Abstract
Background & Aims
Sirtuin 1 (SIRT1), the most conserved mammalian NAD+-dependent protein deacetylase, is an important metabolic sensor in many tissues. However, little is known about its role in the small intestine, which absorbs and senses nutrients. We investigated the functions of intestinal Sirt1 in systemic bile acid and cholesterol metabolism in mice.
Methods
Sirt1 was specifically deleted from intestines of mice using the Flox-villin-Cre system (Sirt1 iKO mice). Intestinal and heptic tissues were collected, and bile acid absorption was analyzed using the everted gut sac experiment. Systemic bile acid metabolism was studied in Sirt1 iKO and Flox control mice placed on standard diets, diets containing 0.5% cholic acid or 1.25% cholesterol, or lithogenic diets.
Results
Sirt1 iKO mice had reduced intestinal Fxr signaling via Hnf1a compared with controls, which reduced expression of the bile acid transporter genes Asbt and Mcf2l (encodes Ost) and absorption of ileal bile acids. Sirt1 regulated Hnf1α–Fxr signaling partially through Dcoh2, which increases dimerization of Hnf1α. Sirt1 was found to deacetylate DCoH2, promoting its interaction with Hnf1α and inducing DNA binding by Hnf1α. Intestine-specific deletion of Sirt1 increased hepatic bile acid biosynthesis, reduced hepatic accumulation of bile acids, and protected animals from liver damage from high-bile acid diets.
Conclusions
Intestinal Sirt1, a key nutrient sensor, is required for ileal bile acid absorption and systemic bile acid homeostasis in mice. We delineated the mechanism of metabolic regulation of Hnf1α–Fxr signaling. Reagents designed to inhibit intestinal SIRT1 might be developed to treat bile acid-related diseases such as cholestasis.
Keywords: ileal bile acid absorption, bile acid synthesis, liver damage, cholestasis
Introduction
Bile acids are the major end products of hepatic cholesterol catabolism. They are essential for intestinal absorption of dietary lipids, cholesterol and fat-soluble vitamins, and play an important role in cholesterol metabolism. Whole body bile acid homeostasis is maintained by efficient enterohepatic cycling of bile acids between liver and small intestine, in which 95% of bile acids released from the liver into the proximal duodenum in response to entering of dietary fats are reabsorbed by the distal ileum and transported back to the liver via the portal circulation 1-3. The unabsorbed 5% of the bile acids eliminated through feces in each enterohepatic cycle will then be supplemented by new hepatic synthesis so that a constant pool of bile acids is maintained. In addition, a small amount of bile acids spilled over into systemic circulation will be excreted from urine. Therefore, bile acid metabolism is tightly regulated by various nutritional and hormonal cues, and its dysregulation has been associated with a number of gastrointestinal and metabolic diseases, including cholestasis, hypercholesterolemia, defective liver regeneration, cholesterol gallstone disease, and diabetes 4-7.
A key regulatory factor for systemic bile acid metabolism is Farnesoid X receptor (FXR), a cellular bile acid sensor and a member of adopted orphan nuclear receptors 2, 8, 9. In enterocytes, activation of FXR by bile acids enhances the transcription of a number of bile acid transporters, particularly the intestinal basolateral organic solute transporters Ostα/Ostβ that are responsible for bile acid export from enterocytes into portal blood 10. Activation of FXR in enterocytes also induces a hormone, fibroblast growth factor 15 (FGF15), which travels to the liver and repress the hepatic bile acid biosynthesis through its receptor, Fgfr4, and the JNK pathway 2, 11. In addition to FXR, Hepatocyte Nuclear Factor 1α (HNF1α), a homeodomain-containing transcription factor, is also critical in regulation of intestinal and systemic bile acid metabolism 12. HNF1α directly binds to the promoter of FXR gene to modulate its expression 12. HNF1α also regulates ileal bile acids uptake through transcriptional induction of an apical sodium-dependent bile acid transporter Asbt 13. Therefore, the intestinal HNF1α and FXR signaling pathways tightly control the ileal bile acid uptake and systemic bile acid homeostasis in response to nutritional and hormonal signals.
SIRT1 is a member of sirtuins, a family of highly conserved NAD+-dependant protein deacetylases and/or ADP-ribosyltransferases 14. Accumulating evidence has indicated that sirtuins are crucial regulators for a variety of cellular processes, ranging from energy metabolism, stress response, to tumorigenesis and aging 15, 16. As the most conserved mammalian sirtuin, SIRT1 couples the deacetylation of numerous transcription factors and co-factors to the cleavage of NAD+, an indicator of cellular metabolic status 17, 18. Therefore, SIRT1 is an important regulator of energy homeostasis in several tissues, including liver, adipose tissues, pancreas, and hypothalamus 18, 19. However, the function of SIRT1 in the small intestine, particularly in intestinal nutrient absorption, has not yet been determined.
In order to investigate the role of SIRT1 in intestinal physiology, we generated an intestine-specific SIRT1 knockout mouse strain (SIRT1 iKO). In this report, we show that intestinal SIRT1 regulates ileal bile acid absorption and feedback impacts systemic bile acid and cholesterol metabolism in mice. Intestinal SIRT1 modulates the DNA binding ability of HNF1α, partially through deacetylation of a dimerization co-factor of HNF1α, pterin 4α carbinolamine dehydratase 2/dimerization cofactor of HNF1α 2 (DCoH2), a novel acetylated protein. Consequently, deletion of intestinal SIRT1 decreases the expression of FXR and Asbt, reducing ileal bile acid absorption. Intriguingly, in contrast to hepatic deficiency of SIRT1, which leads to decreased HNF1α/FXR signaling pathways, diminished hepatic bile acid excretion and increased liver damage 20, deletion of intestinal SIRT1 reduces hepatic accumulation of bile acids and blunts the inhibition of hepatic bile acid synthesis under high bile acid diets, protecting animals from liver damage.
Materials and Methods
Animal experiments
The intestinal specific SIRT1 knockout mice (SIRT1 iKO) in a C57BL/6 background were generated by crossing mice carrying a SIRT1 exon 4 floxed allele 21 with Villin-cre mice (Jackson laboratory 22). Three to four-month old SIRT1 iKO mice (Villin-Cre+, SIRT1 flox/flox) and their littermate Flox controls (Villin-Cre-, SIRT1 flox/flox), as well as age-matched wild type and Villin-Cre mice, were fed either a standard laboratory chow diet, a chow diet containing 0.5% cholic acid (Research Diets, custom made), a chow diet containing 1.25% Cholesterol (Research Diets, custom made), or a lithogenic diet (D12383, Research Diets) for indicated times. All animal experiments were approved by the NIEHS/NIH Animal Care and Use Committee.
Bile acids analysis
To determine the fecal bile acid outputs, feces were collected from individually housed mice over 48 h and fecal bile acids were extracted with 75% ethanol at 50 °C for 2 h. To determine the total bile acid pool size, liver, gall bladder, and full-length small intestine were dissected and homogenized in water and extracted with 75% ethanol. Bile acids in the resulting supernatants were measured with the total bile acid kit (Diazyme Laboratories). Gallbladder bile acid profiles were analyzed by the ultra performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) as described 23.
Everted gut sac experiment
The everted gut sac experiments were carried out essentially as described 10.
Chromatin immunoprecipitation (ChIP) analysis
Chromatin immunoprecipitation (ChIP) analysis was performed essentially as described by the manufacturer (Millipore) with some modifications. Briefly, Flox and SIRT1 iKO mice were perfused with 1% paraformaldehyde in PBS at room temperature via heart perfusion to cross-link protein-DNA complexes. After 10 min, the cross-linking was stopped by perfusion with 0.125 M glycine. The sonicated cross-linked chromatin was subjected to immunoprecipition with antibodies against FXR, HNF1α (Santa Cruz biotechnology), or normal rabbit IgG. DNA fragments were analyzed by qPCR using primers flanking indicated promoter regions.
Protein acetylation analysis
To analyze the deacetylation of DCoH2 by SIRT1, HEK293T SIRT1RNAi cells were transfected with constructs expressing HA-DCoH2, with or without FLAG-p300 and SIRT1 as indicated. 48 h after transfection, cells were treated with 5 μM TSA for 2 hours, harvested, and homogenized. HA-DCoH2 was immuno-purified with anti-HA beads (Santa Cruz biotech), subjected to SDS-PAGE, and analyzed with anti-acetyl-lysine polyclonal antibodies (Cell Signaling Technology).
Identification of lysine acetylation sites in DCoH2 protein
The two acetylated sites (K124 and K131) in DCoH2 (Figure S4) were identified by a proteomics screening as described 24, 25.
Statistical analysis
Values are expressed as mean ± standard error of mean (SEM). Significant differences between the means were analyzed by the two-tailed, unpaired, nonparametric Mann-Whitney test, and differences were considered significant at p< 0.05.
Results
Deletion of SIRT1 in intestine reduces ileal bile acid absorption
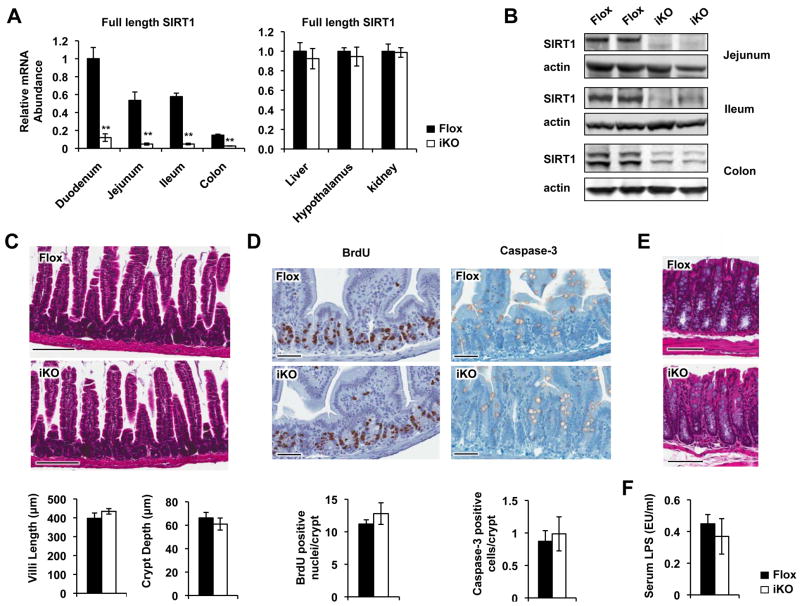
To study the function of SIRT1 in the intestine, we generated a SIRT1 iKO mouse strain as described in the Materials and Methods. The resulting SIRT1 iKO mice efficiently deleted the exon 4 of the SIRT1 gene down the length of the intestine, yet the expression of SIRT1 was normal in other selected tissues (Figure 1A, 1B, and Figure S1A).
Figure 1.
Generation of intestine-specific SIRT1 KO mice (SIRT1 iKO mice). (A) mRNA levels of full-length SIRT1 in intestine and other control tissues in Flox and SIRT1 iKO mice (n=6, **p<0.01). (B) SIRT1 protein in the jejunum, ileum, and colon of Flox and SIRT1 iKO mice. (C) SIRT1 iKO mice have normal morphology of jejunum under standard feeding conditions (n=6). Bars, 200 μm. (D) SIRT1 iKO mice display normal rates of proliferation and apoptosis in the jejunum of small intestine (n=6). Bars, 60 μm. (E) SIRT1 iKO mice have normal morphology of colon under standard feeding conditions (n=4-5). Bars, 200 μm. (F) SIRT1 iKO mice have normal levels of serum LPS levels on the chow diet (n=5-6).
SIRT1 iKO mice were phenotypically normal on the chow diet, with no signs of defects in morphology, proliferation, and apoptosis of intestinal epithelial cells (Figure 1C-E). They also had normal expression levels of different intestinal epithelial cell markers (Figure S1B-C), and maintained normal intestinal barrier functions, as indicated by normal serum LPS levels (Figure 1F) and a normal paracellular transport rate of [14C] inulin, a routinely used “non-absorbable” polysaccharide, across everted sacs of the small intestine segments in vitro (Figure S1D).
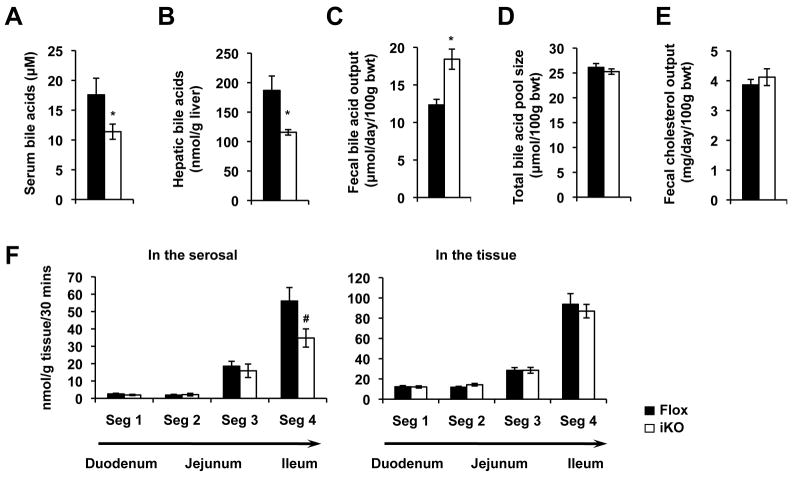
Additionally, SIRT1 iKO mice had normal levels of the most tested serum metabolites (Figure S2A-D). However, their serum and hepatic levels of bile acids were significantly reduced compared to those from control mice (Figure 2A and 2B), suggesting a role of intestinal SIRT1 in bile acid metabolism. Consistent with this possibility, SIRT1 iKO mice displayed increased fecal bile acid excretion (Figure 2C) without changes in their total bile acid pool size (Figure 2D). In contrast, their fecal total cholesterol output was normal (Figure 2E), indicating a selective effect on bile acid metabolism.
Figure 2.
Loss of intestinal SIRT1 alters systemic bile acid metabolism under normal feeding conditions. (A-B) Deletion of SIRT1 in the intestine decreases serum bile acid levels (A, n=20) and hepatic bile acid contents (B, n=6-7). (C) Deletion of SIRT1 in intestine increases the fecal bile acid outputs. (n=6). (D) SIRT1 iKO mice display a normal total bile acid pool size (n=7). (E) SIRT1 iKO mice have normal fecal cholesterol outputs (n=6). (F) Intestinal SIRT1 deficiency leads to decreased mucosal-to-serosal transport of taurocholate in ileal gut sacs. The transport of taurocholate was measured as described in Materials and Methods (n=9). # p=0.07, *p<0.05.
The decreased serum and hepatic bile acid levels, and increased fecal bile acid loss suggest a partial reduction in ileal bile acid transport in SIRT1 iKO mice. To test this hypothesis, we determined the mucosal-to-serosal transport of [3H] taurocholate, a conjugated bile acid, across the small intestinal segments using a well establish everted gut model 26. As shown in Figure 2F, in agreement with previous observations 10, 27, the mucosal-to-serosal transport of taurocholate was largely restricted to the ileum in both control and SIRT1 iKO mice. Deletion of SIRT1 in the intestine led to a 40-50% decrease of trans-ileal transport of taurocholate (Figure 2F, left panel, p=0.07), whereas the amount of tissue-associated taurocholate was normal (Figure 2F, right panel), suggesting that both apical uptake and basolateral export of bile acids are reduced in SIRT1 deficient intestine. Collectively, our observations indicate that intestinal deficiency of SIRT1 reduces the transport of bile acids in the distal ileum, resulting in increased fecal elimination and decreased circulating and hepatic levels of bile acids.
Deletion of intestinal SIRT1 reduces the HNF1α/FXR signaling pathways
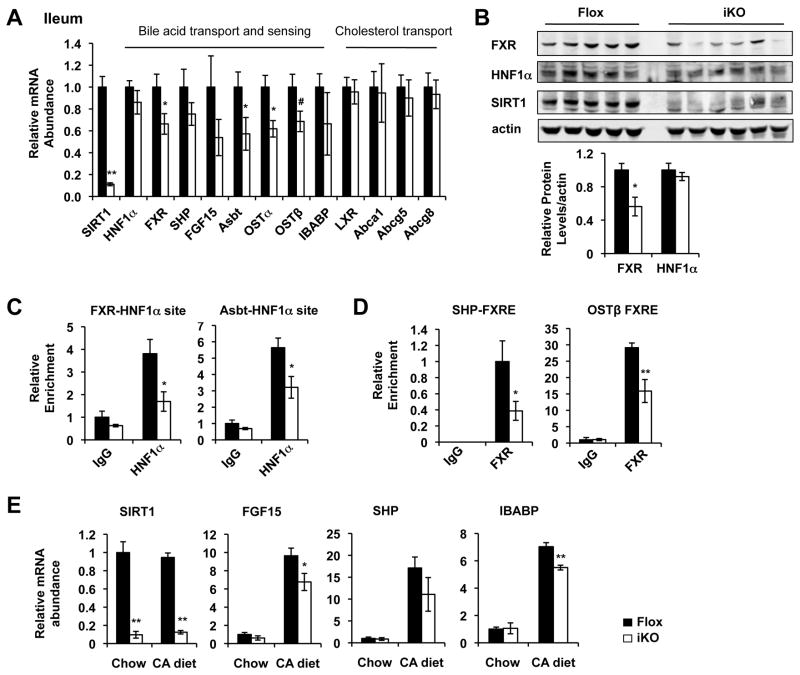
The ileal bile acid absorption/transport is regulated by the HNF1α/FXR pathways 2, 12. To dissect molecular mechanisms underlying the ileal bile acid transport defect in SIRT1 iKO mice, we analyzed ileal expression levels of genes in the HNF1α/FXR signaling pathway by quantitative real-time PCR. As shown in Figure 3A and Figure S3, deletion of intestinal SIRT1 led to reduced ileal expression of many HNF1α/FXR target genes involved in bile acid absorption, transport and sensing, including FXR, Asbt, Ostα/Ostβ, and FGF15 (when compared between paired littermates). FXR protein levels were also significantly decreased in the ileum of SIRT1 iKO mice (Figure 3B). As a control, mRNA levels of genes in the LXR signaling pathway in the ileum were normal (Figure 3A), suggesting that SIRT1 specifically modulates the ileal bile acid metabolism.
Figure 3.
Loss of intestinal SIRT1 reduces the HNF1α/FXR mediated bile acid transport in the small intestine. (A) Deletion of intestinal SIRT1 results in reduced expression levels of HNF1α/FXR target genes in the ileum (n=8). (B) SIRT1 deficiency in the ileum leads to decreased levels of FXR protein (n=5-6). (C) SIRT1 deficiency in the ileum leads to decreased association of HNF1α with HNF1α binding sites on its target promoters (n=4). (D) SIRT1 deficiency in the ileum leads to decreased association of FXR with the FXR binding sites on the SHP and Ostβ promoters (n=4). (E) Deletion of intestinal SIRT1 leads to reduced induction of FGF15 and IBABP in the ileum in response to a high bile acid diet feeding (cholic acid diet, CA diet) (n=6-9). # 0.05<p<0.10, *p<0.05, **p<0.01.
To elucidate the mechanisms underlying the reduced expression of Asbt, FXR, and FXR target genes in SIRT1 iKO mice, we examined the recruitment of HNF1α, an upstream transcription factor for both FXR and Asbt 12, 20, to the FXR and Asbt promoters in the ileum using chromatin-immunoprecipitation (ChIP). As shown in Figure 3C, the HNF1α levels associated with these two promoters were significantly reduced in the SIRT1 deficient ileum, but the total HNF1α protein levels were normal (Figure 3B), indicating that intestinal SIRT1 deficiency decreases the DNA binding of HNF1α. In line with the reduced FXR levels, SHP and Ostβ promoters-associated FXR levels were also significantly decreased in the SIRT1 deficient ileum (Figure 3D). Additionally, when challenged with a high bile acid diet containing 0.5% cholic acid (CA diet), SIRT1 iKO mice had a reduced induction of a couple of FXR target genes in the ileum, such as FGF15 and an intestinal bile acid binding protein IBABP (Figure 3E). Together, our data indicate that deletion of intestinal SIRT1 reduces the recruitment of HNF1α to the FXR and Asbt promoters, thereby decreasing the FXR signaling pathway and ileal bile acid transport.
SIRT1 regulates the HNF1α/FXR signaling pathways partially through deacetylation of HNF1α dimerization co-factor DCoH2
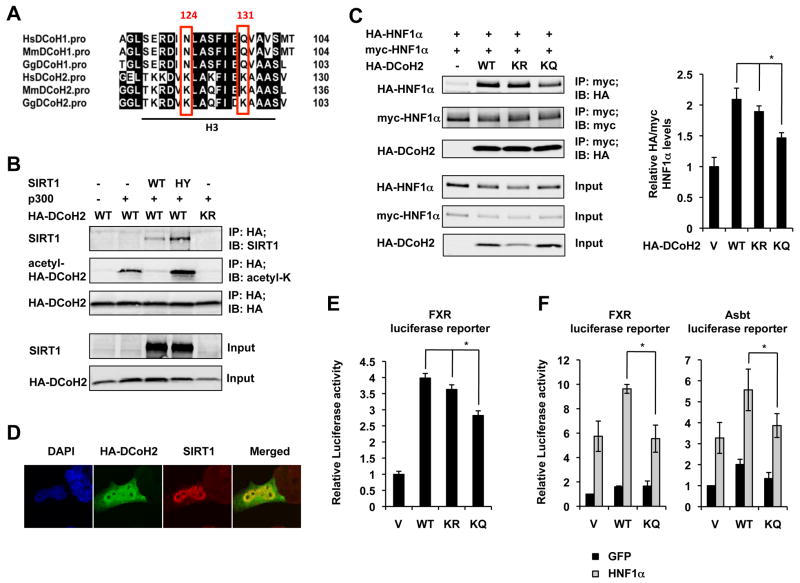
To further dissect the molecular mechanisms by which SIRT1 regulates the DNA binding ability of HNF1α, we investigated whether HNF1α and/or some of its cofactors are SIRT1 deacetylation substrates. Mass spectrometry analyses revealed that neither endogenous nor overexpressed HNF1α protein had detectable acetylation modifications. However, a dimerization co-factor of HNF1α, DCoH2, was found to be acetylated at K124 and K131 residues, two residues located at the DCoH2 protein surface (Figure 4A, Figure S4 and Figure S5A).
Figure 4.
SIRT1 regulates the dimerization of HNF1α through deacetylation of DCoH2. (A) DCoH2 is acetylated at K124 and K131 in the helix 3 (H3) domain. The K124 and K131 residues in DCoH2 and the corresponding N and Q residues in DCoH1 were highlighted. (B) SIRT1 interacts with and deacetylates DCoH2 in HEK293T cells. (C) Acetylation status of DCoH2 affects HNF1α dimerization in HEK293T cells (n=5). (D) DCoH2 is localized in both cytosol and nucleus in Hepa1-6 cells. (E) Acetylation status of DCoH2 affects its co-activation activity on a HNF1α target gene, FXR, in Hepa1-6 cells in a luciferase reporter assay (n=3). (F) Acetylation of DCoH2 reduces HNF1α mediated transcriptional activation on FXR and Asbt promoters (n=3). *p<0.05.
To test whether DCoH2 could be a substrate of SIRT1, we examined whether SIRT1 could physically interact with DCoH2 and induce its deacetylation. As shown in Figure 4B, in HEK293T SIRT1 RNAi cells, the wild-type (WT) SIRT1 protein was co-immunoprecipitated with HA-DCoH2 (IP-HA; IB-SIRT1). Consistent with the idea that DCoH2 is a substrate of SIRT1, the association between SIRT1 and HA-DCoH2 was increased in cells expressing the SIRT1 H355Y (HY) protein, a mutant that is known to stall the deacetylation reaction at the ternary enzyme:ADP-ribose:acetyl-substrate intermediate stage 29. Furthermore, expression of an acetyltransferase, p300, induced the acetylation levels of WT DCoH2 but not an acetylation defective mutant (K124R/K131R, KR), and co-expression of WT but not catalytically inactive HY mutant SIRT1 decreased the acetylation levels of DCoH2 (IP-HA; IB-acetyl-K). Taken together, these results indicate that DCoH2 is a substrate of SIRT1.
To test the possibility that the acetylation status of DCoH2 regulates the dimerization of HNF1α thereby modulating its DNA binding ability, we investigated whether the deacetylation mimetic (K124R/K131R, KR) and acetylation mimetic (K124Q/K131Q, KQ) of DCoH2 display distinct abilities to regulate HNF1α dimerization. As shown in Figure 4C and Figure S5B, the dimerization of HNF1α was significantly reduced in cells co-expressing the DCoH2 KQ mutant protein, as indicated by a decreased association between HA- and myc-tagged HNF1α proteins. Therefore, this observation demonstrates that acetylation of K124 and K131 residues in DCoH2 decreased the dimerization of HNF1α.
Finally, to test whether the acetylation status of DCoH2 indeed affects the transactivation of HNF1α in cells, we first analyzed the co-activation activities of WT, KR, and KQ DCoH2 proteins in Hepa1-6 cells, a mouse hepatic cell line that has significant expression levels of both HNF1α and FXR. DCoH2 was distributed in both the nucleus and cytoplasm in this hepatic cell line, with a strong nuclear staining that closely resembled the pattern of SIRT1 (Figure 4D). As shown in Figure 4E, the DCoH2 KQ mutant protein displayed a significantly reduced ability to activate a luciferase reporter driven by the mouse FXR promoter in Hepa1-6 cells. We then investigated the co-activation activity of DCoH2 proteins in a rat small intestinal epithelial cell line, IEC-6 cells (Figure 4F). Because the endogenous HNF1α levels in IEC-6 cells were low, we infected them with lentiviruses expressing control GFP proteins or HNF1α proteins. As shown in Figure 4F, the DCoH2 KQ mutant protein showed significantly decreased co-activation activity on HNF1α-mediated activation of both FXR and Asbt promoters. Collectively, our findings suggest that SIRT1 stimulates the transcriptional activity of HNF1α in part through deacetylation of its dimerization co-factor DCoH2.
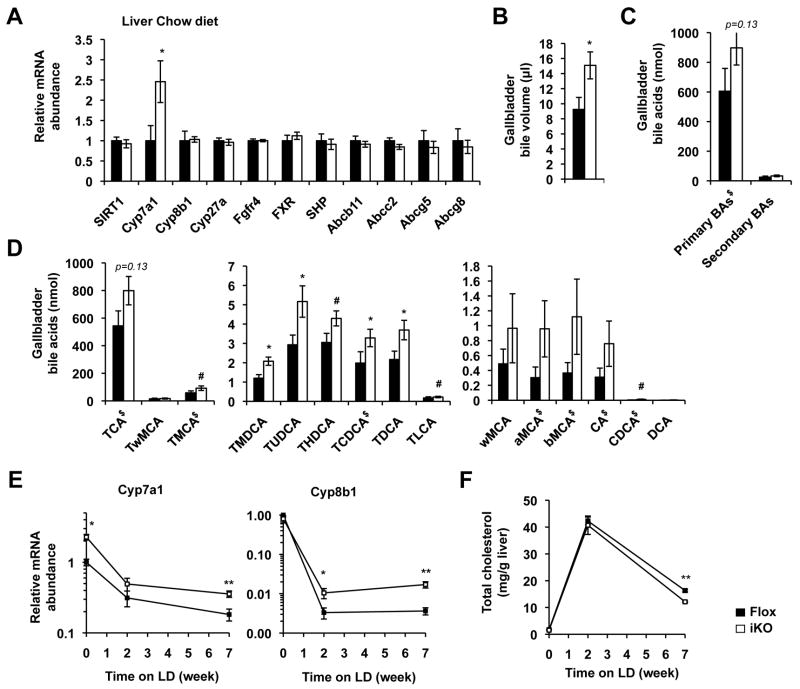
Intestine-specific deletion of SIRT1 increases hepatic bile acid biosynthesis, reduces hepatic accumulation of bile acids, and protects animals from high bile acid diet induced liver damage
As important regulators of bile acid metabolism, deficiency of HNF1α and FXR in humans and mice has been associated with a number of hepatic disorders, including an increased rate of hepatic bile acid synthesis 12, 30, 31. To test whether intestinal SIRT1 deficiency induced reduction of HNF1α/FXR pathways has any impacts on hepatic bile acid synthesis in vivo, we measured the hepatic expression levels of bile acid and cholesterol metabolism genes. As shown in Figure 5A, Cyp7a1, a rate limiting hepatic bile acid synthesis gene, was significantly elevated in SIRT1 iKO mice, whereas other genes involved in bile acid metabolism and cholesterol transport were not significantly altered, indicating that deletion of intestinal SIRT1 specifically induces hepatic bile acid synthesis. Consistent with this notion, SIRT1 iKO mice had increased gallbladder bile excretion (Figure 5B) and a trend of elevation in the gallbladder primary bile acid content (Figure 5C and 5D, Table S1) on the chow diet. Furthermore, when challenged with a high bile acid diet containing high-fat, high-cholesterol, and 0.5% cholic acid (lithogenic diet, LD), the repression of hepatic Cyp7a1 and Cyp8b1 was significantly attenuated in SIRT1 iKO mice compared to Flox control mice (Figure 5E). Since bile acids are synthesized from hepatic cholesterol, this increased residual hepatic bile acid synthesis in SIRT1 iKO mice may also account for the decreased hepatic cholesterol content (Figure 5F) and the reduced fecal cholesterol output (Figure S7E) on the LD diet. The fecal bile acid output, on the other hand, was not altered in SIRT1 iKO mice after 7 weeks on the LD diet (Figure S7F), probably due to saturation of the ileal bile acid transporters by the extremely high levels of dietary bile acids. The increase in hepatic bile acid synthesis and decrease in hepatic cholesterol content were also observed in SIRT1 iKO mice fed with a high-cholesterol-only diet (Figure S6). Altogether, these data suggest that deficient ileal bile acid absorption, possible also reduced expression of ileal FGF15, attenuates inhibition of hepatic bile acid biosynthesis from cholesterol in SIRT1 iKO mice.
Figure 5.
Deletion of SIRT1 in the intestine results in increased hepatic bile acid biosynthesis. (A) Deletion of SIRT1 in intestine results in increased expression of Cyp7a1 in liver (n=6-9). (B) SIRT1 iKO mice have increased gallbladder bile volume (n=8). (C-D) SIRT1 iKO mice have increased amount of biliary primary bile acids (n=8, labeled by $ in D). (E) Intestinal SIRT1 deficiency results in decreased inhibition of hepatic bile acid synthesis genes when fed with LD (n=7-8). (F) SIRT1 iKO mice have reduced hepatic levels of cholesterol after 7 weeks of LD feeding (n=7-8). # 0.05<p<0.10, *p<0.05, **p<0.01.
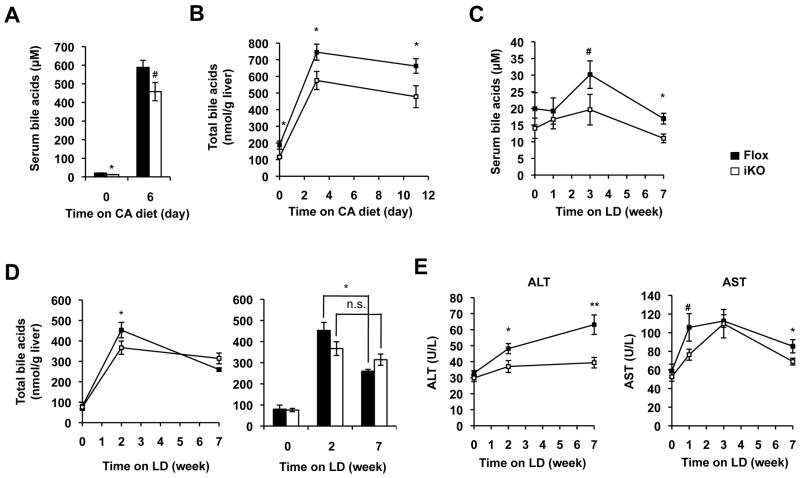
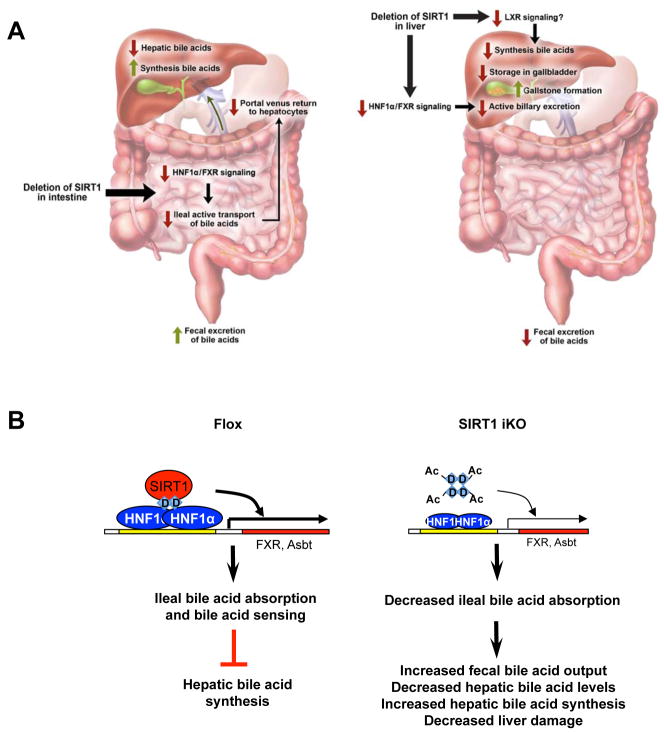
To further examine the pathophysiological effects of blunted HNF1α and FXR signaling in SIRT1 deficient mice, we challenged Flox and SIRT1 iKO mice with high bile acid diets. In line with the reduced ileal bile acid transport, SIRT1 iKO had decreased levels of serum and hepatic bile acids when fed with the CA diet (Figure 6A and 6B), and maintained lower serum bile acid levels during the 7-week feeding period of the lithogenic diet (LD) (Figure 6C), with normal body weight, food intake, and other serum lipid levels (Figure S7A and S7B). Again, consistent with the observation that SIRT1 iKO mice decreased their ileal bile acid absorption, the liver of SIRT1 iKO mice displayed significantly reduced levels of hepatic bile acids 2 weeks after LD feeding (Figure 6D). However, this reduction was lost in SIRT1 iKO mice with prolonged feeding of the LD diet due to a significant decrease of the hepatic bile acid content in Flox but not SIRT1 iKO mice (Figure 6D, right panel), confirming a less inhibition of hepatic bile acid synthesis in SIRT1 iKO mice. Interestingly, it appears that this decreased accumulation of hepatic bile acids partially protects SIRT1 iKO mice from LD-induced liver damage. As shown in Figure 6E, the serum ALT and AST levels were significantly reduced in SIRT1 iKO mice during 7 weeks of LD feeding without significant alterations in the hepatic histological lesions (Figure S7C). Together, these findings support the hypothesis that intestinal deletion of SIRT1 leads to defective HNF1α/FXR signaling pathways, resulting in reduced ileal bile acid absorption, increased fecal bile acid loss, enhanced hepatic bile acid synthesis, decreased hepatic bile acid accumulation, and reduced risk of liver damage (Figure 7A, left panel).
Figure 6.
Intestine-specific deletion of SIRT1 reduces intestinal bile acid uptake and hepatic bile acid accumulation upon feeding with high bile acid diets. (A-B) SIRT1 iKO mice have reduced serum (A) and hepatic (B) levels of total bile acids during 11-day feeding with the CA diet (n=6-9). (C-D) SIRT1 iKO mice display reduced levels of serum (C) and hepatic (D) levels of bile acids during 7-week feeding with LD diet (n=6-8). (E) Deletion of SIRT1 in the intestine leads to reduced serum Alanine transaminase (ALT) and Aspartate transaminase (AST) activities upon LD feeding (n=6-8). # 0.05<p<0.10, *p<0.05, **p<0.01.
Figure 7.
Intestinal SIRT1 is an important regulator of systemic bile acid homeostasis. (A) Deletion of intestinal SIRT1 or hepatic SIRT1 has distinct impacts on systemic bile acid homeostasis. Left, deletion of intestinal SIRT1 impairs the transcriptional activity of HNF1α, leading to a decreased FXR signaling pathway and thereby reduced bile acid absorption in the distal ileum and increased elimination of bile acid from feces. The blunted ileal bile acid absorption further feedback increases hepatic bile acid synthesis, leading to decreased liver damage under lithogenic condition. Right, deletion of hepatic SIRT1 decreases the HNF1α/FXR signaling pathways and reduces active biliary excretion of bile acids. Hepatic deletion of SIRT1 also impairs hepatic bile acid synthesis, probably through the LXR signaling pathway, further reducing biliary bile acid contents. The reduced storage of bile acids in gallbladder leads to hypersaturation of biliary cholesterol and formation of cholesterol gallstones. Please note that red arrows designate decreased activities whereas green arrows designate increased activities. (B) Intestinal SIRT1 regulates ileal bile acid absorption and systemic bile acid metabolism through the DCoH2/HNF1α/FXR signaling pathways. Our data suggests that deletion of SIRT1 leads to hyperacetylation of DCoH2, which in turn results in decreased dimerization and DNA binding of HNF1α as well as reduced the transcription levels of HNF1α downstream genes such as FXR and Asbt.
Discussion
Bile acids not only play an important role in the regulation of lipid absorption and cholesterol homeostasis, but also function as hormones to modulate systemic energy metabolism in various metabolic tissues 32-34. Defects in bile acid homeostasis have been associated with a number of metabolic diseases, including cholestasis, hypercholesterolemia, defective liver regeneration, and diabetes 4-7. We and others have showed that hepatic SIRT1 is a crucial regulator of the HNF1α/FXR signaling pathway and hepatic bile acid metabolism, and that SIRT1 modulates the FXR signaling pathway at multiple levels, including transcriptional regulation through HNF1α and posttranslational deacetylation 20, 28. In the present study, using an intestine-specific SIRT1 knockout mouse model (with the Villin-Cre strain as an additional control (Figure S8)), we demonstrate that the intestinal HNF1α/FXR signaling and the bile acid enterohepatic circulation are also under control of intestinal SIRT1 through a similar multi-level mechanism. This interaction between SIRT1 and HNF1α/FXR signaling pathways plays an essential role in the regulation of systemic bile acid homeostasis. However, in contrast to hepatic SIRT1 deficiency (Figure 7A, right panel) 20, deletion of intestinal SIRT1 reduced ileal bile acid transport and FGF15 expression, increasing hepatic biosynthesis of bile acids yet protecting mice from high bile acid diets induced cholestasis and liver damage (Figure 5 and 6). Therefore, our findings suggest that the impacts of pharmacological modulation of SIRT1 activity on bile acid and cholesterol homeostasis are tissue-specific and more complex than previously expected.
Our studies further indicate that intestinal SIRT1 deficiency impairs systemic bile acid homeostasis through multiple pathways, including both HNF1α transcriptional network and FXR signaling, and possibly additional factors that may affect bile acid metabolism directly or indirectly. As a result, SIRT1 iKO mice did not phenocopy any single knockout mouse models. For instance, even though SIRT1 iKO mice have reduced ileal FGF15 expression and increased hepatic bile acid synthesis (Figure 4 and 5) just like intestinal-specific FXR KO mice 30, 31, deletion of SIRT1 in intestine protects liver from cholestasis and damage (Figure 6), whereas activation of intestinal FXR yields similar phenotypes 35. Moreover, FXR intestinal KO mice have a slight increase in total bile acid pool size 30, 31, whereas SIRT1 iKO mice have normal bile acid pool size (Figure 2D). A reasonable explanation for these discrepancies is that SIRT1 deficiency primarily impairs the activity of HNF1α other than FXR. It is likely that the increased fecal bile acid loss (resulted from the decreased HNF1α-Asbt axis) dominates the increased in hepatic bile acid synthesis in SIRT1 iKO mice, resulting in less accumulation of bile acids in liver (cholestasis) and reduced liver damage. This dominant effect may also account for the normal bile acid pool size of SIRT1 iKO mice, as the increased hepatic bile acid synthesis was offset by the increased fecal bile acid loss. In addition, tissue-specificity may also be a contributing factor to the apparent inconsistency between SIRT1 iKO mice and reported whole body HNF1α null 12 and/or FXR null mice 31. Since FXR iKO mice and FXR whole body null mice do not have identical bile acid phenotypes, particularly the serum levels of bile acids and fecal bile acid output 31, further studies that compare SIRT1 iKO with intestine-specific HNF1α or FXR KO mouse models will be helpful to further resolve these inconsistencies.
Our current study also revealed that SIRT1 modulates the HNF1α/FXR pathway through deacetylation of DCoH2 and subsequent dimerization and DNA binding of HNF1α, therefore uncovering a previously unknown link between DCoH2, SIRT1, and HNF1α (Figure 4 and Figure 7B). We discovered that DCoH2 is an acetylated protein and a substrate of SIRT1 (Figure 4). Interestingly, two lysine residues that are acetylated in DCoH2, K124 and K131, are not conserved in DCoH1, a closely related protein. Instead, they are replaced by asparagine (N) and glutamine (Q), which are acetyl-lysine mimetics (Figure 4A). These observations suggest that DCoH1 could be an acetyl mimetic of DCoH2, and DCoH1 and DCoH2 might have different abilities to co-activate HNF1α. Previous studies indicate that this is indeed the case. It has been shown that the DCoH1 protein forms stable tetramers, and cannot efficiently promote the dimerization of HNF1α and subsequent DNA binding 36. In contrast, the purified DCoH2 tetramers are less stable, can efficiently promote the formation of the DCoH2:HNF1α/DNA complex 36. Therefore, it is likely that deletion of SIRT1 leads to hyperacetylation of DCoH2 and stabilization of the DCoH2 homo-tetramers, thereby a decreased formation of the DCoH2:HNF1α hetero-tetramers (Figure 7B). Additional biochemical and structural studies with WT, KR and KQ DCoH2 proteins are needed to investigate how the acetylation status of DCoH2 affects the interaction between DCoH2 and HNF1α as well as HNF1α dimerization.
In summary, we have shown that intestinal SIRT1 plays a vital role in the regulation of ileal bile acid absorption and systemic bile acid homeostasis. Intestinal deletion of SIRT1 decreases the HNF1α/FXR signaling through hyperacetylation of its cofactor DCoH2, leading to reduced intestinal bile acid transport and altered systemic bile acid homeostasis. Our findings point out that the same molecular mechanism can yield distinct physiologies in different tissues, and suggest that therapeutic strategies based on the SIRT1 small molecule modulators against bile acid and cholesterol diseases have to consider tissue specificity as a complicating factor.
Supplementary Material
Acknowledgments
We thank Drs. Anton Jetten, Stavros Garantziotis, and members of the Li laboratory for critical reading of the manuscript; Dr. Frederic Alt at Harvard Medical School for providing SIRT1 exon 4 floxed allele; and Dr. P. Kay Lund at University of North Carolina-Chapel Hill for IEC-6 cells. We also thank Qing Xu and Ashley Kang for technical support; Dr. Arun Pandiri, Beth Mahler and Eli Ney from the National Toxicology Program for pathological analyses of liver sections; NIEHS Laboratory of Experimental Pathology for histological staining and serum biochemistry, and Dr. Grace Kissling from the NIEHS Biostatistics Branch for advice on statistical methodology. This research was supported by the Intramural Research Program of National Institute of Environmental Health Sciences of the NIH, to X.L. (Z01 ES102205), the National Institute of Diabetes and Digestive Diseases of the NIH to P.A.D. (DK047987), and the National Cancer Institute of the NIH to Y.Z. (CA126832).
Footnotes
Author Contributions
N. K. designed experiments, carried out experiments, analyzed data, and wrote the manuscript. M. R. M., A. P., J. L., A. R., S. L., M. P.-H., A. L., and I. C. carried out experiments and analyzed data, Y. Z. and P. A. D. designed experiments, analyzed data, and critically reviewed the manuscript. X. L. designed experiments, carried out experiments, analyzed data, and wrote the manuscript.
Conflict of Interest
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–74. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 2.Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31:572–80. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Dawson PA, Hubbert ML, Rao A. Getting the mOST from OST: Role of organic solute transporter, OSTalpha-OSTbeta, in bile acid and steroid metabolism. Biochim Biophys Acta. 2010;1801:994–1004. doi: 10.1016/j.bbalip.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–44. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 5.Cariou B, Staels B. FXR: a promising target for the metabolic syndrome? Trends Pharmacol Sci. 2007;28:236–43. doi: 10.1016/j.tips.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–6. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 7.Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–9. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt DR, Mangelsdorf DJ. Nuclear receptors of the enteric tract: guarding the frontier. Nutr Rev. 2008;66:S88–97. doi: 10.1111/j.1753-4887.2008.00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. The organic solute transporter alpha-beta, Ostalpha-Ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc Natl Acad Sci U S A. 2008;105:3891–6. doi: 10.1073/pnas.0712328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–25. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, Stoffel M. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–82. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 13.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–35. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 15.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Kazgan N. Mammalian sirtuins and energy metabolism. Int J Biol Sci. 2011;7:575–87. doi: 10.7150/ijbs.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–74. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 18.Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Annals of Medicine. 2011;43:198–211. doi: 10.3109/07853890.2010.547211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X. SIRT1 and energy metabolism. Acta Biochim Biophys Sin (Shanghai) 2013;45:51–60. doi: 10.1093/abbs/gms108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purushotham A, Xu Q, Lu J, Foley JF, Yan X, Kim DH, Kemper JK, Li X. Hepatic deletion of SIRT1 decreases hepatocyte nuclear factor 1alpha/farnesoid X receptor signaling and induces formation of cholesterol gallstones in mice. Mol Cell Biol. 2012;32:1226–36. doi: 10.1128/MCB.05988-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003 doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–83. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Limaye PB, Lehman-McKeeman LD, Klaassen CD. Dysfunction of organic anion transporting polypeptide 1a1 alters intestinal bacteria and bile acid metabolism in mice. PLoS One. 2012;7:e34522. doi: 10.1371/journal.pone.0034522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- 26.Wilson TH, Wiseman G. The use of sacs of everted small intestine for the study of the transference of substances from the mucosal to the serosal surface. J Physiol. 1954;123:116–25. doi: 10.1113/jphysiol.1954.sp005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lack L, Weiner IM. In vitro absorption of bile salts by small intestine of rats and guinea pigs. Am J Physiol. 1961;200:313–7. doi: 10.1152/ajplegacy.1961.200.2.313. [DOI] [PubMed] [Google Scholar]
- 28.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith BC, Denu JM. Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–82. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–43. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim I, Ahn SH, Inagaki T, Choi M, Ito S, Guo GL, Kliewer SA, Gonzalez FJ. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–72. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–91. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 33.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–25. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–91. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- 35.Modica S, Petruzzelli M, Bellafante E, Murzilli S, Salvatore L, Celli N, Di Tullio G, Palasciano G, Moustafa T, Halilbasic E, Trauner M, Moschetta A. Selective activation of nuclear bile acid receptor FXR in the intestine protects mice against cholestasis. Gastroenterology. 2012;142:355–65. e1–4. doi: 10.1053/j.gastro.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 36.Rose RB, Pullen KE, Bayle JH, Crabtree GR, Alber T. Biochemical and structural basis for partially redundant enzymatic and transcriptional functions of DCoH and DCoH2. Biochemistry. 2004;43:7345–55. doi: 10.1021/bi049620t. [DOI] [PubMed] [Google Scholar]
- 37.Moschetta A, Bookout AL, Mangelsdorf DJ. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat Med. 2004;10:1352–8. doi: 10.1038/nm1138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.