Abstract
The laboratory mouse is an ideal model organism for studying disease because it is physiologically similar to human and also because its genome is readily manipulated. Genetic engineering allows researchers to introduce specific loss-of-function or gain-of-function mutations into genes and then to study the resulting phenotypes in an in vivo context. One drawback of using traditional transgenic and knockout mice to study human diseases is that many mutations passed through the germline can profoundly affect development, thus impeding the study of disease phenotypes in adults. New technology has made it possible to generate conditional mutations that can be introduced in a spatially and/or temporally restricted manner. Mouse strains carrying conditional mutations represent valuable experimental models for the study of human diseases and they can be used to develop strategies for prevention and treatment of these diseases. In this article, we will describe the most widely used DNA recombinase systems used to achieve conditional gene mutation in mouse models and discuss how these systems can be employed in vivo.
INTRODUCTION
In the last two decades, gene targeting in embryonic stem (ES) cells has been used extensively as a tool to generate predesigned mouse mutants for studying gene function in vivo. In its original form, gene targeting allowed for the disruption of a specific gene in the mouse germline via insertion of a selectable marker (Capecchi 1989). The vast majority of the >3000 currently available knockout mouse strains have been created following this design strategy and many of these mouse models have provided valuable insights into the biological function of the genes studied. Nevertheless, because these “conventional” knockout mutations typically result in complete loss of function and are transmitted through the germline, they frequently result in embryonic or early postnatal lethality.
In 1994, site-specific recombinase systems (e.g., Cre, FLP, Dre) emerged as a technology that would allow for the design of more refined mouse models of human disease. These methodologies, along with gene targeting techniques, now make it possible for researchers to modify the mouse genome in almost any desired manner, for example by making loss-of-function or gain-of-function mutations (Lakso et al. 1992) that are spatially and temporally restricted (Lobe and Nagy 1998; Nagy and Rossant 2001). Consequently, gene function in adult mice can be precisely studied at a specific developmental stage or in a specific cell/tissue type of choice. Human diseases that arise late in life or in specific tissues (e.g., neurodegeneration or cancer) can now be studied effectively in an experimental organism.
SITE-SPECIFIC RECOMBINASES
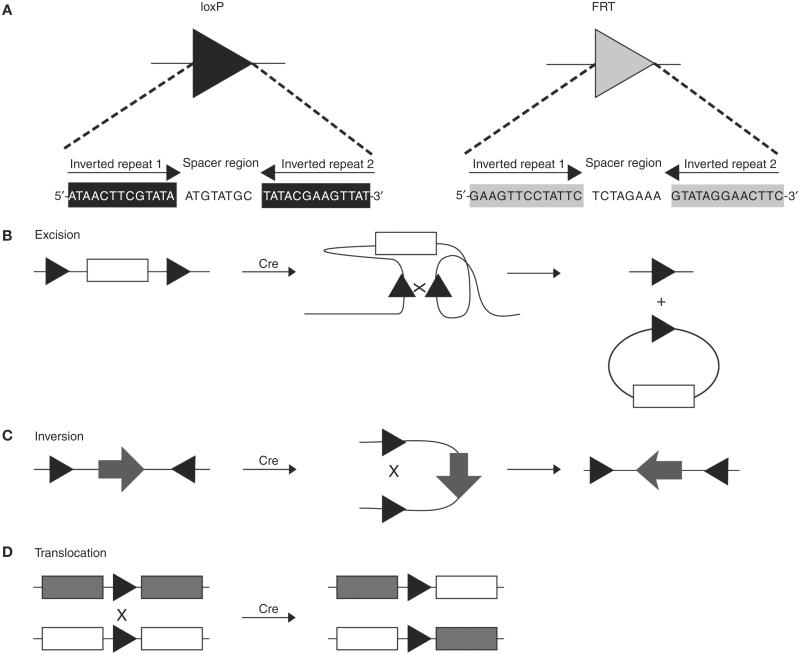
Cre (cyclization recombination) is a 38 kDa, site-specific DNA tyrosine recombinase derived from P1 bacteriophage. It catalyzes DNA recombination between two 34-bp recognition sequences referred to as loxP sites (locus of crossing over {X} of P1). Since Cre does not require high-energy cofactors or accessory proteins and exhibits optimal recombinase activity at 37°C (Buchholz et al. 1996), the system is optimal for genetic engineering in cultured mammalian cells and in mice. Each loxP site is composed of two 13-bp palindromic sequences that are separated by an 8-bp asymmetric core region that determines the orientation (Fig. 1A, Table 1). Owing to its size, the loxP site is unlikely to be F1 T1 found at random in the mouse genome (Nagy 2000). Upon binding to the loxP sites, Cre-mediated recombination results in cleavage of the DNA and reciprocal strand exchange between the two loxP sites (Grindley et al. 2006). The orientation of the two-loxP sites determines the different DNA products that are produced (Fig. 1B–D). When two loxP sites are arranged in the same orientation on a linear DNA fragment, Cre-mediated recombination results in excision of the loxP-flanked DNA fragment in a circular form, leaving behind a single loxP site on the linear DNA. When two loxP sites are arranged in opposite orientation to each other on a linear DNA fragment, Cre-mediates the inversion of the loxP-flanked DNA fragment. Cre can also exchange DNA sequences that are distal to the loxP sites on two different linear DNA fragments, such as a pair of nonhomologous chromosomes (Branda and Dymecki 2004).
FIGURE 1.
Cre and FLP recognition sequences and mechanism of recombination. (A) The Cre recombinase recognition sequence is termed loxP (black arrow), while the Flp recombinase recognition sequence is termed FRT (gray arrow). Both loxP and FRT contain two 13-bp inverted repeats that are separated by an 8-bp spacer region. (B) Cre mediates excision or circularization of a DNA segment (white rectangle) when flanked by two loxP sites (black triangles) in a cis (same DNA strand) arrangement and oriented in the same direction. (C) Cre mediates inversion of a DNA segment (black arrow) when flanked by two loxP sites (black triangles) in a cis arrangement and oriented in the opposite direction. (D) Cre mediates translocation of a DNA segment (rectangles) when the loxP sites (black triangles) are located on different strands of DNA (trans arrangement) and are oriented in the same direction.
TABLE 1.
Cre, FLP, and Dre recognition sites: classical and variant sites
| Recombinase | Recognition Site | Inverted Repeat 1 | Spacer | Inverted Repeat 2 |
|---|---|---|---|---|
| Cre | loxP | ATAACTTCGTATA | ATGTATGC | TATACGAAGTTAT |
| Cre | lox2272 | ATAACTTCGTATA | ATGTGTAC | TATACGAAGTTAT |
| Cre | lox511 | ATAACTTCGTATA | ATGTATAC | TATACGAAGTTAT |
| Cre | lox m2 | ATAACTTCGTATA | AGAAACCA | TATACGAAGTTAT |
| Cre | lox66 | ATAACTTCGTATA | ATGTATGC | TATACGAACGGTA |
| Cre | lox71 | TACCGTTCGTATA | ATGTATGC | TATACGAAGTTAT |
| Cre | lox72 | TACCGTTCGTATA | ATGTATGC | TATACGAACGGTA |
| FLP | FRT | GAAGTTCCTATTC | TCTAGAAA | GTATAGGAACTTC |
| FLP | F3 | GAAGTTCCTATTC | TTCAAATA | GTATAGGAACTTC |
| FLP | F5 | GAAGTTCCTATTC | TTCAAAAG | GTATAGGAACTTC |
| Dre | rox | TAACTTTAAATAAT | GCCA | ATTATTAAAGTTA |
Nucleotides, in the variant sites, that have been altered from the classical sites are underlined.
A drawback of the Cre/lox system is that DNA methylation, transcriptional activity, and chromatin structure at the locus of interest, and the distance between the loxP sites, can influence the efficiency of recombination (Vooijs et al. 2001). To increase expression and enhance the enzymatic activity of Cre protein, the cre gene has been modified to create a new recombinase variant. These changes include a reduced CpG content, several silent mutations, and an improved Kozak translation initiation consensus sequence. The encoded iCre (improved Cre) demonstrated a 1.6-fold increase in expression over the conventional Cre when expressed from the same vector in mammalian cells. iCre was also found to be 1.8-fold more efficient at DNA recombination than conventional Cre (Shimshek et al. 2002).
FLP (Flippase) is a tyrosine recombinase that was originally isolated from Saccharomyces cerevisiae (Sadowski 1995). Similar to Cre, FLP alters the arrangement of DNA by strand cleavage, exchange, and ligation. The FLP recombinase mediates DNA recombination between two 34-bp recognition sites referred to as FRT (Flippase Recognition Target), which have a similar structure to loxP sites (Fig. 1A, Table 1). Like Cre, FLP does not require high-energy cofactors or accessory proteins to catalyze recombination, but unlike Cre, it exhibits optimal activity at 30°C and is unstable at 37°C (Buchholz et al. 1998). Mutational analysis of FLP resulted in the identification of FLPe, which exhibits improved thermostability at 37°C, but this mutant is still less stable compared to Cre (Buchholz et al. 1998). FLPe exhibits a 4-fold increase in recombinase activity over wild-type FLP when analyzed in culture. In mice, FLPe can readily mediate recombination between FRTs separated by as little as a few kilobases or as much as 150 kb (Farley et al. 2000; Awatramani et al. 2003). Despite this improvement, the recombination efficiency of FLPe still remains low in cells (Farley et al. 2000). To improve the translational efficiency in mammalian cells, FLPe was re-engineered de novo according to the native amino acid sequence, but with mouse codon usage. The base composition of the codon-optimized FLPe gene (FLP optimized or FLPo) was modified to prolong mRNA half-life, and two stop codons were also included to ensure efficient translational termination. FLPo was shown to exhibit enhanced recombinase activity comparable to Cre (Raymond and Soriano 2007).
Other recombinases have also been identified for genome engineering in mice. For example, comparative analysis of P1-like phages identified Dre recombinase from the bacteriophage D6. Dre is also a tyrosine recombinase that is able to catalyze site-specific recombination in a similar manner to Cre, but it recognizes a recombination target site that is distinct from loxP. The rox site is a 32-bp DNA sequence that consists of two 14-bp inverted repeats that are separated by a 4-bp asymmetric core region (Table 1). Owing to a difference in amino acid sequence between Cre and Dre at the DNA recognition site, Dre is not able to catalyze recombination at loxP and Cre cannot catalyze recombination at rox (Sauer and McDermott 2004). The Dre-rox system has been shown to function both in mammalian cells and in mice, and therefore may be used in conjunction with the Cre-lox system for genome engineering. The Dre-rox system is relatively new with only three ubiquitous mouse lines developed based on the CAGGs promoter and the Rosa26 locus (Anastassiadis et al. 2009).
TRANSGENIC EXPRESSION OF CRE
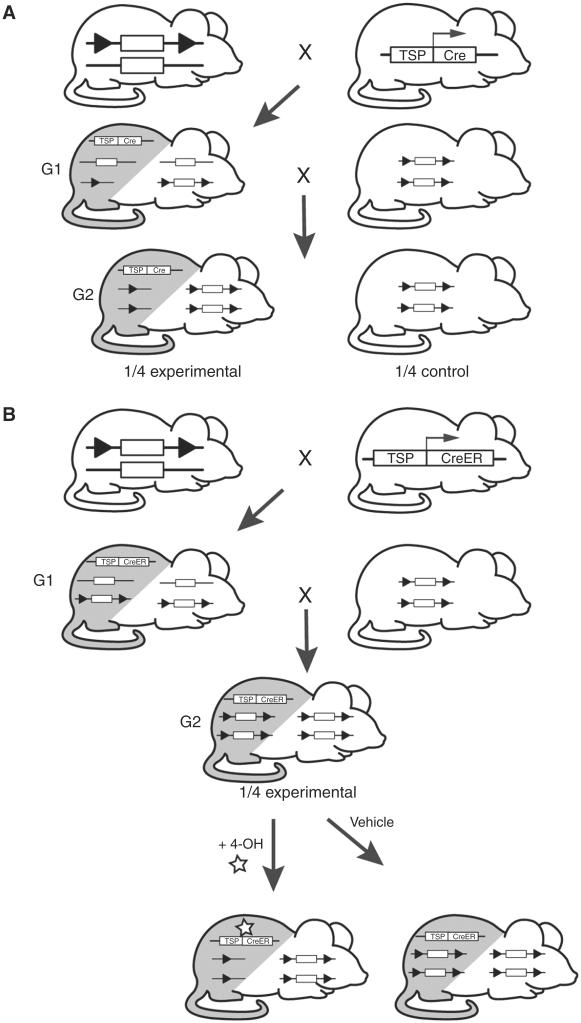
The high efficiency of the Cre-loxP system in mammalian cells (Gu et al. 1994; Sauer 1994) has led to its common use for genetic engineering in vivo. The in vivo delivery of Cre to cells in a mouse can be achieved in several ways. For example, animals carrying a conditional allele of a gene, typically flanked by two loxP sites (“floxed” allele) in the desired orientation, can be crossed with animals carrying a transgene or knock-in that expresses Cre under the control of a promoter that restricts its expression to a particular developmental stage, a particular tissue, or a particular cell type (Fig. 2A, Table 2). In the absence of Cre recombinase, mice carrying two floxed alleles will express wild-type protein at endogenous levels. Cross-breeding with a Cre strain will lead to inactivation of the targeted gene within the expression domain of the recombinase, while the target gene remains intact in other cells where the recombinase is not expressed.
FIGURE 2.
Breeding schemes for generating Cre-lox conditional, tissue-specific mutant mice. (A) Classical Cre-lox strategy to obtain conditional gene knockout in a tissue-specific manner. Mice harboring the conditional allele are crossed with Cre transgenic mice, in which Cre recombinase is under the control of a tissue-specific promoter (TSP). Breeding of these two mice will produce heterozygous offspring with excision of the floxed gene of interest only in the Cre-expressing cells/tissue (gray), while the gene of interest remains functional in other cell/tissue types (white). Offspring that are heterozygous for the conditional allele and also contain the Cre transgene (G1) are crossed to homozygous conditional mutant mice. Offspring that are homozygous for the conditional allele and heterozygous for the Cre transgene (G2) represent the conditional deletion mice, which can be generated at a frequency of 25%. Littermates that are homozygous for the conditional allele but do not carry the Cre transgene can be used as experimental controls. (B) Ligand-inducible strategy to obtain conditional gene knockout. Mice harboring the conditional allele are crossed with mice carrying a CreER fusion protein that is under the control of a tissue specific promoter (TSP). Breeding of these two mice will produce heterozygous offspring that contain the floxed gene of interest and the CreER fusion transgene. These animals are crossed to homozygous conditional mutant mice (G1). Offspring that are homozygous for the conditional allele and heterozygous for the CreER transgene (G2) can be generated at a frequency of 25% and represent the experimental animals. In the presence of ligand (tamaxofin [4-OH]) the floxed gene of interest will be excised only in the CreER-expressing cells/tissue (gray), while the gene of interest remains functional in other cell/tissue types (white). Control animals are treated with vehicle and the floxed gene of interest remains functional in all cell/tissue types since the CreER transgene is not activated by the ligand.
TABLE 2.
Tissue-Specific Cre-Expressing Mouse Lines
| Specificity | Promoter | Transgene | Reference |
|---|---|---|---|
| Ubiquitous | Human cytomegalovirus (CMV) | CMV-Cre CreERT2 CMV |
Schwenk et al. (1995), Feil et al. (1996) |
| Ubiquitous | Rosa26 | Rosa26-Cre | Soriano (1999) |
| Adipocytes | aP2 | aP2-Cre | Barlow et al. (1997) |
| B-lymphocytes | CD-19 gene albumin | Cd19-Cre Cd-19-CreERT2 |
Rickert et al. (1997), Boross et al. (2009) |
| Brain (neurons) | Calbindin 2 | Calb2-CreERT2 | Taniguchi et al. (2011) |
| Cardiac muscle | α-Myosin heavy chain (α-MHC) | α-MHC-Cre | Gupta et al. (1998) |
| Central nervous system (hippocampus) |
Mouse pro-opiomelanocortin-alpha (POMC) | POMC-Cre | McHugh et al. (2007) |
| Colonic epithelium | Human caudal type homeo-box 2 (CDX2) | CDX2-Cre | Hinoi et al. (2007) |
| Epiblast cells | Mouse SRY-box containing gene 2 (Sox2) | Sox2-Cre | Hayashi and McMahon (2002) |
| Kidney epithelium | Gamma-glutamyltransferase 1(Ggt1) | Ggt1-Cre | Iwano et al. (2002) |
| Liver (hepatocytes) | Albumin promoter/enhancer (Alb) | Alb-Cre Alb-CreERT2 |
Postic and Magnuson (1999), Schuler et al. (2004) |
| Lung (endoderm) | Human surfactant pulmonary-associated protein C (SFTPC) |
SFTPC-Cre | Okubo and Hogan (2004) |
| Mammary gland | Whey acidic protein (Wap) | Wap-Cre | Wagner et al. (1997) |
| Midbrain/hindbrain | Mouse engrailed 2 promoter/enhancer (En2) | En2-Cre | Sgaier et al. (2005) |
| Pancreatic epithelium | Pancreatic and duodenal homeobox1 (pdx1) | Pdx1-Cre | Gannon et al. (2000a) |
| Pancreatic β-cells | Rat insulin II | Ins2-Cre | Gannon et al. (2000b) |
| Skeletal muscle | Mouse muscle creatine kinase (MCK) | MCK-Cre | Levin et al. (2007) |
| Skin (keratinocytes) | Human keratin 14 | K14-Cre K14-CreERT2 |
Dassule et al. (2000), Vasioukhin et al. (1999) |
| Small and large intestine (crypts) |
Mouse villin 1 | Vil-Cre Vil-CreERT2 |
Madison et al. (2002), Vasioukhin et al. (1999) |
| Small and large intestine (epithelium) |
Fatty acid binding protein (Fabp) | Fabpl-Cre | Saam and Gordon (1999) |
| Small and large intestine (epithelium) |
Leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5) |
Lgr5-EGFP-IRES- CreERT2 |
Barker et al. (2007) |
The Cre-lox system can also be used to obtain gain-of-function mutations to generate new models of human cancer. Mice harboring a conditionally activatable allele of oncogenic K-ras have been used to generate various mouse models of human cancer, including lung adenocarcinoma (Jackson et al. 2001). Expression of oncogenic K-ras is controlled by a removable transcriptional stop element (Lox-STOP-LOX). In the presence of Cre, the STOP element is removed and oncogenic K-ras is expressed only in cells where the recombinase is expressed (Tuveson et al. 2004). Combining gain-of-function and loss-of-function mutations has led to the development of more advanced cancer models (Babaei-Jadidi et al. 2011; Young et al. 2011).
Using this standard approach, recombinase expression, and theoretically, the timing and pattern of gene mutation are entirely dependent on the endogenous activity of the chosen promoter. Practically, however, the desired tissue specificity and the level of induction are difficult to control in a transgenic line, often resulting in mosaic expression of the recombinase or expression in unwanted tissues (Schwenk et al. 1998; Rossant and McMahon 1999) (Fig. 3). Despite this limitation, this approach has been utilized effectively for modeling human diseases such as cancer. For example, specific mutation of Apc and Rb1 in the intestinal epithelium effectively induces adenocarcinoma (Kucherlapati et al. 2008).
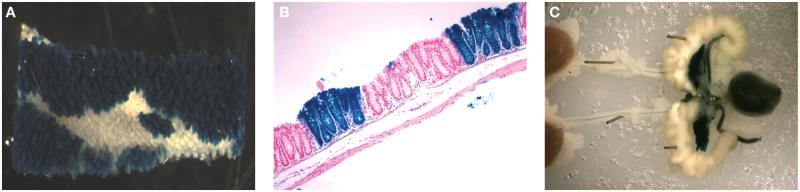
FIGURE 3.
Cre expression mediated by the “tissue-specific” liver fatty acid binding protein (Fabpl) promoter. (A) High magnification of β-galactosidase staining in whole mount of the distal small intestine from Fabpl-Cre/+; R26R/+ mice. Cre-positive portions of the tissue stain blue when exposed to X-Gal. The Fabpl-Cre transgene directs expression of Cre recombinase to the distal small intestine and colon. (B) β-galactosidase expression in a paraffin section of the distal colon from Fabpl-Cre/+; R26R/+ mice. The expression of the Cre transgene is mosaic and therefore normal (white) and mutant (blue) crypts can be analyzed in the same mouse. (C) High magnification of β-galactosidase staining in whole mount of the urinary tract from Fabpl-Cre/+; R26R/+ mice. The positive staining (blue) illustrates the off-target effects that can be attained using this system.
While many successful Cre transgenic mice have been generated and used in laboratories, several caveats have been reported. For example, because the mammalian genome does not contain endogenous loxP sites, expression of Cre in mice was expected to be harmless. Nevertheless, several incidences of Cre toxicity both in vitro and in vivo have been reported (Loonstra et al. 2001; Forni et al. 2006). To minimize the toxic effect of Cre, a self-deleting retroviral vector that incorporates a negative feedback loop limiting the duration and intensity of Cre expression has been engineered. The vector carries the Cre cDNA and a single loxP site in its U3 region of the 3’LTR, which allows for transient expression of Cre in the target cell and subsequent deletion from the genome in a Cre-dependent manner (Pfeifer et al. 2001; Silver and Livingston 2001). Additionally, even tissue-restricted mutation of a gene can be lethal when it occurs early during development (Braun et al. 2004). There are two ways to overcome these limitations: (1) by using transgenic mouse strains in which the expression or activity of Cre can be regulated acutely and (2) through exogenous delivery of Cre, for example, by using viruses.
ACUTE REGULATION OF CRE ACTIVITY
There are several ways to regulate recombinase activity acutely, for example, by driving Cre expression from a promoter that is induced by treatment of mice with a particular chemical. In the AhCre strain, Cre expression is under the control of the rat cytochrome P450 1A1 (CYP1A1) promoter element that is transcriptionally up-regulated in response to β-napthanoflavone. As such, Cre-mediated recombination is only induced following injection with β-napthanoflavone (Ireland et al. 2004). This approach has been used to study the effects of deleting Apc and c-Myc in the intestinal epithelium, where the CYP1A1 promoter is strongly induced (Sansom et al. 2007). In Mx1-Cre mice, Cre is under the control of the Mx1 promoter, which is strongly induced in hematopoietic cells and in the liver by administration of interferon α and β, or polyriboinosinic acid/polyribocytidylic acid (poly I:C) (Kuhn et al. 1995). Mx1-Cre mice were crossed with animals carrying conditional mutations in Kras (Braun et al. 2004) and Nras (Li et al. 2011) to model myeloproliferative disease and myeloid leukemia.
One particularly successful strategy has involved fusing a mutant estrogen receptor (ER) ligand binding domain (LBD) to the C terminus of Cre (Metzger et al. 1995; Schwenk et al. 1998). Addition of the ligand (4-OH tamoxifen {4-OH}) allows CreER to access the nucleus in an active conformation leading to Cre-mediated recombination (Indra et al. 1999). A more powerful approach to study human disease is the development of inducible gene expression systems with tissue-restricted expression of these fusions using specific enhancer elements to gain both temporal and spatial control over recombinase and gene activity (Logie and Stewart 1995; Danielian et al. 1998).
Another approach used for temporal regulation in transgenic mouse models is the reverse tetracycline-regulated gene expression system (Gossen and Bujard 1992). In this system a transgenic mouse line is produced in which the promoter driving Cre expression contains a tetracycline-responsive promoter element (tetO). In the presence of doxycycline, the inducer, Cre is rapidly expressed, thus allowing gene deletion (Lewandoski 2001). While this system has been successfully used, the main disadvantage of this approach is the complexity of the tripartite system, which complicates mouse husbandry due to the mating of the three different transgenic lines.
Although ligand-inducible recombinase systems have opened new avenues in experimental biology, temporal control of recombinase activity is limited by the time required for cell/tissue permeation. They also require systemic administration of the inducer, making it difficult to restrict Cre induction to a small area. Recently developed light-switchable transgene systems could provide precise and robust spatiotemporal control over gene expression in both cultured cells and mice (Kennedy et al. 2010; Wang et al. 2012). This system takes advantage of genetically encoded dimerization modules based on plant photoreceptors that control protein–protein interactions by light (Shimizu-Sato et al. 2002; Levskaya et al. 2009). Arabidopsis light oxygen voltage (LOV) domain-containing proteins (e.g., CIB1 and CRY2) form dimers upon exposure to blue-light (Zoltowski and Crane 2008). Artificial dimerizers can be engineered to allow inducible control of a “split” protein, in which two inactive fragments are brought together to reconstitute functional protein activity. Using this system a split Cre recombinase can be fused to CIB-CRY modules, allowing light-dependent control of DNA recombination (Jullien et al. 2003; Kennedy et al. 2010). These modules are transferred into mice using a hydrodynamic procedure and can be activated by two-photon stimulation, which is ideal when using mice, illustrating high induction with fast kinetics (Wang et al. 2012).
VIRAL DELIVERY OF CRE
A major limitation of transgenic approaches to providing Cre to cells is that it is extremely difficult to limit expression to a small population of cells. In many cases (e.g., investigations of immune responses), it is desirable to mutate a gene broadly within a specific tissue or cell type. For modeling certain disease states (e.g., cancer), however, it is actually preferable to target only a small population of cells. Adenoviruses and lentiviruses can be used to effectively deliver genes to both dividing and nondividing cells in vivo (Wang et al. 1996) and, because the viruses can be delivered directly (e.g., by injection) into specific anatomic locations in animals carrying conditional alleles, the extent of the conditional genetic modification can be restricted to a small area of viral spread. Adenoviral delivery is advantageous because the viral DNA does not integrate into the host genome. As such, expression of genes carried by the recombinant virus is transient. Adenovirus is disadvantageous, however, because it elicits a host immune response. Lentiviruses, by contrast, stably integrate into the genome, providing sustained transgene expression and they generate little or no immune response (Naldini et al. 1996; Lai et al. 2002). Since these viruses integrate into the host genome, however, they can induce mutation of host genes, which is a concern for experimental studies. In comparison to other viral systems, lentiviral vectors have a larger insert capacity, thus large gene inserts, longer (e.g., tissue-specific) promoters, and more than one gene (poly-cistronic cassettes) can be subcloned into the vectors (Heldt and Ressler 2009). The production of Cre-containing lentiviruses in tissue culture cells and subsequent purification are detailed in Producing and Concentrating Lenti-Cre for Mouse Infections (Gierut et al. 2014a).
Viral delivery approaches have been used extensively to model cancer in mouse models. For example, acute infection of the respiratory epithelium via intranasal instillation (see In vivo Delivery of Lenti-Cre or Adeno-Cre using Intranasal Instillation [Gierut et al. 2014b]) or intratracheal injection of Adeno-Cre induces non-small-cell lung cancer in animals carrying conditional alleles Kras and Trp53 (Jackson et al. 2005) and small-cell lung cancer in animals carrying conditional alleles Rb1 and Trp53 (Vooijs et al. 1998; Jonkers et al. 2001; Meuwissen et al. 2003). Similarly lentiviral vectors have been engineered to introduce both Cre recombinase and exogenous antigens in order to mimic tumor neoantigens. For example, intranasal instillation of Lenti-Cre vectors that also express T-cell antigens into animals carrying a conditional allele of Kras led to tumors that had large numbers of infiltrating lymphocytes, including both T and B cells, several weeks after tumor initiation (DuPage et al. 2011). Intracranial injection of Adeno-Cre into mice carrying conditional alleles of Egfr and Arf or Pten generates a rapid-onset high-grade malignant glioma phenotype that is molecular and pathologically similar to glioblastoma multiforme (GBM) in humans.
VALIDATION OF CRE ACTIVITY
A useful exercise when generating new transgenic Cre strains, or when analyzing phenotypes associated with conditional mutation of a specific gene, is to characterize the pattern of expression of the recombinase using Cre reporter strains. The reporter strain contains a floxed transcriptional stop sequence that prevents the expression of the downstream reporter gene (usually β-galactosidase or GFP) in the absence of Cre (Soriano 1999). When crossed with a Cre transgenic mouse strain or after infection with the virus, the stop element is removed and the reporter gene is expressed in tissues where Cre is expressed (Fig. 3). This procedure is detailed in Whole Mount X-Gal Staining of Mouse Tissues (Gierut et al. 2014c). If the expression pattern of the Cre strain is unknown, one should first perform whole mount staining of all the major mouse tissues in order to assess the Cre activity throughout the entire body of the mouse. When using an established Cre strain or viral infection, histological sections of the specific tissue(s) should be analyzed in order to obtain detailed information about the regional and cell type-specific expression of the recombinase.
DUAL RECOMBINASE TECHNOLOGY
Because many diseases result from the combined effects of multiple mutations, it is often desirable to combine conditional alleles together. This creates a potential problem because Cre can promote both intragenic and intergenic recombination (Fig. 1A). As a result, the combination of multiple conditional alleles can lead to undesired genomic arrangements. To overcome this limitation, variant Cre and Flp target sites can be used in different conditional alleles (Table 1) (Hoess et al. 1986; Senecoff et al. 1988). In this situation, recombination is proficiently mediated between pairs of homotypic (loxP/loxP or lox2272/lox2272) but not heterotypic (loxP/lox2272) sites (Senecoff and Cox 1986). Likewise, for Flp-mediated recombination, the spacer variants F3 and F5 have been shown to be insensitive to recombination with wild-type FRT sites (Schlake and Bode 1994). In contrast, the Cre target site lox511 (Bethke and Sauer 1997) has been found to recombine (1%–5%) with a wild-type loxP site (Kolb 2001) and therefore it is less effective for this type of gene inactivation strategy (Table 1).
While Cre-mediated deletion is essentially unidirectional, inversion is reversible and therefore not used for stable gene alternations in cells that express Cre. Site-directed mutagenesis of wild-type loxP sites has led to the identification of variant loxP pairs that display a favorable forward reaction and for unidirectional Cre-mediated inversion (Albert et al. 1995). Cre-mediated recombination of the two asymmetric, mutant loxP sites, lox66 (right) and lox71 (left), yields one loxP site with two mutated inverted repeats (lox72) and one wild-type loxP site. Although successful application of lox66/71 mediated recombination has been observed in mouse ES cells (Araki et al. 1997; Zhang and Lutz 2002), an in-depth characterization of lox66/lox71 recombination in Cre-transgenic mouse strains has yet to be determined.
As an alternative to using distinct recognition sites for the same recombinase, conditional alleles for different recombinases can be combined. Dual recombinase technology has become a major area of study and interest because (1) it enables distinct gene mutations to be directed to different cell types and (2) it allows for sequential mutation of genes. For example, a Cre-dependent allele of Trp53 and a FRT-dependent allele of Kras were combined to examine the effects of sequential mutations on the development of soft tissue sarcoma (Young et al. 2011).
FUTURE DIRECTIONS
The completion of the human and mouse genome sequencing projects, together with advances in site-specific recombinase technology, herald a new era of genetic analysis in the mouse. The original Cre and FLP recombinases demonstrated their utility in developing conditional gene targeting. Now, other analogous recombinases are ready to be used, in the same way or in combined strategies, to achieve more sophisticated experimental schemes for addressing complex biological questions. The properties of site-specific recombinases, in combination with other tools (tet on/off system, siRNA mediating gene silencing, fluorescent proteins, viral vectors, etc.), make them useful instruments to induce precise mutations in specific cells/tissues and in a temporally controlled manner. This spatially and temporally controlled activity can be applied to functional genomics in many ways: from conditional and inducible gene targeting to controlled expression of transgenes and recombination-mediated cassette exchange in mouse models. Indeed, with the tools of site-specific recombinase technology in hand, the field of mouse genetics is primed to determine the contribution of the vast majority of genes to development and disease.
REFERENCES
- Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Disease Models Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- Araki K, Araki M, Yamamura K. Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res. 1997;25:868–872. doi: 10.1093/nar/25.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awatramani R, Soriano P, Rodriguez C, Mai JJ, Dymecki SM. Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet. 2003;35:70–75. doi: 10.1038/ng1228. [DOI] [PubMed] [Google Scholar]
- Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R, Abuzinadah M, Davis H, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barlow C, Schroeder M, Lekstrom-Himes J, Kylefjord H, Deng CX, Wynshaw-Boris A, Spiegelman BM, Xanthopoulos KG. Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic Acids Res. 1997;25:2543–2545. doi: 10.1093/nar/25.12.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke B, Sauer B. Segmental genomic replacement by Cre-mediated recombination: Genotoxic stress activation of the p53 promoter in single-copy transformants. Nucleic Acids Res. 1997;25:2828–2834. doi: 10.1093/nar/25.14.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boross P, Breukel C, van Loo PF, van der Kaa J, Claassens JW, Bujard H, Schonig K, Verbeek JS. Highly B lymphocyte-specific tamoxifen inducible transgene expression of CreER T2 by using the LC-1 locus BAC vector. Genesis. 2009;47:729–735. doi: 10.1002/dvg.20549. [DOI] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Braun BS, Tuveson DA, Kong N, Le DT, Kogan SC, Rozmus J, Le Beau MM, Jacks TE, Shannon KM. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF. Different thermostabilities of FLP and Cre recombinases: Implications for applied site-specific recombination. Nucleic Acids Res. 1996;24:4256–4262. doi: 10.1093/nar/24.21.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. The new mouse genetics: Altering the genome by gene targeting. Trends Genet. 1989;5:70–76. doi: 10.1016/0168-9525(89)90029-2. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- DuPage M, Cheung AF, Mazumdar C, Winslow MM, Bronson R, Schmidt LM, Crowley D, Chen J, Jacks T. Endogenous T cell responses to antigens expressed in lung adenocarcinomas delay malignant tumor progression. Cancer Cell. 2011;19:72–85. doi: 10.1016/j.ccr.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni PE, Scuoppo C, Imayoshi I, Taulli R, Dastru W, Sala V, Betz UA, Muzzi P, Martinuzzi D, Vercelli AE, et al. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. J Neurosci. 2006;26:9593–9602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, Herrera PL, Wright CV. Mosaic Cre-mediated recombination in pancreas using the pdx-1 enhancer/promoter. Genesis. 2000a;26:143–144. doi: 10.1002/(sici)1526-968x(200002)26:2<143::aid-gene13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Gannon M, Shiota C, Postic C, Wright CV, Magnuson M. Analysis of the Cre-mediated recombination driven by rat insulin promoter in embryonic and adult mouse pancreas. Genesis. 2000b;26:139–142. doi: 10.1002/(sici)1526-968x(200002)26:2<139::aid-gene12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Gierut JJ, Jacks TE, Haigis KM. Producing and concentrating lenti-Cre for mouse infections. Cold Spring Harb Protoc. 2014a doi: 10.1101/pdb.prot073437. doi: 10.1101/pdb. prot073437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierut JJ, Jacks TE, Haigis KM. In vivo delivery of lenti-Cre or adeno-Cre using intranasal instillation. Cold Spring Harb Protoc. 2014b doi: 10.1101/pdb.prot073445. doi: 10.1101/ pdb.prot073445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierut JJ, Jacks TE, Haigis KM. Whole mount X-Gal staining of mouse tissues. Cold Spring Harb Protoc. 2014c doi: 10.1101/pdb.prot073452. doi: 10.1101/pdb.prot073452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Gupta M, Zak R, Libermann TA, Gupta MP. Tissue-restricted expression of the cardiac alpha-myosin heavy chain gene is controlled by a downstream repressor element containing a palindrome of two ets-binding sites. Mol Cell Biol. 1998;18:7243–7258. doi: 10.1128/mcb.18.12.7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Heldt SA, Ressler KJ. The use of lentiviral vectors and Cre/loxP to investigate the function of genes in complex behaviors. Front Mol Neurosci. 2009;2:22. doi: 10.3389/neuro.02.022.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, Fearon ER. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 2007;67:9721–9730. doi: 10.1158/0008-5472.CAN-07-2735. [DOI] [PubMed] [Google Scholar]
- Hoess RH, Wierzbicki A, Abremski K. The role of the loxP spacer region in P1 site-specific recombination. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland H, Kemp R, Houghton C, Howard L, Clarke AR, Sansom OJ, Winton DJ. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: Effect of loss of beta-catenin. Gastroenterology. 2004;126:1236–1246. doi: 10.1053/j.gastro.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre Q4 recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res. 2003;31:e131. doi: 10.1093/nar/gng131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb AF. Selection-marker-free modification of the murine beta-casein gene using a lox2272 [correction of lox2722] site. Anal Biochem. 2001;290:260–271. doi: 10.1006/abio.2000.4984. [DOI] [PubMed] [Google Scholar]
- Kucherlapati MH, Yang K, Fan K, Kuraguchi M, Sonkin D, Rosulek A, Lipkin M, Bronson RT, Aronow BJ, Kucherlapati R. Loss of Rb1 in the gastrointestinal tract of Apc1638N mice promotes tumors of the cecum and proximal colon. Proc Natl Acad Sci. 2008;105:15493–15498. doi: 10.1073/pnas.0802933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lai Z, Han I, Park M, Brady RO. Design of an HIV-1 lentiviral-based gene-trap vector to detect developmentally regulated genes in mammalian cells. Proc Natl Acad Sci. 2002;99:3651–3656. doi: 10.1073/pnas.062032499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B, Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, Westphal H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin MC, Monetti M, Watt MJ, Sajan MP, Stevens RD, Bain JR, Newgard CB, Farese RV, Sr, Farese RV., Jr Increased lipid accumulation and insulin resistance in transgenic mice expressing DGAT2 in glycolytic (type II) muscle. Am J Physiol Endocrinol Metab. 2007;293:E1772–1781. doi: 10.1152/ajpendo.00158.2007. [DOI] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Li Q, Haigis KM, McDaniel A, Harding-Theobald E, Kogan SC, Akagi K, Wong JC, Braun BS, Wolff L, Jacks T, et al. Hematopoiesis and leukemogenesis in mice expressing oncogenic NrasG12D from the endogenous locus. Blood. 2011;117:2022–2032. doi: 10.1182/blood-2010-04-280750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobe CG, Nagy A. Conditional genome alteration in mice. BioEssays. 1998;20:200–208. doi: 10.1002/(SICI)1521-1878(199803)20:3<200::AID-BIES3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Logie C, Stewart AF. Ligand-regulated site-specific recombination. Proc Natl Acad Sci. 1995;92:5940–5944. doi: 10.1073/pnas.92.13.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem. 2002;277:33275–33283. doi: 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S. Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science. 2007;317:94–99. doi: 10.1126/science.1140263. [DOI] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–189. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- Nagy A. Cre recombinase: The universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- Nagy A, Rossant J. Chimaeras and mosaics for dissecting complex mutant phenotypes. Int J Dev Biol. 2001;45:577–582. [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of non-dividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Okubo T, Hogan BL. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J Biol. 2004;3:11. doi: 10.1186/jbiol3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A, Brandon EP, Kootstra N, Gage FH, Verma IM. Delivery of the Cre recombinase by a self-deleting lentiviral vector: Efficient gene targeting in vivo. Proc Natl Acad Sci. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Magnuson MA. [Role of glucokinase (GK) in the maintenance of glucose homeostasis. Specific disruption of the gene by the Cre-loxP technique] Journees annuelles de diabetologie de l’Hotel-Dieu. 1999:115–124. [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PloS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, McMahon A. “Cre”-ating mouse mutants—a meeting review on conditional mouse genetics. Genes Dev. 1999;13:142–145. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- Saam JR, Gordon JI. Inducible gene knockouts in the small intestinal and colonic epithelium. J Biol Chem. 1999;274:38071–38082. doi: 10.1074/jbc.274.53.38071. [DOI] [PubMed] [Google Scholar]
- Sadowski PD. The Flp recombinase of the 2-microns plasmid of Saccharomyces cerevisiae. Progr Nucleic Acid Res Mol Biol. 1995;51:53–91. [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676–679. doi: 10.1038/nature05674. [DOI] [PubMed] [Google Scholar]
- Sauer B. Recycling selectable markers in yeast. BioTechniques. 1994;16:1086–1088. [PubMed] [Google Scholar]
- Sauer B, McDermott J. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res. 2004;32:6086–6095. doi: 10.1093/nar/gkh941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33:12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- Schuler M, Dierich A, Chambon P, Metzger D. Efficient temporally controlled targeted somatic mutagenesis in hepatocytes of the mouse. Genesis. 2004;39:167–172. doi: 10.1002/gene.20039. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Kuhn R, Angrand PO, Rajewsky K, Stewart AF. Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res. 1998;26:1427–1432. doi: 10.1093/nar/26.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senecoff JF, Cox MM. Directionality in FLP protein-promoted site-specific recombination is mediated by DNA-DNA pairing. J Biol Chem. 1986;261:7380–7386. [PubMed] [Google Scholar]
- Senecoff JF, Rossmeissl PJ, Cox MM. DNA recognition by the FLP recombinase of the yeast 2 mu plasmid. A mutational analysis of the FLP binding site. J Mol Biol. 1988;201:405–421. doi: 10.1016/0022-2836(88)90147-7. [DOI] [PubMed] [Google Scholar]
- Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL. Morphogenetic and cellular movements that shape the mouse cerebellum; insights from genetic fate mapping. Neuron. 2005;45:27–40. doi: 10.1016/j.neuron.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vooijs M, van der Valk M, te Riele H, Berns A. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene. 1998;17:1–12. doi: 10.1038/sj.onc.1202169. [DOI] [PubMed] [Google Scholar]
- Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krushel LA, Edelman GM. Targeted DNA recombination in vivo using an adenovirus carrying the cre recombinase gene. Proc Natl Acad Sci. 1996;93:3932–3936. doi: 10.1073/pnas.93.9.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Meth. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71:4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lutz B. Cre recombinase-mediated inversion using lox66 and lox71: Method to introduce conditional point mutations into the CREB-binding protein. Nucleic Acids Res. 2002;30:e90. doi: 10.1093/nar/gnf089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry. 2008;47:7012–7019. doi: 10.1021/bi8007017. [DOI] [PMC free article] [PubMed] [Google Scholar]