Abstract
1H-detected solid-state nuclear magnetic resonance (NMR) experiments are recorded on both intact and trypsin-cleaved sedimented measles virus (MeV) nucleocapsids under ultra-fast magic-angle spinning. High-resolution 1H,15N-fingerprints allow probing the degree of molecular order and flexibility of individual capsid proteins, providing an exciting atomic-scale complement to electro microscopy (EM) studies of the same systems.
Introduction
The atomic-level characterization of large viral particles represents one of the largest challenges of modern structural biology, as well as a fundamental step for the design of effective antiviral treatments. In viruses, the viral genome (double- or single-stranded RNA or DNA) is associated to multiple copies of a capsid protein, forming predominantly icosahedral or helical architectures. The description of the interactions among capsid proteins, and between capsid proteins and nucleic acids, requires the determination of structural details at atomic resolution. These complex superstructures are often studied by x-ray crystallography and electron microscopy (EM) (1). However, only information at low resolution is usually available from EM, and extended and flexible architectures such as those encountered for helical nucleocapsids do not provide single crystals amenable to diffraction studies, and the determination of atomic-scale models requires modeling by homology (2) or the use of extensive large-scale molecular dynamics simulations (3).
Major technical as well as methodological developments have recently extended the applicability of solid-state NMR under magic-angle spinning (MAS) to access the structure in more and more challenging biological solids, ranging from microcrystalline domains (4) to insoluble membrane proteins (5,6), heterogeneous assemblies (7), or fibrillar systems (8–10). In particular, these approaches have proven to be a powerful tool to study the structure and dynamics of polyethylene glycol- (PEG-)precipitated model viral nucleocapsids, such as the Pf1 and fd bacteriophages (11–13). However, resolution and sensitivity become an issue when the approach is applied to systems composed of large coat proteins, as in the case of HIV1 conical assemblies (14,15).
Suitable deuteration strategies (16–18) and the development of MAS probes capable of spinning small rotors at the so-called fast (30 to 40 kHz) (19–23) and ultra-fast (60 kHz) (24–26) regimes have opened a new avenue in solid-state NMR, enabling the detection of resolved 1H resonances. This advancement results in a sensitivity boost that has been shown to considerably accelerate the resonance assignment procedure as well as the acquisition of structurally important restraints, on both deuterated and fully protonated crystalline and noncrystalline biological targets (23,24,26–31).
In this study, we combine 1H-detection at ultra-fast MAS and high magnetic field with a sedimentation protocol, to provide access to high-resolution proton solid-state NMR spectra of large nucleocapsid-like particles. Sedimentation, either via regular ultracentrifugation, or directly in the MAS rotor (32–34), is emerging as a sample preparation method to obtain high-quality solid-state NMR spectra. On the one hand, the large size of the viral particles makes them a perfect substrate for sedimentation techniques, which represent a common intrinsic step for their purification. On the other hand, the repetitive positioning of the nucleoproteins within the nucleocapsids provides the relative degree of order necessary for well-resolved solid-state NMR spectra, as recently shown by Polenova and coworkers for HIV capsid assemblies (35). In our research we apply this methodology to the study of recombinant nucleocapsids of measles virus (MeV), a member of the Paramyxoviridae family of the Mononegavirales order, for which no crystal structure is currently available.
In MeV, the single-stranded viral RNA genome is encapsidated by multiple copies of the nucleoprotein (N), forming a helical nucleocapsid with each N subunit binding six nucleotides (36–39). The MeV nucleoprotein consists of two distinct domains, a globular N-terminal domain (NCORE, residues 1 to 400) and a C-terminal domain (NTAIL, residues 401 to 525). NCORE contains all the regions necessary for self-assembly and RNA binding (40). NTAIL is intrinsically disordered and is responsible for the interaction with the polymerase complex (41,42).
Low-resolution cryo-EM maps of MeV capsids show a left-handed helical arrangement with an outer diam. of 20 nm (D in Fig. 1) and a diam. of the inner channel of 6.5 nm (d in Fig. 1) (2,43,44). Solution NMR and small angle x-ray scattering data showed that NTAIL is disordered in the context of the intact nucleocapsids, and suggested that it can exfiltrate through the spacing between the turns of the supramolecular helix (45). Removal of NTAIL by trypsin digestion results in a major structural rearrangement, yielding more compact and regular assemblies, with reduced diameters and shorter pitch (Fig. 2, in brown) (2). Capsid morphology is a key viral property, but the relation between atomic-level structure and morphology remains elusive. No atomic level information is available for NCORE and for the N-terminal portion of NTAIL in the intact assemblies, and it is unknown to what extent the structure of NCORE monomers differs between intact and cleaved nucleocapsids.
Figure 1.
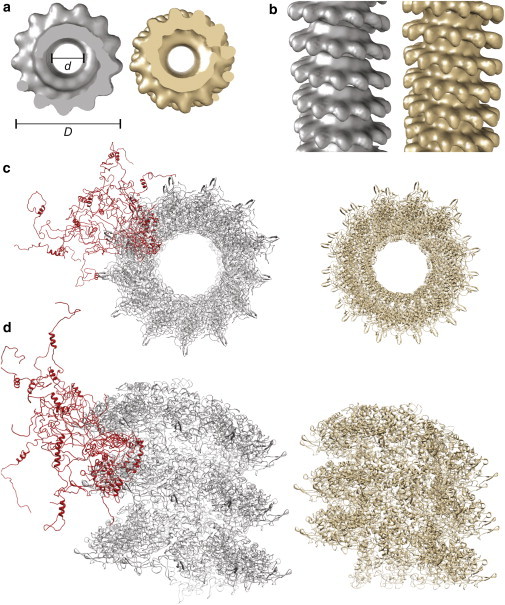
(A) Top view and (B) side view of the cryo-EM maps of intact (gray) and cleaved (brown) measles nucleocapsids (2). (C) Top view and (D) side view of atomic-level ribbon models of the nucleocapsids, obtained by modeling the coordinates of NCORE by homology with the N monomer of the closely related respiratorial syncytial virus (RSV) (46), and fitting them into cryo-EM maps of the capsids. In the intact nucleocapsid, different conformations of the disordered NTAIL are shown in red (45). To see this figure in color, go online.
Figure 2.
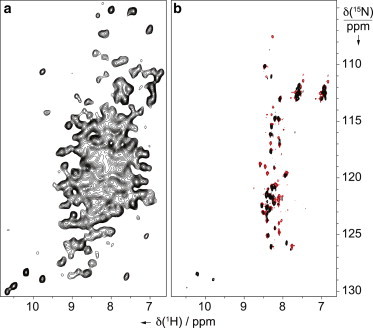
(A) Dipolar-based 1H-15N CP-HSQC and (B) scalar-based 1H-15N J-HSQC spectra (black contours) of intact sedimented MeV nucleocapsids; (B) 1H-15N HSQC spectrum (red contours) of the intact MeV nucleocapsid-like particles in solution (45). The experiments were carried out on a 1 GHz spectrometer equipped with a 1.3 mm triple-channel probe at 60 kHz MAS (black spectra) or equipped with a triple-channel cryoprobe (red spectrum). The pulse sequences of all solid-state NMR experiments are given in the Supporting Material. Note that in the red spectrum only the last 75 residues of NTAIL can be detected (45) and that comparison with the solution NMR spectrum of free NTAIL, where all 125 signals could be assigned (57), suggests that the first 50 amino acids of NTAIL experience restricted dynamics through interaction with NCORE. To see this figure in color, go online.
We have recorded a set of high-resolution 1H-detected solid-state NMR experiments on both intact and trypsin-cleaved sedimented MeV nucleocapsids that allowed us to probe their relative molecular order and flexibility within the supramolecular assemblies, as well as their level of hydration.
Materials and Methods
Sample preparation
Expression and purification of recombinant [U-13C, 15N]-labeled MeV nucleocapsids were performed as described elsewhere (2,45). For solid-state NMR analysis, suspensions containing ∼ 3 mg of nucleocapsids were centrifuged at 165,000 × g for 15 h at 12°C directly into a 1.3 mm rotor using a device similar to one previously described (47), and manufactured in collaboration with Bruker Biospin (Rheinstetten, Germany) (48).
Fig. S1 in the Supporting Material shows Transmission Electron Microscopy (TEM) images of intact and cleaved nucleocapsids, before and after the NMR measurements.
Solid-state NMR spectroscopy
All experiments were run on a 1 GHz Bruker NMR spectrometer, equipped with a 3-channel HCN 1.3 mm probe with an external lock.
For all the proton-detected experiments, the MAS rate was 60 kHz and the target temperature on the spectrometer was set to 240 K, corresponding to an actual sample temperature of 300 K. 1H and 15N 90° pulses were 2.5 and 5 μs, respectively. For the dipolar-based CP-HSQC experiments (Fig. S1 A), cross-polarization (CP) steps from 1H to 15N and from 15N to 1H were performed using a constant Radio Frequency (RF) amplitude of 35 kHz applied to 15N and linearly ramped pulses from 70% to 100% (direct CP) and 100% to 70% (back CP) with a maximum RF amplitude of 25 kHz on 1H. CP contact times of 1000 and 400 μs were used, respectively. For the 15N-1H scalar-based J-HSQC (Heteronuclear Single-Quantum Coherence) experiment (Fig. S1 B), the J evolution delays were set to 1/(4JNH) = 2.7 ms. WALTZ-16 (49) decoupling at 10 kHz was used on the 1H channel during indirect 15N detection and on the 15N and 13C channels during direct 1H acquisition. All 15N-1H correlation spectra were recorded with 200 t1 points to total indirect 15N acquisition times of 20 ms, using 192 transients per increment with direct 1H acquisition times of 30 ms and total experimental times of 10.7 h each. The States-Two-Phase Pulse Incrementation (TPPI) (50) method was employed for quadrature detection in the indirect dimension.
For 15N T1rho measurement (Fig. S1 C), relaxation delays of 0, 0.005, 0.01, 0.025, 0.05, 0.1, 0.15, 0.2, 0.25, 0.3, and 0.5 s were used, during a spin-lock pulse at 20 kHz applied to 15N (31,51).
For the water-edited experiments, a 1D 15N-1H CP-HSQC sequence was implemented in analogy to previously introduced 13C-detected approaches (52,53) (see Fig. 4 A). A selective 90° pulse was applied on the water resonance using a Q5 Gaussian (54) cascade shape of 1000 points with a duration of 6 ms. The total dephasing echo delay was 0.8 ms. The direct 1H acquisition time was 30 ms, and the number of scans was 128 for each diffusion delay τmix. 48 diffusion delays were sampled in the range from 0 to 470 ms.
Figure 4.
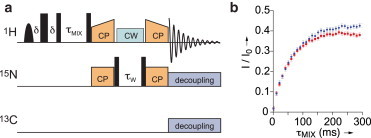
(A) Pulse sequence for the proton-detected water-edited 1D 15N-1H CP-HSQC experiment. Narrow and broad black rectangles indicate π/2 and π pulses, respectively, and the bell shape represents a band-selective water excitation pulse. (B) Buildup curves representing the relative intensities of the water-edited CP-HSQC spectra compared with the standard 15N-1H CP-HSQC spectra of amide protons. Curves obtained on intact and cleaved MeV nucleocapsids are represented in blue and red, respectively. Experiments were carried out on a 1 GHz spectrometer equipped with a triple-channel 1.3 mm probe at 60 kHz MAS. To see this figure in color, go online.
The MISSISSIPI scheme (55) at 15 kHz during a saturation time of 300 ms was used for water suppression in all of the above experiments.
CP-PDSD (proton-driven spin-diffusion) 13C-13C correlation experiments were carried out under 10 kHz MAS. The target temperature on the spectrometer was set to 285 K, corresponding to an actual sample temperature of 290 K. 1H and 13C 90° pulses were 2.5 and 3.3 μs, respectively. CP from 1H to 13C was performed using a constant RF amplitude of 20 kHz applied to 13C and a linearly ramped pulse from 90% to 100% with a maximum RF amplitude of 30 kHz on 1H for a contact time of 1500 μs. The PDSD mixing time was set to 100 ms. Spinal-64 (56) decoupling at 80 kHz was applied in the proton channel during 13C direct and indirect evolutions. The spectra were recorded with 1000 t1 points to total indirect acquisition times of 9 ms, using 64 transients per increment with direct 13C acquisition times of 15 ms and total experimental times of 42.7 h. The States-TPPI (50) method was employed for quadrature detection in the indirect dimension.
Results and Discussion
Intact [13C,15N]-labeled nucleocapsids from MeV incorporating random cellular RNA were obtained by overexpressing the N-protein in Escherichia coli (E. coli), and purified by differential sedimentation (2,45). Removal of NTAIL to obtain cleaved nucleocapsids can be achieved afterward by trypsin digestion. These sedimented samples can be directly transferred into a 1.3 mm NMR rotor and studied under ultra-fast MAS. In contrast to its solution counterpart, solid-state NMR under MAS does not suffer from molecular weight limitations, and spectra from both rigid and flexible regions can be obtained by the application of dipolar- and scalar-based experiments, respectively.
Fig. 2 A shows a dipolar-based 15N-1H correlation spectrum (“CP-HSQC”) (We use here the HSQC acronym to refer to a 1H-detected heteronuclear SQ-SQ correlation experiment. Note however that in the dipolar version of the experiment, there is no heteronuclear two-spin coherence during any evolution period. Rather a population is transferred from 1H to 15N during the CP step, it evolves as 15Nx magnetization, and then it is transferred back to 1H for detection.) recorded under ultra-fast MAS on intact nucleocapsids that allows access to the rigid portion of NCORE. A large number of signals from this region are visible, with 1H linewidths of isolated peaks on the order of 100 to 150 Hz, similar to those observed for microcrystalline solids (26) or amorphous materials such as protein precipitates or fibrillar aggregates (17,23).
Complementarily, the scalar-based 15N-1H correlation spectrum (“J-HSQC”) recorded on the same sample (Fig. 2 B, black contours) displays resonances from NTAIL and superimposes reasonably well with the 1H-15N HSQC spectrum recorded on capsids resuspended in solution (Fig. 2 B, red contours, and reference (45)). This indicates that the solid sample retains the structural properties of the intact nucleocapsids in solution with a flexible NTAIL, opening the way to more detailed structural investigations, as illustrated in Fig. 2.
Fig. 3 A shows a comparison of 15N-1H dipolar correlation spectra of intact and cleaved nucleocapsids. We note that most resolved signals are present in both spectra, showing little or no chemical shift changes, which suggests that there is no major reorganization of the secondary structure of NCORE in the two samples. This is also supported by a comparison of 13C-13C dipolar correlation spectra (CP-PDSD (58)) of intact and cleaved nucleocapsids, which show no obvious 13C chemical shift changes between the two samples (Fig. S2). Interestingly, a few more signals can be detected in the 15N-1H correlation spectrum of the intact sample (in blue). This may be attributable to a portion of the N-terminal stretch of NTAIL being sufficiently rigid and ordered in the intact nucleocapsids to be observed in dipolar correlation spectra.
Figure 3.
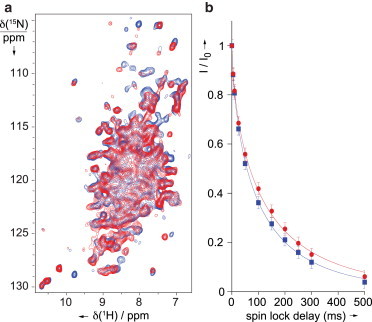
(A) Comparison of dipolar-based 15N-1H HSQC correlation spectra acquired on intact (blue contours) and trypsin-cleaved (red contours) MeV nucleocapsids, at pH7. Experiments were run on a 1 GHz spectrometer with a triple-channel 1.3 mm probe under 60 kHz MAS. (B) Bulk 15N R1rho decays measured on intact (blue points) and cleaved (red points) samples. The data were fitted using a stretched exponential function I(t) = I0 [exp(-t/T1rho)β], yielding average T1rho values of 99 ± 2 ms (β = 0.66) and 119 ± 2 ms (β = 0.65) for intact and cleaved samples, respectively. The stretching factors account for a distribution of T1rho values in the samples; their similarity between intact and cleaved nucleocapsids permits direct comparison of resultant T1rho values. To see this figure in color, go online.
Also, we observe that for most resolved signals in NCORE, the resolution is similar in the two samples, but their bulk 15N rotating-frame relaxation rates (T1rho) (31,51) are significantly different with an average value of 99 ± 2 and 119 ± 2 ms for the intact and cleaved assemblies, respectively (see Fig. 3 B), indicative of differential local dynamics on a submicrosecond timescale. These two experiments indicate that NCORE is generally well ordered in both samples but exhibits increased local dynamics on the submicrosecond timescale in the intact capsids. We speculate that this differential flexibility may be attributable to either residual domain motions caused by the presence of NTAIL that prevents extensive contacts between the interpitch regions of NCORE,, or to perturbations caused by transient interaction with flexible NTAIL segments.
Additionally, the availability of 1H-detected spectra from NCORE provides a sensitive method to evaluate molecular-level order by measuring the capsid hydration level. Reduced hydration is a direct indicator of close intermolecular packing, which itself requires a more rigid structure. In solid-state NMR experiments on hydrated biosolids, signals from at least two water pools are usually observed, namely the interstitial water (e.g., water in contact with the protein), which can exchange magnetization with the protein during the NMR experiment, and the bulk water, which can exchange magnetization only over a much longer timescale (47,59). Although in a standard dipolar-based 15N-1H CP-HSQC experiment, the polarization is first driven from the protein protons to the neighboring 15N spins, a water-edited scheme can be designed that selects the water magnetization before a transfer via chemical exchange and spin diffusion during the mixing period τmix to the protein protons. The corresponding experimental scheme is shown in Fig. 4 A and is similar to water-edited 13C-detected experiments described in the literature (52,53).
At long diffusion times, a plateau is reached indicating that all the interstitial water magnetization has equilibrated with the whole protein 1H bath. On this timescale, there is no contact with the bulk water and the plateau value is proportional to the amount of interstitial water. This measurement is therefore complementary to the atomic level measurements from Fig. 3 and provides a view at the molecular level of overall compactness of both the NCORE monomers themselves, and their packing within the nucleocapsid particles.
A series of water-edited 1D 15N-1H CP-HSQC experiments was acquired on intact and cleaved nucleocapsids to assess the solvent accessibility of NCORE. For a diffusion time of 200 ms, the relative water-edited CP-HSQC signal of the cleaved nucleocapsids is 15% smaller than that of the intact nucleocapsids. These experiments are based on dipolar transfers, thus only the rigid parts of the nucleoprotein contribute to the observed signals. The presence of flexible parts of NTAIL in the intact nucleocapsids is hence not responsible for the differences in signal intensities, although immobilized regions of NTAIL would contribute. This difference confirms that the intact nucleocapsid is on average surrounded by a larger amount of interstitial water. In agreement with EM studies, this provides further evidence that in the presence of NTAIL the capsids adopt a less-ordered supra-molecular arrangement. This could be attributable to larger inner cavities, increased pitch, heterogeneous shapes, or less compact packing of the particles.
Conclusions
In conclusion, we have applied state-of-the-art 1H-detected solid-state NMR to the recombinant viral nucleocapsids of MeV. These complexes are high molecular weight noncrystalline protein-RNA assemblies representing challenging systems in the field of molecular structural biology. We show that, despite their lack of crystalline order, sedimented precipitates of these systems yield high-resolution solid-state NMR spectra, while preserving a viable environment in which the dynamical conformational behavior are retained. The experiments carried out in this study probe the degree of molecular order and flexibility of individual N proteins on length scales from Angstroms to microns and timescales from nanoseconds to microseconds, and thereby provide an exciting complement to EM studies of the same systems.
Acknowledgments
We acknowledge support from the Agence Nationale de la Recherche (ANR 08-BLAN-0035-01 and 10-BLAN-713-01), from Joint Research Activity and Access to Research Infrastructures in the 7th Framework program of the EC (EAST-NMR n.228461, BioNMR n. 261863), from the CNRS (TGIR-RMN-THC FR3050), from the Human Frontier Science Program (long-term fellowship LT000322/2011-L to R.S.), and from Finovi.
Supporting Material
References
- 1.Prasad B.V., Schmid M.F. Principles of virus structural organization. Adv. Exp. Med. Biol. 2012;726:17–47. doi: 10.1007/978-1-4614-0980-9_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desfosses A., Goret G., Gutsche I. Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J. Virol. 2011;85:1391–1395. doi: 10.1128/JVI.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao G., Perilla J.R., Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDermott A. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu Rev Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 5.Cady S.D., Schmidt-Rohr K., Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S.H., Das B.B., Opella S.J. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature. 2012;491:779–783. doi: 10.1038/nature11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jehle S., Rajagopal P., Oschkinat H. Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers. Nat. Struct. Mol. Biol. 2010;17:1037–1042. doi: 10.1038/nsmb.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasmer C., Lange A., Meier B.H. Amyloid fibrils of the HET-s (218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen J.T., Bjerring M., Nielsen N.C. Unique identification of supramolecular structures in amyloid fibrils by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2009;48:2118–2121. doi: 10.1002/anie.200804198. [DOI] [PubMed] [Google Scholar]
- 10.Loquet A., Sgourakis N.G., Lange A. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. doi: 10.1038/nature11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldbourt A., Day L.A., McDermott A.E. Intersubunit hydrophobic interactions in Pf1 filamentous phage. J. Biol. Chem. 2010;285:37051–37059. doi: 10.1074/jbc.M110.119339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abramov G., Morag O., Goldbourt A. Magic-angle spinning NMR of a class I filamentous bacteriophage virus. J. Phys. Chem. B. 2011;115:9671–9680. doi: 10.1021/jp2040955. [DOI] [PubMed] [Google Scholar]
- 13.Park S.H., Yang C., Mueller L.J. Resolution and measurement of heteronuclear dipolar couplings of a noncrystalline protein immobilized in a biological supramolecular assembly by proton-detected MAS solid-state NMR spectroscopy. J. Magn. Reson. 2013;237:164–168. doi: 10.1016/j.jmr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y., Ahn J., Polenova T. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2010;132:1976–1987. doi: 10.1021/ja908687k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Byeon I.J., Hou G., Polenova T. Motions on the millisecond time scale and multiple conformations of HIV-1 capsid protein: implications for structural polymorphism of CA assemblies. J. Am. Chem. Soc. 2012;134:6455–6466. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chevelkov V., Rehbein K., Reif B. Ultrahigh resolution in proton solid-state NMR spectroscopy at high levels of deuteration. Angew. Chem. Int. Ed. Engl. 2006;45:3878–3881. doi: 10.1002/anie.200600328. [DOI] [PubMed] [Google Scholar]
- 17.Linser R., Dasari M., Reif B. Proton-detected solid-state NMR spectroscopy of fibrillar and membrane proteins. Angew. Chem. Int. Ed. Engl. 2011;50:4508–4512. doi: 10.1002/anie.201008244. [DOI] [PubMed] [Google Scholar]
- 18.Asami S., Rakwalska-Bange M., Reif B. Protein-RNA interfaces probed by 1H-detected MAS solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2013;52:2345–2349. doi: 10.1002/anie.201208024. [DOI] [PubMed] [Google Scholar]
- 19.Paulson E.K., Morcombe C.R., Zilm K.W. High-sensitivity observation of dipolar exchange and NOEs between exchangeable protons in proteins by 3D solid-state NMR spectroscopy. J. Am. Chem. Soc. 2003;125:14222–14223. doi: 10.1021/ja037559u. [DOI] [PubMed] [Google Scholar]
- 20.Paulson E.K., Morcombe C.R., Zilm K.W. Sensitive high resolution inverse detection NMR spectroscopy of proteins in the solid state. J. Am. Chem. Soc. 2003;125:15831–15836. doi: 10.1021/ja037315+. [DOI] [PubMed] [Google Scholar]
- 21.Zhou D.H., Graesser D.T., Rienstra C.M. Sensitivity and resolution in proton solid-state NMR at intermediate deuteration levels: quantitative linewidth characterization and applications to correlation spectroscopy. J. Magn. Reson. 2006;178:297–307. doi: 10.1016/j.jmr.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 22.Zhou D.H., Shah G., Rienstra C.M. Proton-detected solid-state NMR spectroscopy of fully protonated proteins at 40 kHz magic-angle spinning. J. Am. Chem. Soc. 2007;129:11791–11801. doi: 10.1021/ja073462m. [DOI] [PubMed] [Google Scholar]
- 23.Zhou D.H., Nieuwkoop A.J., Rienstra C.M. Solid-state NMR analysis of membrane proteins and protein aggregates by proton detected spectroscopy. J. Biomol. NMR. 2012;54:291–305. doi: 10.1007/s10858-012-9672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knight M.J., Webber A.L., Pintacuda G. Fast resonance assignment and fold determination of human superoxide dismutase by high-resolution proton-detected solid-state MAS NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2011;50:11697–11701. doi: 10.1002/anie.201106340. [DOI] [PubMed] [Google Scholar]
- 25.Lewandowski J.R., Dumez J.-N., Oschkinat H. Enhanced resolution and coherence lifetimes in the solid-state NMR spectroscopy of perdeuterated proteins under ultrafast magic-angle spinning. J. Phys. Chem. Lett. 2011;2:2205–2211. [Google Scholar]
- 26.Marchetti A., Jehle S., Pintacuda G. Backbone assignment of fully protonated solid proteins by 1H detection and ultrafast magic-angle-spinning NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2012;51:10756–10759. doi: 10.1002/anie.201203124. [DOI] [PubMed] [Google Scholar]
- 27.Asami S., Schmieder P., Reif B. High resolution 1H-detected solid-state NMR spectroscopy of protein aliphatic resonances: access to tertiary structure information. J. Am. Chem. Soc. 2010;132:15133–15135. doi: 10.1021/ja106170h. [DOI] [PubMed] [Google Scholar]
- 28.Huber M., Hiller S., Meier B.H. A proton-detected 4D solid-state NMR experiment for protein structure determination. Chem. Phys. Chem. 2011;12:915–918. doi: 10.1002/cphc.201100062. [DOI] [PubMed] [Google Scholar]
- 29.Linser R., Bardiaux B., Reif B. Structure calculation from unambiguous long-range amide and methyl 1H-1H distance restraints for a microcrystalline protein with MAS solid-state NMR spectroscopy. J. Am. Chem. Soc. 2011;133:5905–5912. doi: 10.1021/ja110222h. [DOI] [PubMed] [Google Scholar]
- 30.Knight M.J., Felli I.C., Pintacuda G. Rapid measurement of pseudocontact shifts in metalloproteins by proton-detected solid-state NMR spectroscopy. J. Am. Chem. Soc. 2012;134:14730–14733. doi: 10.1021/ja306813j. [DOI] [PubMed] [Google Scholar]
- 31.Knight M.J., Pell A.J., Pintacuda G. Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR. Proc. Natl. Acad. Sci. USA. 2012;109:11095–11100. doi: 10.1073/pnas.1204515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertini I., Luchinat C., Turano P. Solid-state NMR of proteins sedimented by ultracentrifugation. Proc. Natl. Acad. Sci. USA. 2011;108:10396–10399. doi: 10.1073/pnas.1103854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertini I., Engelke F., Turano P. NMR properties of sedimented solutes. Phys. Chem. Chem. Phys. 2012;14:439–447. doi: 10.1039/c1cp22978h. [DOI] [PubMed] [Google Scholar]
- 34.Gardiennet C., Schütz A.K., Meier B.H. A sedimented sample of a 59 kDa dodecameric helicase yields high-resolution solid-state NMR spectra. Angew. Chem. Int. Ed. Engl. 2012;51:7855–7858. doi: 10.1002/anie.201200779. [DOI] [PubMed] [Google Scholar]
- 35.Han Y., Hou G., Polenova T. Magic angle spinning NMR reveals sequence-dependent structural plasticity, dynamics, and the spacer peptide 1 conformation in HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 2013;135:17793–17803. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egelman E.H., Wu S.S., Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J. Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calain P., Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolakofsky D., Pelet T., Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J. Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruigrok R.W., Crépin T., Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Bankamp B., Horikami S.M., Moyer S.A. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology. 1996;216:272–277. doi: 10.1006/viro.1996.0060. [DOI] [PubMed] [Google Scholar]
- 41.Longhi S., Receveur-Bréchot V., Canard B. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 2003;278:18638–18648. doi: 10.1074/jbc.M300518200. [DOI] [PubMed] [Google Scholar]
- 42.Kingston R.L., Baase W.A., Gay L.S. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 2004;78:8630–8640. doi: 10.1128/JVI.78.16.8630-8640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhella D., Ralph A., Yeo R.P. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J. Mol. Biol. 2004;340:319–331. doi: 10.1016/j.jmb.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 44.Schoehn G., Mavrakis M., Ruigrok R.W. The 12 A structure of trypsin-treated measles virus N-RNA. J. Mol. Biol. 2004;339:301–312. doi: 10.1016/j.jmb.2004.03.073. [DOI] [PubMed] [Google Scholar]
- 45.Jensen M.R., Communie G., Blackledge M. Intrinsic disorder in measles virus nucleocapsids. Proc. Natl. Acad. Sci. USA. 2011;108:9839–9844. doi: 10.1073/pnas.1103270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tawar R.G., Duquerroy S., Rey F.A. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 47.Böckmann A., Gardiennet C., Lesage A. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR. 2009;45:319–327. doi: 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- 48.Bertini I., Engelke F., Ravera E. On the use of ultracentrifugal devices for sedimented solute NMR. J. Biomol. NMR. 2012;54:123–127. doi: 10.1007/s10858-012-9657-y. [DOI] [PubMed] [Google Scholar]
- 49.Shaka A.J., Keeler J., Freeman R. An improved sequence for broadband decoupling: WALTZ-16. J. Magn. Reson. 1983;52:335–338. [Google Scholar]
- 50.Marion D., Ikura M., Bax A. Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J. Magn. Reson. 1989;85:393–399. [Google Scholar]
- 51.Lewandowski J.R., Sass H.J., Emsley L. Site-specific measurement of slow motions in proteins. J. Am. Chem. Soc. 2011;133:16762–16765. doi: 10.1021/ja206815h. [DOI] [PubMed] [Google Scholar]
- 52.Lesage A., Emsley L., Böckmann A. Investigation of dipolar-mediated water-protein interactions in microcrystalline Crh by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2006;128:8246–8255. doi: 10.1021/ja060866q. [DOI] [PubMed] [Google Scholar]
- 53.Ader C., Schneider R., Baldus M. Structural rearrangements of membrane proteins probed by water-edited solid-state NMR spectroscopy. J. Am. Chem. Soc. 2009;131:170–176. doi: 10.1021/ja806306e. [DOI] [PubMed] [Google Scholar]
- 54.Emsley L., Bodenhausen G. Gaussian pulse cascades: new analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem. Phys. Lett. 1990;165:469–476. [Google Scholar]
- 55.Zhou D.H., Rienstra C.M. High-performance solvent suppression for proton detected solid-state NMR. J. Magn. Reson. 2008;192:167–172. doi: 10.1016/j.jmr.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fung B.M., Khitrin A.K., Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 57.Gely S., Lowry D.F., Longhi S. Solution structure of the C-terminal X domain of the measles virus phosphoprotein and interaction with the intrinsically disordered C-terminal domain of the nucleoprotein. J. Mol. Recognit. 2010;23:435–447. doi: 10.1002/jmr.1010. [DOI] [PubMed] [Google Scholar]
- 58.Szeverenyi N.M., Sullivan M.J., Maciel G.E. Observation of spin exchange by two-dimensional fourier-transform 13C cross polarization-magic-angle spinning. J. Magn. Reson. 1982;47:462–475. [Google Scholar]
- 59.Lesage A., Gardiennet C., Böckmann A. Polarization transfer over the water-protein interface in solids. Angew. Chem. Int. Ed. Engl. 2008;47:5851–5854. doi: 10.1002/anie.200801110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


