Abstract
N6-methyladenosine (m6A) is a common modification of mRNA, with potential roles in fine-tuning the RNA life-cycle. Here, we identify a dense network of proteins interacting with METTL3, a component of the methyltransferase complex, and show that three of them, WTAP, METTL14 and KIAA1429, are required for methylation. Monitoring m6A levels upon WTAP depletion allowed the definition of accurate and near single-nucleotide resolution methylation maps, and their classification into WTAP-dependent and independent sites. WTAP-dependent sites are located at internal positions in transcripts, are topologically static across a variety of systems we surveyed, and are inversely correlated with mRNA stability, consistent with a role in establishing ‘basal’ degradation rates. WTAP-independent sites form at the first transcribed base as part of the cap structure, and are present at thousands of sites, forming a previously unappreciated layer of transcriptome complexity. Our data sheds new light on proteomic and transcriptional underpinnings of this epitranscriptomic modification.
Introduction
DNA, RNA and proteins are all subjected to biochemical modifications following synthesis, which can alter and fine-tune their function by diverse regulatory mechanisms. N6-methyladenosine (m6A) is a highly prevalent base modification occurring on mammalian mRNA. Recent studies used immunoprecipitation of methylated RNA fragments followed by sequencing (m6A–Seq) to globally map transcript regions enriched in m6A in mammalian cells, finding that it is strongly enriched near stop codons and in long exons (Dominissini et al., 2012; Meyer et al., 2012).
Conceptually, m6A in mammals has the potential of fine-tuning RNA function in different ways. One possibility is that genes are subjected to methylation only under specific conditions or in specific tissues (‘condition specific methylation’), as appears to be the case in yeast meiosis (Agarwala et al., 2012; Clancy et al., 2002; Schwartz et al., 2013). A non-mutually exclusive scenario is that m6A may mark and regulate a specific sets of transcripts (‘transcript specific methylation’), for instance by affecting their stability (Wang et al., 2013). To explore potential roles for m6A, it is necessary to investigate the extent to which m6A varies across physiologically relevant conditions. To date, mammalian methylated sites have been mapped and characterized in only a small number of mammalian cell lines/tissues (Dominissini et al., 2012; Meyer et al., 2012; Wang et al., 2013) limiting the ability to evaluate methylation dynamics. Moreover, the resolution of these maps was limited, with sites typically being >20 nt away from the nearest consensus signal, potentially reflecting a non-negligible amount of false positives.
Obtaining accurate maps of mRNA methylation requires identification of the proteins involved in catalyzing them. We recently found in yeast that available protocols for m6a–Seq identified both true methylated sites and false positive sites, and that these two classes could be distinguished by mapping methylations that remain after knockout of the methyltransferases (Schwartz et al., 2013). Until recently, only one protein – METTL3 (Methyl-transferase-like 3) – was implicated in m6A methylation of mammalian mRNA (Bokar et al., 1997). However, it had been recognized that additional components were crucial for methylation (Bokar et al., 1997). While this manuscript was under preparation, two additional proteins, WTAP and METTL14, were identified as required for methylation (Liu et al., 2013a; Ping et al., 2014; Wang et al., 2014). These studies, however, did not study the extent to which individual sites were dependent on these proteins, which is important both to eliminate false positives (Schwartz et al., 2013) and to identify sites that are methylated using orthogonal pathways.
Here, we have employed an unbiased proteomic approach to characterize the components of the methyltransferase complex, allowing us to identify and validate known and novel components required for methylation. By mapping sites upon experimental depletion of these components, we were able to classify and characterize methylated sites based on their dependency on these proteins. Our analyses provide important resources at the proteomic and transcriptomic levels towards understanding the regulators (‘who’) and targets (‘where’) of RNA methylations, two crucial milestones towards addressing the function (‘why’) of this epitranscriptomic modification.
Results
Proteomic screens identify novel components of the methyltransferase complex
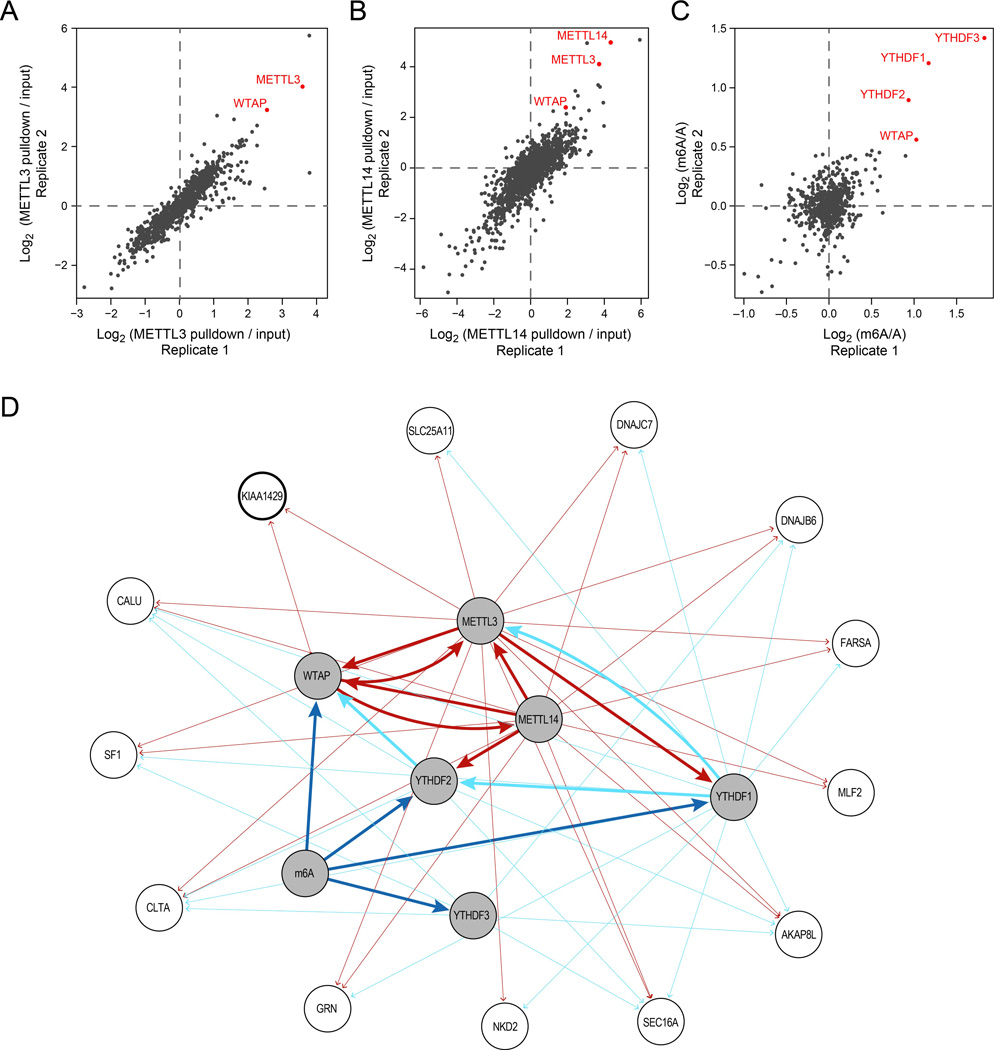
To identify components of the human m6A methyltransferase complex, we performed co-immunoprecipitation (co-IP) experiments using an overexpressed C-terminus HIS-tagged METTL3 in HEK293 cells, followed by LC-MS/MS (Methods). After filtering out background contaminants using the CRAPOME database (Mellacheruvu et al., 2013) (Methods), one of the most enriched proteins was WTAP, the human ortholog of Mum2, a crucial component of the yeast methyltransferase complex (Agarwala et al., 2012; Schwartz et al., 2013) (Fig. 1A). Proteins interacting with WTAP have recently been characterized in a proteomic screen (Horiuchi et al., 2013). Analysis of this data revealed reciprocal binding of WTAP to both METTL3 and to METTL14, a close paralog of METTL3 (Bujnicki et al., 2002). Confirming this association, both METTL3 and WTAP were enriched when we performed mass-spectrometry following pulling down on a 3’ terminus V5-tagged version of METTL14, (Fig. 1B). The physical association of METTL14 and WTAP with METTL3, and our previous findings that all three proteins have tightly coevolved (Schwartz et al., 2013), thus strongly implicated them in mRNA methylation.
Figure 1. Proteomic identification of methyltransferase complex components.
(A-B) Proteins associated with HIS-tagged METTL3 (A) and V5-tagged METTL14 (B). Fold-changes quantifying enrichment versus control (Methods) across two biological replicates. (C) Proteins associated with biotinylated, methylated RNA baits, compared to biotinylated, non-methylated counterparts, in two biological replicates. (D) Network of associations with the bait proteins (shaded grey nodes) identified in the different mass-spec experiments. An edge from bait A to target B indicates that B was enriched (fold change > 1.5) when performing IPs on A. Only target nodes with an incoming degree ≥2 are displayed. Edge width distinguishes association between baits (thick lines) and with non-baits (thin lines). Edges are colored in dark red if they were enriched by a protein bait involving an m6a ‘writer’ (METTL3 / METTL14 / WTAP), cyan if they were enriched by a ‘reader’ (YTHDF1 / YTHDF2 / YTHDF3), or dark blue if they were enriched with a methylated bait. The node for KIAA1429, further analyzed functionally, is marked by a thicker border.
To explore additional proteins involved in the methylation program, we used in vitro transcribed biotinylated baits, which were either methylated or non-methylated at a single position, to capture proteins from HEK293 protein lysates, followed by quantitative LC-MS/MS (Fig. 1C). YTHDF1, YTHDF2 and YTHDF3, three proteins from the YTH domain family, were the top three enriched proteins in this screen. This enhances our previous results (Dominissini et al., 2012), where two of these three proteins were identified as m6A binders (Dominissini et al., 2012), and is consistent with very recent biochemical results showing that these three proteins all selectively bind m6A (Wang et al., 2013). Interestingly, the fourth most enriched protein in this assay was WTAP, suggesting that WTAP may be involved not only in mediating methylation, but also in binding it.
Finally, we used a V5-tagged version of each of three YTH proteins in affinity proteomics. These experiments highlighted potential associations between different YTH proteins (e.g., between YTHDF1 and YTHDF2) or between the YTH proteins and other components of the methyltransferase machinery identified here (e.g., YTHDF2 and WTAP, YTHDF1 and METTL3). The full proteomics datasets are available in Supplemental Table S1.
Integrating the proteomic data into a network (Fig. 1D; Methods), highlights the centrality of WTAP and METTL14 in this complex, consistent with the three recent reports (Liu et al., 2013a; Ping et al., 2014; Wang et al., 2014). The density and topology of the network suggest that the conceptual distinction between m6A ‘writers’ and ‘readers’ may not be clear-cut, as the putative writers (METTL3, WTAP, METTL14) and readers (YTH proteins) may physically interact with each other. Finally, 13 proteins with diverse functions interact with two or more of the 7 core components studied here, and may therefore form part of the methyltransferase complex as well. However, we cannot rule out that some of these associations may be spurious, and each of these candidates will therefore have to be individually tested. Below we validate three of these proteins.
WTAP is necessary for m6A mRNA methylation
To determine whether WTAP is required for mRNA methylation in vivo, we depleted it in p53−/− mouse embryonic fibroblasts (MEFs) using WTAP-targeting shRNAs to 10–15% of WT levels (Fig. S1A–D). We used anti-hnRPDL and anti-GFP shRNAs as negative controls. We immunoprecipitated and sequenced methylated RNA from these samples using an m6a–Seq procedure, optimized for enhanced resolution and scalability using decreased starting material (Schwartz et al., 2013). Analyzing these profiles (Methods) we identified 16,487 putative m6A sites, present in at least two of the 8 profiled conditions.
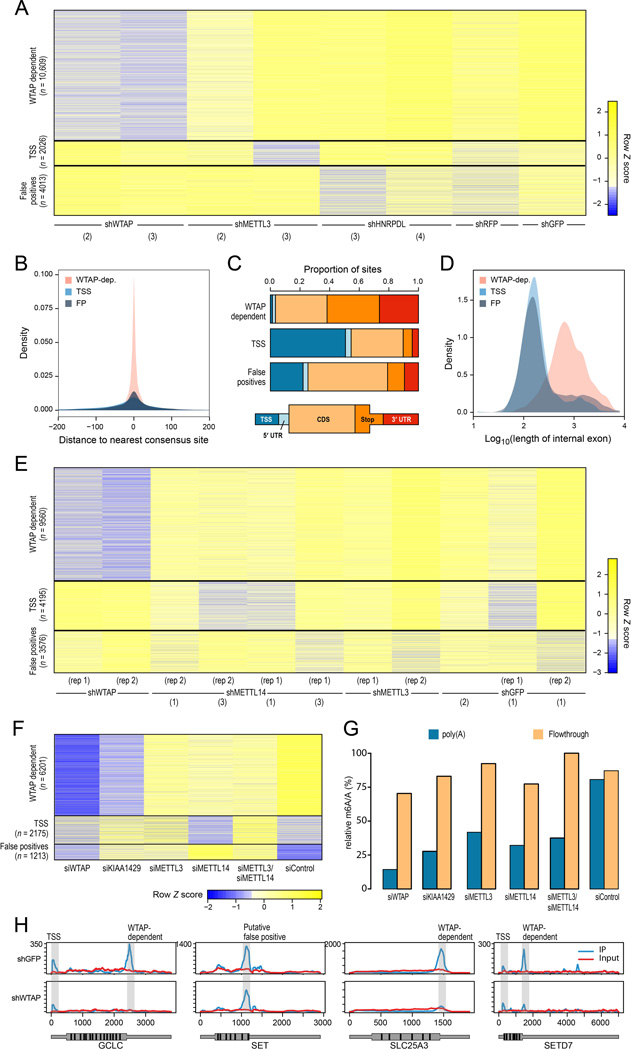
Most sites (10,609/16,487, 64.3%) were WTAP-dependent (‘WTAP-dependent’ cluster), showing dramatically decreased methylation Peak Over Input (POI) scores compared to any of the controls (P<2.2×10−16 for all comparisons, Mann-Whitney) following knockdown with either of two shRNAs targeting different regions of WTAP (Fig. 2A). WTAP-dependent sites were strongly enriched for hallmarks of m6A methylation: sites were within a median distance of 5 nt from the nearest consensus site (Fig. 2B, Fig. S1E–F); 34.6% of sites were within 200 nt of the stop codon (Fig. 2C); and median internal exons harboring sites in this cluster tended to be ∼7-fold longer than non-methylated exons (Fig. 2D).
Figure 2. Perturbation of methyltransferase complex components identifies complex-dependent and -independent methylation sites.
(A) K-means clustering of log2 transformed Peak Over Input scores (yellow: high, blue: low) across 16,487 sites (rows) detected in mouse embryonic fibroblasts (MEFs). Only sites detected in 2 or more of the presented samples (columns), with a maximal Peak Over Input score greater than 8, and within genes expressed above the 60th percentile are shown. Sites were initially clustered into 5 clusters using k-means, and three of the clusters were subsequently manually merged with existing clusters, based on similarity of characteristics, to yield 3 clusters (separated by black lines). For visualization purposes, the log2(POI) scores were capped at 4.5, and then transformed into Z scores. The internal shRNA identifiers are indicated in parentheses. (B) Distributions of distance from the nearest consensus site of the sites in the three clusters in (A). (C) Distribution of the sites in (A) grouped by the clusters in (A) across five gene segments, defined as in (Dominissini et al., 2012). (D) Distribution of length of internal exons comprising the sites identified in (A) across the identified clusters. (E) K-means clustering of log2 transformed Peak Over Input scores across 17,331 sites (rows) detected in human A549 cells, visualized as in A. (F) K-means clustering of log2 transformed Peak Over Input scores for set of experiments involving perturbations of A549 cells with siRNAs, as indicated. Only WTAP-dependent sites (rows) are plotted within genes expressed above the 60th percentile across all conditions. (G) Relative m6A/A ratios across A549 cells treated with siRNA as indicated, as measured by HPLC-M/S. Values are plotted for both the poly(A) fraction (blue) and the flowthrough (orange). (H) Examples of coverage from IP (blue) and input (red) experiments across selected genes in A549 cells treated with shGFP (top row) or shWTAP (bottom row).
The remaining sites formed two distinct clusters (Fig. 2A), neither of which was enriched for the methylation consensus signal (median distances to consensus motif were 27 and 38 nt, respectively), or for any of the other hallmarks of m6A methylation (Fig. 2B–D). One of these two clusters showed dramatic enrichment for sites near the TSS, with ∼50% of the sites within 200 nt of the TSS (‘TSS-enriched cluster’); we show below that the sites in this cluster represent methylations on the first transcribed nucleotide as part of the cap structure. The other cluster did not exhibit such dramatic enrichment for the TSS segment, but was pervasive across all sampled conditions, and showed no enrichment for the m6A consensus motif. These properties were reminiscent of a set of false positive sites that we found in yeast when performing the same m6a–Seq protocol to strains lacking a functional methyltransferase (Schwartz et al., 2013). Indeed, sequence analysis revealed that the sites in the cluster (‘False-positive cluster’) were highly enriched for the same degenerate purine-rich motif (‘AGAAGAA’) that we found at the yeast false positive sites (Fig. S1G–H). In yeast, we have previously shown that these sites were false positive sites enriched in a non-specific manner during the immunoprecipitation process, since they are enriched even when we perform m6A–seq with in vitro synthesized sites without any methylations (Schwartz et al., 2013); Thus, we considered it highly likely that these sets of sites are false positives, and conservatively opted to either analyze them separately, or eliminate them altogether, in the subsequent analyses.
We confirmed these results in human A549 cell lines where WTAP was perturbed using either shRNAs (Fig. 2E) or siRNAs (Fig. 2F). We observed a similarly strong reduction of methylation upon WTAP-depletion in both sets of experiments, and clustering of the detected sites yielded very similar clusters to those obtained in the mouse studies above (Fig. 2E–F), with similar characteristics (Fig. S1I–K).
KIAA1429 is required for mRNA methylation
Of the other 13 candidates associating with methyltransferase components in our proteomics screen (Fig. 1D), we focused our attention on KIAA1429, since its Drosophila ortholog was biochemically shown to interact with Drosophila WTAP in the context of sex-specific splicing (Ortega et al., 2003). We used siRNAs to deplete KIAA1429 in human A549 cells to ∼6% of WT levels (Fig. S1C), and performed m6A–Seq on the resulting cells. We observed a median ∼4-fold decrease (Mann-Whitney, P<2.2×10−16) in peak scores compared to cells treated with non-targeting siRNAs (Fig. 2F). Although these decreases were less dramatic than those observed upon knockdown of WTAP (median decrease: 6.25 fold), they were substantially and significantly (Mann-Whitney, P<2.2×10−16) more prominent than observed upon knockdown of either METTL3 or METTL14 (see below), demonstrating that KIAA1429 is required for the full methylation program in mammals.
Knockdown of METTL3 or/and METTL14 leads to milder decreases in methylation levels
We next used shRNAs and siRNA to test the effect of depleting METTL3 and METTL14 in mouse fibroblasts (METTL3, Fig. 2A) and in human A549 cells (both, Fig. 2E–F). In mouse, we observed a moderate depletion of m6A levels for one of the hairpins (shMETTL3-2, where METTL3 transcript was reduced to 10% of WT levels), but not for another (shMETTL3-3, where knockdown efficiency was lower) (Fig. S1A). In human, shRNA mediated knockown of METTL3 levels to ∼10% of WT levels did not reveal a discernible effect on m6A methylation, but use of siRNAs, which enabled even higher knockdowns (∼5% of WT levels, Fig. S1C–D), yielded POI scores that were reduced to 60% of control (Mann-Whitney, P<2.2×10−16, Fig. 2F). Similarly, for METTL14 knockdown, we failed to observe an effect when knocking it down using shRNAs to ∼20% of WT levels (Fig. S1B), but upon knocking it down to ∼6% using siRNAs (Fig. S1C) we observed reductions to similar levels as for METTL3 (median decrease: 51% of control). Similar levels were achieved also with dual knockdown of METTL3 and METTL14 (median decrease: 53% of control, Fig. S1C). Our results, which are consistent with (Liu et al., 2013a), demonstrate a strong threshold dependence for both METTL3 and METTL14, which is likely due to adequacy of even low levels of METTL3 and METTL14 to mediate mRNA methylations.
Bulk-measurements of m6A–levels highly consistent with m6A–Seq
To further confirm our results, we used HPLC-M/S to measure bulk levels of m6A in poly(A) mRNA from A549 cells upon siRNA mediated knockdown of the above genes (Fig. 2G). Consistently, highest levels of depletion in m6A content in the oligo-dT selected poly(A) fraction were obtained for WTAP (5.6 fold depletion), followed by KIAA1429 (∼3 fold), followed by single or combined knockdown of METTL3 and METTL14 (1.9–2.5 fold). M6A levels in the flowthrough fraction were roughly equal for all samples (Fig. 2G). These results were expected, as the flowthrough is dominated by rRNA, which harbors m6A at non-consensus positions (Liu et al., 2013b; Machnicka et al., 2013) that are likely deposited by proteins distinct from the ones identified here.
A high-quality, high-resolution catalogue of the m6A methylome across four dynamic systems
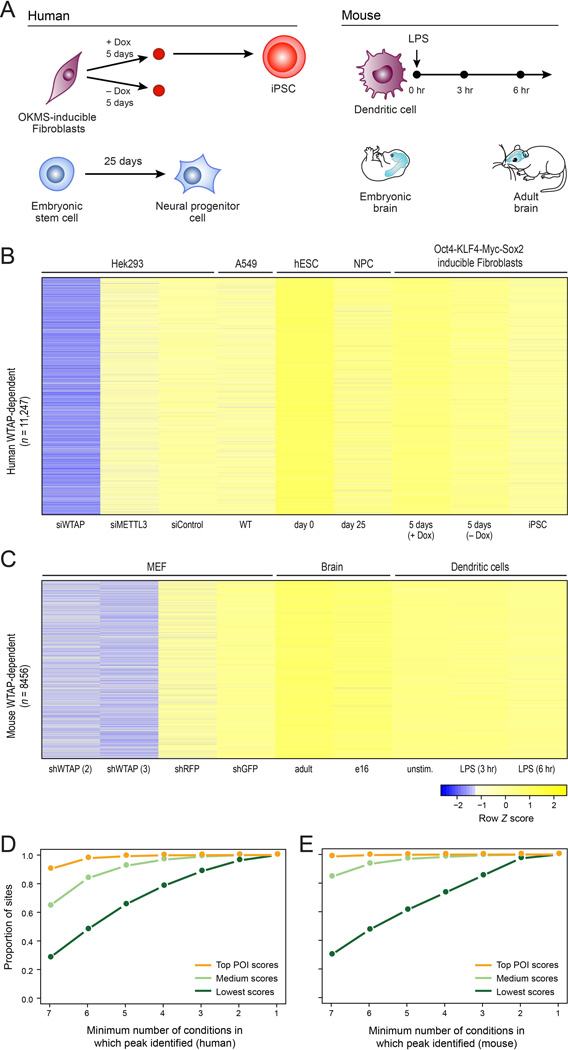
To date, mammalian methylated sites have been mapped and characterized in only a limited number of cell lines and tissues. To determine the extent to which m6A methylation may be modulated in physiologically relevant contexts, in both mitotic and post-mitotic cells, we mapped methylation across four distinct dynamic systems in human and mouse: (1) mouse bone-marrow derived dendritic cells (BMDCs) responding to lipopolysaccharide (LPS), (2) mouse embryonic (e16) and adult brains, a system chosen based on previous reports of lower levels of bulk m6A in embryonic brain (Meyer et al., 2012), (3) human fibroblasts undergoing reprogramming into induced pluripotent stem cells (iPSC) following doxycycline-induced expression of polycistronic OCT4-KLF4-MYC-SOX2, and (4) human embryonic stem cells undergoing differentiation into NPCs (Fig. 3A).
Figure 3. A human and mouse catalogue of methylation dynamics.
(A) Scheme depicting the four experimental systems profiled in this study. Top left: Doxycycline-inducible OKMS (OCT4, KLF4, MYC and SOX2) fibroblast reprogramming to iPSCs. Bottom left: Human embryonic stem cells undergoing differentiation to neural progenitor cells. Top right: Bone marrow derived dendritic cells stimulated by lipopolysaccharide (LPS). Bottom right: Embryonic and adult mice brain. (B,C) Heatmaps of POI scores (visualized as in Fig. 1A) of all WTAP-dependent sites (rows) identified in one or more sample (columns), within genes expressed above the 60th percentile across all conditions (Methods) in human (B) or mouse (C). (D, E) Number of conditions (X axis, of a total of 7, as in B and C respectively, excluding any genetic perturbations) in human (D) and mouse (E), in which sites are identified as present (POI > 4). Sites were grouped into three equally sized groups, based on POI score.
Integrating all m6A profiles yielded 40,742 sites in human and 31,423 in mouse, present in at least two conditions, tripling the reported number of detected putative methylation sites compared to previous studies. We classified each site as high, intermediate, or low confidence based on a linear combination of three features: (1) number of samples in which peak was identified, (2) extent of dependency on WTAP, and (3) maximal POI score observed across any of the samples. The features were selected as informative based on distance to nearest consensus site (Fig. S2A; Methods), and the resulting ranking was independently consistent with enrichment of sites in long exons (Fig. S2B) and near stop codons (Fig. S2C). The sites identified here are at a dramatically increased resolution, compared with previous studies, with 50% of the top 15,000 sites being within 5–6 nt from the nearest consensus site, in both datasets. The full set of sites is provided (Supplemental Table S2) allowing custom filtration of the data based on user-defined criteria.
WTAP-dependent sites have a static topology across multiple dynamic systems
While initial examination suggested the presence of many condition-specific methylations, closer examination indicated that observed differences were largely due to changes in underlying gene expression. For example, in the mouse dataset, 3,629 sites classified as ‘high’ or ‘intermediate’ are present only in the two brain samples, and 1,312 such sites are present exclusively in the DCs. However, these differences clearly reflected the expression levels of the genes harboring the sites (Fig. S2D,E), which indeed were enriched for genes involved in neural processes and immune response, respectively (Fig. S2F,G).
To reliably compare sites across samples despite different expression levels, we therefore conservatively limited our analysis to a subset of WTAP-dependent sites within genes expressed above the 60th percentile in all samples, resulting in 11,247 sites in human (Fig. 3B) and 8,456 sites in mouse (Fig. 3C). Dividing these into three equally sized sets based on their maximal POI scores across all samples, we found that among the highest scoring sites, >90% of the sites in human (Fig. 3D), and >98% of the sites in mouse (Fig. 3E) were detected as enriched (POI > 4) across all samples. Among lower scoring sites, cases of peaks being called in one condition but not in another were more common (Figs. 3D,E); however, upon manual inspection of such putative differential sites, it was difficult to identify convincing sites present in one condition and absent from another. Rather, such differences were typically due to sub-threshold peaks not being called in a particular sample or insufficient coverage in a particular region. Indeed, the variability of POI scores across the different dynamic sets of conditions was similar to the variability across different experiments performed in the same cell line (MEFs in mouse, A549 in human) (Fig. S2H,I). Thus, overall methylation profiles appear to be similar across diverse conditions, at least under our conservative criteria, suggesting that in mammals m6A plays a basal, ubiquitous role, shared across different cell types and systems. However, as m6A–seq cannot directly quantify methylation levels, our approach may be blind to quantitative differences in methylation levels between samples (see Discussion).
WTAP-dependent methylation inversely correlates with mRNA stability, and is depleted in housekeeping genes
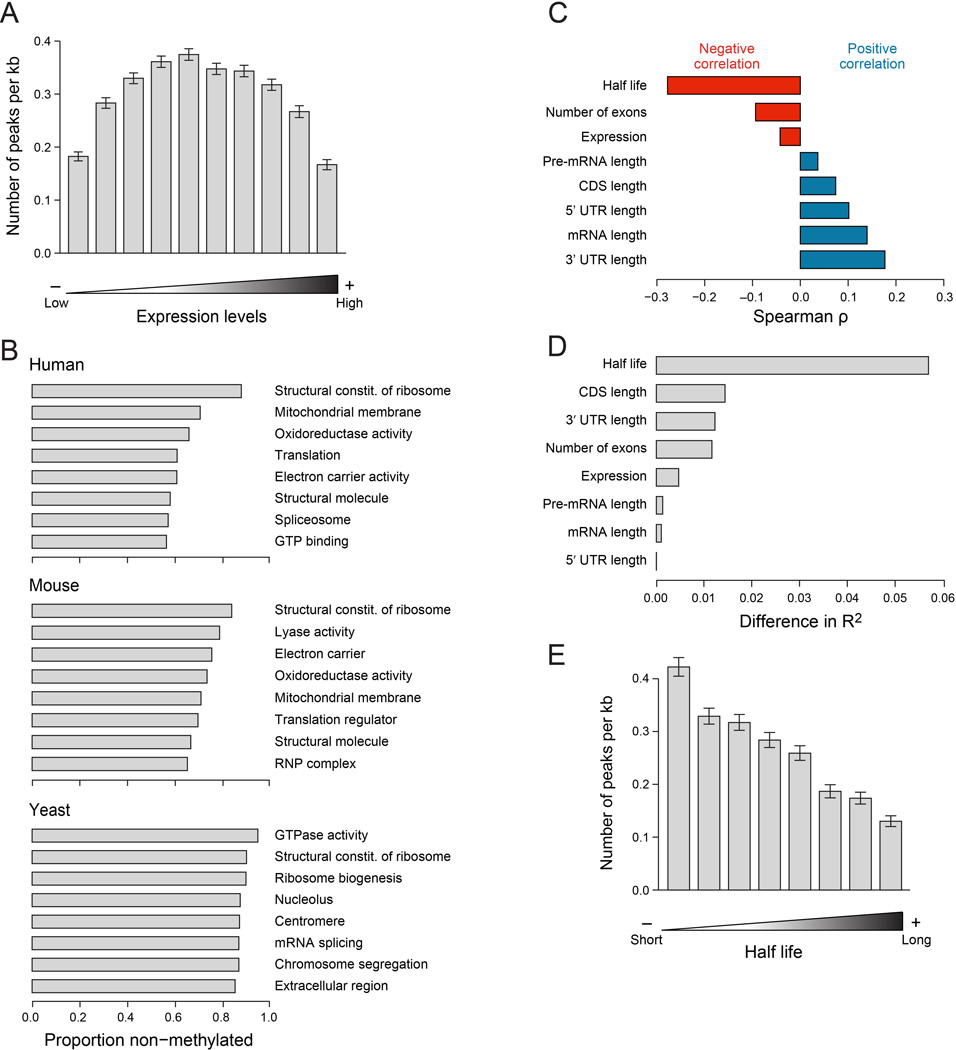
As we found little evidence for condition-specific differences in methylation, we next sought to analyze the potential of a transcript-specific role. Examining the relationship between gene-level attributes and methylation densities (number of WTAP-dependent methylation sites normalized by gene length) in mouse, we noted that the top 20% of expressed genes were significantly less likely to be methylated compared to more lowly expressed genes (Fig. 4A). Indeed, genes completely lacking methylation sites are highly enriched for those involved in ‘housekeeping’ cellular processes like translation, mitochondrial related processes, chromatin regulation, and splicing. To study this further, we examined the proportion of genes lacking methylations across all Gene Ontology (GO) categories in human, mouse and yeast. This analysis revealed that the functional group ‘structural constituents of ribosome’ ranked highest in proportion of genes lacking methylation in both human and mouse, and second highest in yeast (Fig. 4B). Other groups of housekeeping genes ranking high in both yeast and mammals include splicing and GTPase activity (Fig. 4B). Thus, lack of methylations among ribosomal proteins, in particular, and specific sets of housekeeping proteins, in general, is conserved between yeast and mammals.
Figure 4. Methylations are depleted from housekeeping genes, and associated with decreased transcript stability.
(A) Association between gene expression and methylation density in MEFs. (B) Top GO categories harboring non-methylated genes in human (top), mouse (middle), and yeast (yeast). Shown is the proportion of non-methylated genes in each category. Only genes expressed >60th percentile and longer than 500nt and only GO categories harboring >30 matches genes were considered for this analysis. (C) Correlations (Spearman ρ, X axis) between different gene attributes (Y axis) and methylation density. Transcript half-lives were obtained from (Schwanhausser et al., 2011). We used only sites defined in A549 cells (Fig. 2E), in protein coding genes longer than 500nt and expressed above the 60th quantile, with a maximum POI score >8, with at least 50% increased POI score in non-perturbed cells compared to WTAP-depleted cells, and which were in segments other than the TSS segment. (D) Analysis examining the difference in variability explained (R2, X axis) upon elimination of each of the indicated variables from a linear regression model predicting methylation density on the basis of all of them. (E) Association between mRNA half lives (Schwanhausser et al., 2011) and methylation densities. Half lives were binned into 10 equally sized bins, and mean methylation densities for each bin were calculated. Error bars - SEM.
Since housekeeping genes generally have shorter mRNAs with shorter CDSs and UTRs and fewer exons (Eisenberg and Levanon, 2003) and longer RNA half-lives (Schwanhausser et al., 2011), we directly examined the correlation between each of these variables and methylation densities (Fig. 4C,D). Strikingly, the variable correlating most strongly with methylation density was mRNA half-life (Fig. 4C–E), as previously estimated in mouse fibroblasts (Schwanhausser et al., 2011) (Spearman ρ=−0.27, P=1.2×10−69), and this correlation remained of similar magnitude (ρ=−0.22) when performing this analysis after eliminating the top 10% or top 20% of genes, ranked by expression levels. Consistently, examining the difference in proportion of variance explained (R2) when predicting methylation densities from all these variables, compared to exclusion of any single one, revealed that mRNA half-life accounted for the greatest difference (Fig. 4D). We obtained similar results when performing these analyses for human transcripts using transcript stability estimates in human lymphoblastoid cell lines (Duan et al., 2013) (Fig. S3A,B), with half-life exhibiting the strongest correlation with methylation density (Fig. S3C), albeit at somewhat reduced levels (ρ=−0.14, P=3.4×10−34). Our results are consistent with reports implicating methylations in mRNA degradation (Wang et al., 2013; Wang et al., 2014), and suggest that m6A methylation may help set a basal degradation program, and that highly-abundant messages might have evolved to maximize their stability by avoiding mRNA methylation.
WTAP-independent m6A methylome at the mRNA cap identifies methylated TSS
Studies in the mid-1970s involving bulk analysis of mRNA caps revealed than when the first nucleotide of a transcript is an adenosine, this base can be methylated at the N6 position (Furuichi et al., 1975; Keith et al., 1978; Wei et al., 1975). However, no study to date has been able to resolve which mRNA caps within which messages are methylated. We hypothesized that the consistently-observed WTAP-independent peaks enriched near the TSS (Fig. 2A,E,F) reflect methylations occurring at the cap. Corroborating this hypothesis, we found that 30.5% of the mouse genes expressed above the 60th percentile with an ‘TSS peak’ (i.e., an m6A peak within the first 200 nt) are annotated to begin with an ‘A’, compared to only 23% of the genes lacking a peak (Fisher’s exact test, P=2.8×10−13), consistent with analyses performed in (Dominissini et al., 2012). While this enrichment was significant, it did not explain the peaks observed in the remaining ∼70% of the genes.
We therefore considered the ∼70% of genes whose RNA transcripts harbored a TSS peak but whose annotated TSS did not contain an ‘A’ at the first base. We suspected that many of these cases might reflect limitations of the genomic annotation, as transcription initiation often occurs from a variety of closely spaced positions (Carninci et al., 2006; Ni et al., 2010; Plessy et al., 2010). We therefore leveraged the fact that our library construction method relies on ligation of adapters to both ends of the captured RNA fragment, such that although each transcript is fragmented ‘randomly’, every transcript should yield some fragment beginning at its 5’ terminus, resulting in a ‘pileup’ of reads beginning at the 5’ terminus of genes. Thus, these pileups harbor information on the precise transcript initiation sites. Moreover, methylated transcription start sites (mTSSs) should have a high ratio between the size of these pileups in the IP sample compared to the input sample; sites with high ratios should therefore be highly enriched towards harboring an adenosine, as opposed to typical transcription start sites which tend to begin with a guanosine (Carninci et al., 2006).
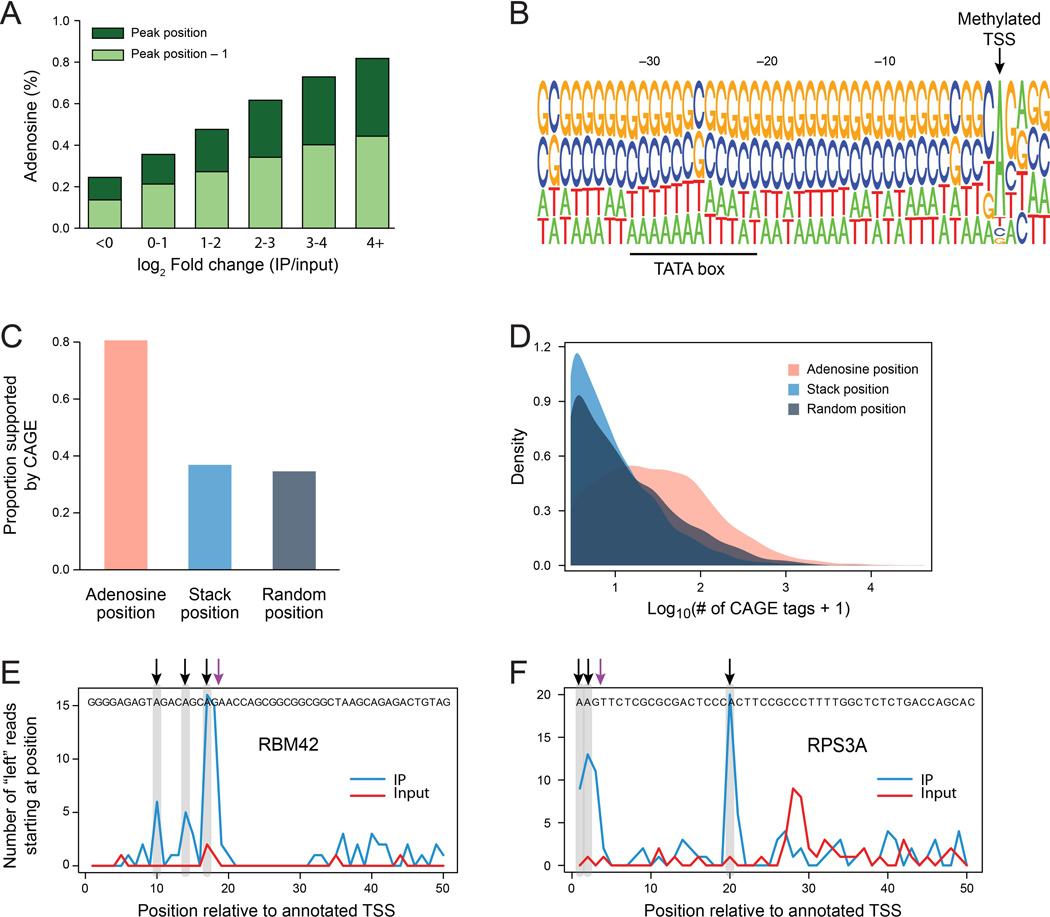
To detect methylated transcription start sites (mTSSs), we compared the number of reads stacks beginning at each of the first 50 annotated positions in the transcript in the IP and Input samples (Methods). We assembled a catalogue of 33,714 sites in mouse with evidence of >10 reads beginning at a specific site (across all conditions), and assigned each site a fold-change corresponding to its enrichment in IP over input. As expected, we found that sites with stronger fold-changes were also dramatically more likely to harbor an adenosine either at the detected site or at the position immediately preceding it, with >80% of the sites harboring an adenosine in the bin of most highly enriched sites (Fig. 5A). (The tendency for adenosines at the position preceding the pileups is likely due reverse transcriptase drop off one base prior to the TSS, as supported by several lines of evidence (Supplemental Note 1, Figures S4A,B, and below). Further corroboration of the validity of the detected sites was obtained from an observation of a strong bias towards pyrimidines at position −1, and a discernible TATA box ∼30 bp upstream of the detected sites, two hallmarks of transcription initiation sites (Carninci et al., 2006) (Fig. 5B).
Figure 5. Identification of methylated transcription start sites (mTSSs).
(A) Proportion of sites harboring an adenosine (Y axis) at the detected position (dark green bars) or the position immediately upstream (light green bars) in 33,714 sites with >10 reads within 50 nt of annotated TSSs. Sites were binned based on fold changes (IP over input; X axis), using thresholds as indicated. (B) Sequence logo for a set of 3,445 putative mTSSs with a log2 fold-change exceeding 4.5. The position of the putative methylated mTSS is indicated, and of a TATA box signal located ∼30 bp upstream. (C) Proportion of putative mTSSs supported by 2 or more CAGE tags (Y axis) for a subset of 4,879 sites, in which the ‘Adenosine’ position precedes the ‘Stack’ position. Proportions are shown both for the adenosine position (pink bar), the stack position (cyan), and 4,879 positions randomly assigned to the same set of genes within the first 50 nt (grey bar). (D) Distributions of the log10 transformed number of CAGE tags observed for the three groups defined in (C). (E, F) Examples of genes with multiple internal mTSSs in human iPSCs. Number of reads beginning at each of the first 50 annotated positions in the IP (blue) and input (red) sample are shown. Detected mTSSs are highlighted in grey, and marked with a black arrow; Putative penultimate-base RT-dropoffs are indicated with a purple arrow.
Our predicted mTSSs also agree well with the results of cap analysis of gene expression (CAGE) data. We performed the above analyses in human (obtaining highly similar results; Fig. S4C) and focused on a subset of 9,757 putatively methylated TSSs (mTSSs) that were independently detected in at least two experiments in A549 cells, exhibiting at least 4-fold enrichment in IP over input, and harboring an adenosine at either the position harboring the read stacks or the one preceding it. We compared these sites to CAGE data in A549 cells, obtained from the ENCODE project (Consortium et al., 2012). Of the 9,757 sites, 7,255 (74%) were supported by at least two CAGE tags, compared to only 34.6% among an equally sized set of random controls (Fisher’s exact test, P <2.2×10−16). To confirm that the positions harboring the adenosines (‘adenosine positions’) were the true TSS even when they preceded the observed stacks by one base (‘stack position) (a phenomenon we attribute to RT-dropoff), we examined a subset of 4,879 sites in which the adenosine positions preceded the stack positions. While 80.5% of the ‘adenosine positions’ were supported by at least two CAGE tags, only 36.8% of the ‘stack positions’ were supported by CAGE data, similar to the 34.5% overlap obtained in the random dataset (Fig. 5C). Moreover, there was >6 fold increase in the mean number of CAGE tags supporting the ‘adenosine positions’, compared to either the ‘stack positions’ or the random positions (Fig. 5D). Finally, when restricting this analysis to positions other than the first annotated position, these results were essentially identical (data not shown). Together, this confirms that our approach can identify transcription initiation sites at single nucleotide resolution and that mTSSs can be reliably detected also within a large number of positions other than the annotated TSS.
Based on these analyses, we compiled a catalogue of putatively methylated transcription start sites (mTSS), comprising all unique adenosine-containing sites enriched >4 fold in at least one condition, encompassing 15,961 sites from 6,454 mouse genes and 12,601 sites from 5,774 human genes (Supplemental Table S4). Many sites – and particularly sites with stronger associated fold-changes - were shared across multiple conditions (Fig. S4D–F), and 51% of the above genes in mouse (23% in human), had more than one detected mTSS across all conditions. Limiting this analysis to a single condition, on average 13% of the transcripts had more than one mTSS in both mouse and human (e.g. Fig. 6E,F). Evaluation of the number of mTSSs found as a function of gene expression, revealed that mTSS detection in our data has not reached saturation (Fig. S4G–H), and thus the numbers reported here likely reflect a lower bound.
To gain insight into the potential role(s) of mTSSs, we examined the extent to which mTSS presence in a gene was associated with gene structure, RNA stability and translational efficiency features. As our ability to detect mTSSs is biased towards more highly expressed genes, we used a randomly sampled control dataset matching the expression patterns of the mTSS harboring genes (but lacking mTSSs). We found that the presence of mTSS is significantly inversely correlated with 5’ UTR length in both mouse and human (Fig. S5A–E), and positively correlated with translation efficiency (Figs. S5A–B,F-H). However, in contrast to internal methylation, mTSSs were not associated with transcript stability (Figs. S5A–B; see also Supplemental Note 2). These results may suggest that mTSSs and internal methylation sites may differ both in the factor(s) mediating their catalysis, and in their function, with internal sites potentially affecting stability and mTSSs potentially affecting translation. However, as these results are based on comparison between different transcripts, which may vary from each other in other respects that could potentially correlate with mTSS state, they must be interpreted with care. The presence and abundance of the mTSS identified here add an additional unappreciated layer of complexity to the mammalian transcriptome, suggesting that a substantial fraction of transcripts exist in distinct isoforms, differing not only in the location of the TSS but also in its methylation state. It will be crucial to develop experimental methodologies to specifically perturb the methylation state of individual transcripts, and to monitor the impact of such perturbations on various stages of an RNA’s life cycle, to conclusively understand the role of mTSSs.
Discussion
The m6A methylation landscape
RNA methylations offer the potential of dynamically modulating characteristics of an mRNA’s life cycle. Such regulation may potentially be applied either in specific conditions, or to specific transcripts, two scenarios which we set out to explore. By mapping methylations across various dynamic systems in human and mouse, we were able to more than triple the number of sites previously identified for either human or mouse (Dominissini et al., 2012; Meyer et al., 2012), while simultaneously enhancing the resolution of the detected sites, to nearly single-nucleotide level. We found that across the monitored conditions methylations appeared to be largely static, suggesting that the role of m6A in mammals is to a large extent condition-independent. These results should best be interpreted with caution, as it is possible that m6a–Seq is unable to capture subtle, quantitative differences in the proportion of transcripts methylated at a specific position, which could still be of functional consequence. To better assess the extent to which m6A is quantitative, measurements by orthogonal methods, such as the recently developed SCARLET (Liu et al., 2013b) will be required for a large number of sites.
While the topology of WTAP-dependent methylations is relatively static across mRNAs, we find that their distribution across genes is not uniform, and that methylations are depleted from abundantly transcribed messages. We moreover find that methylation densities correlate inversely with mRNA half-life. These findings are consistent with recent findings that YTHDF2 binds to methylated mRNAs and decreases messenger stability (Wang et al., 2013), and with the fact that transcript half-lives are generally constant across different conditions. Indeed, in DCs, we do not see any change in m6A densities for those ∼15% of transcripts where degradation rates change dynamically in response to LPS (data not shown). Collectively, our results suggest that internal m6A sites may play a ‘basal’ role in controlling the half-lives of the methylated transcripts, and that this role is relatively constant across the different surveyed conditions.
The multi-component methyltransferase complex
Initial characterization of the mammalian methyltransferase complex revealed that at least two separable complexes (875 and 200 kDa in size) were required for restoring full m6A activity in vitro (Bokar et al., 1994). Subsequent analysis identified METTL3 as one of the components in the smaller complex (Bokar et al., 1997). Here, we show that both WTAP, KIAA1429 and METTL14 are required for full m6A activity in vivo. The multi-unit structure of the m6A methyltransferase is unique with respect to other characterized nucleic acid methyltransferases (Bokar et al., 1994), and offers the potential for complex regulation; Such is the case in yeast where onset and offset of methylations in meiosis were governed by up/down-regulation of different members of this complex at different time points in meiosis (Schwartz et al., 2013).
Our results regarding WTAP and METTL14 are consistent with recently published results (Liu et al., 2013a; Ping et al., 2014; Wang et al., 2014), whereas KIAA1429 was not identified by these studies. The physical association of KIAA1429 with the methyltransferase complex is supported by the physical association of Drosophila homologs of KIAA1429 (virilizer) and WTAP (fl(2)d) in the context of female-specific alternative splicing (Granadino et al., 1990). Moreover, KIAA1429 in human localizes to the nuclear speckles (Horiuchi et al., 2013), as do METTL3 (Bokar, 2005) and WTAP (Horiuchi et al., 2013; Little et al., 2000), further supporting their association with each other. We identified another 13 proteins interacting with two or more of the above components. This collection may contain false positives, and it will therefore be important to experimentally validate them via similar strategies.
The expression patterns of the different methyltransferase components offer important clues about the processes in which they are implicated. The yeast WTAP homolog, Mum2 is expressed in a meiosis-specific manner (Agarwala et al., 2012; Schwartz et al., 2013) and required for proper progression of meiosis (Davis et al., 2001). In Drosophila, fl(2)d and Virilizer are both expressed at highest levels in the ovaries and brain (Robinson et al., 2013). Analysis of METTL14 expression levels across the Illumina Human Body Map (Farrell et al., 2014) a;sp reveals particularly high levels of expression in testis and ovaries. These observations, combined with previous findings that ALKBH5, an m6A demethylase, is expressed at highest levels in the testis and required for spermatogenesis (Zheng et al., 2013), suggest that the methyltransferase complex may have an evolutionary conserved role in mammalian gametogenesis. Nonetheless, the ubiquitous expression of mammalian WTAP and KIAA1429 across tissues and ubiquitous presence of m6A suggests that this modification has acquired additional roles in mammals.
Rich landscape of m6A at the mRNA cap
In higher eukaryotes, but not in yeast, the 5’ cap structure contains 2′-O-ribose methylations at either the first nucleotide or the first and second nucleotide (Wei et al., 1975). When the first nucleotide of the transcript is an adenosine, this base can be methylated at the N6 position (Furuichi et al., 1975; Keith et al., 1978; Wei et al., 1975). However, no studies to date have resolved the methylation states of the caps of specific transcripts. Here, we simultaneously measured transcription initiation sites and their methylation status at thousands of sites, and found that the transcription initiation landscape is highly complex, with transcript isoforms differing from each other in their methylation states. Our findings that mTSS state at the cap is correlated negatively with 5’ UTR length and positively with translation efficiencies suggest that the process most likely to be affected by mTSSs is translation. To investigate this more thoroughly, it will be necessary to better understand the enzymatic components involved in cap methylation (Keith et al., 1978), and to develop targeted methodologies for monitoring and perturbing methylation status at the cap.
The ubiquity of m6A across dramatically different cell types and systems, the conservation of of the different components involved in mediating and binding it across most studied systems, and the phenotypes associated with its depletion all suggest a fundamental role in mammalian cell biology. The rich resources proteomic and transcriptomic resources we provide in this study will help advance our understanding on the function of this epitranscriptomic modification.
Methods
Cell culture
Human 293T cells were transfected with plasmids encoding 3’ tagged proteins (Table S7) using Lipofectamine 2000 (Life Technologies). Pulldown was performed using Anti-HIS (GenScript) or Anti-V5 (Life Technologies) antibodies. siRNAs (Thermo Scientific) were delivered using Lipofectamine RNAiMAX (Life Technologies); shRNAs (Broad RNAi Consortium) were delivered in polybrene supplemented media and selected for 72–96 hours. Details regarding derivation/culturing of HUES9, neural progenitors, mice brain samples, BMDCs, and hTERT immortalized fibroblasts reprogrammed into iPSCs are in Extended Methods.
Mass Spectrometry data analysis
Details pertaining mass-spectrometry data acquisition are in Extended Methods. All mass spectra were processed using the Spectrum Mill software package v4.0 beta (Agilent Technologies, Santa Clara, CA) according to (Mertins et al., 2012). For each peptide, a log2 fold change was calculated between its intensities in the pulldown sample compared to the control. The peptides were mapped to genes based on the Uniprot database, and the median fold-change was assigned to each gene. We then subtracted the median of the distribution of the log2 transformed values (across all genes) from the individual fold-changes of each gene, to center the fold-change distribution around 0. To filter out background contaminants, we used the CRAPOME database (Mellacheruvu et al., 2013), which summarizes the number of peptides identified for each protein across 343 control experiments. A protein was considered a contaminant if it was present (based on ≥ 2 peptides) across ≥20 control experiments, and filtered from subsequent analysis. We supplemented our data with published proteomics data of WTAP pulldown, obtained from (Horiuchi et al., 2013). We then generated a network, whereby a directed edge existed between bait A (METTL3, WTAP, METTL14, YTHDF1, YTHDF2, YTHDF3, or m6A bait) and target B if the interaction between the two, in any of the experiments, was associated with a fold change >1.5.
M6A-Seq and analysis of internal m6A methylation sites
Isolation of total RNA, preparation of poly(A) RNA, the m6A pull-down procedure, and library preparation were performed as detailed in (Schwartz et al., 2013). The computational analysis for internal sites is based on (Schwartz et al., 2013), with some modifications.
Analysis of putative sites in the TSS
To identify putative mTSSs, we first counted the number of ‘left’ reads originating at the first 50bp of all annotated transcripts, and recorded all positions in which a stack of 5 or more reads originated. An initial collection of all sites matching these criteria, and identified across any of the perturbations or dynamic systems was generated. To account for the presumed RT dropoff, each site was assigned an ‘effective position’, which corresponded to the stack position, unless that position did not harbor an adenosine and the position immediately preceding it did, in which case it was assigned the position preceding it. Sites were then aggregated based on genes and effective positions, and for each position we summarized (1) total number of reads originating from all input samples at each effective site, (2) total number of reads from all IP samples, and (3) a fold change between (1) and (2).
Statistical analysis
All statistical analyses and visualizations were performed in R: Sequence logos were prepared using the SeqLogo package (Bembom, 2011) and heatmaps were generated using the gplots package (Warnes, 2012).
Accession Numbers
Sequencing data have been deposited into the Gene Expression Omnibus (GEO, accession number GSE54365).
Supplementary Material
Highlights.
-
-
METTL3, METTL14, WTAP and KIAA1429 are required for mRNA methylation
-
-
Methylation maps, upon depletion of WTAP, reveal two classes of methylation
-
-
WTAP-dependent sites are mostly static and correlate with mRNA stability
-
-
Thousands of WTAP-independent sites at first transcribed nucleotide
Acknowledgments
This work was supported by NHGRI CEGS P50 HG006193 (A.R.), U54 HG003067 (E.S.L) and Broad Institute Funds. AR was supported by an NHGRI Pioneer Award and HHMI. SS was supported by a European Molecular Biology Organization fellowship, and S.S. and D.C. were supported by Human Frontier Science Program fellowships. M.J. was supported by fellowships of the Swiss National Science Foundation for advanced researchers (SNF) and the Marie Sklodowska-Curie IOF.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwala S, Blitzblau H, Hochwagen A, Fink G. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS genetics. 2012:8. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bembom O. seqLogo: Sequence logos for DNA sequence alignments. 2011 [Google Scholar]
- Bokar J, Rath-Shambaugh M, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. The Journal of biological chemistry. 1994;269:17697–17704. [PubMed] [Google Scholar]
- Bokar J, Shambaugh M, Polayes D, Matera A, Rottman F. RNA. Vol. 3. New York, NY: 1997. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase; pp. 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Bokar JA. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. In: Grosjean Henri., editor. Fine-Tuning of RNA Functions by Modification and Editing. Vol. 12. 2005. pp. 141–177. [Google Scholar]
- Bujnicki J, Feder M, Radlinska M, Blumenthal R. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. Journal of molecular evolution. 2002;55:431–444. doi: 10.1007/s00239-002-2339-8. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature genetics. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Clancy M, Shambaugh M, Timpte C, Bokar J. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic acids research. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Barbera M, McDonnell A, McIntyre K, Sternglanz R, Jin Q, Loidl J, Engebrecht J. The Saccharomyces cerevisiae MUM2 gene interacts with the DNA replication machinery and is required for meiotic levels of double strand breaks. Genetics. 2001;157:1179–1189. doi: 10.1093/genetics/157.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A–seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Duan J, Shi J, Ge X, Dolken L, Moy W, He D, Shi S, Sanders AR, Ross J, Gejman PV. Genome-wide survey of interindividual differences of RNA stability in human lymphoblastoid cell lines. Scientific reports. 2013;3:1318. doi: 10.1038/srep01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends in genetics : TIG. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Farrell CM, O’Leary NA, Harte RA, Loveland JE, Wilming LG, Wallin C, Diekhans M, Barrell D, Searle SM, Aken B, et al. Current status and new features of the Consensus Coding Sequence database. Nucleic acids research. 2014;42:D865–D872. doi: 10.1093/nar/gkt1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, Morgan M, Shatkin AJ, Jelinek W, Salditt-Georgieff M, Darnell JE. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci U S A. 1975;72:1904–1908. doi: 10.1073/pnas.72.5.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- Granadino B, Campuzano S, Sanchez L. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. The EMBO journal. 1990;9:2597–2602. doi: 10.1002/j.1460-2075.1990.tb07441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proc Natl Acad Sci U S A. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T, Hamakubo T. Identification of Wilms’ Tumor 1-associating Protein Complex and Its Role in Alternative Splicing and the Cell Cycle. J Biol Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith JM, Ensinger MJ, Mose B. HeLa cell RNA (2’-O-methyladenosine-N6-)-methyltransferase specific for the capped 5’-end of messenger RNA. J Biol Chem. 1978;253:5033–5039. [PubMed] [Google Scholar]
- Little NA, Hastie ND, Davies RC. Identification of WTAP, a novel Wilms’ tumour 1-associating protein. Human molecular genetics. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N-adenosine methylation. Nat Chem Biol. 2013a doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. RNA. Vol. 19. New York, NY: 2013b. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA; pp. 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic acids research. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Udeshi ND, Clauser KR, Mani DR, Patel J, Ong SE, Jaffe JD, Carr SA. iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Molecular & cellular proteomics : MCP 11, M111. 2012:014423. doi: 10.1074/mcp.M111.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Saletore Y, Zumbo P, Elemento O, Mason C, Jaffrey S. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3’ UTRs and near Stop Codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods. 2010;7:521–527. doi: 10.1038/nmeth.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A, Niksic M, Bachi A, Wilm M, Sanchez L, Hastie N, Valcarcel J. Biochemical function of female-lethal (2)D/Wilms’ tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem. 2003;278:3040–3047. doi: 10.1074/jbc.M210737200. [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell research. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SW, Herzyk P, Dow JA, Leader DP. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic acids research. 2013;41:D744–D750. doi: 10.1093/nar/gks1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis. Cell. 2013 doi: 10.1016/j.cell.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2013 doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nature cell biology. 2014 doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR. gplots: Various R programming tools for plotting data. 2012 [Google Scholar]
- Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4:379–386. doi: 10.1016/0092-8674(75)90158-0. [DOI] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.