Abstract
Structural understanding of eukaryotic translation lags behind that of translation on bacterial ribosomes. Here we present two subnanometer-resolution structures of S. cerevisiae 80S ribosome complexes formed with either one or two tRNAs, bound in response to an mRNA fragment containing the Kozak consensus sequence. The ribosomes adopt two globally different conformations, related to each other by the rotation of the small subunit. Comparison with bacterial ribosome complexes reveals that the global structures and modes of intersubunit rotation of the yeast ribosome significantly differ from those in the bacterial counterpart, most notably in the regions involving the tRNA, small ribosomal subunit and conserved helix 69 of the large ribosomal subunit. The structures provide insight into ribosome dynamics implicated in tRNA translocation and help elucidate the role of the Kozak fragment in positioning an open reading frame during translation initiation in eukaryotes.
Introduction
Translation of genetic information to protein sequence is performed by ribosomes in all organisms. Whereas the functional sites of the ribosome, such as the decoding center and the peptidyl transferase center, are universally conserved, there are substantial differences between bacterial and eukaryotic translation, as reflected in distinct mechanisms of initiation, termination and ribosome recycling (Aitken and Lorsch, 2012; Dever and Green, 2012).
Translation initiation plays an important role in gene expression regulation in homeostasis, cell stress, development and disease (Sonenberg and Hinnebusch, 2009). In bacteria, translation initiation depends on three initiation factors, and results in the formation of a 70S ribosome complex with initiator tRNAfMet bound in the P site in response to the AUG codon (Myasnikov et al., 2009). The Shine-Dalgarno sequence upstream of the open reading frame (Dalgarno and Shine, 1973) forms base-pairing interactions with the complimentary anti-Shine-Dalgarno region of the ribosomal 16S RNA. Formation of this specific contact results in positioning of the downstream AUG start codon in the P site of the small 30S subunit, thus determining the open reading frame of the mRNA for translation (Kaminishi et al., 2007; Korostelev et al., 2007; Yusupova et al., 2006). By contrast, initiation in eukaryotes depends on at least a dozen initiation factors (Aitken and Lorsch, 2012). An mRNA region termed the Kozak consensus sequence flanking the AUG start codon is required for efficient translation initiation (Kozak, 1986). The context around the start codon is thus considered critical for selection of the correct AUG start codon among several possible AUG trinucleotides at the 5′ end of an mRNA. Although the Kozak sequence is variable among groups of eukaryotes, in vertebrates the sequence is strongly biased toward containing purines at positions −3 and +4, relative to A+1 of the AUG start codon (Cavener and Ray, 1991). Mutation of a purine to pyrimidine at position −3 was shown to have the most detrimental effect on the efficiency of translation, compared to mutations at other positions (Kozak, 1986). In non-vertebrate eukaryotes, the most stringent requirement is that a nucleotide at position −3 is a purine, whereas the conservation and functional importance of the identity of the nucleotide at position +4 is less pronounced (Cavener and Ray, 1991). Despite suggestions that there is some analogy of the Kozak consensus and Shine-Dalgarno sequences based on their location, the Kozak sequence likely does not act by forming base pairing interactions with 18S ribosomal RNA. First, variability of the Kozak sequence among eukaryotes does not correlate with the high conservation of the eukaryotic 18S ribosomal RNA. Second, the 3′ end of 18S rRNA of most species does not contain sequences that would be strongly complementary to the Kozak consensus (Verrier and Jean-Jean, 2000). Third, introducing complementarity between the untranslated 5′ region and the 3′ end of 18S rRNA leads to inhibition of translation rather than enhancement (Verrier and Jean-Jean, 2000). As such, the molecular mechanism of the Kozak sequence function remains unknown.
Upon initiation, elongation of the polypeptide chain takes place. The cycle of elongation is accompanied by consecutive movement (translocation) of tRNAs and respective mRNA codons from the A (aminoacyl-) to P (peptidyl-) to E (exit) site. In the studies of bacterial ribosomes, significant progress has been made in structural and functional characterization of translocation intermediates reflecting the key steps of translocation. Upon peptidyl transfer from the peptidyl-tRNA to aminoacyl-tRNA, translocation occurs in two consecutive steps. These are (1) the movement of the acceptor arms of the peptidyl- and deacyl-tRNAs from the A and P sites into the P and E sites on the large subunit (resulting in tRNAs adopting hybrid A/P and P/E conformations) followed by (2) movement of the anticodon stem loops into the P and E sites on the small subunit, catalyzed by elongation factor G (in bacteria) or eEF-2 (in eukaryotes). Crystallographic and electron cryo-microscopy (cryo-EM) structures as well as solution experiments helped establish that during translation the ribosome undergoes large-scale conformational changes. The most pronounced is the intersubunit rotation (Frank and Agrawal, 2000) coupled with formation of hybrid states by tRNAs (Ermolenko et al., 2007; Moazed and Noller, 1989; Spiegel et al., 2007). The ribosome adopts a non-rotated state when tRNAs are located in the classical A, P and E sites. The small subunit rotates relative to the large subunit by ~9 degrees when tRNAs adopt hybrid A/P and P/E states. Importantly, the ribosome and tRNAs fluctuate spontaneously between the non-rotated (classical) and rotated (hybrid) states, independent of elongation factors, emphasizing that formation of translocation intermediates is an inherent property of the ribosome (Cornish et al., 2008). In addition to intersubunit rotation, large-scale intrasubunit rearrangements take place, including: (a) rotation of the head of the small subunit (Guo and Noller, 2012; Ratje et al., 2010), which allows widening of the mRNA-tRNA corridor between the P and E sites (Schuwirth et al., 2005), and (b) movement of the L1 stalk, which interacts with the tRNA elbow as tRNA progresses from the P/E to E states (Cornish et al., 2009; Fei et al., 2008).
Structural understanding of eukaryotic translation lags behind that of bacterial translation. Some studies suggest that the dynamic behavior of eukaryotic ribosomes and tRNAs as well as their structural details may differ from those of bacterial counterparts. Specifically, cryo-EM reconstructions of vacant eukaryotic ribosomes revealed that eukaryotic ribosomes predominantly adopt a rotated conformation, unlike vacant bacterial ribosomes, which predominantly sample the non-rotated state (Cornish et al., 2008; Spahn et al., 2004). Furthermore, the predominant conformations of eukaryotic ribosomes bound with elongation factor eEF-2 and endogenous tRNA found near the E site (Anger et al., 2013) differ from those of bacterial ribosomes bound with EF-G and tRNA sampling the hybrid P/E states (Brilot et al., 2013; Frank and Agrawal, 2000; Pulk and Cate, 2013; Tourigny et al., 2013; Zhou et al., 2013) or classical P state (Gao et al., 2009). To understand the mechanism of translocation on eukaryotic ribosomes, a detailed structural reconstruction of 80S translocation intermediates is required. Whereas a number of tRNA- and elongation factor eEF2-bound eukaryotic complexes have been visualized by cryo-EM, the structural details of 80S ribosome dynamics and interactions with tRNA and mRNA remain poorly characterized due to several reasons. These include insufficient resolution, compositional heterogeneity of complexes containing endogenous mixtures of mRNA and tRNA, and until recently the absence of a high-resolution crystal structure of the eukaryotic ribosome that could be used for structural comparisons. A 3 Å crystal structure of the yeast 80S ribosome in the absence of tRNA and mRNA was recently determined (Ben-Shem et al., 2011), allowing for detailed model building and interpretation of cryo-EM structures. Concurrently, pre-translocation mammalian ribosomes assembled with defined mRNA and tRNA were visualized by cryo-EM at 9 to 10.6 Å resolution (Budkevich et al., 2011). The latter structures revealed that in the presence of the A-site tRNA, the pre-translocation ribosome adopts the rotated and non-rotated states, coupled with tRNAs adopting the hybrid and classical states, respectively. The positions of the P and E-site tRNAs in the non-rotated ribosome were reported to significantly differ from those in the bacterial ribosomes. It remained unclear, however, whether the A-site tRNA plays a role in inducing such differences. In order to reconstruct the structural pathway of translocation on the 80S ribosome, additional complexes are required to be visualized, including the post-translocation ribosome containing tRNA in the P and E sites, as well as other tRNA-bound intermediates that may bring insights in ribosome dynamics.
In this work, we present ~6 Å resolution cryo-EM structures for two compositionally homogeneous tRNA-bound Saccharomyces cerevisiae 80S complexes formed with a defined mRNA construct containing the S. cerevisiae Kozak consensus fragment. Improvements in cryo-EM data classification (Lyumkis et al., 2013) and the use of the 80S ribosome crystal structure (Ben-Shem et al., 2011) refined against our cryo-EM maps allow us to present detailed structural models of the 80S•2tRNA and 80S•tRNA complexes. Comparison of the 80S structures with those of the bacterial counterparts provides new insights into the kingdom-specific structural aspects of translocation. In addition, our structures bring new insights into the role of the Kozak fragment in positioning the open reading frame on the ribosome. Specifically, the structures suggest that the Kozak sequence works at least in part as a molecular ruler, allowing placement of the AUG codon in the P site, due to interactions of the Kozak sequence elements with the conserved structures of the small ribosomal subunit.
Results and Discussion
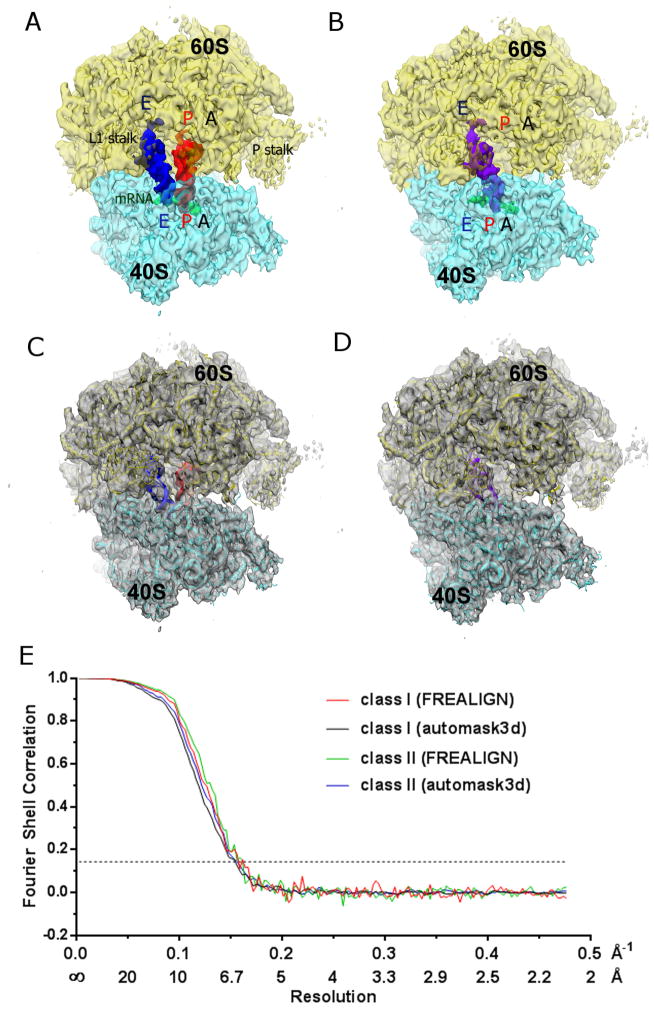
We have assembled S. cerevisiae 80S ribosome complexes in the presence of initiator tRNAfMet and an mRNA fragment that includes an authentic S. cerevisiae Kozak fragment preceding the AUG start codon, as described in Experimental Procedures. To ensure compositional homogeneity, vacant ribosomes free of endogenous tRNA (Ben-Shem et al., 2011) were purified prior to complex assembly. Cryo-EM classification of 80S particles (Fig. S1) revealed two main 80S classes at ~6 Å resolution, accounting for nearly 60% of particles (Fig. 1). The quality of both maps allowed interpretation of protein and RNA secondary structure. In the core of the ribosome, which is most highly ordered, it is possible to interpret phosphate backbone and individual bulged or stacked nucleotides. We used real-space refinement to accurately fit the 3 Å crystal structure of S. cerevisiae ribosome into the cryo-EM maps essentially as described (Koh et al., 2014) (Figs. 1C–D). Class I comprises non-rotated 80S ribosomes, bound with two tRNA molecules, in the P and E sites (80S•2tRNA; Fig. 1A). Class II represents rotated 80S ribosomes bound with a single tRNA in the hybrid P/E state (80S•tRNA; Fig. 1B).
Figure 1.
Cryo-EM maps of the ribosome bound with (A) two tRNAs and (B) single tRNA. Large 60S subunit is shown in yellow-orange, small 40S subunit in cyan, E-site tRNA molecule in blue, P-site tRNA molecule in red, hybrid P/E tRNA in purple. (C–D) Molecular structures of the ribosome refined into the maps shown in panels A and B. Colors are as in panels A and B. (E) Fourier shell correlation (FSC) curves for classes representing the 80S•2tRNA and 80S•tRNA complexes (I and II, respectively). FSC calculated using automask3d and FREALIGN for each class were generated as described (Experimental Procedures). For convenience, the X-axis is labeled with spatial frequency Å−1 and with Å. The resolution stated in the text corresponds to a FSC = 0.143 cut-off value shown as a dotted line, for FREALIGN-derived FSC.
Structure of the non-rotated 80S ribosome bound with two tRNAs
The complex with two tRNAs bound in the P and E sites of the non-rotated ribosome represents a post-translocation-like state, which to our knowledge has not been characterized for eukaryotic ribosomes in detail. Comparison of our 80S•2tRNA ribosome structure with the bacterial 70S ribosome bound with two tRNAs (Jenner et al., 2010) reveals two global differences. First, the small subunit is significantly tilted relative to the large 60S subunit (Fig. 2A). The tilt of the subunit as a whole places the head of the small 40S subunit by up to 7 Å farther from the core of the large subunit than in the bacterial ribosome. The tilt is coupled with displacement of the intersubunit bridge B1b (Yusupov et al., 2001) formed by the proteins S18 (S13 in bacteria) at the head of the 40S subunit and L11 (L5 in bacteria) at the central protuberance of the large subunit (Fig. 2B). Here, the position of the central protuberance, whose core is formed by 5S ribosomal RNA, relative to the core of the large subunit is different from that in the bacterial ribosome. This protrusion of the central protuberance as a whole results from eukaryote-specific expansion segments 31ES9 (stemming from helix 31), 38ES12 (stemming from helix 38, also known as the A-site finger) and elongated helix 30 (Petrov et al., 2014). These extensions displace 5S ribosomal RNA and the associated L11 toward the small subunit (Figs. 2B–C). In summary, the addition of the eukaryote-specific ribosomal RNA expansion segments next to 5S rRNA results in displacement of the central protuberance toward the small subunit, resulting in the tilting of the latter.
Figure 2.
Structural differences between tRNA-bound non-rotated 80S yeast ribosomes (this work) and ribosomes from other kingdoms of life. (A) The tilt of the small subunit relative to the large subunit, induced by eukaryote-specific expansion segments of 25S ribosomal RNA and resulting in the shift of the head of the small subunit away from the core of the large subunit by up to 7 Å. Ribosomal proteins are not shown for clarity. Comparison of the position of 18S ribosomal RNA in the non-rotated 80S (this work) with that of 16S ribosomal RNA in Thermus thermophilus 70S ribosome (Jenner et al., 2010) bound with two tRNAs (PDB ID 3I9B) was obtained by structural alignment of 25S rRNA (yellow) and 23S rRNA (not shown). (B) Displacement of bridge B1b, formed between the central protuberance of the large subunit and head of the small subunit, in the 80S ribosome relative to that in the 70S ribosome (gray). The superposition was achieved by structural alignment of 25S rRNA of the 80S•2tRNA ribosome (this work) and 23S rRNA of T.th. 70S•2tRNA ribosome structure (Jenner et al., 2010). (C) Differences between the positions of P-site tRNAs in the yeast (red) and Thermus thermophilus (gray) ribosome structures, induced by different architectures of the central protuberance. The superposition was obtained as in panel B, by aligning large-subunit rRNAs of corresponding structures (this work and Jenner et al., 2010). (D) Conformational and positional differences between tRNAs bound to yeast (red and blue, this work) and bacterial T. th. 70S ribosomes (gray; Jenner et al., 2010). Positions of E- and P-site tRNAs were compared by structural alignment of small-subunit rRNAs. (E) Conformational difference between E-site tRNAs bound to yeast (blue, this work) and bacterial (gray; Jenner et al., 2010) ribosomes, obtained by structural alignment of tRNA anticodon stem loops. (F–H) Conformations of the 3′-CCA terminus of E-tRNAs bound to yeast (F, this work) and T. th. (G, Jenner et al., 2010) ribosomes, and an acceptor-arm analog bound to the archaeal H. ma. 50S subunit (H, (Schmeing et al., 2003). Large subunit is shown in yellow (rRNA) and orange (proteins), small subunit in cyan (18S) and marine (proteins), E-site tRNA in blue, P-site tRNA in red, mRNA in green, eukaryote specific elements of 25S rRNA in yellow-green. Elements of the 70S ribosome are shown in grey when compared with those of the 80S ribosome.
The second significant difference between eukaryotic and bacterial ribosomes is in the positions of the tRNAs in the classical P and E sites. In the yeast ribosome, the P-site tRNA is located closer to the E site, whereas the E-site tRNA is closer to the P site (Fig. 2D). This brings the elbows of tRNAs in the 80S complex ~15 Å closer together than in the bacterial ribosome (Korostelev et al., 2006; Selmer et al., 2006). Our analysis of the kingdom-specific structural differences suggests that the positioning of the P-site tRNA is influenced by the placement of the central protuberance, with which the tRNA interacts via L11 (L5 in bacteria) (Fig. 2C). In the E site, the elbow of the tRNA interacts with the L1 stalk (Fig. S2). The difference in bacterial and eukaryotic E-site tRNA positions is likely caused by differences in the L1 stalk structures (Petrov et al., 2014). The L1 stalk of the yeast ribosome (Figs. S2A and S2B) lacks helix 78, which in bacterial ribosomes approaches small subunit protein S11 (Figs. S2C and S2D). Because the contact between the L1 stalk and the small subunit is absent in the non-rotated yeast ribosome, the L1 stalk and E-site tRNA elbow adopt different positions than in the bacterial ribosome.
We also investigated whether the distinct global conformations of the yeast and bacterial complexes are specific to the yeast ribosome or to the post-translocation-like state of the 80S ribosome. To this end, we compared our structure with the ~10 Å resolution pre-translocation-like rabbit 80S ribosomes, in which all the three tRNA sites are occupied by tRNAs (Budkevich et al., 2011). We found that the positions of tRNAs in the P and E sites of these complexes are nearly identical, indicating that the occupancy of the A site does not influence the placement of the P and E tRNAs. The similar positions of tRNAs in the yeast and mammalian ribosomes suggest that the tRNA positioning is conserved among eukaryotes.
The relative positions of ribosomal functional centers in the A, P and E sites are nearly identical in bacterial and eukaryotic ribosomes. Our observation of different tRNA positions in the bacterial and eukaryotic ribosomes prompted us to look also for conformational differences within the tRNAs that could account for different positioning of the tRNAs relative to the similarly positioned functional centers. tRNAs are known to be highly flexible, and their conformational dynamics are thought to be critical for tRNA translocation through the ribosome, during which the acceptor arm and the CCA end sample different conformations relative to the anticodon stem loop (Caulfield et al., 2011). Superposition of tRNAs from our 80S structures with those of the bacterial 70S counterpart (Jenner et al., 2010) revealed that the classical P-site tRNA adopts nearly identical conformations. By contrast, the E-site tRNA adopts significantly different conformations in the 70S and 80S complexes. Specifically, the CCA end in the 80S complex is bent by ~20° relative to that in the bacterial complex (Fig. 2E). The E-site tRNA interacts with the E site of the large subunit (Fig. 2F), which in eukaryotes is compositionally and conformationally different from that in bacteria. In bacterial tRNA-bound ribosomes, the CCA 3′ end of the tRNA interacts with helix 82 of 23S rRNA and protein L28 (Korostelev et al., 2006; Selmer et al., 2006). The two cytosines of the CCA trinucleotide form a system of stacked nucleotides with the acceptor arm (Fig. 2G). In our structure of the non-rotated 80S ribosome, the CCA end interacts with the conserved h82 (in the vicinity of C2765 and G2793) and with protein L44e, which is structurally distinct from bacterial L28. In contrast to the bacterial E-site tRNA structure, the penultimate nucleotide C75 of the tRNA appears unstacked from C74 and instead interacts with L44e in the vicinity of Tyr43 (Fig. 2F). In this conformation, the 3′ terminus of the tRNA and its interactions closely resemble those of a hairpin loop mimicking the tRNA acceptor arm in the 3.1 Å crystal structure (Fig. 2H) of the archaeal 50S subunit (Schmeing et al., 2003), consistent with the phylogenetic conservation of L44e between eukaryotes and archaea. In summary, the phylogenetic differences in the composition of the E sites between bacteria and eukaryotes contribute to the diverged conformations of the tRNA bound in the E site.
Structure of the rotated 80S ribosome bound with the P/E tRNA
Upon classification of cryo-EM images, the second most populated class contained a single deacylated tRNA. Structural studies of bacterial ribosomes bound with a deacylated tRNA have revealed a variety of inter- and intra-subunit rearrangements, providing important insights into the dynamics of the 70S ribosome (Agirrezabala et al., 2012). This prompted us to characterize the 80S•tRNA complex in detail. In the 80S•tRNA structure, the ribosome adopts a single rotated conformation, in which the small subunit is rotated relative to that in the 80S•2tRNAs complex by ~8.5° (Fig. 1B).
As in the analyses described above, we also compared the rotated 80S•tRNA ribosome and the crystal structure of its bacterial counterpart (Dunkle et al., 2011). We found significant differences in the positions of the small subunit, tRNA and helix 69 of the large subunit, which forms an intersubunit bridge next to the decoding center of the small subunit. These differences arise from the tilted position of the small subunit relative to the large subunit, resulting from the interaction between the protruding central protuberance and the 40S head (Fig. 3A). The tilt of the small subunit in the rotated ribosome is more pronounced than that in the non-rotated structure (~6° vs. ~3°). The tilt results in shifted positions of tRNA binding sites on the 40S subunit relative to the core of the 60S subunit in comparison to the sites in bacterial ribosomes. Specifically, the anticodon stem loop of P/E tRNA and helix 69 (h69) of the rotated 80S ribosome are shifted together with the small subunit by almost 6 Å toward to the E site in comparison with those in the bacterial ribosomes (Fig. 3B). The rearrangement of h69 relative to the large subunit is thus coupled with the movement of the small subunit, with which h69 forms intersubunit bridge B2a (Yusupov et al., 2001).
Figure 3.

Differences in the conformations of rotated yeast and bacterial ribosomes bound with P/E tRNA. (A) The tilt of the small 40S subunit relative to the large 60S subunit, resulting in the shift of the head of the small subunit away from the core of the 60S subunit by up to 15 Å. Proteins are not shown for clarity. Comparison of the positions of 18S ribosomal RNA in the rotated 80S ribosome (this work) with E. coli 16S ribosomal RNA (Dunkle et al., 2011) was obtained by structural alignment of 25S rRNA (yellow) and 23S rRNA (not shown). (B) Difference in positions of the tRNA, mRNA and helix 69 between the rotated yeast (this work) and E. coli (Dunkle et al., 2011) ribosomes. The superposition was obtained by structural alignment of large-subunit rRNA of the 80S and 70S complexes, respectively (not shown). (C) Intersubunit rotation axes for yeast (brown) and E. coli (black) ribosomes. The rotation axis for each ribosome was calculated in Chimera (Pettersen et al., 2004), using the small subunit rRNA from non-rotated (bound with two tRNAs) and rotated (bound with single tRNA) ribosome structures. The superposition of yeast and bacterial (Dunkle et al., 2011) ribosome structures was obtained by structural alignment of large-subunit rRNAs. Head and body domains of the small subunit, and the central protuberance (CP) of the large subunit are labeled. 60S is in yellow and orange, 40S in cyan and marine, 70S in grey, 80S rotation axis in brown, 70S rotation axis in black.
We next asked whether the 80S ribosome with a single deacylated P/E tRNA is different from the rotated rabbit 80S ribosome, in which two tRNAs are bound in the P/E and A/P states, respectively. Superposition of the large subunit ribosomal RNAs revealed that the global conformations of the yeast and mammalian ribosomes are similar, as well as the position of the P/E tRNA. This indicates that the occupancy of the A/P hybrid state does not affect the overall global conformation of the ribosome. The position of h69 in the mammalian ribosome structure (Budkevich et al., 2011) appears to be different from that in the yeast ribosome, perhaps due to the occupancy of the A site of the 40S subunit, which is located near h69. It therefore remains to be clarified whether h69 moves passively as the ribosome rearranges between the rotated and non-rotated states or is involved in A-site tRNA repositioning in the course of translocation.
Intersubunit rotation of the 80S ribosome
To gain insights into the mechanism of rotation of the small subunit between the non-rotated and rotated conformations, we compared our 80S•2tRNA and 80S•tRNA structures. Superposition of the large ribosomal subunit rRNA reveals a ~8.5° rotation of the small subunit, reminiscent of the range of rotation for bacterial ribosomes (Agirrezabala et al., 2008; Julian et al., 2008). The axis of rotation (Fig. 3C) crosses the penultimate helix (h44), similarly to that in the bacterial system (Korostelev and Noller, 2007). Notably, the position of the rotation axis in the eukaryotic ribosome is distinct from that of the bacterial ribosome. In particular, the tilt of the upper part of the small subunit caused by the protruded central protuberance results in the tilt of the rotation axis by ~23° (Fig. 3C), indicating that the detailed mode of rotation and rearrangement of intersubunit contacts is likely different in bacterial and eukaryotic ribosomes. Visualization of additional intermediates of the intersubunit rotation is required to further address the mechanism of intersubunit rearrangements.
In summary, the comparison of the rotated and non-rotated eukaryotic ribosome complexes with bacterial counterparts suggests that while the subunits of ribosomes from both kingdoms undergo a large-scale rotation, the detailed mode of the rotation is different. Our structures reveal kingdom-specific differences, including the previously unreported distinct dynamics of helix 69, coupled with rearrangements of the small subunit. It is possible that in the globally similar pathways of bacterial and eukaryotic tRNA and mRNA translocation, the intermediates of ribosome rearrangements may be distinct between the two kingdoms. In turn, the similarity between the E-site architectures of eukaryotic and archaeal ribosomes suggests that the archaeal ribosomes and tRNAs sample the conformational states that are similar to eukaryotic but distinct from bacterial ribosomes. These kingdom-specific differences should be taken into account when parallels are drawn between the well-studied bacterial and understudied archaeal and eukaryotic translation mechanisms.
Kozak sequence as a molecular ruler
The sequence context of the AUG start codon was shown to be of key importance for the efficiency of translation initiation in eukaryotes and thus for the selection of the open reading frame initiation (Kozak, 1986). Whereas the sequence requirements (Kozak consensus) differ among eukaryotic species, the presence of a purine nucleotide at position −3 is the most stringent requirement in all eukaryotes (Cavener and Ray, 1991). Adenosine is thought to occupy this position in more than 70% of mRNAs, and guanosine in more than 25% (Cavener and Ray, 1991). The structural role of this stringent requirement is not clear as it could not be addressed by previous structural studies of the 80S complexes due to low resolution and/or compositional heterogeneity of endogenous mRNA found in the ribosome. In our work, we have used a fragment of mRNA, which contains an authentic S. cerevisiae Kozak sequence AAAA (Hamilton et al., 1987) immediately upstream of the AUG codon. The presence of the initiator tRNA base paired with the AUG codon at the 40S P site in our maps indicates that the position of the mRNA fragment within the ribosome reflects that of a tRNA-bound initiation state of the 80S ribosome.
The density for mRNA in the A, P and E sites is well resolved, allowing us to build and refine an mRNA model at the three tRNA-mRNA binding sites of the 40S subunit. The structures of mRNA are similar in the 80S•tRNA and 80S•2tRNA complexes. The three nucleotides immediately preceding the AUG codon are located in the E site (Fig. 4A). Density in the E site is characteristic of stacked nucleotides and contains sufficient detail to identify phosphate groups. The positions and conformations of the nucleotides in the E site (Fig. 4B) are similar to those found in the 3.1-Å resolution crystal structure of the bacterial 70S ribosome (PDB ID 3I8G) (Jenner et al., 2010), in which the mRNA in the E site was well resolved (Fig. 4C). The similarity between our mRNA model and the mRNA in the crystal structure of the 70S ribosome, as well as the stereochemically plausible packing of nucleotides within the density suggest the following interactions. The adenosine of interest (A-3) is positioned to form stacking interactions with the universally conserved G904 (G693 in E. coli) located at the tip of the hairpin loop 23 of the small ribosomal RNA. At the other side, A-3 stacks on A-2, as suggested by the similar contact in the high-resolution structure of the bacterial ribosome (Fig. 4C). The bases of A-3 and A-2 are positioned next to the glycine-rich tip of a beta hairpin of protein S5 (amino acids 152–154, Fig. 4A). Our structural model is consistent with results of biochemical experiments showing that the nucleotide in position −3 cross-links with G904 (Demeshkina et al., 2000) and S5 (Pisarev et al., 2006) in mammalian ribosomes.
Figure 4.
Interactions of mRNA with the E and P sites of the 40S subunit. (A) Fit of the molecular structure of the 80S bound with single tRNA into cryo-EM density. (B) Interactions of mRNA with the elements of the 40S subunit and the anticodon-stem loop of tRNA in the P site (this work). (C) Interactions of mRNA in the 3.1 Å crystal structure of Thermus thermophilus 70S ribosome (PDB ID 3I8G, Jenner et al., 2010). Small-subunit rRNA is in cyan, mRNA in green, P/E-site tRNA in magenta, P-site tRNA in red, ribosomal proteins in marine.
Visualization of the AAA trinucleotide preceding the AUG codon provides insights into the critical role that nucleotide A-3 plays in selecting an authentic start codon during initiation. Interactions of a purine residue with G904 and S5 during initiation likely stabilize the position of the mRNA in this region. Purine-purine stacking may provide up to 10-fold stronger binding than pyrimidine-purine stacking (Friedman and Honig, 1995), suggesting that a pyrimidine at −3 would result in less stable interactions with G904. Thus, the conserved G904 of the 18S rRNA forms a stacking foundation to support the placement of A-3 or G-3 in the E site, resulting in proper positioning of downstream nucleotides. Interactions of S5 with A-3 may also contribute to stabilization of the Kozak sequence (Fig. 4A). Together, the three nucleotides in the E site likely act as a molecular ruler to position the following AUG start codon in the P site, allowing the stable initiation complex to form.
Experimental procedures
Vacant 80S ribosomes were purified from Saccharomyces cerevisiae strain W303 as described (Ben-Shem et al., 2011). 80S ribosome complexes were assembled in the presence of initiator E. coli tRNAfMet (ChemBlock) and an mRNA fragment (AAAAAUGUAAAAAA, IDT) containing the start codon AUG (underlined) and adenosine at position −3 (bold and underlined) characteristic for the S. cerevisiae Kozak fragment. Cryo-EM imaging was performed essentially as previously described (Koh et al., 2014). Image processing was performed essentially as described (Brilot et al., 2013; Grigorieff, 2007). The initial dataset consisted of 86866 particles. Particles belonging to the two highest-resolution classes were refined using FREALIGN and optimal filtering (Sindelar and Grigorieff, 2012). 25136 particles were extracted from the larger dataset for further refinement to yield the reconstruction occupied with one tRNA. 23136 particles were extracted from the larger dataset for further refinement to yield the reconstruction with two tRNAs. The 80S•tRNA and 80S•2tRNA structures were independently refined against the reconstructions (maps) employing stereochemically-restrained real-space refinement essentially as described (Koh et al., 2014). Real-space R-factors are 0.2 for both refined all-atom structures, indicating good fit of the models to the map. Details of ribosome sample formation, cryo-EM data collection and processing, fitting and refinement of structural models are described in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Yeast 80S ribosome adopts a non-rotated state in the presence of E and P-site tRNAs
Small subunit of the 80S ribosome bound with single P/E tRNA is rotated by ~8.5°
Conformations of 80S ribosomes differ from bacterial 70S in respective states
Conserved −3 purine of Kozak sequence helps position the AUG codon in the P site
Acknowledgments
We thank Allan Jacobson for providing a S. cerevisiae strain; Rohini Madireddy for help with protein purification; Zhiheng Yu and Jason de la Cruz for help with collecting data on the Titan Krios microscope; Sarah Stumper for help with picking particles and initial steps of image processing. The study was supported by grants from Worcester Foundation for Biomedical Research, UMMS Center for AIDS Research (awarded to A.A.K) and the National Institutes of Health grants R01 GM106105 (awarded to A.A.K.) and P01 GM62580 (awarded to N.G.), and from NSERC (awarded to A.F.B.). Accession codes: cryo-EM maps and structural models have been deposited to EMDataBank (ID 5976 and 5977) and RCSB (PDB ID 3J77 and 3J78), respectively.
Footnotes
Author contributions
E. S., A.F.B, N.G and A.A.K. designed the project; C.S.K. prepared purified ribosomes; E. S. prepared ribosome complexes; A.F.B. collected cryo-EM images; A.F.B and N.G. analyzed cryo-EM data; E. S. and A.A.K. built and refined structural models. All authors contributed to structure analysis and manuscript writing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirrezabala X, Liao HY, Schreiner E, Fu J, Ortiz-Meoz RF, Schulten K, Green R, Frank J. Structural characterization of mRNA-tRNA translocation intermediates. Proc Natl Acad Sci U S A. 2012;109:6094–6099. doi: 10.1073/pnas.1201288109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell. 2011;44:214–224. doi: 10.1016/j.molcel.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield TR, Devkota B, Rollins GC. Examinations of tRNA Range of Motion Using Simulations of Cryo-EM Microscopy and X-Ray Data. J Biophys. 2011;2011:219515. doi: 10.1155/2011/219515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106:2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgarno L, Shine J. Conserved terminal sequence in 18SrRNA may represent terminator anticodons. Nat New Biol. 1973;245:261–262. doi: 10.1038/newbio245261a0. [DOI] [PubMed] [Google Scholar]
- Demeshkina N, Repkova M, Ven’yaminova A, Graifer D, Karpova G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P, and E sites: a crosslinking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA. 2000;6:1727–1736. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- Friedman RA, Honig B. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys J. 1995;69:1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. Journal of Structural Biology. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton R, Watanabe CK, de Boer HA. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15:3581–3593. doi: 10.1093/nar/15.8.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- Julian P, Konevega AL, Scheres SH, Lazaro M, Gil D, Wintermeyer W, Rodnina MV, Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminishi T, Wilson DN, Takemoto C, Harms JM, Kawazoe M, Schluenzen F, Hanawa-Suetsugu K, Shirouzu M, Fucini P, Yokoyama S. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure. 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Koh CS, Brilot AF, Grigorieff N, Korostelev AA. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA–like structural element in the ribosomal decoding center. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1406335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Noller HF. Analysis of structural dynamics in the ribosome by TLS crystallographic refinement. J Mol Biol. 2007;373:1058–1070. doi: 10.1016/j.jmb.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Asahara H, Laurberg M, Lancaster L, Noller HF. Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome. Proc Natl Acad Sci U S A. 2007;104:16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Brilot AF, Theobald DL, Grigorieff N. Likelihood-based classification of cryo-EM images using FREALIGN. J Struct Biol. 2013;183:377–388. doi: 10.1016/j.jsb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr Opin Struct Biol. 2009;19:300–309. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Gulen B, Waterbury CC, Hershkovits E, Hsiao C, Harvey SC, Hud NV, Fox GE, Wartell RM, Williams LD. Secondary Structures of rRNAs from All Three Domains of Life. PLoS One. 2014;9:e88222. doi: 10.1371/journal.pone.0088222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA. 2003;9:1345–1352. doi: 10.1261/rna.5120503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Sindelar CV, Grigorieff N. An adaptation of the Wiener filter suitable for analyzing images of isolated single particles. J Struct Biol. 2012;176:60–74. doi: 10.1016/j.jsb.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel PC, Ermolenko DN, Noller HF. Elongation factor G stabilizes the hybrid-state conformation of the 70S ribosome. RNA. 2007;13:1473–1482. doi: 10.1261/rna.601507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourigny DS, Fernandez IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier SB, Jean-Jean O. Complementarity between the mRNA 5′ untranslated region and 18S ribosomal RNA can inhibit translation. RNA. 2000;6:584–597. doi: 10.1017/s1355838200992239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.