
Keywords: Parasite diversity, Virus, Helminths, Ectoparasites, Chiroptera, Asia, Distribution range, Fragmentation
Highlights
-
•
We investigated the parasite and viral richness in bats from Southeast Asia.
-
•
We tested determinants of parasite richness using phylogenetic comparative analyses.
-
•
Viral and parasite richness were explained by area distribution shape of bats.
-
•
Future changes in bat distribution may alter their viral and parasite richness.
Abstract
Interest in bat-borne diseases and parasites has grown in the past decade over concerns for human health. However, the drivers of parasite diversity among bat host species are understudied as are the links between parasite richness and emerging risks. Thus, we aimed at exploring factors that explain macro and microparasite species richness in bats from Southeast Asia, a hotspot of emerging infectious diseases. First, we identified bat species that need increased sampling effort for pathogen discovery. Our approach highlights pathogen investigation disparities among species within the same genus, such as Rhinolophus and Pteropus. Secondly, comparative analysis using independent contrasts method allowed the identification of likely factors explaining parasite and viral diversity of bats. Our results showed a key role of bat distribution shape, an index of the fragmentation of bat distribution, on parasite diversity, linked to a decrease for both viral and endoparasite species richness. We discuss how our study may contribute to a better understanding of the link between parasite species richness and emergence.
1. Introduction
Incidence of emerging infectious diseases (EIDs) has dramatically increased in recent decades (Jones et al., 2008). A majority of EIDs are zoonoses and most of them originate in wildlife (zoonotic spillover). EID events are often due to environmental alteration, including agricultural intensification and habitat modification, global trade and travel (Cunningham, 1996; Daszak et al., 2000). Several studies and reviews have suggested that the risk of disease transmission from wildlife to humans should increase with biodiversity loss and the expansion of human populations (Dobson et al., 2006; Keesing et al., 2009), as humans will get into contact with a large pool of known and unknown zoonotic pathogens from wildlife (Mahy and Brown, 2000; Murray and Daszak, 2013). Bacteria and rickettsia represent the majority of EIDs, viral and prion pathogens cause 25.4% and other parasites 20.3% of EID events (Jones et al., 2008).
Among known reservoir species of viral EIDs, bats, which represent 20% of mammal species (Simmons, 2005), play an important role in the maintenance and spread of various viral diseases (Sulkin and Allen, 1974; Ghatak et al., 2000; McColl et al., 2000; Olival et al., 2012; Luis et al., 2013), including members of the alphaviruses, flaviviruses, paramyxoviruses, rhabdoviruses, coronaviruses and arenaviruses among others. Several notable bat-borne viruses are a public health concern in Southeast Asia (SEA) including Nipah virus, lyssaviruses and Severe Acute Respiratory Syndrome coronavirus (Mackenzie et al., 2003). While bats are increasingly viewed as a threat to human health, these mammals have important roles in ecosystems such as pollination, seed dispersal and predation on insects. They are excellent bioindicators of environmental changes as they are sensitive to a wide range of anthropogenic disturbances such as urbanization, agricultural intensification, habitat loss and fragmentation (Clarke et al., 2005; Jones et al., 2008). Understanding the ecology of these potential reservoirs of zoonotic pathogens is needed for improving management of bats and their habitats, ultimately ensuring the health of humans, livestock and wildlife species, while keeping their functional roles in the ecosystems (Breed et al., 2006).
The potential of human–bat interactions, either direct or indirect, may be underappreciated and greater than expected for some species. While bats rarely seek direct contact with humans, they often roost in or near human dwellings, which can lead to accidental contact or exposure to bat excreta. For example, Nipah virus has been transmitted to people who have eaten bat-contaminated date palm sap (Luby et al., 2006).
Wild animals are known to host different pathogens at the same time (Petney and Andrews, 1998; Drake and Bundy, 2000; Bordes et al., 2008; Bordes and Morand, 2011). Polyparasitism (or multiple infections) and infection dynamics in host species are important features to understand the mechanisms of EID; the risk of disease transmission may depend on the pathogen richness found in natural reservoir species (Wolfe et al., 2005). Morse (1993) termed this pathogen diversity in wildlife hosts the ‘zoonotic pool’. In the literature, most studies emphasized the links between biodiversity loss and the risk of pathogen emergence and transmission (Keesing et al., 2009; Derne et al., 2011), whereas very few investigated the role of parasite diversity, or the size of the ‘zoonotic pool’, in the risk of emergence.
Investigating parasitic and microbial diversity in bat species may help to identify species that are reservoir sources of a greater diversity of pathogens and to understand factors influencing this richness, particularly those related to life or ecological traits. The determinants of parasite diversity in wildlife may be linked to their biogeography (e.g., latitude, distribution area), ecology (e.g., density, migration), life-history traits (e.g., longevity and fecundity) or immunity (e.g., white blood cell counts, spleen size, immune gene diversity) (Morand and Poulin, 2000; Wegner et al., 2003; Nunn et al., 2003b; Guernier et al., 2004; Ezenwa, 2004; Bordes et al., 2007; Šimková et al., 2008; Pedersen and Grieves, 2008; Turmelle and Olival, 2009; Bordes and Morand, 2011; Nunn, 2012; Luis et al., 2013). Some of these determinants can also influence the probability of contact of bats with humans and thus the potential risk of contamination and transmission; a widely distributed bat species living at high density has a greater probability of repeated contacts with humans than a species living in low density with a restricted distribution. This may be particularly true for synanthropic species that appear to be generalist in their ecology and rich in the parasite diversity they harbour (Herbreteau et al., 2012). Finally, a question rarely investigated about species richness is: “is there any correlation between microparasite (viruses, parasitic bacteria, protists, fungi) richness and macroparasite (helminths and arthropods) richness in bats?” (Bordes et al., 2008; Turmelle and Olival, 2009). The diversity of microparasites (e.g., virus and bacteria) may then depend on the diversity of macroparasites (e.g., helminths) through the activation and maintenance of different pathways of the immune system (Bordes and Morand, 2011; Ezenwa and Jolles, 2011). A positive correlation may suggest that hosts with high macroparasite diversity also harbour high viral and bacterial richness.
We focused our study on Southeast Asia (SEA), a hotspot of biodiversity and EIDs with pandemic potential (Myers et al., 2000; Jones et al., 2008; Coker et al., 2011). It is also a natural laboratory to study the evolutionary history (Guillén et al., 1997) and the impact of high human environmental pressures (Sodhi and Brook, 2006; Clements et al., 2006; Stibig et al., 2007; Wilcove et al., 2013). For our study, we defined parasite species richness (PSR) as the total number of parasite species such as microparasites as well as macroparasites identified in a given host (Poulin and Morand, 2004; Bordes et al., 2007) at the regional scale of SEA. In the present study we aimed at investigating the likely factors that may explain PSR in bats from SEA. From the literature we compiled information on parasitic and infectious agents in SEA bats found, as well as information available on their life-history and ecological traits. Then, we tested potential factors that may explain the whole pool of parasite diversity in bats using phylogenetic comparative analyses and model selection.
We tested hypotheses related to (1) influences of the size and shape of the geographic distribution (index of the fragmentation of bat distribution, defined as the ratio of the surface of the distribution area to the edge length); (2) the size of bat colonies and the number of breeding seasons. We hypothesized that a large area should favor the accumulation of parasites and that an increase of fragmentation of bat distribution, potentially linked with increased habitat diversity, lead to an increase of the overall parasite diversity. We also hypothesized that bats species living in large colonies with a high number of breeding seasons will support a larger pool of parasite species due to large susceptible populations.
2. Materials and methods
2.1. Data on bats and their parasites
Information on bats and their parasites were compiled from the literature. Only Southeast Asian countries were selected: Brunei; Cambodia; Indonesia; Lao People’s Democratic Republic; Malaysia; Myanmar; Philippines; Singapore; Thailand; Timor; and Vietnam.
A total of 292 species bats inventoried in SEA were included in the database. Several variables were documented for each host species: distribution size; distribution shape (border edge, as a measure of the fragmentation of bat distribution area estimated by the ratio of the distribution circumference to the distribution area size); breeding seasons per year; colony size and average adult body mass (Table 1). The distribution shape ranges from 0 (cylindrical and compact distribution area) to 1 (fragmented distribution area) (Fortin et al., 2005). Information was gathered from several sources: IUCN Red List (http://www.iucnredlist.org/); Wilson and Reeder «Mammal Species of the World» (http://www.bucknell.edu/msw3/); and Harrison Institute (http://www.sc.psu.ac.th/batdb/index.asp). In addition, published articles were obtained through match searches using Google Scholar (http://scholar.google.com) and the Web of Knowledge (http://apps.webofknowledge.com/).
Table 1.
List of bat species with information on parasite richness (endoparasitic helminths, ectoparasitic arthropods, virus species, virus genus and virus family), sampling effort (SE Para = number of individual bats investigated for a parasite, SE virus = number of individual bats investigated for a virus), investigation effort (Pub para = number of publications citing a parasitic of microbial agent in a given bat species, Pub bats = number of publication of a bat species), body mass (g), distribution size (km2), distribution shape (area range/circumference), colony size (0 = ⩽10; 1 = ⩽100; 2 = ⩽1000; 3 = >1000), roosting behavior (0 = solitary; 1 = depend; 2 = gregarious), breeding seasons (1 = one season; 2 = two seasons; 3 = all year long) and IUCN status (0 = least concern; 1 = near threatened; 2 = vulnerable).
| Bat species | Endoparasite | Ectoparasite | Virus sp. | Virus genus | Virus fam | SE Para | SE Virus | Pub para | Pub bats | Body mass | Distribution size | Distribution shape | Colony size | Roost | Breed season | IUCN status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acerodon leucotis | 1 | 2 | 400 | 12832 | 0.1601 | 1 | 2 | 2 | ||||||||
| Aethalops alecto | 5 | 5 | 3 | 23.1 | 686808 | 0.0195 | 0 | 0 | 0 | |||||||
| Aselliscus stoliczkanus | 1 | 1 | 1 | 8 | 6.1 | 1368853 | 0.0069 | 2 | 2 | 1 | 0 | |||||
| Balionycteris maculata | 3 | 50 | 5 | 4 | 13.5 | 574183 | 0.0161 | 0 | 2 | 0 | ||||||
| Cheiromeles torquatus | 1 | 10 | 15 | 6 | 181.5 | 1338608 | 0.0162 | 2 | 2 | 0 | ||||||
| Chironax melanocephalus | 4 | 4 | 1 | 18 | 178954 | 0.0298 | 0 | 2 | 2 | 0 | ||||||
| Coelops frithii | 2 | 2 | 1 | 3.7 | 2505073 | 0.0159 | 1 | 2 | 0 | |||||||
| Cynopterus brachyotis | 1 | 11 | 6 | 3 | 3 | 93 | 79 | 35 | 54 | 32.8 | 2715326 | 0.0145 | 0 | 1 | 2 | 0 |
| Cynopterus horsfieldii | 1 | 4 | 2 | 2 | 2 | 2 | 13 | 1 | 59 | 1495518 | 0.0153 | 2 | 2 | 2 | 0 | |
| Cynopterus sphinx | 1 | 1 | 1 | 81 | 81 | 1 | 128 | 47.7 | 6489257 | 0.0088 | 0 | 2 | 2 | 0 | ||
| Dobsonia exoleta | 1 | 2 | 0 | 249 | 181117 | 0.0468 | 0 | 2 | 0 | |||||||
| Dyacopterus spadiceus | 2 | 2 | 3 | 80 | 447874 | 0.0174 | 0 | 2 | 1 | |||||||
| Emballonura monticola | 5 | 5 | 0 | 35 | 1701771 | 0.0204 | 1 | 2 | 1 | 0 | ||||||
| Eonycteris major | 1 | 8 | 737545 | 0.0113 | 1 | 2 | ||||||||||
| Eonycteris spelaea | 3 | 20 | 5 | 4 | 4 | 246 | 44 | 33 | 25 | 58.4 | 3546214 | 0.0199 | 2 | 2 | 3 | 0 |
| Eptesicus dimissus | 1 | 1 | 1 | 4087 | 0.0673 | |||||||||||
| Eptesicus serotinus | 2 | 1 | 1 | 1 | 1 | 27 | 128 | 25 | 12129638 | 0.0087 | 0 | 1 | 0 | |||
| Glischropus tylopus | 1 | 1 | 2 | 3 | 4.5 | 2466827 | 0.0117 | 0 | 2 | 1 | 0 | |||||
| Harpyionycteris whiteheadi | 1 | 1 | 1 | 120.3 | 172958 | 0.0722 | 0 | 1 | 0 | |||||||
| Hesperoptenus blanfordi | 1 | 1 | 4 | 6.4 | 463170 | 0.0220 | 0 | 2 | 0 | |||||||
| Hipposideros armiger | 5 | 11 | 7 | 5 | 4 | 113 | 113 | 40 | 29 | 47 | 4496220 | 0.0093 | 0 | 1 | 1 | 0 |
| Hipposideros bicolor | 9 | 74 | 10 | 15 | 47.7 | 1594983 | 0.0187 | 2 | 2 | 1 | 0 | |||||
| Hipposideros cervinus | 14 | 2 | 3 | 9.3 | 1978116 | 0.0241 | 1 | 2 | 0 | |||||||
| Hipposideros cineraceus | 6 | 6 | 5 | 4.3 | 2214839 | 0.0124 | 0 | 2 | 1 | 0 | ||||||
| Hipposideros diadema | 1 | 7 | 2 | 2 | 2 | 2 | 10 | 9 | 39 | 3340330 | 0.0261 | 3 | 2 | 1 | 0 | |
| Hipposideros galeritus | 4 | 6 | 2 | 7.9 | 2358398 | 0.0166 | 0 | 2 | 1 | 0 | ||||||
| Hipposideros larvatus | 15 | 5 | 5 | 5 | 156 | 143 | 30 | 14 | 20.4 | 3306719 | 0.0119 | 2 | 2 | 1 | 0 | |
| Hipposideros lylei | 1 | 1 | 3 | 36 | 1018679 | 0.0116 | 1 | 2 | 1 | 0 | ||||||
| Hipposideros pomona | 1 | 2 | 2 | 2 | 388 | 388 | 9 | 9 | 6.5 | 2855362 | 0.0092 | 2 | 2 | 1 | 0 | |
| Hipposideros pratti | 1 | 1 | 9 | 37.5 | 2251718 | 0.0062 | 1 | 2 | 0 | |||||||
| Ia io | 1 | 1 | 1 | 1 | 3 | 50 | 53.2 | 1512964 | 0.0078 | 1 | 2 | 0 | ||||
| Kerivoula hardwickii | 1 | 15 | 2 | 7 | 3.9 | 4070770 | 0.0141 | 0 | 0 | 0 | ||||||
| Kerivoula minuta | 1 | 4 | 2 | 2 | 2.2 | 144409 | 0.0282 | 1 | ||||||||
| Kerivoula pellucida | 1 | 2 | 1 | 5 | 6 | 1221402 | 0.0170 | 0 | 2 | 1 | ||||||
| Macroglossus minimus | 6 | 1 | 1 | 1 | 3 | 1 | 7 | 10 | 16.5 | 3613476 | 0.0285 | 0 | 1 | 0 | ||
| Macroglossus sobrinus | 2 | 19 | 2 | 5 | 22.9 | 2449534 | 0.0120 | 0 | 1 | 3 | 0 | |||||
| Megaderma lyra | 3 | 7 | 10 | 113 | 50 | 6070972 | 0.0082 | 0 | 1 | 1 | 0 | |||||
| Megaderma spasma | 7 | 10 | 5 | 26.9 | 4082464 | 0.0193 | 0 | 1 | 1 | 0 | ||||||
| Miniopterus magnater | 5 | 3 | 2 | 189 | 164 | 50 | 6 | 13.4 | 3179606 | 0.0128 | 2 | 2 | 1 | 0 | ||
| Miniopterus medius | 11 | 11 | 0 | 9.5 | 1258546 | 0.0167 | 2 | 2 | 0 | |||||||
| Miniopterus pusillus | 6 | 2 | 2 | 474 | 425 | 27 | 10 | 8.4 | 1113321 | 0.0166 | 2 | 2 | 1 | 0 | ||
| Murina aurata | 1 | 1 | 1 | 1 | 1 | 2 | 5.1 | 314718 | 0.0262 | 0 | 1 | 0 | ||||
| Murina cyclotis | 1 | 1 | 7 | 7.6 | 2032608 | 0.0133 | 0 | 2 | 0 | |||||||
| Murina suilla | 1 | 4 | 1 | 4 | 3.8 | 1451218 | 0.0142 | 0 | 1 | 0 | ||||||
| Myotis adversus | 1 | 2 | 19 | 47.7 | 244183 | 0.0352 | 0 | 2 | 0 | |||||||
| Myotis chinensis | 1 | 1 | 1 | 1 | 1 | 3 | 6 | 27.8 | 2272978 | 0.0059 | 0 | 0 | 0 | |||
| Myotis hasseltii | 1 | 1 | 0 | 10 | 953379 | 0.0173 | 0 | 1 | 1 | 0 | ||||||
| Myotis horsfieldii | 6 | 2 | 1 | 1 | 8 | 8 | 9 | 3 | 8 | 2576798 | 0.1454 | 0 | 1 | 1 | 0 | |
| Myotis muricola | 1 | 1 | 5 | 3.8 | 4668165 | 0.0132 | 0 | 1 | 1 | 0 | ||||||
| Nyctimene minutus | 1 | 1 | 0 | 26128 | 0.0580 | 2 | ||||||||||
| Penthetor lucasi | 10 | 10 | 13 | 2 | 1296841 | 0.0126 | 2 | 2 | 0 | |||||||
| Philetor brachypterus | 1 | 1 | 1 | 13 | 13 | 1 | 0 | 8.2 | 1325800 | 0.0242 | 1 | 2 | 0 | |||
| Pipistrellus abramus | 5 | 2 | 2 | 203 | 203 | 6 | 36 | 4.5 | 3854526 | 0.0133 | 1 | 2 | 0 | |||
| Pipistrellus javanicus | 1 | 1 | 4 | 4.2 | 3727958 | 0.0168 | 1 | 2 | 0 | |||||||
| Pipistrellus stenopterus | 7 | 7 | 4 | 15.5 | 644892 | 0.0176 | 2 | 2 | 0 | |||||||
| Ptenochirus jagori | 1 | 1 | 1 | 16 | 16 | 1 | 6 | 85 | 272675 | 0.0569 | 0 | 0 | 2 | 0 | ||
| Pteropus giganteus | 2 | 5 | 3 | 3 | 184 | 184 | 9 | 96 | 1021 | 4021279 | 0.0065 | 2 | 2 | 0 | ||
| Pteropus hypomelanus | 1 | 3 | 4 | 4 | 3 | 689 | 689 | 22 | 37 | 438 | 527562 | 0.0625 | 1 | 2 | 1 | 0 |
| Pteropus vampyrus | 2 | 3 | 2 | 1 | 1 | 475 | 475 | 34 | 52 | 911.3 | 1953294 | 0.0283 | 1 | 0 | 1 | |
| Rhinolophus acuminatus | 1 | 9 | 1 | 3 | 11.6 | 1083857 | 0.0199 | 1 | 2 | 0 | ||||||
| Rhinolophus affinis | 6 | 8 | 2 | 2 | 2 | 292 | 266 | 25 | 13 | 12.9 | 5534663 | 0.0095 | 2 | 2 | 2 | 0 |
| Rhinolophus lepidus | 6 | 6 | 9 | 7.4 | 3526223 | 0.0076 | 0 | 2 | 1 | 0 | ||||||
| Rhinolophus macrotis | 2 | 3 | 4 | 2 | 2 | 51 | 46 | 9 | 12 | 5.6 | 2348803 | 0.0093 | 0 | 2 | 0 | |
| Rhinolophus malayanus | 1 | 1 | 7 | 2 | 6 | 6.7 | 1547550 | 0.0095 | 2 | 2 | 0 | |||||
| Rhinolophus marshalli | 1 | 3 | 5 | 6.3 | 656136 | 0.0099 | 0 | 2 | 0 | |||||||
| Rhinolophus pearsonii | 5 | 3 | 3 | 2 | 95 | 95 | 8 | 2 | 13 | 3191107 | 0.0076 | 1 | 2 | 1 | 0 | |
| Rhinolophus pusillus | 1 | 1 | 1 | 1 | 6 | 6 | 3 | 15 | 3.5 | 3989176 | 0.0144 | 1 | 2 | 1 | 0 | |
| Rhinolophus sinicus | 1 | 6 | 3 | 2 | 1993 | 1993 | 28 | 10 | 11.4 | 2177356 | 0.0097 | 0 | 1 | 0 | ||
| Rhinolophus stheno | 1 | 6 | 9 | 4 | 6.3 | 2216461 | 0.0129 | 2 | 2 | 0 | ||||||
| Rhinopoma microphyllum | 4 | 5 | 13 | 31 | 7183340 | 0.0060 | 2 | 2 | 1 | 0 | ||||||
| Rousettus amplexicaudatus | 9 | 3 | 3 | 3 | 51 | 51 | 14 | 9 | 64.5 | 4308473 | 0.0217 | 2 | 2 | 0 | ||
| Rousettus leschenaultii | 4 | 4 | 3 | 3 | 16 | 16 | 8 | 7 | 79.8 | 6798300 | 0.0093 | 2 | 2 | 0 | ||
| Saccolaimus saccolaimus | 3 | 3 | 14 | 57.5 | 1969538 | 0.0321 | 0 | 2 | 0 | |||||||
| Scotophilus kuhlii | 7 | 10 | 7 | 6 | 6 | 284 | 284 | 44 | 12 | 21.6 | 5254460 | 0.0173 | 1 | 2 | 1 | 0 |
| Tadarida jobensis | 1 | 1 | 0 | 25 | 2349741 | 0.0127 | 0 | |||||||||
| Tadarida mops | 6 | 7 | 9 | 21.9 | 616082 | 0.0147 | 1 | |||||||||
| Tadarida plicata | 11 | 4 | 4 | 4 | 524 | 524 | 20 | 3 | 13.9 | 2477230 | 0.0178 | 2 | 2 | 2 | 0 | |
| Taphozous longimanus | 1 | 5 | 6 | 24 | 22.5 | 4817713 | 0.0113 | 0 | 2 | 3 | 0 | |||||
| Taphozous melanopogon | 3 | 9 | 3 | 2 | 2 | 102 | 102 | 51 | 25 | 26 | 5654376 | 0.0147 | 3 | 2 | 1 | 0 |
| Taphozous theobaldi | 3 | 2 | 2 | 2 | 157 | 157 | 5 | 3 | 33.5 | 1047949 | 0.0140 | 1 | 2 | 0 | ||
| Thoopterus nigrescens | 1 | 1 | 0 | 84.5 | 184093 | 0.0469 | 0 | |||||||||
| Tylonycteris pachypus | 9 | 2 | 1 | 1 | 123 | 123 | 13 | 12 | 3.2 | 4194084 | 0.0152 | 1 | 2 | 1 | 0 | |
| Tylonycteris robustula | 1 | 7 | 10 | 12 | 5.8 | 2721850 | 0.0152 | 1 | 2 | 1 | 0 |
Data on parasite species richness (PSR), which is the number of parasite species described infecting a given host species, were obtained through searches on Web of Science. To identify relevant information binomial names of every Southeast Asian bat species were combined with related terms (parasite, ectoparasite, endoparasite, virus, bacteria, fungi, helminths, pathogen and disease). All resulting abstracts and available full texts were examined. We examined 964 publications from 1959 to July 2012 and identified 637 species of parasites in SEA.
In addition we used specific databases for SEA, such as the database of Armed Forces Research Institute of Medical Sciences (http://www.afpmb.org/content/welcome-literature-retrieval-system); the Liverpool database (http://www.zoonosis.ac.uk/eid2) for additional records of viral richness; and the British Natural History Museum database (http://www.nhm.ac.uk/research-curation/research/projects/host-parasites/database/index.jsp) for helminths.
2.2. Sampling effort and investigation effort
We measured sampling effort, or ascertainment bias, in two ways. First, we defined the ‘sampling effort’ as the sample size or number of individual bats tested for a given parasite or microbe screened (positive or negative). Second, the ‘investigation effort’ is the number of publications about a given parasite or microbe for a given host species, which represents the research effort for a given parasite investigation (usually corresponds to the number of pathogens’ screened). Parasite species richness (PSR), viral richness, investigation effort and sampling effort were log transformed in order to stabilize variance.
Residual values of the linear regression between log of PSR and sampling effort were calculated. We investigated residual variations among bat hosts in order to identify bat species that harbour more pathogens than expected by the regression model (i.e., bat species with positive residual values), or conversely, to identify bat species that harbour fewer pathogens than the model predicts (negative residual values). This approach can be used to target taxa for future pathogen discovery, as was proposed by Herbreteau et al. (2012) for rodents and rodent-borne diseases. Of the 292 species of bats inventoried in SEA, information on parasite species richness and sampling effort was obtained for 41 bat species, whereas information on viral species richness and sampling effort was obtained for 33 bat species.
2.3. Comparative analyses on parasite species richness
Phylogenetically related species tend to share common characteristics and are not independent observations (Harvey and Pagel, 1991). Comparative analysis using the independent contrasts method allows studying relationships between species traits by limiting false statistical results due to phylogenetically pseudo-replications (Type I and II errors) (Felsenstein, 1985). We computed a working phylogenetic tree of bat species investigated in this analysis using phylogenetic trees from published studies (see Supplementary data).
Of the 292 species of bats inventoried in SEA, we obtained complete information on 81, for which we documented all factors (independent variables in the subsequent analyses) including a fully resolved phylogenetic tree. Analyses were then performed on 20 bat species for viruses, 17 bat species for endoparasites (helminths) and 28 bat species for ectoparasites.
We calculated independent contrasts for each of the investigated variables with the package APE (Paradis et al., 2004) implemented in R (R Development Core Team, 2008). Independent contrasts were calculated for three groups of parasites (viruses, endoparasitic helminths and ectoparasite arthropods) for which we had a full set of explanatory variables: sampling effort or investigation effort; bat body mass; bat distribution range size; distribution shape (fragmentation); bat colony size; number of breading seasons; and bat gregarious behavior. Parasite species richness (PSR), viral richness, investigation effort, sampling effort, range, body weight were log transformed in order to stabilize variance. Distribution shape was transformed using arcsine of square root transformation. To confirm the proper standardisation of contrasts, the absolute values of standardised contrasts were regressed against their standard deviations (Garland et al., 1992). Then contrasts were analyzed using standard multiple regressions, with all intercepts forced through the origin (Garland et al., 1992).
We selected the model using a backward procedure and due to potential co-linearity among variables we performed a Principal Component Analysis in order to select variables. We used Akaike’s Information Criteria (AIC) to select the best models.
3. Results
3.1. Identification of highly parasitized bat species
Linear regression between sampling effort and PSR showed a positive relationship (P < 0.0001, R2 = 0.54). Species were ordered by residuals of the sampling effort regression (Fig. 1A). Macroglossus minimus was identified as the host species with the largest positive residual value, carrying more parasite species than expected by linear regression between species richness and sampling effort, whereas Rhinolophus acuminatus appeared to host fewer parasites than expected (i.e., highest negative residual). Similarly, the regression between virus diversity, a subset of PSR, and sampling effort significantly explained 46% of the virus diversity variation (P < 0.0001). Species were ordered by residual values (Fig. 1B). Hipposideros armiger carried the greatest number of viruses above predictions by the linear regression model, whereas Cynopterus sphinx hosted the fewest viruses.
Fig. 1.

Distribution of residual values from the linear relation between (A) Parasite species richness (PSR) and sampling effort and between (B) viral richness and sampling effort.
3.2. Correlation of parasites types per host species
We found one positive correlation between ectoparasite, endoparasite and virus richness using raw data (Table 2). Ectoparasite species richness was weakly positively correlated with virus species richness in bat hosts (P = 0.04). The relationship between ecto- and endo-parasites was not significant (P = 0.06); nor was the relationship between endoparasite and viral species richness (P = 0.16).
Table 2.
Pearson-moment correlations between richness values of ectoparasite, endoparasite and virus species in bats (with P-value).
| Endoparasite species richness | Ectoparasite species richness | Viral species richness | |
|---|---|---|---|
| Endoparasite species richness | – | 0.45 (0.06) | 0.35 (0.16) |
| Ectoparasite species richness | – | 0.47 (0.04) | |
| Viral species richness | – |
3.3. Multivariate analysis
We performed a Principal Component Analysis on potential determinants of parasite and viral species richness of bats (Fig 2): bat body size; number of publications for each bat species; number of publications about parasites of each bat species; bat range distribution; distribution shape or fragmentation; colony size, breeding seasons; roosting site; and ICUN status. The two first dimensions accounted for 43.1% of the variance. IUCN status appeared correlated with bat distribution shape (fragmentation of distribution area). The number of publications on bat species was related to both bat body mass and bat distribution range, whereas the number of publications on parasites was related to bat distribution range. Bat roosting site was related to colony size.
Fig. 2.
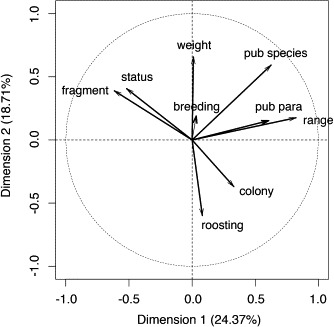
Principal Component Analysis performed on the following potential determinants of parasite species and viral diversities: bat body size (weight); number of publications for each bat species (pub species); number of publications of parasites of each bat species (pub para); bat range distribution (range); distribution shape or fragmentation (fragmentation); colony size (colony); breeding seasons (breeding); roosting site (roosting); and IUCN status (status).
3.4. Comparative analyses using independent contrasts
The best model explaining virus richness included distribution shape, size of bat colony and investigation effort (Table 3). Both distribution shape and colony size were negatively correlated with viral richness (Table 4). Bat species with fragmented distributions and living in large colonies harbour less virus than bat species with continuous distributions and living in small colonies.
Table 3.
Comparison of models used to explain viral richness, ectoparasite and endoparasite species richness in Southeast Asian bats using independent contrasts. The initial model included the following variables: sampling effort (number of hosts sampled for a given virus: hsv); investigating effort (number of publications on parasites for a given host: pubn); host body mass (body mass); colony size (colony); breeding seasons (breeding); geographical distribution range size (range); and shape of the geographical distribution (fragmentation). Models are ranked from the least to the most supported according to corrected Akaike information criteria (AIC).
| Dependent variables | Model ranks | AIC |
|---|---|---|
| Viral richness | Fragmentation + colony + pubn | 31.0 |
| Fragmentation + colony + breeding + pubn | 31.1 | |
| Range + fragmentation + colony + breeding + pubn | 32.6 | |
| Range + fragmentation + colony + breeding + hsv + pubn | 34.3 | |
| Body mass + range + fragmentation + colony + breeding + pubn | 36.2 | |
| Ectoparasite species richness | Body mass + colony + pubn | 98.4 |
| Body mass + colony + breeding + pubn | 99.7 | |
| Body mass + fragmentation + colony + breeding + pubn | 100.8 | |
| Body mass + range + fragmentation + colony + breeding + pubn | 102.3 | |
| Endoparasite species richness | Fragmentation + pubn | 41.3 |
| Body mass + fragmentation + pubn | 42.7 | |
| Body mass + fragmentation + breeding + pubn | 44.6 | |
| Body mass + fragmentation + colony + breeding + pubn | 46.6 | |
| Body mass + range + fragmentation + colony + breeding + pubn | 48.6 | |
Table 4.
Best model explaining viral richness, ectoparasite and endoparasite species richness in Southeast Asian bats using independent contrasts (initial model with investigating effort, host body mass, colony size, number of breeding seasons, geographical distribution size and fragmentation of the geographical distribution) using the AIC criteria (with SD = standard deviation of the slope).
| Dependent variables | Independent variables | Slope (SD) | P | R2, F(P) |
|---|---|---|---|---|
| Virus richness | Fragmentation of distribution | −24.0 (6.8) | 0.003 | |
| Colony size | −1.1 (0.2) | <0.0001 | ||
| Investigating effort (number of publications on parasites) | 3.6 (0.3) | <0.0001 | ||
| 0.87, F3,16 = 41.77 (<0.0001) | ||||
| Ectoparasite richness | Body mass | −2.8 (0.9) | 0.004 | |
| Colony size | 0.5 (0.4) | 0.18 | ||
| Investigating effort (number of publications on parasites) | 8.7 (1.0) | <0.0001 | ||
| 0.76, F2,25 = 38.89 (<0.0001) | ||||
| Endoparasite richness | Fragmentation of distribution | −72.1 (22.0) | 0.006 | |
| Investigating effort | 3.3 (0.7) | <0.001 | 0.64, F2,14 = 12.61 (0.0007) | |
The best model explaining ectoparasite richness identified bat body mass, colony size and investigation effort (Table 3). Body mass was negatively correlated with ectoparasitic arthropod species richness, whereas colony size was positively, but not significantly, correlated with ectoparasite species richness (Table 4).
Finally, investigation effort was positively, and the distribution shape negatively, correlated with endoparasitic helminth species richness (Table 4).
4. Discussion
This study is the first investigation of the likely determinants of parasite and viral species richness in SE Asian bats, using a large data set on parasitic and microbial diversity and ecological traits of their bat hosts. The results of comparative analyses showed that, in addition to sampling effort, factors related to fragmentation of distribution (distribution shape), colony size and body mass seem to explain the variability of parasitic (endoparasitic helminths and ectoparasitic arthropods) and viral species richness.
4.1. Relationship between parasites
We found a positive and significant correlation between ectoparasite and virus species richness in bat species in our data set. This correlation may be related to the vector-borne transmission of some viruses by ectoparasites (e.g., Aznar-Lopez et al., 2013). For example, ectoparasites, including highly adapted and host-specific bat flies (Order Diptera), may play a role as vectors for bacteria and protozoa in bats, e.g., Bartonella (Morse et al., 2012) and possibly Hepatocystis (Garnham, 1951; Olival et al., 2007). However, the feeding behavior of ectoparasites (blood-sucking or not) was not documented as there are few publications, but this information could be added in our analyses when available. Also due to a lack of data, it was not possible to test the correlation between viral richness and bacterial richness. An immunologically-driven mechanism of viral and bacterial diversity in bats may explain their apparent resistance to virus-induced diseases and would be worth testing further. For example in humans, herpes virus latency confers a surprising resistance to infection with bacterial pathogens (Barton et al., 2007).
4.2. Sampling effort, parasite and viral diversity
Sampling and investigation effort were positively correlated with parasite or viral species richness, as observed in many comparative studies (Walther et al., 1995; Poulin, 1995; Guégan and Kennedy, 1996; Nunn et al., 2003a; Turmelle and Olival, 2009; Luis et al., 2013). Based on this pattern, we used residual values of the linear relationship between PSR and sampling effort as a way to draw attention to species with greater than expected numbers of parasites (Herbreteau et al., 2012).
The association found between sampling investigation (i.e., number of publications investigating parasites) and bat distribution area, using multivariate analysis, confirms that parasitologists mostly screen for parasites of common bat species (i.e., living over a large distribution area).
The ordination of residual values of PSR obtained from the linear regression between PSR and sampling effort identified bat species with higher values than expected by the linear regression model: M. minimus; Penthetor lucasi; and Rousettus leschenaultii, etc. Of the 41 bat species studied, half of species with positive residual values are known to carry emerging or potentially emerging viruses: Nipah virus; Australian bat lyssavirus; Phnom Penh bat virus; Kaeng Khoi virus; and Coronaviruses. It may reveal a high level of virus screening in bat species of the same genus. For example, the genus Rhinolophus was considered as a reservoir of a huge diversity of bat-SARS-like Coronaviruses in both Asia and Europe (Wang et al., 2011; Balboni et al., 2012). Rhinolophus was the target of many investigations for Coronavirus discovery (e.g., Lau et al., 2005; Li et al., 2005; Tang et al., 2006). However, disparities in sampling effort among species of a same genus were noted. Rhinolophus sinicus, Rhinolophus malayanus and R. acuminatus seem over-investigated and an increase in sampling effort may not improve pathogen discovery, whereas other species of the genus need more investigation (e.g., Rhinolophus macrotis, Rhinolophus pearsonii, Rhinolophus pusillus and Rhinolophus affinis).
-
1.
Similar ordination of residual values performed for virus species richness showed that three bat species have higher residual values: H. armiger; Scotophilus kuhlii; Cynopterus brachyotis. Of the 33 species studied, half of positive residual values belong to families Hipposideridae, Vespertilionidae and Pteropodidae. Genera belonging to these families are known to carry viruses in the families Paramyxovidae and Coronaviridae. For example, the genus Pteropus is considered as the natural reservoir of the Paramyxovirus, Nipah virus (De Jong et al., 2011; Field and Epstein, 2011) and was well investigated in SEA. In contrast, it seems that several species need more investigation, including H. armiger and all the other species with strongly positive residual values. This prioritization method highlights the over-investigation of some bat species of the family Pteropodidae such as C. sphinx, Pteropus hypomelanus, Pteropus vampyrus, Ptenochirus jagori, Balionycteris maculata, whereas others need more sampling efforts for parasites (e.g., P. lucasi, C. brachyotis, Eonycteris spelaea).
Our analysis of residual values between sampling effort and parasite and viral richness among bat species can be used to target hosts for cost-effective pathogen discovery and also to identify hosts that have been well sampled for pathogen discovery. The genus Pteropus represents one third of the species with positive residual values, i.e., with greater number of parasite and viral species richness than expected by the linear correlation with sampling effort. However, species of the genus Pteropus represent 40% of all species included in our prioritization analysis and more balanced taxonomic sampling in the future will improve the representativeness of analyses similar to ours. Only a few bat species from SEA were well documented and thus integrated to the analysis (41 for PSR and 33 for viruses). As the number of individual hosts tested is often missing in published papers, the number of publications related to a given parasite in a given host species may be used for future residual analyses. This information has the advantage of being more easily documented and was correlated with PSR in our study.
4.3. Ecto- and endoparasite species richness
Ectoparasite species richness was negatively correlated with body mass and positively, but not significantly, correlated with colony size of bat species. A negative correlation of ectoparasite species richness with bat body mass was unsuspected as it was found positively correlated in bats from South America (Bordes et al., 2008). However, using multivariate analysis, body mass appeared to be negatively associated with colony size and roosting behavior (Fig. 2) and these two last factors could explain ectoparasite species richness. The statistical power of our comparative analysis was limited by the number of SEA bat species investigated for ectoparasites. Moreover, our results may suggest that social behavior should be defined in a more complex way in order to better understand factors affecting contact rates, transmission and diversity of parasites (Altizer et al., 2003). Consistent with Bordes et al. (2008), the distribution area did not seem to explain ectoparasite species richness in bats of Southeast Asia. It differs from other studies on PSR in terrestrial mammals (Krasnov et al., 2004; Lindenfors et al., 2007), where it is assumed that widely distributed mammal species accumulate parasites as they increase their chances of contact with parasite species (Lindenfors et al., 2007).
Endoparasite richness was negatively correlated with the distribution shape, an index of the fragmentation of bat distribution. One hypothesis is that endoparasites need specific environmental conditions to survive and infect bats, mostly linked to the diversity of their epidemiological cycles including intermediate hosts and external environment stage. Moreover, parasites are known to suffer from habitat fragmentation that destabilized host-parasite interactions (Kruess and Tscharntke, 2000). A bat carrying a wide diversity of endoparasites may need a variety of environmental conditions to maintain this richness.
4.4. Viral richness
The best model explaining viral richness identified investigation effort, distribution shape and colony size (both negatively correlated). The negative correlation observed for colony size does not correspond to what would be predicted by classical epidemiological models where a large colony should harbour more viruses because of a greater pool of susceptible (larger critical community size). One explanation for this finding may be that our measure of colony size does not consider the structural complexity of bat communities. Some bat species live in close proximity with other species. For example, R. leschenaultii can share its roosting site with Miniopterus schreibersii, Rhinolophus rouxii, Rhinolophus lepidus and other fauna such as rock pigeon (Korad and Gaikwad, 2008). An important variable to take into account is then the size of the whole community of bat species and the rates of interaction among these species. A large colony size does not also equate with high density in term of epidemiological transmission. The social complexity of bats within a colony may limit the overall contacts between individuals leading to a decrease of parasite diversity, such as observed for the ectoparasite species richness in rodents (Bordes et al., 2007). However, information on bat sociality or bat community structure is mostly lacking for the investigated bats of SEA, and is an area for future research.
4.5. Effect of the bat distribution shape
Our analysis highlights the significantly negative effect of bat distribution shape on endoparasite and viral species richness. The negative correlation between the distribution shape and viral or endoparasite species richness corroborate results from Turmelle and Olival (2009), who found greater viral richness in bat species with more genetically structured populations. However, the relationship between genetic diversity and distribution shape (i.e., fragmentation) is not well known but should be a negative as an increase in fragmentation should reduce interactions between bats and thus increase genetic divergence. This hypothesis remains to be tested as more information on genetic diversity of bats from SEA becomes available. Although the distribution shape is not an index of the fragmentation of habitat, it may reflect a border effect of bat species’ distribution area and thus could be considered as a bat species characteristic. This index may originate from the evolutionary biogeographical history of each species (expansion and reduction of population range) (Hampe and Petit, 2005). An increase of distribution shape may reveal species vulnerability, characterized by low population size and patchy distribution leading to a decrease in parasite diversity. The importance of the distribution shape draws attention on environmental factors that may affect border edge, particularly in the face of the ongoing global change.
Parasite richness could be considered as a zoonotic pool involving a risk for human health. According to this view the negative effect of bat distribution shape on endoparasite and viral species richness could be consider as protective for humans. However, fragmentation, due to changes in land use by humans, increase rates of contact between humans and animals (Plowright et al., 2011). This contact may be a critical factor underlying spillover (Keesing et al., 2009). Moreover, habitat fragmentation may favour emergence as the decline of PSR reduces interspecific competitive interactions that could benefits some pathogens.
Future studies should examine consequences of forest/habitat fragmentation impacts on bat distribution and prevalence of EIDs (Murray and Daszak, 2013; Olival et al., 2013). Very few studies have investigated the effect of distribution shape or fragmentation on parasite diversity. Our results are an important first step to understand links between PSR and other host and environmental traits that may influence disease emergence. Thus, future studies should examine effects of habitat fragmentation on the distribution shape (with better information on bat distribution) and PSR of bats, which may be an important determinant of viral richness and PSR more generally.
4.6. Targeting bats as source of potential viruses
We are aware that the results of our comparative study were strongly dependent on the quality of the published literature, and are based purely on correlation analyses. Ordination of residual values of the linear relationship between viral richness and sampling effort has helped prioritization of several species belonging to families Hipposideridae, Vespertilionidae and Pteropodidae for further viral screening (see above). Ordination of the bat distribution shape could be used as a way to target the bat species as source of potential emerging viruses. Among the first ten species that are ordered according to their distribution shape (see Table 1) appear bat species of the family Pteropodidae (such as C. sphinx, Pteropus giganteus or R. leschenaultii), H. armiger (Hipposideridae) and some bat species belonging to Vespertilionidae (e.g. Myotis chinensis, R. pearsonii, Eptesicus serotinus). However, much work should be done to confirm that these species should be targeted and to use this kind of comparative analysis as a tool for predicting bat species as potential sources of emerging viral diseases.
5. Conclusion
We examine determinants of PSR (endoparasite, ectoparasite and viral) in SEA bats. First, we used model residuals as a novel prioritization method to target bat species for a cost-effective pathogen discovery. Secondly, results of comparative analyses suggest that the distribution shape is a significant determinant of PSR, as well as colony size for viruses and distribution size for ectoparasites. Several potential mechanisms may explain the correlation with bat distribution shape through a border effect, although the life-cycle and ecology of these parasites may be of importance, with opposite trends in different parasite groups. Environmental habitat and distribution of bats seem to play a central role in shaping species diversity of parasites and viruses. Ongoing global environmental change is affecting the distribution of bats through the modification of borders, i.e., by decreasing the distribution size and increasing the area shape. Our results suggest that changes in bat distribution shape will alter parasite diversity, with a decrease for endoparasites and viruses. Our study is an important first step in understanding parasite and pathogen species richness in bats from an emerging disease hotspot. However, accurate investigation of emerging risks will require additional information on the role of ecological changes in PSR and contact rates between bats, humans or their domestic animals. Moreover, our study highlights the deficiency of ecological and parasitological data on bat species. Of the 292 species inventoried in SEA only 81 species could be included in our comparative analysis.
Acknowledgments
This study was funded by the French ANR CEP&S, Grant ANR 11 CPEL 002 BiodivHealthSEA (Local impacts and perceptions of global changes: Biodiversity, health and zoonoses in Southeast Asia) (www.biodivhealthsea.org) and by the ATPd CIRAD “Emergences”. KJO is supported by the USAID Emerging Pandemic Threats Program PREDICT project Cooperative Agreement Number (GHN-A-OO-09-00010-00) and an NIH NIAID non-biodefense EID Research Opportunities Award (1 R01 AI079231-01). We thank two anonymous referees and the Editor for helpful comments. The authors declare no conflict of interest.
Appendix A. Supplementary data
References
- Altizer S., Nunn C., Thrall P.H., Gittleman J.L., Antonovics J., Cunningham A.A., Dobson A.P., Ezenwa V., Jones K.E., Pedersen A.B., Poss M., Pulliam J.R.C. Social organization and parasite risk in mammals: integrating theory and empirical studies. Annu. Rev. Ecol. Syst. 2003;34:517–547. [Google Scholar]
- Aznar-Lopez C., Vazquez-Moron S., Marston D.A., Juste J., Ibáñez C., Berciano J.M., Salsamendi E., Aihartza J., Banyard A.C., McElhinney L., Fooks A.R., Echevarria J. Detection of rhabdovirus viral RNA in oropharyngeal swabs and ectoparasites of Spanish bats. J. Gen. Virol. 2013;94:69–75. doi: 10.1099/vir.0.046490-0. [DOI] [PubMed] [Google Scholar]
- Balboni A., Battilani M., Properi S. The SARS-like coronaviruses: the role of bats and evolutionary relationships with SARS coronavirus. New Microbiol. 2012;35:1–16. [PubMed] [Google Scholar]
- Barton E.S., White D.W., Cathelyn J.S., Brett-McClellan K.A., Engle M., Diamond M.S., Miller V.L., Virgin H.W. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- Bordes F., Blumstein D.T., Morand S. Rodent sociality and parasite diversity. Biol. Lett. 2007;3:692–694. doi: 10.1098/rsbl.2007.0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordes F., Morand S., Ricargo G. Bat fly species richness in Neotropical bats: correlations with host ecology and host brain. Oecologia. 2008;158:109–116. doi: 10.1007/s00442-008-1115-x. [DOI] [PubMed] [Google Scholar]
- Bordes F., Morand S. The impact of multiple infections on wild animal hosts: a review. Infect. Ecol. Epidemiol. 2011;1 doi: 10.3402/iee.v1i0.7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed A.C., Field H.E., Epstein J.H., Daszak P. Emerging henipaviruses and flying foxes – conservation and management perspectives. Biol. Conserv. 2006;131:211–220. doi: 10.1016/j.biocon.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke F.M., Rostant L.V., Racey P.A. Life after logging: post-logging recovery of a neotropical bat community. J. Appl. Ecol. 2005;42:409–420. [Google Scholar]
- Clements R., Sodhi N.S., Schilthuizen M. Limestone karsts of Southeast Asia: imperiled arks of biodiversity. Bioscience. 2006;56:733–742. [Google Scholar]
- Coker R.J., Hunter B.M., Rudge J.W., Liverani M., Hanvoravongchai P. Emerging infectious diseases in southeast Asia: regional challenges to control. Lancet. 2011;377:599–609. doi: 10.1016/S0140-6736(10)62004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham A.A. Disease risks of wildlife translocations. Conserv. Biol. 1996;10:349–353. [Google Scholar]
- Daszak P., Cunningham A.A., Hyatt A.D. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- De Jong C., Field H., Newman S., Epstein J.H. Emerging infectious diseases. In: Newman S., Field H.E., De Jong C.E., Epstein J.H., editors. Vol. 12. FAO Animal Production and Health Manual; Rome: 2011. pp. 1–13. (Investigating the Role of Bats in Emerging Zoonoses. Balancing Ecology, Conservation and Public Health Interests). [Google Scholar]
- Derne B.T., Fearnley E.J., Lau C.L., Paynter S., Weinstein P. Biodiversity and leptospirosis risk: a case of pathogen regulation? Med. Hypotheses. 2011;77:339–344. doi: 10.1016/j.mehy.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Dobson A., Cattadori I., Holt R.D., Ostfeld R.S., Keesing F., Krichbaum K., Rohr J.R., Perkins S.E., Hudson P.J. Sacred cows and sympathetic squirrels: the importance of biological diversity to human health. PLoS Med. 2006;3:e231. doi: 10.1371/journal.pmed.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake L.J., Bundy D.A. Multiple helminth infections in children: impacts and control. Parasitology. 2000;122:73–81. doi: 10.1017/s0031182000017662. [DOI] [PubMed] [Google Scholar]
- Ezenwa V.O. Host social behavior and parasitic infection: a multifactorial approach. Behav. Ecol. 2004;15:446–454. [Google Scholar]
- Ezenwa V.O., Jolles A.E. From host immunity to pathogen invasion: the effects of helminth coinfection on the dynamics of microparasites? Integr. Comp. Biol. 2011;51:540–551. doi: 10.1093/icb/icr058. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Field H., Epstein J.H. Henipavirus. In: Newman S., Field H.E., De Jong C.E., Epstein J.H., editors. Vol. 12. FAO Animal Production and Health Manual; Rome: 2011. p. 64. (Investigating the Role of Bats in Emerging Zoonoses. Balancing Ecology, Conservation and Public Health Interests). [Google Scholar]
- Fortin M.-J., Keitt T.H., Maurer B.A., Taper M.L., Kaufman D.M., Blackburn T.M. Species’ geographic ranges and distributional limits: pattern analysis and statistical issues. Oikos. 2005;108:7–17. [Google Scholar]
- Garland T., Harvey P.H., Ives A.R. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 1992;41:18–32. [Google Scholar]
- Garnham P.C.C. An attempt to find the vector of Hepatocystis (=Plasmodium) Kochi (Levaditi and Schoen) Exp. Parasitol. 1951;1:94–107. [Google Scholar]
- Ghatak S., Banerjee R., Agarwal R.K., Kapoor K.N. Zoonoses and bats: a look from human health viewpoint. J. Commun. Dis. 2000;32:40–48. [PubMed] [Google Scholar]
- Guégan J.F., Kennedy C.R. Parasite richness/sampling effort/host range: the fancy three-piece jigsaw puzzle. Parasitol. Today. 1996;12:367–369. doi: 10.1016/0169-4758(96)10054-5. [DOI] [PubMed] [Google Scholar]
- Guernier V., Hochberg M.E., Guégan J.-F. Ecology drives the worldwide distribution of human diseases. PLoS Biol. 2004;2:e141. doi: 10.1371/journal.pbio.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén A., Francis C.M., Salivong K. Wildlife Conservation Society; Vientiane: 1997. Preliminary Survey of Bats in Phou Khao Khouay National Biodiversity Conservation Area. [Google Scholar]
- Harvey P., Pagel M. Oxford University Press; Oxford: 1991. The Comparative Method in Evolutionary Biology. [Google Scholar]
- Hampe A., Petit R.J. Conserving biodiversity under climate change: the rear edge matters. Ecol. Lett. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- Herbreteau V., Bordes F., Jittapalapong S., Supputamongkol Y., Morand S. Rodent-borne diseases in Thailand: targeting rodent carriers and risky habitats. Infect. Ecol. Epidemiol. 2012;2 doi: 10.3402/iee.v2i0.18637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Streygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F., Belden L.K., Daszak P., Jones G., Jacobs D.S., Hunz T.H., Willig M.R., Racey P. Carpe noctem: the importance of bats as bioindicators. Endanger. Species Res. 2009;8:93–115. [Google Scholar]
- Korad, V.S., Gaikwad, M.C., 2008. About frugivorous bats of northern Western Ghats. BAT NET CCINCA Newslett. 9, 9–10.
- Krasnov B.R., Shenbrot G.I., Khokholva I.S., Degen A.A. Flea species richness and parameters of host body, host geography, and host “milieu”. J. Anim. Ecol. 2004;73:1121–1128. [Google Scholar]
- Kruess A., Tscharntke T. Species richness and parasitism in a fragmented landscape experiments and field studies with insects on Vicia sepium. Oecologia. 2000;122:129–137. doi: 10.1007/PL00008829. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.-F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lindenfors P., Nunn C.L., Jones K.E., Cunningham A.A., Sechrest W., Gittleman J.L. Parasite species richness in carnivores: effects of host body mass, latitude, geographical range and population density. Global Ecol. Biogeogr. 2007;16:496–509. [Google Scholar]
- Luby S.P., Rahman M., Hossain M.J., Blum L.S., Husain M.M., Gurley E., Khan R., Ahmed B.-N., Rahman S., Nahar N., Kenah E., Comer J.A., Ksiazek T.G. Foodborne transmission of Nipah virus, Bangladesh. Emerg. Infect. Dis. 2006;12:1888–1894. doi: 10.3201/eid1212.060732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis A.D., Hayman D.T.S., Shea T.J.O., Cryan P.M., Gilbert A.T., Juliet R.C., Mills J.N., Timonin M.E., Willis C.K.R., Cunningham A.A., Fooks A.R., Rupprecht E., Wood J.L.N., Webb C.T. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc. R. Soc. London B. 2013 doi: 10.1098/rspb.2012.2753. [ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie J.S., Field H.E., Guyatt K.J. Managing emerging diseases borne by fruit bats (flying foxes), with particular reference to henipaviruses and Australian bat lyssavirus. J. Appl. Microbiol. 2003;(94 Suppl.):59S–69S. doi: 10.1046/j.1365-2672.94.s1.7.x. [DOI] [PubMed] [Google Scholar]
- Mahy B.W.J., Brown C.C. Emerging zoonoses: crossing the species barrier. Rev. Sci. Tech. OIE. 2000;19:33–40. doi: 10.20506/rst.19.1.1212. [DOI] [PubMed] [Google Scholar]
- McColl K.A., Tordo N., Aguilar Setién A.A. Bat lyssavirus infections. Rev. Sci. Tech. OIE. 2000;19:177–196. doi: 10.20506/rst.19.1.1221. [DOI] [PubMed] [Google Scholar]
- Morand S., Poulin R. Nematode parasite species richness and the evolution of spleen size in birds. Can. J. Zool. 2000;78:1356–1360. [Google Scholar]
- Morse S.F., Olival K.J., Kosoy M., Billeter S., Patterson B.D., Dick C.W., Dittmar K. Global distribution and genetic diversity of Bartonella in bat flies (Hippoboscoidea, Streblidae, Nycteribiidae) Infect. Genet. Evol. 2012;12:1717–1723. doi: 10.1016/j.meegid.2012.06.009. [DOI] [PubMed] [Google Scholar]
- Morse S.S. Oxford University Press; New York: 1993. Emerging Viruses. [Google Scholar]
- Murray K.A., Daszak P. Human ecology in pathogenic landscapes: two hypotheses on how land use change drives viral emergence. Curr. Opin. Virol. 2013;3:79–83. doi: 10.1016/j.coviro.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers N., Mittermeier R.A., Mittermeier C.G., Da Fonseca G.B., Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nunn C.L., Altizer S., Jones K.E., Sechrest W. Comparative tests of parasite species richness in primates. Am. Nat. 2003;162:597–614. doi: 10.1086/378721. [DOI] [PubMed] [Google Scholar]
- Nunn C.L., Gittleman J.L., Antonovics J. A comparative study of white blood cell counts and disease risk in carnivores. Proc. R. Soc. Lond. B Biol. Sci. 2003;270:347–356. doi: 10.1098/rspb.2002.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn C.L. Phylogenetic comparative methods and sleep. In: McNamara P., Barrett D., editors. ABC-CLIO; Greenwood: 2012. pp. 495–504. (The Encyclopedia of Sleep and Dreams: The Evolution, Function, Nature, and Mysteries of Slumber). [Google Scholar]
- Olival K.J., Stiner E.O., Perkins S.L. Detection of Hepatocystis sp. in Southeast Asian flying foxes (Pteropodidae) using microscopic and molecular methods. J. Parasitol. 2007;93:1538–1540. doi: 10.1645/GE-1208.1. [DOI] [PubMed] [Google Scholar]
- Olival K.J., Epstein J.H., Wang L.F., Field H.E., Daszak P. Are bats unique viral reservoirs? In: Aguirre A.A., Ostfeld R.S., Daszak P., editors. New Directions in Conservation Medicine: Applied Cases of Ecological Health. Oxford University Press; Oxford: 2012. pp. 195–212. [Google Scholar]
- Olival K.J., Hoguet R.L., Daszak P. Linking the historical roots of environmental conservation with human and wildlife health. EcoHealth. 2013;10:224–227. doi: 10.1007/s10393-013-0862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pedersen A.B., Grieves T. The interaction of parasites and resources cause crashes in wild mouse population. J. Anim. Ecol. 2008;77:370–377. doi: 10.1111/j.1365-2656.2007.01321.x. [DOI] [PubMed] [Google Scholar]
- Petney T.N., Andrews R.H. Multiparasite communities in animals and humans: frequency, structure and pathogenic signification. Int. J. Parasitol. 1998;28:377–393. doi: 10.1016/s0020-7519(97)00189-6. [DOI] [PubMed] [Google Scholar]
- Plowright R.K., Foley P., Field H.E., Dobson A.P., Foley J.E., Eby P., Daszak P. Urban habituation, ecological connectivity and epidemic dampening: the emergence of Hendra virus from flying foxes (Pteropus spp.) Proc. R. Soc. Lond. B Biol. Sci. 2011;278:3703–3712. doi: 10.1098/rspb.2011.0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin R. Phylogeny, ecology, and the richness of parasites communities in vertebrates. Ecol. Monogr. 1995;5:283–302. [Google Scholar]
- Poulin R., Morand S. Smithsonian Institution Press; Washington DC: 2004. The Parasite Biodiversity. [Google Scholar]
- Šimková A., Lafond T., Ondracková M., Jurajda P., Ottová E., Morand S. Parasitism, life history traits and immune defence in cyprinid fish from Central Europe. BMC Evol. Biol. 2008;8:29. doi: 10.1186/1471-2148-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons N.B. Order chiroptera. In: Wilson D., Reeder D., editors. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd ed. Johns Hopkins University Press; Washington: 2005. pp. 312–529. [Google Scholar]
- Sodhi N.S., Brook B.W. Cambridge University Press; Cambridge, U.K.: 2006. Southeast Asian Biodiversity in Crisis. [Google Scholar]
- Stibig H.-J., Stolle F., Dennis R., Feldkötter C. Office for Official Publications of the European Communities; Luxembourg: 2007. Forest Cover Change in Southeast Asia – The Regional Pattern. [Google Scholar]
- Sulkin S.E., Allen R. Virus infections in bats. Monogr. Virol. 1974;8:1–103. [PubMed] [Google Scholar]
- Tang X.C., Zhang J.X., Zhang S.Y., Wang P., Fan X.H., Li L.F., Li G., Dong B.Q., Liu W., Cheung C.L., Xu K.M., Song W.J., Vijaykrishna D., Poon L.L., Peiris J.S., Smith G.J., Chen H., Guan Y. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006;80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmelle A.S., Olival K.J. Correlates of viral richness in bats (order Chiroptera) EcoHealth. 2009;6:522–539. doi: 10.1007/s10393-009-0263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther B.A., Cotgreave P., Price R.D., Gregory R.D., Clayton D.H. Sampling effort and parasite species richness. Parasitol. Today. 1995;11:306–310. doi: 10.1016/0169-4758(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Wang L.-F., Kuzmin I.V., Tong S. Virus discovery. In: Newman S., Field H.E., De Jong C.E., Epstein J.H., editors. Vol. 12. FAO Animal Production and Health Manual; Rome: 2011. (Investigating the Role of Bats in Emerging Zoonoses. Balancing Ecology, Conservation and Public Health Interests). pp. 197–150. [Google Scholar]
- Wegner K.M., Reusch T.B.H., Kalbe M. Multiple parasites are driving major histocompatibility complex in the wild. J. Evol. Biol. 2003;16:224–232. doi: 10.1046/j.1420-9101.2003.00519.x. [DOI] [PubMed] [Google Scholar]
- Wilcove D.S., Giam X., Edwards D.P., Fisher B., Koh L.P. Navjot’s nightmare revisited: logging, agriculture, and biodiversity in Southeast Asia. Trends Ecol. Evol. 2013 doi: 10.1016/j.tree.2013.04.005. [ahead of print] [DOI] [PubMed] [Google Scholar]
- Wolfe N.D., Daszak P., Kilpatrick A.M., Burke D.S. Bushmeat hunting, deforestation, and prediction of zoonotic disease. Emerg. Infect. Dis. 2005;11:1822–1827. doi: 10.3201/eid1112.040789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


