Abstract
Genetic variation in FFAR1 modulates insulin secretion dependent on non-esterified fatty acid (NEFA) concentrations. We previously demonstrated lower insulin secretion in minor allele carriers of PPARG Pro12Ala in high-NEFA environment, but the mode of action could not been revealed. We tested if this effect is mediated by FFAR1 in humans. Subjects with increased risk of diabetes who underwent oral glucose tolerance tests were genotyped for 7 tagging SNPs in FFAR1 and PPARG Pro12Ala. The FFAR1 SNPs rs12462800 and rs10422744 demonstrated interactions with PPARG on insulin secretion. FFAR1 rs12462800 (p = 0.0006) and rs10422744 (p = 0.001) were associated with reduced insulin secretion in participants concomitantly carrying the PPARG minor allele and having high fasting FFA. These results suggest that the minor allele of the PPARG SNP exposes its carriers to modulatory effects of FFAR1 on insulin secretion. This subphenotype may define altered responsiveness to FFAR1-agonists, and should be investigated in further studies.
Keywords: FFAR1, Free fatty acid receptor 1, G-protein coupled receptor 40, GPR40, PPARG, Insulin secretion
Abbreviations: FFAR1, free fatty acid receptor 1; GPR40, G-protein coupled receptor 40; NEFA, non-esterified fatty acids; OGTT, oral glucose tolerance test; TUEF, Tuebingen Family Study
1. Introduction
In addition to glucose, which is the foremost stimulator of insulin secretion in beta-cells, other nutrients such as free fatty acids (also called non-esterified fatty acids, NEFA) contribute to glucose-stimulated insulin secretion (GSIS). Glucose triggers the processes leading to insulin secretion by its metabolism within beta-cells [1,2]. NEFA also act as fuels in beta-cells and they are able to modulate insulin secretion by influencing intracellular metabolism [3]. It was not until 2003, the discovery of the role of a previously orphaned receptor, the G-protein-coupled receptor 40 (GPR40), now called free fatty acid receptor 1 (FFAR1), that an additional important coupling mechanism between NEFA and insulin secretion was revealed [4]. The acute increase of insulin secretion after raising NEFA levels was shown to be ∼50% in healthy volunteers [5,6], and the FFAR1-pathway is thought to be involved in at least 50% of NEFA-mediated insulin secretion [7].
FFAR1-agonists have been developed to enhance GSIS, some of which could be candidates for a new class of antidiabetic drugs [8]. We recently demonstrated that FFAR1-agonism may be protective against beta-cell apoptosis and provided evidence that common variation near the FFAR1 gene modulates insulin secretion dependent on free-fatty acid levels [9].
Recent data from an investigation conducted with beta-cell cultures and animals suggest that FFAR1 gene expression is modulated by stimulation of peroxisome proliferator-activated receptor gamma (PPARG) [10]. Intriguingly, in 2001 we had demonstrated effects of the PPARG Pro12Ala single nucleotide polymorphism (SNP) on insulin secretion in an elevated NEFA milieu. In this investigation, carriers of the minor allele of the Pro12Ala polymorphism (rs1805192) had lower insulin secretion during hyperglycemic clamp studies conducted with a concomitant intravenous lipid infusion, but no difference was seen between the genotypes without increasing NEFA [11].
Given these data supporting a crosstalk between PPARG and FFAR1 signaling, we set out to look for evidence of a PPARG × FFAR1 interaction in humans. For this purpose, we analyzed insulin secretion effects of interactions between previously described tagging SNPs in FFAR1 [9] and the aforementioned diabetes-related SNP in PPARG, also accounting for NEFA levels.
2. Methods
2.1. Study population
Subjects analyzed in the current study are participants of the Tuebingen Family Study (TUEF). TUEF is a cross-sectional observational study originally designed to characterize the phenotype of increased risk for type 2 diabetes. Persons with either a positive family history, or prior gestational diabetes or known glucose intolerance or overweight from Southern Germany were enrolled. For this analysis, we selected 1928 participants without incident diabetes who had fasting NEFA and full 5-point OGTT measurements for glucose and insulin, and who were also genotyped for the PPARG and FFAR1 SNPs. Of the study population, 3% had prior gestational diabetes, 43% had a family history of diabetes in at least one first degree relative and 40% were obese according to the WHO definition (BMI > 30 kg/m2) (Table 1).
Table 1.
Demographic and metabolic characteristics of the investigated population (N = 1928).
| Trait | Median | IQR |
|---|---|---|
| Age | 39 | (29, 50) |
| BMI (kg/m2) | 28 | (24, 35) |
| Glucose 0′ (fasting, mmol/l) | 5.1 | (4.8, 5.4) |
| Glucose 120′ (post-challenge, mmol/l) | 6.2 | (5.2, 7.3) |
| HbA1c (%) | 5.4 | (5.1, 5.7) |
| Insulin 0′ (fasting, pmol/l) | 51.0 | (34.0, 87.0) |
| Insulin sensitivity index (AU) | 12.4 | (7.3, 20.5) |
| Insulinogenic index (AU) | 124 | (76, 200) |
| Non-esterified fatty acids (fasting, μmol/l) | 561 | (424, 723) |
| Triglycerides (fasting, mg/dl) | 100 | (71, 147) |
| Cholesterol (fasting, mg/dl) | 190 | (167, 215) |
| LDL (fasting, mg/dl) | 116 | (96, 139) |
| HDL (fasting, mg/dl) | 52 | (44, 62) |
2.2. OGTT and laboratory measurements
Following an overnight fast, all participants ingested a 75 g glucose dose at 8 am (OGTT). Plasma glucose, insulin, C-peptide and NEFA concentrations were determined after 0, 30, 60, 90 and 120 min. A bedside glucose analyzer (glucose-oxidase method, Yellow Springs Instruments, Yellow Springs, OH, USA) was used to determine plasma glucose. Plasma insulin and NEFA were measured with commercial chemiluminescence assays for ADVIA Centaur (Siemens Healthcare Diagnostics, Eschborn, Germany).
2.3. Selection of tagging SNPs and genotyping
Selection of the tagging SNPs in FFAR1 was performed as described earlier [9]. Genotyping was carried out on the MassARRAY platform from Sequenom (Sequenom, San Diego, CA, USA). Initially 9 SNPs were selected as tagging SNPs in FFAR1 covering all other common SNPs within the locus with an r2 > 0.8 (100% coverage), but genotype calling was unsuccessful for rs12459138 and Hardy–Weinberg equilibrium was rejected for rs10423648 (p < 0.05), therefore 7 SNPs went into analysis.
2.4. Calculations and statistics
Insulin sensitivity was assessed from 5-point OGTT glucose and insulin values using the Matsuda index (ISI), calculated as
where g and i denote glucose and insulin levels, respectively, at the given time-point (minutes) of OGTT [12]. Insulin secretion was assessed by the insulinogenic index (IGI) which was originally proposed by Seltzer et al. in 1967 [13], and has been since then validated several times as a reliable measure of insulin secretion utilizing insulin and glucose measurements during the first 30 min of an OGTT [14,15]. IGI can be calculated as (i30 − i0)/(g30 − g0). The disposition index (DI), another measure of insulin secretion which also accounts for insulin sensitivity, was calculated as IGI × ISI.
Variables with skewed distributions were loge-transformed prior to linear regression analyses. To increase statistical power in the interaction analyses, genotypes were coded according to the dominant inheritance model. Given the 7 investigated SNPs, to reduce the risk of false positive findings with multiple testing, the limit of statistical significance was defined at 0.05/7 = 0.007, and a p between 0.05 and 0.007 was termed nominally significant. Sex, age and insulin sensitivity were used as covariates of IGI. Sex and age were used as covariates of DI. Interaction of the FFAR1 SNPs with the PPARG SNP and with NEFA was tested adding all combinations of possible interaction terms (FFAR1 × NEFA, PPARG × NEFA, FFAR1 × PPARG, FFAR1 × PPARG × NEFA) to the models. Effect estimates are given as standardized beta (β). All statistical analyses were conducted with JMP11 (SAS).
3. Results
As expected, PPARG genotypes alone did not impact insulin secretion in the study population (a table on the genotype-dependent distribution of key cohort phenotypes is provided as Supplementary Table 1). To test the hypothesis that there is an interaction between genetic variation in PPARG and FFAR1 on insulin secretion, we analyzed gene × gene interactions between rs1805192 in PPARG and 7 tagging SNPs of known frequent variants in FFAR1. Since the modulatory effect of PPARG on insulin secretion manifested only in a high-NEFA environment [11] and FFAR1 effects on insulin secretion were seen only in interaction with fasting NEFA [9], we additionally adjusted for fasting NEFA and added interaction terms of fasting NEFA with the FFAR1 SNPs and the PPARG SNP to the models.
We adjusted insulin secretion as determined by the insulinogenic index for sex, age, insulin sensitivity, fasting NEFA, PPARG Pro12Ala genotype, FFAR1 genotypes and 4 interaction terms: between FFAR1 and PPARG genotypes, between PPARG genotype and NEFA level, between FFAR1 genotype and NEFA and, finally, a 3-way interaction term with PPARG × NEFA × FFAR1-SNP. We found a significant gene × gene interaction between the FFAR1 SNP rs10422744 and the PPARG Pro12Ala SNP (p = 0.005) with a concomitant nominally significant interaction between NEFA level and the PPARG SNP (p = 0.03). Additionally, a nominally significant interaction was found between rs12462800 and PPARG (p = 0.01) with a concomitant PPARG × NEFA interaction (p = 0.03). The 3-way interaction term did not reach statistical significance, although the p-value of 0.0592 suggested a trend for nominal significance in the case of rs12462800. Detailed results of the interaction tests are provided in Supplementary Table 2. An additional adjustment for obesity by adding BMI to the interaction models did not relevantly change the results (data not shown). Alternatively using DI as outcome variable, the gene × gene interaction term had a p-value of 0.01 and 0.001 for rs12462800 and rs10422744, respectively.
Next, we conducted subgroup analyses with stratification along interaction variables, namely the PPARG SNP and fasting NEFA. The study population was thus stratified into high and low NEFA (along the median NEFA, 561 μmol/L), and PPARG minor allele (Ala) carriers. This stratification resulted in 4 subgroups (I: low NEFA and PPARG Pro homozygotes, n = 718, II: low NEFA and PPARG Ala carriers, n = 237, III: high NEFA and PPARG Pro homozygotes, n = 744, IV: high NEFA and PPARG Ala carriers, n = 229). An effect of the FFAR1 genotype on insulin secretion was only evident in the subgroup IV comprising participants with at least one PPARG Ala allele and high fasting NEFA (Figure 1A and B bottom right diagrams [diagrams IV]). In this subpopulation, carriers of the minor allele of rs10422744 and rs12462800 in FFAR1 had lower insulin secretion (β = −0.18, p = 0.001 and β = −0.19, p = 0.0006, respectively) than homozygotes of the major allele, after controlling for sex, age and insulin sensitivity.
Figure 1.
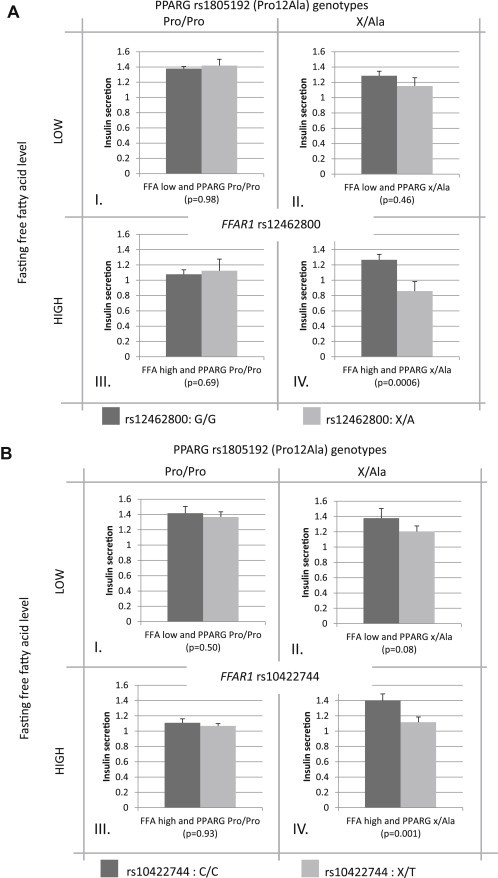
Insulin secretion per rs12462800 (A) and rs10422744 (B) genotype in FFAR1 in interaction with the PPARG Pro12Ala genotype and fasting free fatty acid levels. The bar charts show insulin secretion as residuals of the insulinogenic index adjusted for sex, age and insulin sensitivity (arbitrary units). Dark shaded bar charts represent homozygous carriers of the major FFAR1 alleles (G/G for rs12462800 and C/C for rs10422744), while light shaded bar charts represent hetero- and homozygous carriers of the minor alleles (X/A for rs12462800 and X/T for rs10422744). The sets of bar charts in the left sided quadrants (I, III) show insulin secretion in the subgroup with the homozygous major PPARG allele (Pro/Pro), while the sets of bar charts in the right sided quadrants (II, IV) show insulin secretion in the subgroup that hetero- or homozygously carries the minor PPARG allele (X/Ala). The upper (I, II) and lower (III, IV) bar chart sets represent the subgroups with low and high fasting non-esterified fatty acid (NEFA) levels, respectively. This stratification was performed along the median NEFA value (561 μmol/l) of the total study population.
4. Discussion
In this work, we provide evidence for a biologically relevant crosstalk between PPARG and FFAR1 signaling in the regulation of insulin secretion in humans. Importantly, this crosstalk was only manifest when fasting NEFA levels were high.
The Pro12Ala variant in PPARG has been one of the first candidate SNPs for type 2 diabetes [16,17]. The rare allele of this variant is associated with a 25% reduced risk for the disease [18]. Given the role of PPARG as both a receptor for NEFA and itself a regulator of fat metabolism, our group had earlier investigated the hypothesis that PPARG-mediated alterations in fatty acid signaling could have an impact on insulin secretion. In those hyperglycemic clamp studies, we had compared insulin secretion between Pro/Pro and X/Ala carriers of the PPARG variant in a control and a high-NEFA setting. Only the clamp condition with a concomitant infusion of Intralipid solution, performed to raise NEFA levels, had unmasked striking differences in insulin secretion between PPARG genotypes [11]. However, no explanation could be given for the mechanism of action by that time. Meanwhile, the NEFA receptor FFAR1 has been established as an important stimulator of fatty-acid mediated insulin secretion [4] and genetic variation in FFAR1 has been shown to influence beta-cell function [9,19]. Therefore, FFAR1 seemed to be a plausible link between PPARG and insulin secretion. By analyzing the previously described 7 FFAR1 tagging SNPs, we now found 2 SNPs, which exert a NEFA and PPARG-dependent effect on insulin secretion.
The SNPs rs12462800 and rs10422744 are located 0.8 kb apart in an intergenic regulatory area between the FFAR1 (GPR40) and FFAR3 (GPR41) genes, 3.5 and 3.8 kb from the 3′ end of the single FFAR1 exon. These SNPs are more distal from the gene than the previously described FFAR1 SNP rs1573611 which directly interacts with fasting NEFA in association with insulin secretion [9].
The molecular mechanisms underlying the PPARG–FFAR1 interaction are still speculative. Although the literature is controversial, there is a wealth of data from basic science and clinical studies involving the use of PPARG agonists (thiazolidinediones) indicating that PPARG activation may have a positive impact on beta-cell function and beta-cell survival (reviewed by [20]). The PPARG–FFAR1 interaction, especially in light of the effect modification by NEFA levels, could possibly explain this still poorly understood link between PPARG action and beta-cell function. A recent study demonstrated that PPARG overexpression leads to an increased expression of FFAR1 in rat INS-1 cells and primary rat islets [10]. A possible interaction scenario could thus be a defective PPARG-induced transcriptional or translational activity of the FFAR1 gene in the presence of the FFAR1 minor allele variants, which would consecutively result in a limited compensatory potential to increase insulin secretion when fasting NEFA are elevated. The fundamental human phenotypic alterations associated with the Pro12Ala variant seem to involve lipolysis and its insulin-dependent inhibition [21–23]. Since FFAR1 is activated by medium and long-chained fatty acids [24], perturbations in NEFA levels or their composition as a consequence of altered PPARG action could modulate insulin secretion through FFAR1.
In summary, PPARG and FFAR1 are connected in several ways, and the presented interaction is mechanistically reasonable. Additional investigations are required to elucidate the exact details of the underlying physiology. However, our findings are of immanent importance for the treatment of type 2 diabetes, because the described genotypic interaction between PPARG and FFAR1 could have bearing on the pharmacologic efficacy of FFAR1 agonists. Although a phase 3 clinical trial of the most promising FFAR1-agonist has been recently halted, several of such agents are still under development [8,25]. We infer that the variants identified in the current study, under the constellation of the minor PPARG Pro12Ala allele, could have relevant impact on the effectiveness of FFAR1 agonists. Identification of variants which are candidates for pharmacogenetic interactions could pave the way for investigating genotype-based therapeutic responses within clinical studies and on the long run in clinical practice. This would eventually lead to an improved selection of individuals who would benefit from specific therapies [26].
Acknowledgments
We thank all the research volunteers for their participation. We gratefully acknowledge the excellent technical assistance of Anna Bury, Anja Dessecker, Dr. Louise Fritsche, Ellen Kollmar, Corinna Sailer and Andreas Vosseler (all University of Tübingen, Germany).
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Brisson G.R., Malaisse-Lagae F., Malaisse W.J. The stimulus-secretion coupling of glucose-induced insulin release. VII. A proposed site of action for adenosine-3′,5′-cyclic monophosphate. The Journal of Clinical Investigation. 1972;51:232–241. doi: 10.1172/JCI106808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meglasson M.D., Matschinsky F.M. Pancreatic islet glucose metabolism and regulation of insulin secretion. Diabetes/Metabolism Reviews. 1986;2:163–214. doi: 10.1002/dmr.5610020301. [DOI] [PubMed] [Google Scholar]
- 3.Nolan C.J., Madiraju M.S.R., Delghingaro-Augusto V., Peyot M.-L., Prentki M. Fatty acid signaling in the beta-cell and insulin secretion. Diabetes. 2006;55(Suppl. 2):S16–S23. doi: 10.2337/db06-s003. [DOI] [PubMed] [Google Scholar]
- 4.Itoh Y., Kawamata Y., Harada M., Kobayashi M., Fujii R., Fukusumi S. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422:173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- 5.Boden G., Chen X., Rosner J., Barton M. Effects of a 48-h fat infusion on insulin secretion and glucose utilization. Diabetes. 1995;44:1239–1242. doi: 10.2337/diab.44.10.1239. [DOI] [PubMed] [Google Scholar]
- 6.Kashyap S., Belfort R., Gastaldelli A., Pratipanawatr T., Berria R., Pratipanawatr W. A sustained increase in plasma free fatty acids impairs insulin secretion in nondiabetic subjects genetically predisposed to develop type 2 diabetes. Diabetes. 2003;52:2461–2474. doi: 10.2337/diabetes.52.10.2461. [DOI] [PubMed] [Google Scholar]
- 7.Prentki M., Matschinsky F.M., Madiraju S.R.M. Metabolic signaling in fuel-induced insulin secretion. Cell Metabolism. 2013;18:162–185. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Poitout V., Lin D.C.-H. Modulating GPR40: therapeutic promise and potential in diabetes. Drug Discovery Today. 2013;18:1301–1308. doi: 10.1016/j.drudis.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Wagner R., Kaiser G., Gerst F., Christiansen E., Due-Hansen M.E., Grundmann M. Reevaluation of fatty acid receptor 1 as a drug target for the stimulation of insulin secretion in humans. Diabetes. 2013;62:2106–2111. doi: 10.2337/db12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H.-S., Hwang Y.-C., Koo S.-H., Park K.S., Lee M.-S., Kim K.-W. PPAR-γ activation increases insulin secretion through the up-regulation of the free fatty acid receptor GPR40 in pancreatic β-cells. PLoS ONE. 2013;8:e50128. doi: 10.1371/journal.pone.0050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefan N., Fritsche A., Häring H., Stumvoll M. Effect of experimental elevation of free fatty acids on insulin secretion and insulin sensitivity in healthy carriers of the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor-γ2 gene. Diabetes. 2001;50:1143–1148. doi: 10.2337/diabetes.50.5.1143. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda M., DeFronzo R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 13.Seltzer H.S., Allen E.W., Herron A.L., Jr., Brennan M.T. Insulin secretion in response to glycemic stimulus: relation of delayed initial release to carbohydrate intolerance in mild diabetes mellitus. The Journal of Clinical Investigation. 1967;46:323–335. doi: 10.1172/JCI105534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albareda M., Rodríguez-Espinosa J., Murugo M., de Leiva A., Corcoy R. Assessment of insulin sensitivity and beta-cell function from measurements in the fasting state and during an oral glucose tolerance test. Diabetologia. 2000;43:1507–1511. doi: 10.1007/s001250051561. [DOI] [PubMed] [Google Scholar]
- 15.Hanson R.L., Pratley R.E., Bogardus C., Narayan K.M.V., Roumain J.M.L., Imperatore G. Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. The American Journal of Epidemiology. 2000;151:190–198. doi: 10.1093/oxfordjournals.aje.a010187. [DOI] [PubMed] [Google Scholar]
- 16.Deeb S.S., Fajas L., Nemoto M., Pihlajamäki J., Mykkänen L., Kuusisto A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature Genetics. 1998;20:284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 17.Altshuler D., Hirschhorn J.N., Klannemark M., Lindgren C.M., Vohl M.C., Nemesh J. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nature Genetics. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 18.Stumvoll M., Häring H. The peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism. Diabetes. 2002;51:2341–2347. doi: 10.2337/diabetes.51.8.2341. [DOI] [PubMed] [Google Scholar]
- 19.Kalis M., Levéen P., Lyssenko V., Almgren P., Groop L., Cilio C.M. Variants in the FFAR1 gene are associated with beta cell function. PLoS ONE. 2007;2:e1090. doi: 10.1371/journal.pone.0001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta D., Kono T., Evans-Molina C. The role of peroxisome proliferator-activated receptor γ in pancreatic β cell function and survival: therapeutic implications for the treatment of type 2 diabetes mellitus. Diabetes, Obesity and Metabolism. 2010;12:1036–1047. doi: 10.1111/j.1463-1326.2010.01299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stumvoll M., Wahl H.G., Loblein K., Becker R., Machicao F., Jacob S. Pro12Ala polymorphism in the peroxisome proliferator-activated receptor-2 gene is associated with increased antilipolytic insulin sensitivity. Diabetes. 2001;50:876–881. doi: 10.2337/diabetes.50.4.876. [DOI] [PubMed] [Google Scholar]
- 22.Stumvoll M., Häring H. Reduced lipolysis as possible cause for greater weight gain in subjects with the Pro12Ala polymorphism in PPARgamma2? Diabetologia. 2002;45:152–153. doi: 10.1007/s125-002-8257-y. [DOI] [PubMed] [Google Scholar]
- 23.Vangipurapu J., Stančáková A., Pihlajamäki J., Kuulasmaa T.M., Kuulasmaa T., Paananen J. Association of indices of liver and adipocyte insulin resistance with 19 confirmed susceptibility loci for type 2 diabetes in 6,733 non-diabetic Finnish men. Diabetologia. 2011;54:563–571. doi: 10.1007/s00125-010-1977-4. [DOI] [PubMed] [Google Scholar]
- 24.Ichimura A., Hirasawa A., Hara T., Tsujimoto G. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins & Other Lipid Mediators. 2009;89:82–88. doi: 10.1016/j.prostaglandins.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Lead GPR40 agonist bites the dust. Nature Review Drug Discovery. 2014;13:91. [Google Scholar]
- 26.Wagner R., Staiger H., Ullrich S., Stefan N., Fritsche A., Häring H.-U. Untangling the interplay of genetic and metabolic influences on beta-cell function: examples of potential therapeutic implications involving TCF7L2 and FFAR1. Molecular Metabolism. 2014;3:261–267. doi: 10.1016/j.molmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


