Abstract
Puffs are local Ca2+ signals that arise by Ca2+ liberation from the endoplasmic reticulum through the concerted opening of tightly clustered inositol trisphosphate receptors/channels (IP3Rs). The locations of puff sites observed by Ca2+ imaging remain static over several minutes, whereas fluorescence recovery after photobleaching (FRAP) experiments employing overexpression of fluorescently tagged IP3Rs have shown that the majority of IP3Rs are freely motile. To address this discrepancy, we applied single-molecule imaging to locate and track type 1 IP3Rs tagged with a photoswitchable fluorescent protein and expressed in COS-7 cells. We found that ∼70% of the IP3R1 molecules were freely motile, undergoing random walk motility with an apparent diffusion coefficient of ∼0.095 μm s−1, whereas the remaining molecules were essentially immotile. A fraction of the immotile IP3Rs were organized in clusters, with dimensions (a few hundred nanometers across) comparable to those previously estimated for the IP3R clusters underlying functional puff sites. No short-term (seconds) changes in overall motility or in clustering of immotile IP3Rs were apparent following activation of IP3/Ca2+ signaling. We conclude that stable clusters of small numbers of immotile IP3Rs may underlie local Ca2+ release sites, whereas the more numerous motile IP3Rs appear to be functionally silent.
Introduction
Inositol trisphosphate receptors (IP3Rs) are Ca2+-permeable channels in the membrane of the endoplasmic reticulum (ER) (1). They liberate Ca2+ sequestered in ER stores to generate cytosolic Ca2+ signals that control ubiquitous cellular functions as diverse as gene expression, secretion, and synaptic plasticity (2). The specific regulation of cellular functions is achieved through an exquisite spatiotemporal patterning of cytosolic Ca2+ signals (2) that arises from the properties of the IP3Rs and their cellular organization. First, the gating of IP3R channels is biphasically regulated by cytosolic Ca2+ itself. Small Ca2+ elevations (in the presence of IP3) induce channel openings via Ca2+-induced Ca2+ release (CICR), whereas higher concentrations cause inactivation, thereby creating positive- and negative-feedback loops (3). Second, functional IP3Rs are arranged in a clustered fashion on the ER membrane so that Ca2+ release through a single channel (known as a blip) can trigger the opening of neighboring channels within a cluster to generate local Ca2+ signals (Ca2+ puffs) (4). In turn, recruitment of multiple puff sites by successive cycles of Ca2+ diffusion and CICR can propagate saltatory Ca2+ waves throughout the cell (5,6). Thus, the localization of IP3Rs is crucial for establishing and optimizing the hierarchical spatiotemporal patterning of cytosolic Ca2+ signals to ensure appropriate regulation of downstream signaling pathways (7).
Ca2+ imaging studies in mammalian cell lines and in Xenopus oocytes have demonstrated that puffs arise repeatedly over many minutes at locations that remain essentially fixed within a cell, suggesting that the underlying clusters of IP3Rs on the ER membrane are relatively stable entities (8–11). Ca2+ blips generated by lone IP3Rs are similarly immotile (12,13). In marked contrast to the immotility of IP3Rs as monitored by the Ca2+ signals they generate, studies of fluorescence recovery after photobleaching (FRAP) of GFP-tagged IP3R proteins indicate that a substantial proportion of IP3Rs are freely diffusible within the ER membrane (14–22). Moreover, IP3R proteins have been reported to aggregate into clusters following sustained (minutes) activation of IP3 signaling and/or cytosolic Ca2+ elevation (14,19–23), and even to undergo clustering in response to IP3 within just a few seconds (24). There is also a marked disparity between immunostaining patterns that show IP3Rs densely distributed throughout the entirety of the ER in mammalian cells (14,20,22) and observations that Ca2+ release arises at just a few discrete puff and blip sites (9–12,25–27).
To account for these apparent differences in the density, distribution, and motility of IP3Rs as assessed by functional Ca2+ imaging versus protein localization, we previously hypothesized that a majority of IP3Rs may be motile but are either functionally unresponsive or mediate Ca2+ liberation only during sustained global elevations of cytosolic [Ca2+] (12,28). We further proposed that local Ca2+ signals arise from a much smaller subset of IP3Rs that are anchored individually or in tight clusters by association with static cytoskeletal structures. Possibly as a consequence of this anchoring, these IP3Rs display a high sensitivity to IP3 and are uniquely able to generate Ca2+ blips and puffs (28).
Here, we sought to examine this hypothesis by using photoconvertible, genetically encoded proteins to track the motility of thousands of individual IP3R molecules with nanometer spatial resolution using single-particle-tracking photoactivation localization microscopy (sptPALM) (29). We expressed type 1 IP3Rs tagged with the photoconvertible fluorescent protein mEos2 in COS-7 cells. By using stochastic activation of mEos2 molecules at low density, we were able to track IP3Rs with high precision by fitting a two-dimensional Gaussian function to determine the centroid of the blurred spots (point-spread function) formed by the microscope in successive image frames. We found that the IP3Rs could be classified into two groups with relatively high or low/zero motility, and that the immotile IP3Rs appeared to be preferentially grouped within tight clusters. Further, we report that agonist activation of IP3/Ca2+ signaling caused no appreciable short-term (seconds) changes in the motility or distribution of IP3Rs.
Materials and Methods
Plasmid construction
The type 1 IP3R construct was a gift from Kevin Foskett. The IP3R1 was cloned into the EcoR1/Not1 restriction sites of pcDNA3.1/Zeo(+), yielding pcDNA3.1- IP3R1. mEos2 was then tagged onto the N-terminus (20,30) of the IP3R1 by cloning into the multiple cloning site of pcDNA3.1- IP3R1 using the NheI/EcoRI restriction site. We named this final construct mEos2-IP3R1.
Cell culture
COS-7 cells (ATCC Manassas, VA) were cultured according to the supplier’s instructions in Dulbecco’s modified Eagle’s medium supplemented with 10% (v/v) fetal bovine serum (FBS). Cells were incubated at 37°C in a humidified incubator gassed with 95% air and 5% CO2, passaged every 3–4 days, and used for up to 20 passages. For expression of mEos2-IP3R1 in COS-7 cells, 5 × 105 cells were harvested and transfected using a Nucleofector (Kit V, program A-024; Lonza, Basel, Switzerland) or Neon Transfection System (Life Technologies, Carlsbad, CA). Transfected cells were plated at 5 × 104 cells/ml in Petri dishes with glass coverslips as the base (MatTek Ashland, MA). Cells were imaged 24–72 h later in HEPES-buffered saline (HBS; (in mM) NaCl 135, KCl 5, MgCl2 1.2, CaCl2 2.5, HEPES 5, glucose 10) at room temperature. For fixed-cell experiments, cells were rinsed in PBS, fixed for 15 min in 4% paraformaldehyde (PFA) at room temperature, and subsequently washed three times in PBS before imaging.
Live-cell microscopy
Cells were imaged at room temperature (∼21°C) using a custom-built total internal reflection (TIRF) microscope system based on an Olympus IX 70 microscope equipped with an Olympus 100× SAPO objective (NA 1.40). Activation and excitation lasers illuminated the sample in semi-TIRF mode, with the angle of light exiting the objective adjusted to create a light sheet restricted to the bottom few micrometers of the specimen. Images of emitted fluorescence were captured by an Evolve 512 electron-multiplied CCD camera (Roper Scientific Tucson, AZ) at a resolution of 256 × 256 pixels (1 pixel = 0.166 μm) at an exposure time of 150 ms (6.67 frames s−1). The camera was operated in quantitative mode, where one count = one detected photon. Cells expressing mEos2-IP3R1 were identified by their green fluorescence using 488 nm excitation and a longpass emission filter. A 561 nm laser was then used to preferentially excite the red, photoswitched form of mEos, which was imaged using a 568 nm sharp-edge longpass filter. Preexisting red fluorescence was bleached by exposing the cell to high-intensity 561 nm excitation and the intensity was then reduced (to ∼100 W/cm2) to track single molecules. In some instances, spontaneous photoswitching of mEos provided a sufficient density of red fluorescent molecules. In other cases, continuous or pulsed activation by a 405 nm laser was used, with the intensity adjusted to give an appropriate density of molecules. Typically, the density of fluorescent spots in a given frame was ∼1 per 30 μm2. Sequential image frames were captured using MetaMorph 7.5 (Molecular Dynamics Pittsburgh, PA) to accumulate continuous image stacks, which typically comprised 5000 frames (∼12½ min).
Superresolution tracking and data analysis
Image stacks were imported into Insight3 (custom software provided by X. Zhuang, Harvard University) to track the motions of individual molecules. The program identified valid fluorescent spots (molecules) within each image frame on the basis of their intensity (>250 photoelectrons) and spatial width (200–600 nm), and localized them with subpixel precision by fitting a two-dimensional circular Gaussian function to the diffraction-limited spot. Molecular tracks were then assigned by linking a spot in one frame to the closest spot within a defined search radius in the following frame. The search radius was generally set to 4 pixels (664 nm) to minimize the possibility of false linkages between different adjacent molecules while being wide enough to encompass almost all Brownian displacements of a single molecule. Insight3 thus produced a data set of tracked molecules, listing sequential x-y coordinates during the track and the corresponding frame numbers (times) when they were observed.
Further analysis of the data set was performed in MATLAB (The MathWorks, Natick, MA) using custom-written routines. For each track, we calculated the individual displacements between successive frames (single-lag displacements (SLDs)), as well as the mean location of the molecule averaged across its track. Data were filtered to exclude continuous tracks that persisted for fewer than four frames or longer than 40 frames. To further account for the blinking behavior of mEos, we applied a custom-written MATLAB algorithm to link together temporally (1–25 frames) and spatially (<167 nm) adjacent track segments of selected immotile molecules so that blinking molecules would not be overcounted.
STORM imaging of native IP3Rs
We performed superresolution localization of native type 1 IP3Rs in COS-7 cells (Fig. 1 G) by employing STORM imaging (31) utilizing an anti-IP3R 1 (AB5882 (Millipore Billerica, MA), a rabbit polyclonal antibody raised to a synthetic peptide corresponding to amino acids 2732–2750 of rat IP3R1)) and a secondary antibody generated by dual labeling of donkey-anti-rabbit IgG (Jackson ImmunoResearch West Grove, PA) with Cy3 (mono-reactive dye pack; GE Healthcare Pittsburgh, PA) and Alexa Fluor 647 (Invitrogen Grand Island, NY). Cells were fixed and reduced by incubation for 15 min at room temperature in PBS containing 3% PFA and 0.1% glutaraldehyde (GA), followed by 7 min in a freshly prepared solution of 0.1% NaBH4 in PBS (without additional Ca2+/Mg2+) The cells were rinsed quickly (×3) with PBS, permeabilized by incubation for 1 h in blocking buffer (3% (m/v) bovine serum albumin (BSA), 0.2% TritonX-100 (v/v) in PBS), and probed with the anti-IP3R 1 antibody (1:100 dilution in blocking buffer) for 1 h. The cells were quickly rinsed with wash buffer (0.2% (m/v) BSA, 0.05% TritonX-100 (v/v) in PBS), washed (×3) for 5 min with PBS, and fluorescently labeled by incubation with the Cy3-Alexa Fluor 647-conjugated secondary antibody (1:100 dilution in blocking buffer) for 1 h. Finally, the cells were rinsed with wash buffer and PBS (as previously), and fixed again by incubation with 3% PFA and 0.1% GA for 5 min. Imaging was carried out on a Nikon N-STORM system based on a Nikon Eclipse Ti microscope equipped with a Nikon 100× APO TIRF objective (NA 1.49). In semi-TIRF mode, a 647 nm laser continually illuminated the cells, driving the Alexa Fluor 647 dye into a nonfluorescent state. Sparse single-molecule reactivation of individual Alexa Fluor 647 molecules was achieved by weak excitation of Cy3 using a 561 nm laser. The imaging buffer was composed of 50 mM Tris, pH 8, 10 mM NaCl, 10% glucose, 0.14 mg/ml glucose oxidase (Sigma), 0.2 mg/ml bovine liver catalase (Sigma), and 0.14 M β-mercaptoethanol. Fifteen thousand frames were recorded from the center quad of an Andor iXon3 electron-multiplying charge-coupled device (EMCCD) camera with an exposure time of 15 ms per frame. Single-molecule fitting and drift correction were performed with the N-STORM software in NIS Elements (version 3.1.0.11901). Localization coordinates were exported into Microcal Origin Version 9.1 (OriginLab, Northampton, MA) for graphing.
Figure 1.
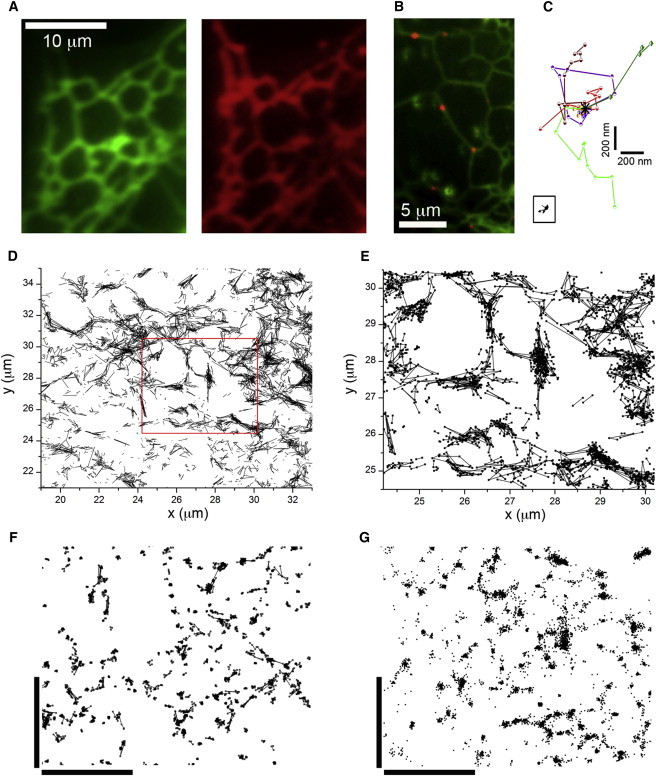
Expression of mEos-tagged IP3R1 in COS-7 cells for single-molecule tracking. (A) Conventional fluorescence image showing the recticular distribution of tagged IP3R1 in the periphery of a COS-7 cell (left) and a pseudocolored image of the same region of the cell showing fluorescence of the dye ER tracker blue/white (right). (B) Overlay showing autofluorescence of the ER (green) with fluorescence of sparsely photoactivated mEos molecules in red. (C) An overlay of seven representative tracks (different colors) of mEos IP3R1s in live COS-7 cells, each localized over nine consecutive image frames (1.35 s) and plotted after aligning their starting positions (marked by asterisk). The inset trace (boxed) shows a representative track of an mEos IP3R1 in a fixed cell. (D) Overlay of >5000 mEos IP3R1 tracks in the periphery of a COS-7 cell. (E) Enlarged view of the region marked in red in (D). (F) Corresponding overlay of mEos IP3R1 localizations in a fixed COS-7 cell. (G) STORM superresolution image of native IP3R1 in the periphery of a control, nontransfected COS-7 cell. The plot depicts drift-corrected fluorophore localizations derived from a cell immunostained with a primary antibody raised against IP3R1 and a secondary antibody labeled with a Cy3-Alexa Fluor 647 dye pair. Bars in F and G = 2 μm. To see this figure in color, go online.
Results
Expression and imaging of fluorescently tagged IP3R molecules
To track single IP3Rs, we employed COS-7 cells, which flatten close to the coverglass, allowing the ER to be visualized in the cell periphery as an extended reticulum forming a nearly two-dimensional, fishnet-like sheet (Fig. 1, A and B). Cells were transfected 24–72 h before imaging with type 1 IP3Rs tagged with mEos2, a photoswitchable fluorescent protein that initially fluoresces green but can be switched to a red fluorescent form by weak illumination with near-UV (405 nm) light. Transfected cells were identified by their green fluorescence when excited by blue (488 nm) laser light. The pattern of green mEos2 fluorescence (Fig. 1 A, left) closely overlapped that of an ER marker (Fig. 1 A, right). After selecting a region of cell to image, we bleached out preexisting red Eos using strong 561 nm laser irradiation, and then visualized the fluorescence of individual red mEos molecules using a lower-power (561 nm) excitation and a 568 nm longpass emission filter (Fig. 1, B and C). The camera exposure time (150 ms) was chosen as a compromise between blurring of the single-molecule fluorescence spot by Brownian motion within a frame and allowing a measurable displacement between successive frames. Movie S1 in the Supporting Material shows the image sequence from which Fig. 1 C was obtained.
We typically acquired image stacks of 5000 sequential frames (750 s) using the center quad of the camera chip (250 × 250 pixels; 167 nm/pixel at the specimen). Spontaneous photoswitching of mEos often provided a sufficient density of fluorescent molecules, and in other cases continuous or pulsed weak illumination with 405 nm light was used to induce photoswitching at a rate controlled by the intensity of the activating light.
Tracking single IP3Rs
Molecules were identified, localized, and tracked by offline analysis of the image stacks using the Insight3 software package to generate a database of unique tracks with x,y,t coordinates for each position of a given molecule. Images were obtained from the cell periphery, where COS-7 cells flatten to a thickness of only ∼800 nm. We thus analyzed the tracking data in only two (x,y) spatial dimensions.
A molecule could typically be tracked for several consecutive frames (mean = 6.34 ± 0.22 frames, n = >30,000 tracks) before it reverted to a dark state. A majority of molecules in live cells showed apparently random displacements, with average track lengths of several hundred nanometers (upper traces, Fig. 1 C). A map obtained by superimposing all tracks from an image stack traced out the reticular pattern of the ER (Fig. 1, D and E). In contrast to this, localizations of individual IP3R molecules in cells after paraformaldehyde fixation were tightly constrained (inset trace, Fig. 1 C), although their distributions again mapped out a reticular pattern (Fig. 1 F).
To confirm that the overexpressed, Eos-tagged IP3Rs showed a subcellular distribution similar to that of native IP3Rs, we further undertook superresolution STORM imaging of IP3Rs immunostained with a type 1-specific primary antibody and utilizing a secondary antibody optimized for single-molecule localization by dual labeling with Cy3 and Alexa Fluor 647 (32).The pattern of IP3R1 staining in fixed, control (nontransfected) COS 7 cells (Fig. 1 G) closely resembled that of expressed Eos-tagged IP3Rs in fixed cells (cf. Fig. 1 F).
To analyze the tracking data, we calculated the displacements d of molecules from any given point during their track to the point reached after a time t (expressed as the number of lags at a frame interval of 150 ms). The SLD is the displacement during a single lag (i.e., from one frame to the next), and the mean SLD is the average SLD across the entire length of a single-molecule track.
Diffusional motility of IP3R
Fig. 2 A shows plots of the mean-squared displacements (MSDs: mean d2) of IP3R molecules in COS-7 cells as a function of time (lags). Square symbols show data from three representative experiments with live cells. Consistent with IP3R molecules undergoing random walk (Brownian) motion, the MSD increased approximately linearly with time (33). The slope of a linear fit to an MSD-versus-time plot provides a measure of the diffusion coefficient D. Assuming that diffusion in the flattened sheet of ER in COS-7 cells approximates a two-dimensional situation, D = <d2>/4t, where <d2> is the MSD at time t. This gave an aggregate mean value of 0.064 ± 0.002 μm2 s−1 for the live-cell data in Fig. 2 A. In contrast to this, data from fixed cells (open symbols, Fig. 2 A) showed little or no consistent increase in MSD with time, yielding an apparent diffusion coefficient of <0.0003 μm2 s−1.
Figure 2.
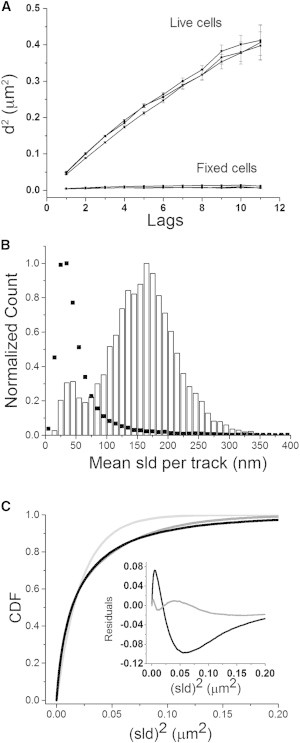
Diffusional characteristics of IP3R1 derived from single-molecule tracking. (A) Plot showing the square of the mean distance (d2) traveled by molecules from any given starting point as a function of lag time (1 lag = 1 frame interval of 150 ms). The data shown are from experiments in three live COS-7 cells (squares), with >2000 tracks in each case. Circles show corresponding data from recordings from three cells after fixation. (B) Distribution of mean SLDs from selected tracks longer than eight lags. Each count (y axis) represents a track with a given mean SLD. Open histogram bars show pooled data from six experiments in live COS-7 cells (3838 tracks). Black symbols show corresponding measurements from four experiments on fixed COS-7 cells (∼25,000 tracks). Both data sets are normalized to the same peak height. (C) Plot showing the CDF of mean-squared SLDs (SLD2) from a representative experiment. The solid black curve shows the cumulative fraction of observations (y axis) for which the SLD2 exceeded a given value (x axis). Curves are single-exponential (light gray) and double-exponential (dark gray) fits to the data (black). The inset shows residual errors remaining after subtracting the single (black trace)- and double (grey trace)-component exponential fits from the data.
Aggregate diffusion coefficients measured in live cells at cooler temperatures (10–12°C, n = 7 cells) were only modestly lower than at 21°C (0.049 ± 0.003 and 0.069 ± 0.005 μm2 s−1, respectively; n = 14). This slight temperature dependence (Q10 = 1.40) is consistent with passive diffusion, whereas an active transport mechanism would be expected to display a much stronger temperature dependence (Q10 ∼3 (34)).
Multiple components of IP3R diffusion
Diffusion coefficients derived as in Fig. 2 A for IP3R in live cells are a population aggregate from all tracked molecules. To determine whether multiple underlying components represent populations of IP3R in live cells with different diffusional motilities, we first analyzed the distribution of mean SLDs of individual molecules and then performed two-component fits to the cumulative density functions (CDFs) of molecular displacements.
Fig. 2 B shows the distribution of mean SLDs (histogram bars), which we pooled from 3838 tracks in six live COS-7 cells after selecting long (9–40 frames) tracks so as to obtain more precise estimates of the mean SLD of each track. The data show two clearly separable components with mean SLDs centered around 46 and 169 nm, corresponding to respective diffusion coefficients of 0.0035 and 0.0476 μm2 s−1. For comparison, we overlay in Fig. 2 B the distribution of mean SLDs measured in fixed cells (solid symbols). This closely overlaps the slow component observed in live cells, indicating that the slow component represents a population of essentially immotile IP3Rs whose SLDs largely reflect jitter in the experimental localizations rather than true movements of the molecules.
We further analyzed the CDF of SLDs and multiple-lag displacements in live COS-7 cells, taking track lengths of 4–40 frames. Fig. 2 C shows representative data from one experiment, illustrating the CDF of SLDs. In the case of a single population of diffusing molecules, the CDF is expected to follow a monoexponential function (35,36) where the exponential fit parameter is a measure of MSD d2 and gives a measure of the effective diffusion coefficient D = d2/4t.
The experimental data could not be fit adequately by a single exponential component, but were fit well (but not perfectly) by two exponential components, indicating two distinct diffusing populations with different diffusion coefficients and relative weights. Two-component exponential fits to the SLD data illustrated in Fig. 2 C corresponded to diffusion coefficients of 0.093 and 0.0075 μm2 s−1, with respective weights of 69% and 31% for these motile and relatively immotile components.
To obtain more precise values, we fitted two-component exponentials to CDFs calculated for all lags between one and 10, and averaged the values obtained within each experiment. Data from six experiments yielded mean diffusion coefficients of 0.095 ± 0.0057 and 0.0075 ± 0.0005 μm2 s−1, with respective weights of 69% ± 1.2% and 31% ± 1.2% for these motile and relatively immotile components. The fraction of motile receptors was similar among all six experiments (range: 64–71%), although the expression level of tagged IP3Rs as assessed by the total number of identified tracks varied ∼3-fold (range: 2835–9136 tracks per experiment).
Spatial segregation of motile and immotile IP3Rs
We were then interested in determining the respective spatial distributions throughout the ER of the populations of IP3R with different motilities. Based on the results in Fig. 2, B and C, we defined immotile molecules as those showing tracks with mean SLDs of <84 nm (i.e., similar to those seen for molecules in fixed cells and within our localization jitter). Correspondingly, we selected rapidly motile molecules as having mean SLDs of >200 nm.
Fig. 3 A shows a representative overlay of the locations (mean position) of immotile IP3Rs (red dots) and rapidly motile IP3Rs (black dots) in the periphery of a COS-7 cell. From visual inspection, it appears the immotile IP3Rs do not closely colocalize with motile IP3Rs. To validate this subjective impression, we measured distances from every immotile IP3R to the closest neighboring motile IP3R, and distances from every motile IP3R to the closest neighboring motile IP3R. The distribution of immotile-to-motile nearest-neighbor distances showed a highly skewed distribution with a pronounced tail at long distances (red, Fig. 3 B). Although roughly two-thirds of immotile molecules were located within <500 nm of a motile IP3R, the remainder were located >500 nm and as far as 5 μm away. For comparison, the nearest-neighbor distances between pairs of motile IP3Rs showed a much tighter distribution (black, Fig. 3 B). Almost all motile IP3Rs were located within <1 μm of a motile neighbor, and only 13.1% were >500 nm away from a neighbor.
Figure 3.
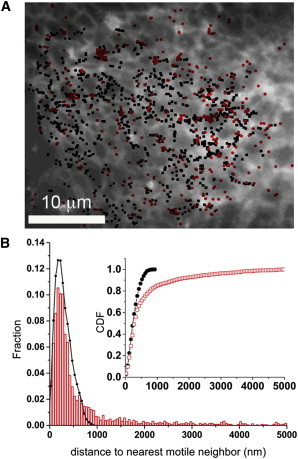
Spatial segregation of immotile and rapidly motile IP3Rs. (A) The background image is the prebleach fluorescence of a COS-7 cell expressing mEos-tagged IP3R1, showing the distribution of IP3Rs across the ER. Superimposed on this are the localizations (mean positions of tracks) of immotile (red dots) and rapidly motile (black dots) IP3R molecules. (B) The main graph plots the distributions of nearest-neighbor distances between each immotile molecule and its nearest motile molecule (red bars), and nearest-neighbor distances between pairs of motile molecules (black circles). The y axis is the fraction of molecules at a given distance from their nearest neighbor. The inset replots the same data as a CDF, i.e., the cumulative fraction of molecules with nearest-neighbor distances shorter than a given value. To see this figure in color, go online.
Clusters of immotile IP3Rs
In light of the immotility of clusters of functional IP3Rs underlying Ca2+ puffs (8,10–12), we sought to determine whether the population of immotile mEos-tagged IP3Rs might show a similarly clustered spatial organization, as distinct from a more diffuse distribution of motile IP3Rs.
This analysis was complicated by the fluorescence blinking behavior of mEos molecules (37). That is to say, after becoming photoactivated, a single mEos molecule could switch repeatedly between fluorescent and dark states within a few seconds before irreversibly bleaching. This would lead to artifactual identification of apparent tight clusters if a single immotile molecule were falsely identified as several distinct, closely localized molecules (38,39). To overcome this problem, we further processed the tracking data from immotile molecules (defined as mean SLD ≤ 84 nm) using a custom-written MATLAB algorithm to link together temporally (0.15–3.75 s) and spatially (<167 nm) adjacent track segments so that blinking molecules would not be overcounted.
After linking the tracks, we calculated the mean position of each molecule and measured the distance from every molecule to every neighboring molecule within an 8 × 8 pixel (1.3 × 1.3 μm) search box. In the case of a uniformly random spatial distribution, the number of neighboring molecules present at a given distance increases linearly with increasing distance (Fig. 4 A). The distribution of intermolecular distances of motile IP3Rs (Fig. 4 B) was closely similar to this linear relationship, as would be expected if their Brownian motion caused them to be randomly distributed throughout the ER. In marked contrast, the spacings between immotile IP3Rs (after linking the tracks) showed a considerable excess at short (<400 nm) distances (Fig. 4 C). This is further illustrated in Fig. 4 D, which shows the difference between the observed distribution of spacings between immotile IP3Rs and the distribution that would expected if they were randomly distributed. Thus, a fraction of the immotile IP3Rs appear to be tightly grouped within <400 nm of neighboring, immotile IP3Rs.
Figure 4.
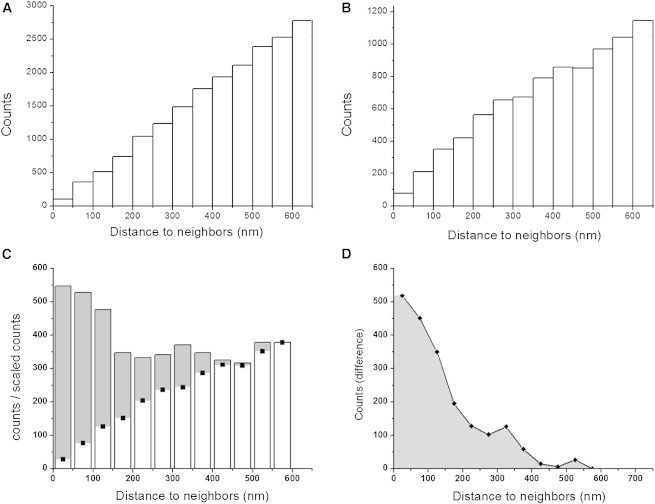
Cluster analysis of IP3R1 distribution. Panels show counts of instances in which two molecules were located within a given distance of one another. (A) Simulation showing the linear distribution of interparticle distances measured from a random spatial spread of 5000 particles. (B) Distribution of distances between motile IP3Rs (mean SLDs >200 nm) in live COS-7 cells. Molecule locations were defined as the mean position along each track, and data are from 9535 tracks in six experiments. (C) Histogram bars show the corresponding distribution of intermolecular distances between immotile IP3Rs. Black squares show the distribution of motile molecules, reproduced from (B) and scaled in amplitude to facilitate comparison. Gray shading indicates the excess numbers of immotile IP3Rs at close interneighbor distances above that expected for a random distribution. (D) Difference between the observed distribution of intermolecular distances of immotile IP3Rs and the scaled distribution of motile IP3Rs.
Nanoarchitecture of immotile IP3R clusters
We then identified putative clusters of immotile IP3R1 by filtering the data set to include only those molecules that met all of the following criteria: 1), IP3Rs with mean SLDs ≤ 84 nm; 2), molecules with ≥2 immotile neighbors within a 700 nm search radius; and 3), a mean distance of ≤ 250 nm to these surrounding neighbors. A representative spatial map of clusters identified in this way within the periphery of a COS-7 cell is shown in Fig. 5 A overlaid on the overall distribution of all IP3R tracks. Pooled over six cells, we identified 70 clusters from a total population of 3331 immotile molecules. To evaluate whether these putative clusters might arise simply from random coincidence, we used the population of rapidly motile IP3Rs as a control, on the premise that their locations (mean positions along their tracks) would be randomized by Brownian motion. Using the same identification criteria and after normalizing for differences in molecule density by randomly sampling equivalent numbers of motile and immotile molecules from each record, we identified 25 apparent clusters, considering the same total number (3331) of rapidly motile molecules in the same cells.
Figure 5.
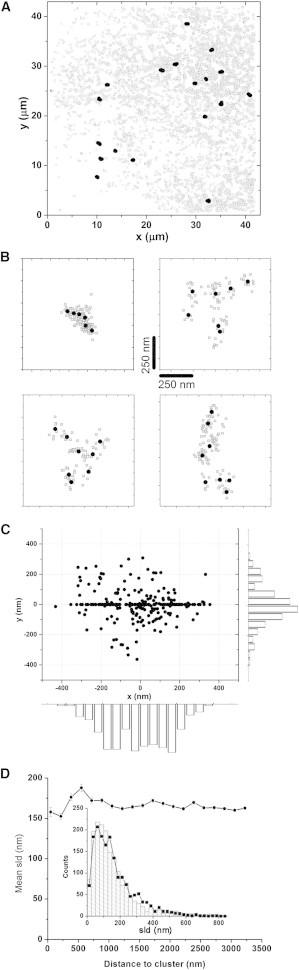
Clusters of immobile IP3Rs. (A) Gray circles denote the mean positions of all IP3R molecule tracks located in a live COS-7 cell. Black dots indicate the positions of clusters of immotile IP3Rs located by criteria as described in the text. (B) Four representative examples of individual IP3R clusters shown at high magnification. Black dots show the positions of individual molecules derived as the mean of all localizations along their tracks (gray circles). (C) The scatterplot is a superposition of IP3R molecules within 70 clusters after aligning the centroids of each cluster and rotating their long axes to lie along the x axis. Bar graphs indicate the numbers of molecules at different distances along the long and short axes to illustrate the mean density distribution of molecules within clusters. The plot along the short axis omits those molecules used to define the long axis of each cluster. (D) Diffusion of motile IP3Rs is not altered in the vicinity of clusters of immotile IP3Rs. The main graph plots mean SLDs (n > 79,000) of all tracks within 3.5 μm of identified clusters (n = 70) as a function of distance from the centroid of the cluster. Inset shows the distributions of SLDs recorded within 334 nm of cluster centroids (histogram bars) and at distances 1.33–1.67 μm away (black symbols and lines).
Fig. 5 B shows magnified views of a representative cluster of immotile IP3Rs identified as above, plotting the mean localizations of each immotile molecule (black dots) and the individual localizations from which the mean positions were obtained (open circles). The distributions of IP3Rs within individual clusters typically were not circularly symmetrical, and instead clusters were elongated at random orientations, as might be expected from the location of IP3Rs along strands of the ER. Fig. 5 C shows an overlay of IP3R localizations obtained from 70 clusters that were formed by aligning their center positions (mean position of molecules within each cluster) and rotating them so that the long axis of each cluster aligned on the x axis. The mean distributions of molecules approximate Gaussian functions of distance from the cluster center, with widths (standard deviations (SDs)) of 195.2 nm along the long axis and 86.84 nm along the short axis.
Motilities of IP3Rs near cluster regions
Clustering of immotile IP3Rs might arise if these molecules are bound to a finite number of fixed anchoring sites, or it might result from a more general environmental influence, such as greatly reduced diffusion within a localized region of ER membrane that has formed a diffusion trap. To discriminate between these possibilities, we examined the motilities of IP3R tracks that passed through or close to (<3.5 μm) clusters, after excluding tracks of those molecules identified as forming the clusters themselves. Fig. 5 D plots SLDs along these tracks as a function of the mean distance of a molecule during that displacement from the centroid of the cluster. The mean data derived from 70 clusters showed little apparent change in SLDs in proximity to a cluster, and the distributions of SLDs within 2 pixels (334 nm) of cluster centroids (Fig. 5 D, inset and histogram) and between 8 and 10 pixels (1.33–1.67 μm) away (Fig. 5 D, inset, line, and symbol) overlapped closely. Thus, with the exception of those immotile IP3Rs that defined a cluster, other IP3Rs in the vicinity of a cluster did not show appreciably different diffusional motilities. A possible caveat to this conclusion is that we analyzed motility only as a single x-y plane, and it is possible that IP3Rs could diffuse freely above or beneath a cluster within the sheet of membrane on the opposite side of the ER lumen from the cluster. However, even if this were the case, we might still expect to detect an overall slowing if the fraction of molecules in the same sheet of membrane as the cluster showed a reduced motility in its vicinity.
Activation of IP3/Ca2+ signaling does not acutely alter IP3R motility or distribution
IP3Rs in the membrane of excised nuclei have been proposed to undergo clustering in response to IP3 within just a few seconds (24). We therefore looked for possible changes in the diffusional motility and spatial distribution of IP3Rs following activation of endogenous IP3 signaling in COS-7 cells. We tracked single-molecule displacements for 750 s at baseline and then for a further 750 s immediately after adding ATP (100 μM) to the bathing solution to activate endogenous IP3-linked purinergic receptors (40). In separate experiments with fluo-8-loaded cells, this concentration of ATP in Ca2+-free medium evoked robust intracellular Ca2+ signals that persisted for ∼1 min (Fig. 6 A). Plots of MSD versus lag from cells before and after addition of ATP were linear and showed no difference in slope (Fig. 6 B). Mean aggregate diffusion coefficients derived from six cells showed no significant difference (before ATP: 0.0956 ± 0.006 μm2 s−1; after ATP: 0.0951 ± 0.007 μm2 s−1; difference not statistically significant, p ≫ 0.05). To examine whether ATP treatment induced clustering of IP3Rs, we performed a near-neighbor distribution analysis of immotile IP3Rs, as in Fig. 4 C. The distributions of intermolecular spacings were closely similar before and after ATP application (Fig. 6 C). Moreover, we found no difference in the numbers of clusters of immotile IP3Rs identified using the same criteria as in Fig. 5 (18.7 clusters per cell in control, 18.3 clusters per cell after ATP; n = 6 cells).
Figure 6.
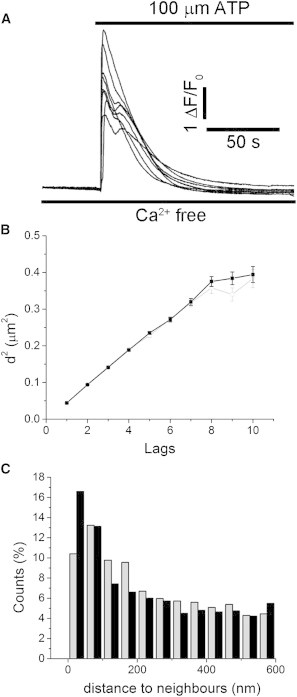
Activation of IP3/Ca2+ signaling has little effect on the motility and spatial distribution of IP3Rs. (A) Superimposed traces show Ca2+-dependent changes in fluo-8 fluorescence from six COS-7 cells in response to bath application of 100 μM ATP in Ca2+-free extracellular solution. (B) The plot shows the MSD of molecules from their origin as a function of increasing lag. The data shown are from a single cell and are representative of results from six cells. Open gray symbols are measurements before application of ATP, and black symbols are measurements after ATP. (C) Distributions of mean interneighbor distances of immotile IP3Rs (mean SLD < 84 nm) before (gray bars) and after (black bars) addition of ATP. Data were pooled from six cells.
Discussion
Ca2+ imaging experiments in numerous cell types have shown that local IP3-mediated Ca2+ signals (Ca2+ puffs) arise at just a few discrete sites within a cell, which are thought to represent tight clusters of several IP3R channels (4,9–11). For example, SH-SY5Y neuroblastoma cells, which express predominantly type 1 IP3R, were estimated to contain <20 discrete Ca2+ release sites, totaling no more than 100 or 200 functional IP3Rs per cell (27). In comparison, estimates of the number of IP3R molecules within a cell range as high as several thousands (41) (E.J.A. Taylor and C.W. Taylor, personal communication), and immunostaining shows an extensive distribution throughout the ER (Fig. 1 G) (11,22). Moreover, several previous studies that applied fluorescence recovery after photobleaching (FRAP) techniques to examine the motility of overexpressed fluorescent protein-tagged IP3Rs in intact cells indicated that a substantial proportion of IP3Rs are freely diffusible within the ER membrane (14–22). This is markedly different from the functional puff sites that remain at essentially fixed locations over a period of minutes (13). Here, we employed single-molecule superresolution imaging to gain insights into how the subcellular localization and motility of individual IP3R proteins might underlie the distribution of functionally active channels and puff sites.
As with all experiments involving heterologous expression of a fluorescently tagged protein, there are caveats as to whether the expressed protein faithfully mimics the behavior of the native protein, and in particular whether overexpression might overwhelm the cellular processes that determine the normal localization and motility of the native protein. However, we found that superresolution snapshots of fixed COS-7 cells showed similar distributions of immunostained native IP3R1s and Eos-tagged, overexpressed IP3R1s, and that the fraction of motile receptors remained approximately constant over a 3-fold range of expression levels. Moreover, GFP-tagged IP3Rs expressed using a construct closely similar to ours have been reported to be functionally active (30), and two reports (16,42) described little difference in FRAP measurements of IP3R motility across a wide range of expression levels, providing reassurance that overexpression would not be a major concern.
From single-molecule tracking, we estimate that ∼70% of the fluorescently tagged IP3R1s we expressed in COS-7 cells were motile, with an apparent two-dimensional diffusion coefficient of 0.095 ± 0.005 μm2 s−1. This diffusion coefficient is well within the extensive range (0.01–0.3 μm2 s−1) reported from FRAP experiments in various cell types (see Table S2 of Pantazaka and Taylor (42)), but is ∼5-fold greater than that derived from previous FRAP measurements of the mobile fraction of EGFP-tagged IP3R1 expressed in COS-7 cells (42). The reason for this discrepancy is not clear, but it may involve both methodological differences and fundamental differences between measurements of single-particle and collective (population) diffusion coefficients in the presence of binding traps (43).
We found that ∼30% of the expressed IP3Rs were essentially immotile, with mean SLDs similar to those measured in fixed cells and thus attributable to localization jitter. The spatial distribution of these immotile IP3Rs showed an excess of closely neighboring molecules over that predicted for a random distribution, and we identified clusters of several molecules spaced within a few hundred nanometers of one another. Representative examples of localizations within individual clusters are shown in Fig. 5 B, although these should not be interpreted as necessarily depicting the true numbers of IP3R proteins present in a cluster. Native, unlabeled IP3R obviously would not be observed, and it is possible that not all of the mEos-tagged molecules were activated during the imaging sequence. Conversely, the tetrameric nature of IP3R and the fact that an mEos molecule may undergo multiple reactivations (38) would tend to result in an overcount, even after correcting for photoblinking artifacts. Nevertheless, the composite overlay Fig. 5 C should provide a representative view of the average dimensions of stationary IP3R clusters at submicron resolution.
In principle, clustering of immotile IP3Rs might result from their binding to a limited number of anchoring sites (e.g., cytoskeleton-associated proteins) or from an environmental effect that greatly slows down diffusional motility, such as a local change in ER membrane lipid composition. We favor the first possibility because tracks of IP3Rs that passed through or nearby identified clusters showed no evidence of a slowing of diffusional motility. A related question concerns the kinetics of IP3R binding to anchoring sites. The very fact that we observe clusters of immotile, fluorescently tagged, exogenous IP3Rs suggests that this binding must be reversible over a timescale of hours; otherwise, the sites would be permanently occupied by native (nonfluorescent) IP3Rs and hence would be inaccessible. However, our ability to discern an interchange of IP3Rs at cluster sites is severely limited by the short time (a few seconds) in which the Eos reporter on a given molecule remains fluorescent.
It has been proposed that IP3R clusters are dynamic and that IP3 regulates the assembly and function of Ca2+ puff sites under physiological conditions (24). We previously argued against this hypothesis on the basis that the low density and restricted motility of functional IP3Rs (i.e., those that give rise to Ca2+ signals) are incompatible with dynamic clustering on a timescale that is fast enough to account for the short latencies (a few hundred microseconds) of puffs evoked by strong flash photorelease of IP3 (12). On the other hand, the results presented here show a high density of freely motile IP3R proteins, suggesting that aggregation of IP3Rs into clusters could occur within hundreds of milliseconds if IP3 were to create a diffusion trap by substantially increasing the affinity between IP3Rs and putative anchoring sites. Nevertheless, we do not believe this to be the case in light of our observations of clusters of immotile IP3Rs in quiescent cells, as well as our failure to observe any changes in diffusional motility or spatial clustering after stimulating cells with ATP to activate the endogenous IP3/Ca2+ signaling pathway.
Given that only a tiny fraction of the IP3Rs in a cell (<5% based on the estimates in SHSY-5Y cells given above) appear to underlie the generation of calcium puffs, a search for those IP3Rs presents a needle-in-a-haystack problem. Although thus far we have been unable to directly correlate the clusters of small numbers of immotile IP3Rs we describe here with active calcium release sites, we believe they are promising candidates for the clusters underlying puff sites. In particular, the IP3R clusters have dimensions (extending over roughly 300–400 nm) that are compatible with estimates of the size of clusters of functional IP3Rs underlying puff sites made via functional Ca2+ imaging, based on simulations of the spatial spread of the Ca2+ fluorescence signal during puffs (44) and on movements of the centroid of the Ca2+ signal during puffs (13). We thus propose the parsimonious explanation that a small fraction of IP3R channels are stably aggregated under resting conditions. Unlike the greater numbers of motile IP3Rs, these clustered IP3Rs are functional. In response to increased [IP3], their openings are concerted via CICR to generate puffs following a brief latency that is determined largely by stochastic Ca2+ activation (45) rather than diffusional aggregation. The functional role of the substantial majority of freely motile IP3Rs remains a mystery.
Acknowledgments
We thank Kevin Foskett (University of Pennsylvania) for providing the type 1 IP3R construct, Xiaowei Zhuang (Harvard University) for providing the Insight3 software and for helping us establish superresolution imaging techniques, and Colin Taylor (Cambridge University) for providing unpublished radioligand data on the number of IP3Rs within a cell.
This work was supported by National Institutes of Health grants GM 048071 and GM 065830 to I.P., and GM 100201 to I.F.S.
Footnotes
Ian F. Smith and Divya Swaminathan contributed equally to this work.
Supporting Material
The ER network was visualized by autofluorescence, and an average of 100 frames is shown in green. Red dots indicate individual red-converted mEos2-IP3R1 molecules.
References
- 1.Foskett J.K., White C., Mak D.O. Inositol trisphosphate receptor Ca2+ release channels. Physiol. Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge M.J., Lipp P., Bootman M.D. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 3.Bezprozvanny I., Watras J., Ehrlich B.E. Bell-shaped calcium-response curves of inositol trisphosphate and calcium-gated channels from endoplasmic reticulum of cerebellum. Nature. 1991;351:751–754. doi: 10.1038/351751a0. [DOI] [PubMed] [Google Scholar]
- 4.Yao Y., Choi J., Parker I. Quantal puffs of intracellular Ca2+ evoked by inositol trisphosphate in Xenopus oocytes. J. Physiol. 1995;482:533–553. doi: 10.1113/jphysiol.1995.sp020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipp P., Niggli E. A hierarchical concept of cellular and subcellular Ca(2+)-signalling. Prog. Biophys. Mol. Biol. 1996;65:265–296. doi: 10.1016/s0079-6107(96)00014-4. [DOI] [PubMed] [Google Scholar]
- 6.Parker I., Choi J., Yao Y. Elementary events of inositol trisphosphate-induced calcium liberation in Xenopus oocytes: hot spots, puffs and blips. Cell Calcium. 1996;20:105–121. doi: 10.1016/s0143-4160(96)90100-1. [DOI] [PubMed] [Google Scholar]
- 7.Shuai J.W., Jung P. Optimal ion channel clustering for intracellular calcium signaling. Proc. Natl. Acad. Sci. USA. 2003;100:506–510. doi: 10.1073/pnas.0236032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dargan S.L., Parker I. Buffer kinetics shape the spatiotemporal patterns of inositol trisphosphate-evoked calcium signals. J. Physiol. 2003;553:775–788. doi: 10.1113/jphysiol.2003.054247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith I.F., Wiltgen S.M., Parker I. Localization of puff sites adjacent to the plasma membrane: Functional and spatial characterization of calcium signaling in SH-SY5Y cells utilizing membrane-permeant caged inositol trisphosphate. Cell Calcium. 2009;45:65–76. doi: 10.1016/j.ceca.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas D., Lipp P., Bootman M.D. Hormone-evoked elementary Ca2+ signals are not stereotypic, but reflect activation of different size channel clusters and variable recruitment of channels within a cluster. J. Biol. Chem. 1998;273:27130–27136. doi: 10.1074/jbc.273.42.27130. [DOI] [PubMed] [Google Scholar]
- 11.Tovey S.C., de Smet P., Bootman M.D. Calcium puffs are generic inositol trisphosphate-activated elementary calcium signals and are downregulated by prolonged hormonal stimulation to inhibit cellular calcium responses. J. Cell Sci. 2001;114:3979–3989. doi: 10.1242/jcs.114.22.3979. [DOI] [PubMed] [Google Scholar]
- 12.Smith I.F., Wiltgen S.M., Parker I. Ca(2+) puffs originate from preestablished stable clusters of inositol trisphosphate receptors. Sci. Signal. 2009;2:ra77. doi: 10.1126/scisignal.2000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiltgen S.M., Smith I.F., Parker I. Superresolution localization of single functional inositol trisphosphate receptor channels utilizing calcium flux as a readout. Biophys. J. 2010;99:437–446. doi: 10.1016/j.bpj.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalmers M., Schell M.J., Thorn P. Agonist-evoked inositol trisphosphate receptor (IP3R) clustering is not dependent on changes in the structure of the endoplasmic reticulum. Biochem. J. 2006;394:57–66. doi: 10.1042/BJ20051130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruttwell C., Bernard J., Mauger J.P. Dynamics of the inositol trisphosphate receptor during polarization of MDCK cells. Biol. Cell. 2005;97:699–707. doi: 10.1042/BC20040503. [DOI] [PubMed] [Google Scholar]
- 16.Ferreri-Jacobia M., Mak D.O., Foskett J.K. Translational mobility of the type 3 inositol 1,4,5-trisphosphate receptor Ca2+ release channel in endoplasmic reticulum membrane. J. Biol. Chem. 2005;280:3824–3831. doi: 10.1074/jbc.M409462200. [DOI] [PubMed] [Google Scholar]
- 17.Fukatsu K., Bannai H., Mikoshiba K. Lateral diffusion of inositol 1,4,5-trisphosphate receptor type 1 is regulated by actin filaments and 4.1N in neuronal dendrites. J. Biol. Chem. 2004;279:48976–48982. doi: 10.1074/jbc.M408364200. [DOI] [PubMed] [Google Scholar]
- 18.Gibson C.J., Ehrlich B.E. Inositol 1,4,5-trisphosphate receptor movement is restricted by addition of elevated levels of O-linked sugar. Cell Calcium. 2008;43:228–235. doi: 10.1016/j.ceca.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwai M., Tateishi Y., Mikoshiba K. Molecular cloning of mouse type 2 and type 3 inositol 1,4,5-trisphosphate receptors and identification of a novel type 2 receptor splice variant. J. Biol. Chem. 2005;280:10305–10317. doi: 10.1074/jbc.M413824200. [DOI] [PubMed] [Google Scholar]
- 20.Tateishi Y., Hattori M., Mikoshiba K. Cluster formation of inositol 1,4,5-trisphosphate receptor requires its transition to open state. J. Biol. Chem. 2005;280:6816–6822. doi: 10.1074/jbc.M405469200. [DOI] [PubMed] [Google Scholar]
- 21.Tojyo Y., Morita T., Tanimura A. The clustering of inositol trisphosphate receptors is triggered by inositol trisphosphate binding and facilitated by depletion of the calcium store. J. Pharmacol. Sci. 2008;107:138–150. doi: 10.1254/jphs.08021fp. [DOI] [PubMed] [Google Scholar]
- 22.Wilson B.S., Pfeiffer J.R., Wojcikiewicz R.J. Calcium-dependent clustering of inositol 1,4,5-trisphosphate receptors. Mol. Biol. Cell. 1998;9:1465–1478. doi: 10.1091/mbc.9.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins S.C., Salter R.D. Functional connectivity between immune cells mediated by tunneling nanotubules. Immunity. 2005;23:309–318. doi: 10.1016/j.immuni.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Taufiq Ur R., Skupin A., Taylor C.W. Clustering of inositol trisphosphate receptors by inositol trisphosphate retunes their regulation by inositol trisphosphate and calcium. Nature. 2009;458:655–659. doi: 10.1038/nature07763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bootman M., Niggli E., Lipp P. Imaging the hierarchical Ca2+ signalling system in HeLa cells. J. Physiol. 1997;499:307–314. doi: 10.1113/jphysiol.1997.sp021928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bootman M.D., Berridge M.J., Lipp P. Cooking with calcium: the recipes for composing global signals from elementary events. Cell. 1997;91:367–373. doi: 10.1016/s0092-8674(00)80420-1. [DOI] [PubMed] [Google Scholar]
- 27.Smith I.F., Parker I. Imaging the quantal substructure of single inositol trisphosphate receptor channel activity during calcium puffs in intact mammalian cells. Proc. Natl. Acad. Sci. USA. 2009;106:6404–6409. doi: 10.1073/pnas.0810799106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker I., Smith I.F. Recording single-channel activity of inositol trisphosphate receptors in intact cells with a microscope, not a patch clamp. J. Gen. Physiol. 2010;136:119–127. doi: 10.1085/jgp.200910390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manley S., Gillette J.M., Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 30.Shinohara T., Michikawa T., Mikoshiba K. Mechanistic basis of bell-shaped dependence of inositol 1,4,5-trisphosphate receptor gating on cytosolic calcium. Proc. Natl. Acad. Sci. USA. 2011;108:15486–15491. doi: 10.1073/pnas.1101677108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bates M., Huang B., Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Codling E.A., Plank M.J., Benhamou S. Random walk models in biology. J. R. Soc. Interface. 2008;5:813–834. doi: 10.1098/rsif.2008.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang K., Osakada Y., Cui B. Single-molecule imaging of NGF axonal transport in microfluidic devices. Lab Chip. 2010;10:2566–2573. doi: 10.1039/c003385e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saxton M.J. Single-particle tracking: the distribution of diffusion coefficients. Biophys. J. 1997;72:1744–1753. doi: 10.1016/S0006-3495(97)78820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weigel A.V., Simon B., Krapf D. Ergodic and nonergodic processes coexist in the plasma membrane as observed by single-molecule tracking. Proc. Natl. Acad. Sci. USA. 2011;108:6438–6443. doi: 10.1073/pnas.1016325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Annibale P., Vanni S., Radenovic A. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS ONE. 2011;6:e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veatch S.L., Machta B.B., Baird B.A. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE. 2012;7:e31457. doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Annibale P., Vanni S., Radenovic A. Identification of clustering artifacts in photoactivated localization microscopy. Nat. Methods. 2011;8:527–528. doi: 10.1038/nmeth.1627. [DOI] [PubMed] [Google Scholar]
- 40.James G., Butt A.M. P2Y and P2X purinoceptor mediated Ca2+ signalling in glial cell pathology in the central nervous system. Eur. J. Pharmacol. 2002;447:247–260. doi: 10.1016/s0014-2999(02)01756-9. [DOI] [PubMed] [Google Scholar]
- 41.Tovey S.C., Dedos S.G., Taylor C.W. Selective coupling of type 6 adenylyl cyclase with type 2 IP3 receptors mediates direct sensitization of IP3 receptors by cAMP. J. Cell Biol. 2008;183:297–311. doi: 10.1083/jcb.200803172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pantazaka E., Taylor C.W. Differential distribution, clustering, and lateral diffusion of subtypes of the inositol 1,4,5-trisphosphate receptor. J. Biol. Chem. 2011;286:23378–23387. doi: 10.1074/jbc.M111.236372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pando B., Dawson S.P., Pearson J.E. Messages diffuse faster than messengers. Proc. Natl. Acad. Sci. USA. 2006;103:5338–5342. doi: 10.1073/pnas.0509576103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuai J., Parker I. Optical single-channel recording by imaging Ca2+ flux through individual ion channels: theoretical considerations and limits to resolution. Cell Calcium. 2005;37:283–299. doi: 10.1016/j.ceca.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 45.Dickinson G.D., Swaminathan D., Parker I. The probability of triggering calcium puffs is linearly related to the number of inositol trisphosphate receptors in a cluster. Biophys. J. 2012;102:1826–1836. doi: 10.1016/j.bpj.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The ER network was visualized by autofluorescence, and an average of 100 frames is shown in green. Red dots indicate individual red-converted mEos2-IP3R1 molecules.


