Abstract
Coyne and Orr found that mating discrimination (premating isolation) evolves much faster between sympatric than allopatric Drosophila species pairs. Their meta-analyses established that this pattern, expected under reinforcement, is common and that Haldane’s rule is ubiquitous in Drosophila species divergence. We examine three possible contributors to the reinforcement pattern: intrinsic postzygotic isolation, dichotomized as to whether hybrid males show complete inviability/sterility; host-plant divergence, as a surrogate for extrinsic postzygotic isolation; and X chromosome size, whether roughly 20% or 40% of the genome is X-linked. We focus on “young” species pairs with overlapping ranges, contrasted with allopatric pairs. Using alternative criteria for “sympatry” and tests that compare either level of prezygotic isolation in sympatry or frequency of sympatry, we find no statistically significant effects associated with X chromosome size or our coarse quantifications of intrinsic postzygotic isolation or ecological differentiation. Although sympatric speciation seems very rare in animals, the pervasiveness of the reinforcement pattern and the commonness of range overlap for close relatives indicate that speciation in Drosophila is often not purely allopatric. It remains to determine whether increased premating isolation with sympatry results from secondary contact versus parapatric speciation and what drives this pattern.
Keywords: reinforcement, Dobzhansky-Muller incompatibilities (DMIs), allopatry, parapatric speciation, extrinsic postzygotic isolation
The biological species concept popularized by Dobzhansky (1937) and Mayr (1942) equates the origin of species with the evolution of reproductive isolation (cf. Coyne and Orr 2004, Ch. 2). Central questions about speciation such as the relative importance of geographic isolation, ecological divergence and sexual selection can be answered by accumulating case studies and by meta-analyses. Coyne and Orr (1989a, 1997) (hereafter, “C&O”) provided the first influential meta-analyses of speciation, using phylogenetic hypotheses, range data, estimates of divergence times, and laboratory estimates of premating isolation and hybrid viability and fertility to extract robust patterns from decades of publications. Their methods and results have been so influential that “Coyne and Orr” has become an adjective describing similar meta-analyses (Funk et al. 2006). The Drosophila data C&O compiled have been repeatedly re-analyzed to address topics including the role of X-chromosome size in determining how long after species divergence Haldane’s rule appears (Turelli and Begun 1997), the correlation of protein evolution versus silent DNA divergence with the evolution of reproductive isolation (Fitzpatrick 2002), the role of ecological divergence in the evolution of prezygotic and intrinsic postzygotic isolation (Funk et al. 2006), the pervasiveness of reinforcement (Yukilevich 2012), and the association of reinforcement with the extent of range overlap (Nosil 2013). Using an expanded Drosophila data set, we address in turn the role of intrinsic postzygotic isolation, ecological differentiation and X chromosome size in speciation, specifically through reinforcement (Dobzhansky 1940), and the geography of speciation (Turelli et al. 2001). Because our sample sizes are small and our measures of ecological divergence and postzygotic isolation are crude, our results are best viewed as guides for future research.
Intrinsic versus extrinsic postzygotic isolation and speciation
We first consider the role of intrinsic postzygotic isolation in Drosophila speciation. Increasing evidence shows that epistatic Dobzhansky-Muller incompatibilities (DMIs), i.e., deleterious combinations of alleles at distinct loci, underlie much of the hybrid inviability and sterility observed under laboratory conditions (Maheshwari and Barbash 2011). These incompatibilities are expected to accumulate with and, less easily, without complete geographical isolation (Orr 1995; Orr and Orr 1996; Orr and Turelli 2001; Kondrashov 2003). Their ubiquity, general recessiveness, and the forces that drive their accumulation, including local adaptation and intragenomic conflict (Meiklejohn and Tao 2010), are likely to explain several broad patterns, including Haldane’s rule (Haldane 1922), the large X-effect (“Coyne’s rule,” Coyne and Orr 1989b), and differences in viability and fertility between F1 hybrids produced from reciprocal crosses (“Darwin’s corollary to Haldane’s rule,” Turelli and Moyle 2007).
However, in some clades, DMIs seem less important to speciation. For instance, reproductive isolation between oscine birds may evolve faster through prezygotic isolation than intrinsic postzygotic isolation (Grant and Grant 1997; Price and Bouvier 2002; Price 2008). For many allopatric Drosophila species pairs, intrinsic postzygotic isolation and prezygotic isolation accumulate at comparable rates (C&O). In contrast, for sympatric pairs (defined as any range overlap), C&O famously showed that prezygotic isolation evolves much faster than intrinsic postzygotic isolation, leading to near-complete reproductive isolation within about 105 years on average, rather than 106 years for allopatric pairs (Coyne and Orr 1997, p. 303; 2004, p. 75).
Many of these sympatric pairs exhibit Haldane’s rule (i.e., inviable or sterile F1 hybrid males, but viable or fertile F1 hybrid females), suggesting that intrinsic postzygotic isolation may provide the impetus for reinforcement, i.e., the evolution of increasing premating isolation driven by hybrid dysfunction (cf. Yukilevich 2012). If so, one might expect pairs with greater intrinsic postzygotic isolation to show more prezygotic isolation (but see our Discussion). Using relatively young species pairs (DNei ≤ 0.5, corresponding to divergence times less than about 106 years, Coyne and Orr 2004, p. 75; Obbard et al. 2012), we test this prediction by comparing premating isolation between pairs having different levels of intrinsic postzygotic isolation. We also test a related prediction. If intrinsic postzygotic isolation facilitates stable co-occurrence (for instance, by driving reinforcement), we expect range overlap to be more common between species pairs showing greater intrinsic postzygotic isolation.
To examine the role of extrinsic postzygotic isolation (i.e., hybrids that are ecologically or behaviorally less fit than their parents), we contrast pairs that do or do not exhibit host-plant differences, as in Funk et al. (2006). As noted by W. J. Etges (pers. comm.) who collected these data and generously shared them, the observed differences in host plants have rarely been directly associated with reduced hybrid fitness (see Soto et al. 2007 and Bono and Markow 2009 for tests). There have been no attempts to assess hybrid performance in nature, where oviposition behavior may be critical to selection against hybrids (e.g., McBride and Singer 2010). Despite the lack of Drosophila evidence, we conjecture that host-plant differences may often be associated with extrinsic postzygotic isolation in nature (Schluter 2001).
X chromosome size and reinforcement
Generally, Drosophila have five chromosome arms, each containing roughly 20% of the nuclear genome (Ashburner et al. 2005, Ch. 4). In many species, such as D. melanogaster, the X chromosome consists of one of these arms; in other species, such as D. pseudoobscura, the X includes two arms, so that roughly 40% of the genome is X-linked. We ask whether X chromosome size affects the level of prezygotic isolation (and presumably the extent of reinforcement) between sympatric pairs or the likelihood of range overlap.
Why might X size matter? We know that X-linked DMIs are central to both the occurrence and timing of Haldane’s rule (Orr 1993; Turelli and Orr 1995, 2000; Turelli and Begun 1997). In particular, large-X pairs show Haldane’s rule at significantly lower levels of genetic divergence (Turelli and Begun 1997); this can reflect either preferential accumulation or preferential expression of X-linked DMIs. Preferential accumulation of X-linked DMIs can occur either because X linkage accelerates molecular evolution (the “faster X” hypothesis of Charlesworth et al. 1987) or because factors such as intra-genomic conflict over sex-ratios preferentially drive the accumulation of X-linked DMIs (Frank 1991; Hurst and Pomiankowski 1991; Meiklejohn and Tao 2010). In Drosophila, comparative genomic data provide only weak support for “faster X” (Vicoso et al. 2008; cf. Mank et al. 2010 for mammals and faster-Z in birds); but there is strong support for differential accumulation of X-linked DMIs (Presgraves 2008; Meiklejohn and Tao 2010). Even without preferential X linkage of DMIs, X-linked DMIs are expected to contribute disproportionately to F1 male hybrid inviability and sterility because of hemizygous expression of recessive X-linked hybrid defects (Muller’s 1940 “dominance theory;” Orr 1993; Turelli and Orr 1995, 2000; Presgraves 2002).
Mathematical theories of reinforcement provide less clear-cut predictions. As shown by Hall and Kirkpatrick (2006) and Lemmon and Kirkpatrick (2006), sex linkage can affect reinforcement via sex-linked loci contributing to hybrid dysfunction, to male traits and/or to female preferences – independent of the intensity of postzygotic isolation. Their mathematical analyses produce complex predictions concerning X linkage; but under many circumstances, X linkage has relatively small effects. One exception is that X-linked (or Z-linked) female preferences for autosomal traits (as documented in Drosophila (Bailey et al. 2011) and flycatchers (Saether et al. 2007)) tend to accentuate reinforcement.
To assess the effects of X linkage, we compare levels of prezygotic isolation and the frequency of range overlap for large-X versus small-X species pairs.
Geography of Speciation
Conventional wisdom among naturalists at the beginning of the 20th century was that allopatric speciation was nearly universal (Jordan 1905; Kellogg 1907, Ch. 9). This consensus was summarized by Mayr (1942, Ch. 7) and emphasized in Mayr (1963, Ch. 16). Late in the 20th century, Schliewen et al. (1994) presented convincing evidence for sympatric speciation by some African crater lake cichlids. Additional examples established the reality of speciation without geographic isolation (e.g., Sorenson et al. 2003; Bolnick and Fitzpatrick 2007; Papadopulos et al. 2011). The initial examples inspired models suggesting that sympatric speciation may be common (Dieckmann and Doebeli 1999; Kondrashov and Kondrashov 1999). However, subsequent theoretical analyses cast doubt on this conclusion (Bolnick 2004; Gavrilets 2005; Polechova and Barton 2005). More importantly, examination of endemism for mobile organisms on small islands, which seemed likely to satisfy the conditions proposed as sufficient for sympatric speciation, suggested that sympatric speciation is rare (Coyne and Price 2000; Losos and Schluter 2000; Kisel and Barraclough 2010). Indeed, for most taxa, at least partial geographic isolation seems necessary for speciation (Kisel and Barraclough 2010). These analyses complement data indicating that purely sympatric speciation requires special conditions only occasionally met (Bolnick 2011; Wagner et al. 2012; Martin and Wainwright 2013). For instance, most lake cichlids do not produce endemic radiations (Seehausen 2006); and even monophagous, host-shifting insects, proposed as prime candidates for sympatric speciation by Mayr (1942, 1963), do not provide ready examples (Feder et al. 2003; Linnen and Farrell 2010).
Although purely allopatric speciation is common and easily documented (e.g., Knowlton et al. 1993; Near and Benard 2004; reviewed by Coyne and Orr 2004, Ch. 3), the biogeographic evidence claimed by Jordan and his peers (Jordan 1903) for pervasive allopatric speciation is far from persuasive (Fitzpatrick & Turelli 2006), even for fishes Jordan studied (Tavera et al. 2012). Part of the problem is that the time scale of climate-induced range changes is typically thousands or tens of thousands of years, whereas speciation is typically slower (Chesser and Zink 1994; Losos and Glor 2003). As emphasized by Barton and Hewitt (1995) and Gavrilets (2003), between the extremes of purely allopatric and purely sympatric speciation lies a continuum of speciation processes with gene flow. We argue that some of the most convincing evidence for non-allopatric speciation is the common reinforcement pattern documented by C&O.
We find no significant effects associated with differences in intrinsic postzygotic isolation, host-plant divergence or X chromosome size, so our Discussion addresses the quality of the data and questions the logic of our analyses.
Materials and Methods
DATA
We primarily used the augmented C&O data compiled by Yukilevich (2012). Yukilevich’s website (http:/www.drosophila-speciation-patterns.com/) provides data on 580 pairs of Drosophila species. For each pair, he gives Nei’s D (DNei, Nei 1972) a protein-based estimate of genetic divergence (and surrogate for divergence time – used because multilocus DNA data are not available for many of these species), laboratory estimates of prezygotic and (intrinsic) postzygotic isolation (denoted Ipre and Ipost, with 0 ≤ I ≤ 1), one or more sources for phylogenetic hypotheses for each clade, and a quantitative estimate of range overlap. While most of Yukilevich’s postzygotic isolation data are quantized following C&O (add 1/4 for each completely inviable or completely sterile sex from the reciprocal crosses), some of Yukilevich’s data did not follow this scoring system. To ensure consistency of all postzygotic isolation measures, we rescored all postzygotic data according to C&O's quantization criteria. W. J. Etges generously provided the Drosophila data used by Funk et al. (2006), including plant hosts (which we use as crude surrogates for ecological differentiation). Despite their limitations and lack of experimental evidence demonstrating extrinsic postzygotic isolation, these qualitative data provide the best available information on ecological divergence for most Drosophila clades. Some discrepancies were found between the data compiled by Yukilevich (2012) and Funk et al. (2006). When these values differ, we analyze the updated data from Yukilevich (2012); however, results are qualitatively equivalent when using Funk et al. (2006) values (Supplementary Material). Information on X chromosome size was taken from sources cited in Turelli and Begun (1997), supplemented with newer references where needed. We provide all of the data analyzed in an online Supplementary Data File.
Miller et al. (2010) found that the maternally inherited bacteria Wolbachia (Werren 1997) contribute to both prezygotic and intrinsic postzygotic isolation of D. paulistorum “semi-species” (Ehrman 1965; Spassky et al. 1971). Their data indicate that five of these six taxa inherited their Wolbachia from the common ancestor of this young clade. Hybrid inviability and sterility between these taxa may, therefore, result from incompatibilities between coevolving Wolbachia and host genomes. These incompatibilities are precisely analogous to DMIs between mtDNA and nuclear genomes (cf., Turelli and Moyle 2007; Bolnick et al. 2008). This is the only Drosophila clade in which Wolbachia are known to contribute to intrinsic postzygotic isolation between species. We present our analyses of intrinsic postzygotic isolation with the paulistorum clade, removing it had no qualitative influence on our results (not shown). From published surveys (Mateos et al. 2006; Bennett et al. 2012), only about 11% (36/319) of sampled Drosophila species harbor Wolbachia. Given this relatively low frequency and the fact that Wolbachia do not contribute to intrinsic postzygotic isolation in most well-studied Drosophila hybridizations, we expect that Wolbachia do not contribute significantly to the patterns observed.
PHYLOGENIES, PHYLOGENETIC CORRECTIONS AND STATISTICAL TESTS
We used specific phylogenetic hypotheses, without attempting to incorporate uncertainty. Incorrect phylogenetic hypotheses are more likely to introduce noise than signal to our comparisons that focus on geography and prezygotic isolation. Following C&O, we “correct” for phylogenetic dependence in patterns of reproductive isolation. Suppose we have several pairwise comparisons of the form Ai vs. Bj, where Ai and Bj are species in sister clades A and B. When comparing allopatric and sympatric pairs, C&O took an unweighted average of all of such comparisons to associate a single observation with the node separating A and B. (This involves using separate phylogenies for the species involved in allopatric versus sympatric comparisons.) This procedure is conservative because it effectively assumes that the reproductive isolation between all (Ai, Bj) pairs evolved between the stem lineages of the sister clades. At the opposite extreme, reproductive isolation between all (Ai, Bj) pairs may evolve only along the terminal branches; so that reproductive isolation between all Ai and Bj represent independent evolutionary events. Following C&O, we collapse all observations across each node to one comparison; but instead of using C&O’s unweighted average, we use the Fitzpatrick (2002) weighting. Because we are unaware of a freely available, user-friendly implementation of this algorithm, we present our R script as a Supplementary File. Figure 1 and Tables 1 and 2 present our data after this phylogenetic correction, Supplementary Tables S1A–S1L present the same comparisons using different cutoffs for range overlap and with and without phylogenetic corrections. Overall, our qualitative results are unaffected by phylogenetic control or sympatry cutoffs. Our statistical analyses and graphics were done with R (R Development Core Team 2008).
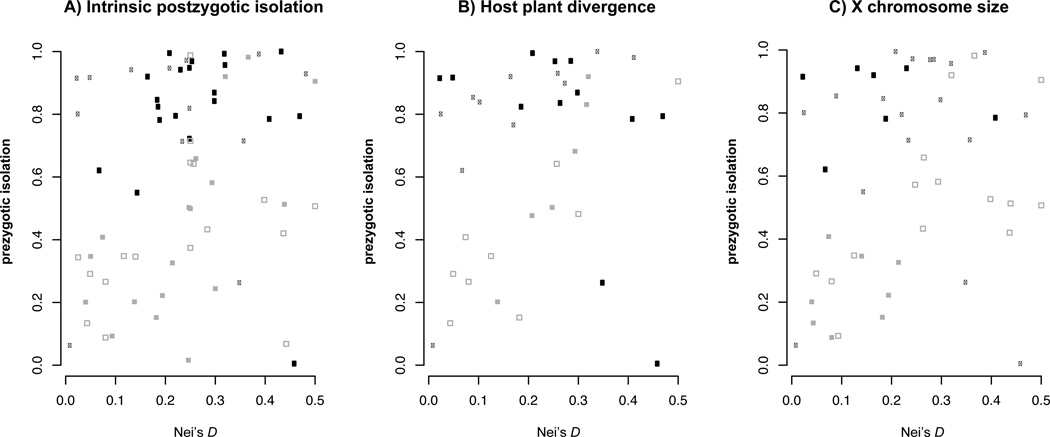
Figure 1.
Patterns of prezygotic isolation as functions of genetic divergence (time) for pairs of Drosophila species with DNei ≤ 0.5. These phylogenetically corrected data are summarized and analyzed in Table 1. Black circles represent sympatric species pairs, grey squares represent allopatric pairs. Panel A. Open and filled symbols represent species contrasts with Ipost = 0 and Ipost > 0, respectively, where Ipost denotes the Coyne and Orr (1989a) index of intrinsic postzygotic isolation. Panel B. Open and filled symbols represent species contrasts with Iext = 0 and Iext > 0, respectively, where Iext denotes whether the pairs show ecological divergence according to Funk et al. (2006). Panel C. Open and filled symbols represent species contrasts with “small” and “large” X chromosomes, respectively.
Table 1.
Levels of prezygotic isolation compared across three dichotomies (intrinsic postzygotic isolation, host plant divergence, and “small” vs. “large” X chromosome size). Our comparisons are for “young” (DNei ≤ 0.5) species contrasts after phylogenetic correction. The entries are mean values of the Coyne and Orr (1989a) index Ipre of prezygotic isolation, the number of comparisons (N), and the average values of DNei. Statistical significance is assessed with Wilcoxon tests. P values in parentheses are for DNei. Following C&O, the criterion for “sympatry” is any range overlap. The data analyzed are from Yukilevich’s web site.
| Dichotomies | Less (N; DNei) | More (N; DNei) | P across dichotomy (for DNei) | |
|---|---|---|---|---|
| Intrinsic postzygotic isolation | Ipost = 0 | Ipost > 0 | ||
| sympatric | 0.77 (13; 0.21) | 0.81 (20; 0.27) | 0.80 (0.34) | |
| allopatric | 0.42 (17; 0.23) | 0.43 (18; 0.23) | 0.88 (0.89) | |
| P for sympatric vs. allopatric | 0.005 (0.529) | < 0.001 (0.43) | ||
| Host plant differences | Ihost = 0 | Ihost > 0 | ||
| sympatric | 0.79 (11; 0.18) | 0.76 (12; 0.27) | 0.88 (0.12) | |
| allopatric | 0.4 (9; 0.18) | 0.6 (6; 0.25) | 0.14 (0.14) | |
| X-linked loci | small X | large X | ||
| sympatric | 0.73 (18; 0.25) | 0.84 (7; 0.17) | 0.98 (0.14) | |
| allopatric | 0.53 (15; 0.29) | 0.23 (8; 0.12) | < 0.001 (0.01) | |
Table 2.
Frequency of range overlap compared across the three dichotomies from Table 1. Our comparisons are for “young” (DNei ≤ 0.5) species contrasts after phylogenetic correction. The entries are fraction of contrasts with overlapping ranges and the number of comparisons (N). Statistical significance is assessed with a Fisher exact test comparing 2×2 tables with the dichotomy on one axis and sympatry versus allopatry on the other. Following C&O, the criterion for “sympatry” is any range overlap. The data analyzed are from Yukilevich’s web site.
| Dichotomies | Less (N) | More (N) | P across dichotomy | |
|---|---|---|---|---|
| Intrinsic postzygotic isolation | DNei ≤ 0.25 | 0.47 (15) | 0.47 (19) | 1 |
| 0.25 ≤ DNei ≤ 0.5 | 0.5 (12) | 0.6 (20) | 0.85 | |
| Host plant differences | DNei ≤ 0.25 | 0.54 (13) | 0.67 (6) | 0.98 |
| 0.25 ≤ DNei ≤ 0.5 | 0.5 (8) | 0.67 (12) | 0.78 | |
| X-linked loci | DNei ≤ 0.25 | 0.58 (12) | 0.5 (12) | 1 |
| 0.25 ≤ DNei ≤ 0.5 | 0.65 (17) | 1 (1) | 1 | |
INTRINSIC AND EXTRINSIC POSTZYGOTIC ISOLATION, X CHROMOSOME SIZE AND REINFORCEMENT
To examine whether intrinsic postzygotic isolation influences reinforcement, we analyzed “young” (DNei ≤ 0.5) species pairs. We compared estimated levels of premating isolation between pairs that show qualitatively different levels of intrinsic postzygotic isolation, namely those that produce at least some fertile males and females from both reciprocal crosses (Ipost = 0) versus those that show complete hybrid inviability or sterility for at least one sex in one of the two reciprocal crosses (Ipost > 0). As a control, we performed this comparison separately for sympatric and allopatric pairs. As a further control, we tested whether pairs with Ipost = 0 vs. Ipost > 0 showed different levels of genetic differentiation as estimated by DNei. Finally, within each class of Ipost values, we compared premating isolation for sympatric vs. allopatric contrasts. As a further test of the role of intrinsic postzygotic isolation in speciation, we compared the frequency of range overlap between pairs of species that do or do not show Ipost > 0. We did this test separately for DNei ≤ 0.25 and 0.25 < DNei ≤ 0.5 to ensure that our analyses are not confounded by the time until sympatry.
Our analysis of extrinsic postzygotic isolation also focused on “young” (DNei ≤ 0.5) species pairs. Using host plant data compiled by Bill Etges and analyzed by Funk et al. (2006), we assigned pairs to “no host difference” (Ihost = 0) if no differences were known; whereas those with any differences were classified as Ihost > 0. We compared the estimates of premating isolation between these groups, as with intrinsic postzygotic isolation. To examine the effect of X chromosome size on reinforcement, we also compared levels of premating isolation for sympatric species pairs with “large” vs. “small” X chromosomes. For both host-use and X-size dichotomies, we also asked whether the frequency of range overlap varied with the categorical differences.
GEOGRAPHY
We calculated the frequency of range overlap between pairs of closely related species in two ways: the fraction of pairs with DNei ≤ 0.5 whose ranges overlap, and the fraction of sister species of any age whose ranges overlap. To account for the fact that reinforcement may require a nontrivial level of range overlap (Sanderson 1989; Noor 1999; Nosil 2013), we used three criteria for “sympatric” species, requiring range overlap >0%, >5%, or >10%, where range overlap is quantified by area of range overlap divided by the area of the smaller range.
Results
INTRINSIC POSTZYGOTIC ISOLATION AND REINFORCEMENT
As shown in Table 1, for sympatric taxa with DNei ≤ 0.5, we find no statistically significant difference in the level of prezygotic isolation between phylogenetically corrected contrasts with Ipost = 0 versus contrasts with Ipost > 0 (Wilcoxon rank sum, P = 0.80, Table 1). This result holds with or without a phylogenetic correction and with alternative sympatry thresholds (> 0%, >5%, or >10% range overlap, Supplementary Tables S1A–L). For contrasts with Ipost = 0, mean Ipre = 0.77 (N = 13); whereas with Ipost > 0, mean Ipre = 0.81 (N = 20). As indicated by Figure 1A, these groups are not significantly different because many contrasts with Ipost = 0 (open circles) show extremely high levels of prezygotic isolation, as originally noted by Coyne and Orr (1989a).
The allopatric contrasts also show no statistically significant difference for either Ipre or DNei when comparing Ipost > 0 versus Ipost = 0 (Table 1 and Tables S1A–L). While the relationship between DNei and Ipre in allopatry is striking (ρ = 0.62, P = 4 × 10−4, N = 38), reinforcement largely flattens the relationship between genetic divergence and prezygotic isolation in sympatry (ρ = 0.32, P = 0.044, N = 40). In light of our negative results relating Ipost to Ipre in sympatry, it is notable that within both the Ipost > 0 and Ipost = 0 groups, we still see C&O’s strong signal of reinforcement (or parapatric speciation, see Discussion and Table 1) when comparing Ipre between sympatric versus allopatric contrasts.
Comparing the frequency of sympatry for the Ipost > 0 vs. Ipost = 0 groups also shows no significant effects for either DNei ≤ 0.25 or 0.25 < DNei ≤ 0.5 (Table 2). Again, this result is robust to: (1) including or excluding paulistorum, (2) using different cutoffs for range overlap, and (3) excluding phylogenetic corrections (Table S2A–L).
EXTRINSIC POSTZYGOTIC ISOLATION
Sympatric pairs that have diverged ecologically (Ihost > 0) according to the Funk et al. (2006) criteria are no older on average that those with Ihost = 0 (DNei = 0.27 vs. 0.18; P = 0.12), and show no more prezygotic isolation (0.76, N = 12 vs. 0.79, N = 11; P = 0.88; Table 1 and Figure 1B). Pooling the Ihost = 0 and Ihost > 0 contrasts, we nevertheless find significantly less premating isolation in allopatry than seen in sympatry (Ipre = 0.49 for all allopatric pairs, N = 42, vs. Ipre = 0.84 for all sympatric pairs, N = 38; P = 10−5). This reinforcement pattern holds for both Ihost > 0 and for Ihost = 0. As in sympatric comparisons, for allopatric there is no difference between mean values of Ipre obtained when Ihost > 0 versus Ihost = 0 (Table 1). We also find no significant differences in the frequency of sympatry for the Ihost > 0 vs. Ihost = 0 groups (Table 2).
As a final attempt to extract a signal associated with increased postzygotic isolation, we considered in our Supplementary Tables two additional categories that combined intrinsic postzygotic isolation and host-plant divergence. We asked whether either prezygotic isolation in sympatry or frequency of range overlap was significantly greater when: (1) either Ipost > 0 or Ihost > 0, or (2) both Ipost > 0 and Ihost > 0. Like our separate analyses of intrinsic and extrinsic postzygotic analyses, neither of these additional comparisons produced a statistically significant result, irrespective of phylogenetic correction, etc. (see Tables S1A–L and S2A–L).
EFFECT OF X-CHROMOSOME SIZE
We found no statistically significant effect of X chromosome size on the level premating isolation in sympatry (P = 0.98, Table 1 and Figure 1C). In contrast, in allopatry we find a significantly lower mean Ipre for large-X contrasts; but this simply reflects the fact that these contrasts also have significantly smaller DNei (0.29 for small X vs. 0.12 for large X, P = 0.01). As noted above, in allopatry Ipre increases significantly with DNei. As with our previous comparisons, the size of the X does not significantly alter the likelihood that closely related species have overlapping ranges (Table 2).
GEOGRAPHY OF SPECIATION
Range overlap is pervasive for closely related Drosophila. Among the 580 species pairs in Yukilevich’s (2012) data, 89 satisfy DNei ≤ 0.5 and have data for percent range overlap; of these, 43, 38, and 36 have greater than 0%, 5%, or 10% range overlap, respectively. Randomly resolving polytomies, we have on average 19.85 sisters pairs with range data, of which 8.3, 7 and 7 have greater than 0%, 5%, or 10% range overlap, respectively. Hence, from both perspectives, range overlap occurs in about a half of closely related Drosophila species.
Discussion
Using updated data of the type originally mined by Coyne and Orr (1989a, 1997), we find no indication that differences in: (1) intrinsic postzygotic isolation, (2) ecological divergence, as indicated by differences in host plants, or (3) X chromosome size are associated with increased prezygotic isolation between sympatric Drosophila species or increased likelihood of range overlap between closely related species. In contrast, we find that (4) non-allopatric speciation is common. We discuss each result in turn.
INTRINSIC (VS. EXTRINSIC) POSTZYGOTIC ISOLATION AND SPECIATION
Coyne and Orr (1989a, 1997) (C&O) showed that reinforcement seems common in Drosophila. They also showed that Haldane’s rule, a specific pattern of intrinsic postzygotic isolation, is a ubiquitous stage of Drosophila divergence. These two results suggest an opportunity for intrinsic postzygotic isolation to drive reinforcement in Drosophila and perhaps other taxa, such as Lepidoptera, in which intrinsic postzygotic isolation, range overlap and speciation occur on commensurate time scales (Presgraves 2002). If intrinsic postzygotic isolation is important to reinforcement, one might expect sympatric species pairs with greater intrinsic postzygotic isolation to show greater evidence of reinforcement, i.e., higher levels of prezygotic isolation. Our analysis does not support this prediction—or the analogous predictions about ecological divergence or X chromosome size (Table 1). Why not?
Poor data or bad predictions?
Our analyses of premating isolation may have produced no statistically significant effects because the data are too crude or because our predictions do not follow from the hypotheses discussed. Our analyses associate three variables, intrinsic postzygotic isolation, ecological divergence and premating isolation, all of which are poorly estimated—and probably systematically underestimated. Development under optimal laboratory conditions may mask significant viability or fecundity defects expressed under more stressful natural conditions. For instance, laboratory assays of fecundity omit host-seeking and oviposition behaviors that are likely to be critical in nature (McBride and Singer 2010). To date, there are no attempts to assess the ecological or behavioral dysfunction of Drosophila hybrids in nature as done, for example, by Hatfield and Schluter (1999) with benthic and limnetic sticklebacks, by Schemske and Bradshaw (1999) with sister species of monkeyflowers, and by McBride and Singer (2010) with conspecific ecomorphs of butterflies. Moreover, standard laboratory measures of hybrid viability and fecundity can miss significant selection against hybrid genotypes, as revealed by more-discriminating fitness assessments in population cages (Fang et al. 2012). Similarly, laboratory assays of mating assortment ignore premating isolation associated with differentiation of mating times or microhabitats. Despite these weaknesses, C&O found a clear signal consistent with reinforcement by comparing sympatric and allopatric species pairs; and they documented the steady accumulation of intrinsic postzygotic isolation.
In the C&O data, range overlap leads to accelerated premating isolation that is evident from laboratory tests. This presumably reflects selection to avoid producing unfit hybrids. Consistent with this, increased premating isolation has also been observed across sympatric versus allopatric populations within species pairs (e.g., Ehrman 1965; Noor 1995). In contrast, by considering only sympatric pairs, we sought a more subtle response. Unlike allopatry versus sympatry, our dichotomies for Ipost, Ihost or X size map less clearly onto more versus less historical selection to avoid maladaptive hybridization. Indeed, for all sympatric species, we expect near-complete premating isolation in nature. Contemporary differences in coarse estimates of intrinsic or extrinsic postzygotic isolation need not reflect differences in the intensity of past selection for assortative mating. Specifically, co-occurring species that now show little intrinsic postzygotic isolation surely experienced other forms of isolation that led to their ability to coexist as distinct taxa. With dubious approximations of historical selection, the shortcomings of lab mating tests may become more important; for instance, laboratory mating assays impose temporal and spatial overlap on species pairs that may not mate at the same times or in the same microhabitats in nature (see Yost and Kay 2009). Thus, the disconnect we find between contemporary indices of intrinsic or extrinsic postzygotic isolation and laboratory mating performance may say little about the forces that drove the pervasive reinforcement pattern identified by C&O.
Our analysis of the frequency of range overlap as a function of each dichotomy was an attempt to circumvent the lack of variation in premating isolation among sympatric pairs expected to rarely hybridize. There may be more discriminatory power in comparing the properties of species whose ranges do or do not overlap (e.g., Davies et al. 2007; Grossenbacher and Whittall 2011). Our negative findings from range-overlap tests may simply reflect the crudeness of our measures of intrinsic and extrinsic postzygotic isolation.
It should also be noted that even the range overlap data may be suspect. For instance, the island endemics D. sechellia and D. mauritiana are tallied as allopatric from their close relative D. simulans, yet D. simulans can now be captured in the Seychelles and on Mauritius (W. J. O. Ballard, pers. comm.) and a recent analysis suggests a long history of range overlap for this pair (Brand et al. 2013). (Counting sechellia and mauritiana as sympatric does not alter any of our conclusions, results not shown.) Many Drosophila, especially from Southeast Asia, have rarely been collected; hence ranges and even species identity may be revised (A. Kopp, pers. comm.).
Measures of intrinsic postzygotic isolation are too coarse?
To obtain reasonable sample sizes, C&O used the most widely available data to measure intrinsic postzygotic isolation. A hybridization that produced many fertile females but only one fertile male was classified as Ipost = 0, even though there is significant intrinsic postzygotic isolation that Haldane (1922) would have counted as an example of Haldane’s rule. A better measure of intrinsic postzygotic isolation might average four values describing the relative fertility/viability of the two sexes obtained from reciprocal crosses, with each value normalized by intraspecific controls. These more quantitative data are generally not available. As a preliminary test, we used the more refined quantitative data on intrinsic postzygotic isolation provided by Yukilevich (2012). These data did not alter our qualitative results; however, he included quantitative data for only a handful of species, and tests with this heterogeneous mix of qualitative and quantitative data are not ideal.
Our measure of ecological differences is far too crude
In their meta-analyses, Funk et al. (2006) found that ecological divergence is associated with greater premating and postzygotic isolation across a wide range of taxa. Combining allopatric and sympatric Drosophila, they found that host-plant differences are associated with greater premating isolation than expected from genetic divergence alone, but the difference was not statistically significant. Our reanalysis of their data also produces no significant effect of host-plant divergence on premating isolation for sympatric pairs. Indeed, at least for sympatric Drosophila, the development of significant intrinsic postzygotic isolation, as indicated by Haldane’s rule, may often follow the evolution of relatively complete reproductive isolation (Mallet 2006). Schemske (2010) summarizes data suggesting that for many taxa, intrinsic postzygotic isolation lags behind premating and extrinsic postzygotic isolation. We obviously need are more systematic assays of ecological differences between Drosophila species and their effects on hybrids—in nature. Encouragingly, novel ecological differences may now be easier to describe using metagenomic analyses of gut contents (e.g., Chandler et al. 2011).
What about Yukilevich’s (2012) more subtle “concordant asymmetries” reinforcement prediction?
Yukilevich (2012) used a subset of the data we analyzed on intrinsic postzygotic isolation versus premating isolation for sympatric species to test a more subtle reinforcement prediction. He found that asymmetries in prezygotic isolation between pairs of sympatric taxa generally agree in direction with reciprocal-cross differences in intrinsic postzygotic isolation. So, for instance, if a cross between taxon A females and taxon B males produces sterile F1 males whereas the reciprocal cross produces fertile males, fewer matings are typically observed between A females and B males than between B females and A males. This suggests that intrinsic postzygotic isolation is central to driving the C&O reinforcement pattern. However, an alternative explanation is that slight asymmetries in gene flow (as indicated by premating isolation in the laboratory) produce asymmetrical accumulation of intrinsic postzygotic isolation. For instance, allopatric populations of the sympatric sister species D. parisiena and D. straubae show asymmetrical Haldane’s rule for sterility, whereas sympatric populations produce fertile males from both reciprocal crosses (Wasserman 1992). Yet, these species, which have diverged in host-plant use, show increased prezygotic isolation in sympatry (Wasserman 1992), as expected under reinforcement driven by extrinsic postzygotic isolation.
A more definitive test of Yukilevich’s (2012) hypothesis that “concordant asymmetries” are produced by asymmetric reinforcement would be to contrast levels of premating asymmetry and intrinsic postzygotic isolation between allopatric versus sympatric populations of sympatric species pairs that show asymmetric intrinsic postzygotic isolation in sympatry (cf. Ehrman 1965; Noor 1995). Reinforcement predicts greater premating isolation asymmetry between sympatric populations, whereas a suppression of DMI accumulation by gene flow would produce less intrinsic postzygotic isolation between sympatric than allopatric populations. Such tests would also help disentangle the contribution of asymmetry in range overlap and asymmetry in offspring viability to observed variation in premating isolation, which were not distinguishable in Yukilevich’s (2012) analysis.
X-CHROMOSOME SIZE AND REINFORCEMENT
We find no evidence that the size of the X chromosome influences the level of premating isolation observed between sympatric species pairs—or the frequency of range overlap. This is consistent with the hypothesis that X-linked DMIs, which play a key role in intrinsic postzygotic isolation, and especially Haldane’s rule, for Drosophila (Muller 1940; Orr 1993; Turelli and Begun 1997; Presgraves 2008; Meiklejohn and Tao 2010), are not central to driving C&O’s reinforcement pattern.
GEOGRAPHY OF SPECIATION
Range overlap is pervasive among closely related Drosophila species, with about half of sister species sympatric. In clades of three species, even if the sisters do not overlap, one of them is often sympatric with the third species. As first documented by C&O and elaborated by Yukilevich (2012), this extensive range overlap is associated with increased premating isolation. This may result from either reinforcement after secondary contact (which Coyne and Orr (2004) dubbed “allo-parapatric” speciation) or parapatric speciation. As argued by Fitzpatrick et al. (2009), the biology is more important than the terminology—some geographic isolation is surely involved and so is the evolution of reproductive isolation in the face of gene flow, assuming that reproductive isolation is not complete when secondary contact occurs. Thus, purely allopatric speciation, envisioned to be nearly universal by Wagner (1873), Jordan (1905) and Mayr (1963), is surely less common than they supposed.
In the Drosophila paulistorum clade of “semispecies,” Ehrman (1965) documented that apparent reinforcement is pervasive. Comparing levels of assortative mating between sympatric versus allopatric population pairs, she found greater premating isolation between sympatric populations in seven of eight tests. Noor (1995) found a similar pattern with D. pseudoobscura and D. persimilis. Reinforcement can influence premating isolation even for allopatric species. As illustrated by Hoskin et al. (2005), if two non-sister groups diverge in response to reinforcement, that divergence can produce premating isolation between the sisters as a byproduct. (We looked for but found no evidence of this effect in our data, analysis not shown.) The potential rapidity of reinforcement is amply documented by laboratory experiments (Rice and Hostert 1993; Etges 1998); and Matute (2010a,b) has demonstrated that reinforcement can occur in the face of gene flow and can affect postmating, prezygotic isolation as well as premating isolation. In light of such evidence, we believe that reinforcement frequently contributes to the evolution of reproductive isolation, and hence speciation in Drosophila (cf. Yukilevich 2012; Nosil 2013).
The papers of Coyne and Orr (1989a, 1997) are landmarks advancing our understanding of speciation through meta-analyses. Our analyses suggest that their data and methods can still provide new insights, but additional progress is likely to require new data from the field.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jerry (“King”) Coyne and H. Allen Orr for their encouragement and comments on early drafts. Bill Etges generously shared his data on Drosophila host-plant use, and Roman Yukilevich kindly answered many questions about his online data set. Graham Coop, Bill Etges, Mohamed Noor, Trevor Price; D. Frank Schemske, FCS; and Roman Yukilevich provided many comments, suggestions and references that greatly improved our analyses and presentation. Bill Ballard and Artyom Kopp provided valuable field expertise. This research was supported by grants from the National Science Foundation: DEB 0815145 to MT, an NSF Bioinformatics postdoctoral fellowship to YB, and a predoctoral fellowship to JRL; and by the National Institute of Health, R01-GM-104325-01 to MT.
LITERATURE CITED
- Ashburner M, Golic KG, Hawley RS. Drosophila: a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Bailey RI, Innocenti P, Morrow EH, Friberg U, Qvarnstrom A. Female Drosophila melanogaster gene expression and mate choice: The X chromosome harbors candidate genes underlying sexual isolation. PLoS One. 2011;6:e17358. doi: 10.1371/journal.pone.0017358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH, Hewitt GM. Adaptation, speciation and hybrid zones. Nature. 1995;341:497–503. doi: 10.1038/341497a0. [DOI] [PubMed] [Google Scholar]
- Bennett G, Pantoja NA, O’Grady PM. Diversity and phylogenetic relationships of Wolbachia in Drosophila and other native Hawaiian insects. Fly. 2012;6:1–11. doi: 10.4161/fly.21161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick DI. Waiting for sympatric speciation. Evolution. 2004;58:895–899. doi: 10.1111/j.0014-3820.2004.tb00421.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI. Sympatric speciation in threespine stickleback: Why not? Int. J. Ecology. 2011;2011:942847. [Google Scholar]
- Bolnick DI, Fitzpatrick BM. Sympatric speciation: models and evidence. Ann. Rev. Ecol. Syst. 2007;38:459–487. [Google Scholar]
- Bolnick DI, Turelli M, López-Fernández H, Wainwright PC, Near TJ. Accelerated mitochondrial evolution and ‘Darwin’s corollary’: asymmetric viability of reciprocal F1 hybrids in centrarchid fishes. Genetics. 2008;178:1037–1048. doi: 10.1534/genetics.107.081364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono JM, Markow TA. Post-zygotic isolation in cactophilic Drosophila: larval viability and adult life-history traits of D. mojavensis/D. arizonae hybrids. J. Evol. Biol. 2009;22:1387–1395. doi: 10.1111/j.1420-9101.2009.01753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand CL, Kingan SB, Wu L, Garrigan D. A selective sweep across species boundaries in Drosophila. Mol. Biol. Evol. 2013;39:2177–2186. doi: 10.1093/molbev/mst123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JA, Morgan Lang J, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: Ecological context of a host-microbe model system. PLoS Genetics. 2011;7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 1987;130:113–146. [Google Scholar]
- Chesser RT, Zink RM. Modes of speciation in birds: a test of Lynch’s method. Evolution. 1994;48:490–497. doi: 10.1111/j.1558-5646.1994.tb01326.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989a;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Two rules of speciation. In: Otte D, Endler JA, editors. Speciation and its consequences. Sunderland, MA: Sinauer Associates; 1989b. pp. 180–207. [Google Scholar]
- Coyne JA, Orr HA. “Patterns of speciation in Drosophila” revisited. Evolution. 1997;51:295–303. doi: 10.1111/j.1558-5646.1997.tb02412.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Coyne JA, Price TD. Little evidence for sympatric speciation in island birds. Evolution. 2000;54:2166–2171. doi: 10.1111/j.0014-3820.2000.tb01260.x. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Meiri S, Barraclough TG, Gittleman JL. Species co-existence and character divergence across carnivores. Ecology Letters. 2007;10:146–152. doi: 10.1111/j.1461-0248.2006.01005.x. [DOI] [PubMed] [Google Scholar]
- Dieckmann U, Doebeli M. On the origin of species by sympatric speciation. Nature. 1999;400:354–357. doi: 10.1038/22521. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. New York: Columbia Univ. Press; 1937. [Google Scholar]
- Dobzhansky T. Speciation as a stage of evolutionary divergence. Am. Nat. 1940;74:312–321. [Google Scholar]
- Ehrman L. Direct observation of sexual isolation between allopatric and sympatric strains of the different Drosophila paulistorum races. Evolution. 1965;19:459–464. [Google Scholar]
- Etges WJ. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. IV. Correlated responses in behavioral isolation to artificial selection in a life-history trait. Am Nat. 1998;152:129–144. doi: 10.1086/286154. [DOI] [PubMed] [Google Scholar]
- Fang S, Yukilevich R, Chen Y, Turissini DA, Zeng K, Boussy IA, Wu C-I. Incompatibility and competitive exclusion of genomic segments between sibling Drosophila species. PLoS Genetics. 2012;8:e1002795. doi: 10.1371/journal.pgen.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Berlocher SH, Roethele JB, Dambrowski H, Smith JL, Perry WL, Gavrilovic V, Filchak KE, Rull J, Aluja M. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. USA. 2003;100:10314–10319. doi: 10.1073/pnas.1730757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick BM. Molecular correlates of reproductive isolation. Evolution. 2002;56:191–198. doi: 10.1111/j.0014-3820.2002.tb00860.x. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick BM, Turelli M. The geography of mammalian speciation: mixed signals from phylogenies and range maps. Evolution. 2006;60:601–615. [PubMed] [Google Scholar]
- Fitzpatrick BM, Fordyce JM, Gavrilets S. Pattern, process and geographic modes of speciation. J. Evol. Biol. 2009;22:2342–2347. doi: 10.1111/j.1420-9101.2009.01833.x. [DOI] [PubMed] [Google Scholar]
- Frank S. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- Funk DJ, Nosil P, Etges WJ. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc. Natl. Acad. Sci. USA. 2006;103:3209–3213. doi: 10.1073/pnas.0508653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilets S. Models of speciation: What have we learned in 40 years? Evolution. 2003;57:2197–2215. doi: 10.1111/j.0014-3820.2003.tb00233.x. [DOI] [PubMed] [Google Scholar]
- Gavrilets S. “Adaptive speciation” — It is not that easy: A reply to Doebeli et al. Evolution. 2005;59:696–699. [Google Scholar]
- Grant PR, Grant BR. Genetics and the origin of species in birds. Proc. Natl. Acad. Sci. USA. 1997;94:7768–7775. doi: 10.1073/pnas.94.15.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossenbacher DL, Whittall JB. Increased floral divergence in sympatric monkeyflowers. Evolution. 2011;65:2712–2718. doi: 10.1111/j.1558-5646.2011.01306.x. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Sex-ratio and unidirectional sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Hall DW, Kirkpatrick M. Reinforcement and sex linkage. Evolution. 2006;60:908–921. [PubMed] [Google Scholar]
- Hatfield T, Schluter D. Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution. 1999;53:866–873. doi: 10.1111/j.1558-5646.1999.tb05380.x. [DOI] [PubMed] [Google Scholar]
- Hoskin CJ, Higgie M, McDonald KR, Moritz C. Reinforcement drives rapid allopatric speciation. Nature. 2005;437:1353–1356. doi: 10.1038/nature04004. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DS. The origin of species through isolation. Science. 1905;22:545–562. doi: 10.1126/science.22.566.545. [DOI] [PubMed] [Google Scholar]
- Kellogg VL. Darwinism to-day. London: George Bell & Sons; 1907. [Google Scholar]
- Kisel Y, Barraclough TG. Speciation has a spatial scale that depends on levels of gene flow. Am. Nat. 2010;175:316–334. doi: 10.1086/650369. [DOI] [PubMed] [Google Scholar]
- Knowlton N, Weigt LA, Solorzano LA, Mills DK, Bermingham E. Divergence in proteins, mitochondrial DNA, and reproductive compatibility across the Isthmus of Panama. Science. 1993;260:1629–1632. doi: 10.1126/science.8503007. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Accumulation of Dobzhansky-Muller incompatibilities within a spatially structured population. Evolution. 2003;57:151–153. doi: 10.1111/j.0014-3820.2003.tb00223.x. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS, Kondrashov FA. Interactions among quantitative traits in the course of sympatric speciation. Nature. 1999;400:351–354. doi: 10.1038/22514. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Kirkpatrick M. Reinforcement and the genetics of hybrid incompatibilities. Genetics. 2006;173:1145–1155. doi: 10.1534/genetics.105.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen CR, Farrell BD. A test of the sympatric host race formation hypothesis in Neodiprion (Hymenoptera: Diprionidae) Proc. Roy. Soc. B. 2010;277:3131–3138. doi: 10.1098/rspb.2010.0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losos JB, Glor RE. Phylogenetic comparative methods and the geography of speciation. Trends Ecol. Evol. 2003;18:220–227. [Google Scholar]
- Losos JB, Schluter D. Analysis of an evolutionary species-area relationship. Nature. 2000;408:847–850. doi: 10.1038/35048558. [DOI] [PubMed] [Google Scholar]
- Maheshwari S, Barbash DA. The genetics of hybrid incompatibilities. Ann. Rev. Genetics. 2011;45:331–355. doi: 10.1146/annurev-genet-110410-132514. [DOI] [PubMed] [Google Scholar]
- Mallet J. What does Drosophila genetics tell us about speciation? TREE. 2006;21:386–393. doi: 10.1016/j.tree.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution. 2010;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- Martin CH, Wainwright PC. Multiple fitness peaks on the adaptive landscape drive adaptive radiation in the wild. Science. 2013;339:208–211. doi: 10.1126/science.1227710. [DOI] [PubMed] [Google Scholar]
- Mateos M, Castrezana SJ, Nankivell BJ, Estes AM, Markow TA, Moran NA. Heritable endosymbionts of Drosophila. Genetics. 2006;174:363–376. doi: 10.1534/genetics.106.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR. Reinforcement of gametic isolation in Drosophila. PLoS Biology. 2010a;8:e1000341. doi: 10.1371/journal.pbio.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute DR. Reinforcement can overcome gene flow during speciation in Drosophila. Curr. Biol. 2010b;20:2229–2233. doi: 10.1016/j.cub.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Systematics and the origin of species. New York: Columbia Univ. Press; 1942. [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge, Mass: Belknap Press; 1963. [Google Scholar]
- McBride CS, Singer MC. Field studies reveal strong postmating isolation between ecologically divergent butterfly populations. PLoS Biology. 2010;8:e1000529. doi: 10.1371/journal.pbio.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Tao Y. Genetic conflict and sex chromosome evolution. Trends Ecol. Evol. 2010;25:215–223. doi: 10.1016/j.tree.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, Ehrman L, Schneider D. Infectious speciation revisited: impact of symbiont-depletion on female fitness and mating behavior in Drosophila paulistorum. PLoS Pathogens. 2010;6:e1001214. doi: 10.1371/journal.ppat.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Bearing of the Drosophila work on systematics. In: Huxley J, editor. The new systematics. Oxford: Clarendon Press; 1940. pp. 185–268. [Google Scholar]
- Near TJ, Benard MF. Rapid allopatric speciation in logperch darters (Percidae: Percina) Evolution. 2004;58:2798–2808. doi: 10.1111/j.0014-3820.2004.tb01631.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Genetic distance between populations. Am. Nat. 1972;106:282–292. [Google Scholar]
- Noor MAF. Speciation driven by natural selection in Drosophila. Nature. 1995;375:674–675. doi: 10.1038/375674a0. [DOI] [PubMed] [Google Scholar]
- Noor MAF. Reinforcement and other consequences of sympatry. Heredity. 1999;83:503–508. doi: 10.1038/sj.hdy.6886320. [DOI] [PubMed] [Google Scholar]
- Nosil P. Degree of sympatry affects reinforcement in Drosophila. Evolution. 2013;67:808–812. doi: 10.1111/j.1558-5646.2012.01817.x. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Maclennan J, Kim K-W, Rambaut A, O’Grady PM, Jiggins FM. Estimating divergence dates and substitution rates in the Drosophila phylogeny. Mol. Biol. Evol. 2012;29:3459–3473. doi: 10.1093/molbev/mss150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. A mathematical model of Haldane’s rule. Evolution. 1993;47:1606–1611. doi: 10.1111/j.1558-5646.1993.tb02179.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Orr LH. Waiting for speciation: the effect of population subdivision on the time to speciation. Evolution. 1996;50:1742–1749. doi: 10.1111/j.1558-5646.1996.tb03561.x. [DOI] [PubMed] [Google Scholar]
- Orr HA, Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution. 2001;55:1085–1094. doi: 10.1111/j.0014-3820.2001.tb00628.x. [DOI] [PubMed] [Google Scholar]
- Papadopulos AST, Baker WJ, Crayn D, Butlin RK, Kynast RG, Hutton I, Savolainen V. Speciation with gene flow on Lord Howe Island. Proc. Natl. Acad. Sci. USA. 2011;108:13188–13193. doi: 10.1073/pnas.1106085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polechova J, Barton NH. Speciation through competition: a critical review. Evolution. 2005;59:1194–1210. [PubMed] [Google Scholar]
- Presgraves DC. Patterns of postzygotic isolation in Lepidoptera. Evolution. 2002;56:1168–1183. doi: 10.1111/j.0014-3820.2002.tb01430.x. [DOI] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends in Genetics. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price T. Speciation in birds. Greenwood, CO: Roberts & Co.; 2008. [Google Scholar]
- Price TD, Bouvier MM. The evolution of F1 postzygotic incompatibilities in birds. Evolution. 2002;56:2083–2089. [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. ISBN 3-900051-07-0, URL http://www.R-project.org. [Google Scholar]
- Rice WR, Hostert EE. Laboratory experiments on speciation: What have we learned in 40 years? Evolution. 1993;47:1637–1653. doi: 10.1111/j.1558-5646.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- Saether SA, Saether G-P, Borge T, Wile C, Svedin N, et al. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science. 2007;318:95–97. doi: 10.1126/science.1141506. [DOI] [PubMed] [Google Scholar]
- Sanderson N. Can gene flow prevent reinforcement? Evolution. 1989;43:1223–1235. doi: 10.1111/j.1558-5646.1989.tb02570.x. [DOI] [PubMed] [Google Scholar]
- Schemske DW. Adaptation and The Origin of Species. Am. Nat. 2010;176:S4–S25. doi: 10.1086/657060. [DOI] [PubMed] [Google Scholar]
- Schemske DW, Bradshaw HD. Pollinator preference and the evolution of floral traits in monkeyflowers (Mimulus) Proc. Natl. Acad. Sci. USA. 1999;96:11910–11915. doi: 10.1073/pnas.96.21.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliewen UK, Tautz D, Pääbo S. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature. 1994;368:629–632. doi: 10.1038/368629a0. [DOI] [PubMed] [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends Ecol. Evol. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Seehausen O. African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. Lond. B. 2006;273:1987–1998. doi: 10.1098/rspb.2006.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson MD, Sefc KM, Payne RB. Speciation by host switch in brood parasitic indigobirds. Nature. 2003;424:928–931. doi: 10.1038/nature01863. [DOI] [PubMed] [Google Scholar]
- Soto EM, Soto IM, Carreira VP, Fanara JJ, Hasson E. Host-related life history traits in interspecific hybrids of cactophilic Drosophila. Entomologia Experimentalis et Applicata. 2007;126:18–27. [Google Scholar]
- Spassky B, Richmond RC, Perez-Salas S, Pavlovsky O, Mourao CA, et al. Geography of the sibling species related to Drosophila willistoni, and of the semispecies of the Drosophila paulistorum complex. Evolution. 1971;25:129–143. doi: 10.1111/j.1558-5646.1971.tb01866.x. [DOI] [PubMed] [Google Scholar]
- Tavera JJ, Acero P A, Balart EF, Bernardi G. Molecular phylogeny of grunts (Teleostei, Haemulidae), with an emphasis on the ecology, evolution, and speciation history of New World species. BMC Evol. Biol. 2012;12:57. doi: 10.1186/1471-2148-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Begun DJ. Haldane’s rule and X chromosome size in Drosophila. Genetics. 1997;147:1799–1815. doi: 10.1093/genetics/147.4.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. The dominance theory of Haldane’s rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Barton NH, Coyne JA. Theory and speciation. Trends Ecol. Evol. 2001;16:330–343. doi: 10.1016/s0169-5347(01)02177-2. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Haddrill PR, Charlesworth B. A multispecies approach for comparing sequence evolution of X-linked and autosomal sites in Drosophila. Genet. Res. Camb. 2008;90:421–431. doi: 10.1017/S0016672308009804. [DOI] [PubMed] [Google Scholar]
- Wagner CE, Harmon LJ, Seehausen O. Ecological opportunity and sexual selection together predict adaptive radiation. Nature. 2012;487:366–369. doi: 10.1038/nature11144. [DOI] [PubMed] [Google Scholar]
- Wagner M. In: The Darwinian theory and the law of the migration of organisms. from the 1868 GermanLaird JL, translator. London: Stanford; 1873. [Google Scholar]
- Wasserman M Cytological evolution of the Drosophila repleta species group. Drosophila inversion polymorphism. Boca Raton, FL: CRC Press; 1992. pp. 455–541. [Google Scholar]
- Werren JH. Biology of Wolbachia. Ann. Rev. Ent. 1997;47:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- Yost JM, Kay KM. The evolution of postpollination reproductive isolation in Costus. Sex. Plant Reprod. 22:247–255. doi: 10.1007/s00497-009-0113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukilevich R. Asymmetric patterns of speciation uniquely support reinforcement in Drosophila. Evolution. 2012;66:1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.