Abstract
Background
The lung microbiome’s contribution to IPF pathogenesisis unknown. Using COMET-IPF (Correlating Outcomes with biochemical Markers to Estimate Time-progression in Idiopathic Pulmonary Fibrosis), the goal of this study was to determine whether unique microbial signatures would associate with disease progression.
Methods
IPF subjects within four years of diagnosis aged 35–80 were eligible for inclusion. Subjects were followed for up to a maximum of 80 weeks. This completed observational study is registered with ClinicalTrials.gov, number NCT01071707. Progression-free survival was defined as death, acute exacerbation, lung transplant, or decline in FVC of 10% or DLCO of 15%.DNA was isolated from 55 bronchoscopic alveolar lavage (BAL) samples. 454 pyrosequencing was used to assign operational taxonomic units (OTUs) based on a 3% sequence divergence. Adjusted Cox models identified OTUs significantly associated with progression-free survival at a p<0·10 level. These OTUs were then used in principal components (PC) analysis. The association between PCs and microbes with high factor loadings from the PC analysis and progression-free survival were examined via Cox regression analyses.
Findings
Mean FVC was 70·1% and mean DLCO 42·3 %predicted. Significant associations with disease progression were noted with increased % relative abundance of two OTUs identified by PC analysis, a Streptococcus OTU. (p<0·0009) and a Staphylococcus OTU(p=0·01). Strength of associations using PCs versus two OTUs alone was similar. Threshold analysis helped define a cut point for % relative abundance for each OTU associated with progression-free survival, >3·9% for the Streptococcus OTU, HR 10·19 (95% CI 2·94, 35·35; p=0·0002) and >1·8% for the Staphylococcus OTU, HR 5·06 (1·71, 14·93; p=0·003).
Interpretation
These preliminary data suggest IPF disease progression is associated with presence of specific members within the Staphylococcus and Streptococcus genera.
Introduction
Established risk factors for idiopathic pulmonary fibrosis (IPF) include male gender and advanced age. Recently genetic factors including MUC5B, TOLLIP1, TERT2 and TLR33 have been implicated in the pathogenesis of IPF.4 Other prognostic biomarkers in IPF have also been identified including specific gene expression profiles of peripheral blood mononuclear cells,5 down-regulation of CD28 on circulating CD4 T-cells6 and peripheral blood protein signatures including MMP7.7 While infectious agents have not been implicated in disease pathogenesis outside of acute exacerbations, accumulating data point to a role for bacteria in the pathobiology of IPF.8 A placebo-controlled clinical trial of co-trimoxazole in IPF recently reported a reduction in mortality for patients randomized to the antibiotic, but an explanation for this effect was not clear.9 The lack of data regarding an association between IPF and bacteria may relate to the difficulty in determining the true microbial composition of the lung as over 70% of the bacterial species inhabiting bodily surfaces cannot be cultured by currently available techniques.10 We hypothesized that a bacterial signature identified via 16 S sequences would associate with rapid disease progression. In order to answer this question, we used samples that had previously been collected as part of the COMET-IPF study, a prospective, observational study to identify biomarkers of disease progression in IPF11.
Methods
Patient Selection
Subjects included in this analysis were participants in COMET-IPF (Correlating Outcomes with biochemical Markers to Estimate Time-progression in Idiopathic Pulmonary Fibrosis), a prospective, observational study correlating biomarkers with disease progression(NCT01071707)11. This multicenter investigation recruited subjects at nine clinical centers in the US. Inclusion criteria required diagnosis of IPF and age 35-80 years. Subjects were excluded if the diagnosis of IPF was >4 years prior to screening or if there was a diagnosis of collagen-vascular disorder, FEV1/FVC <0·60, evidence of active infection at screening, or comorbid conditions other than IPF likely to result in death within one year. Subjects were followed for up to a maximum of 80 weeks. No limitation was placed on therapeutic approaches during the observational period. Informed consent was obtained from all participants. The study protocol was reviewed by the institutional review board of each participating center. The first subject was enrolled March 12, 2010 and the last subject was enrolled March 10, 2011. AssL While COMET-IPF was a prospective study, the decision to analyze samples for the purposes of microbiome analysis was not prespecified and performed after the study had been completed.
Diagnosis of IPF and Endpoints
IPF diagnosis was confirmed using a multidisciplinary diagnostic approach perinternational guidelines12 using expertise from clinicians, radiologists, and pathologists at the local, enrolling clinical center13,14. Our combined endpoint was progression-free survival defined by time until death, acute exacerbation of IPF15, lung transplant, or relative drop in FVC of 10% or DLCO of 15%.
Sample acquisition and processing
Per the COMET protocol, bronchoscopy was performed at the enrollment visit only in patients with clinical stability without evidence of active infection. Patients received conscious sedation and nebulized lidocaine. The bronchoscope was advanced via the mouth or nose and through the vocal cords. The bronchoscope was wedged into a right middle lobe or lingular segmental airway with the location determined by the physician performing the procedure based on the extent of imaging abnormality on HRCT. BAL was performed with 4 instillations of 50 ml of sterile isotonic saline aliquots for a total of 200 ml with all possible return collected and pooled. Samples for microbiome analysis were transported on ice and 5 ml subsequently centrifuged in 2 ml dolphin-nosed Eppendorf tubes at 13,000 rpm in a microcentrifuge at 4°C for 30 min. Supernatants were discarded, the pellets snap frozen in liquid nitrogen and stored at −80°C until the time of DNA extraction. Microbial cultures were not performed on these samples.
Data from two lung explants are also presented in this manuscript. These two lungs were separate from the COMET protocol and were obtained immediately following surgical removal at the time of clinically indicated lung transplant. These specimens were sterilely collected in the hospital and transported to the lab for dissection in a laminar flow hood, using appropriate biosafety precautions. After harvest, a protected, sterile brush was inserted into the main stem and advanced distally where an airway brushing was performed. A second, protected sterile brush was advanced more proximally where the procedure was repeated. Brushes were then placed in sterile 5 ml cryogenic vials and the excess wire trimmed
DNA isolation
Genomic DNA was extracted from BAL pellets using a previously published modification of the QiagenDNeasy protocol16. Brushes were treated similarly with the only difference being the addition of the frozen brush to the UltraClean fecal DNA bead tubes (MO-BIO, Carlsbad, CA) followed by the addition of 360μl ATL buffer (QiagenDNeasy Blood & Tissue kit). From homogenization on, samples were treated identically.
DNA Genotyping
Genotyping of DNA extracted from peripheral blood samples was conducted using the iPLEX Gold™ platform following manufacturer’s protocols (Sequenom, San Diego, CA).
454 Pyrosequencing
The V3-V5 hyper variable regions of the bacterial 16S rRNA gene were sequenced in the reverse direction using barcoded primer sets corresponding to 357F and 929R. Primary PCR cycling conditions were 95°C for two minutes, followed by 20 cycles of touchdown PCR (95°C 20 sec, followed by an annealing for 30 sec beginning at 60°C and decreasing one degree every two cycles until 50°C, and an elongation of 72°C 45 sec.), then 20 cycles of standard PCR (95°C for 20 sec, 50°C for 30 sec., and 72°C for 45 sec.), and finished with 72°C for 5 min.Quality control and sequencing was carried out at the University of Michigan Pyrosequencing Core, using the Roche 454 GS Junior according to established protocols17.
Data analysis
Sequence data were processed and analyzed using the software mothur v.1.27.0 according to the Schloss 454 Standard Operating Procedure (http://www.mothur.org,4) through the generation of the shared community file where the operational taxonomic units (OTUs) were binned at 97% identity and the phylotyped(genus-level grouping) file. OTUs detected in non-template controls were subtracted from all specimens prior to analysis.Count data was normalized to the percent of total reads for each individual. These files, along with the files containing the taxonomic information for the OTUs, were imported and further analyzed using the R-package vegan 2.0–4 for diversity analyses and ordinations, and a custom R script for sorting classification results into tables (ClassifierSorter v.2). Classification of OTUs was carried out using the mothur implementation of the Ribosomal Database Project (RDP) Classifier and the RDP taxonomy training set (http://rdp.cme.msu.edu).We restricted the principal components and regression analyses to OTUs that were present at greater than 0·5% of a given sample’s population and taxonomically identifiable.
Statistical Analysis
All analyses were performed in R version 2.15.2. Data were first normalized using vegan package for R decostand function. Using an approach for conducting a supervised principal components (PC) analysis similar to that recommended by Hastie, Tibshirani and Friedman18, we first identified OTU’s that met p<0·10 significance in a Cox proportional hazards analysis associated with the combined endpoint adjusted for baseline FVC%, DLCO%, desaturation <88% during six minute walk test (6MWT), age, gender and smoking status. These 43 OTU’s were then studied using a principal components analysis, giving us PC1 and PC2 risk factors based on linear combinations of weighted OTU’s that showed signal with regard to our combined endpoint. Upon viewing weights for PC1 and PC2, two OTUs classified as Streptococcussp. OTU 1345 and Staphylococcus sp. OTU 1348 had weights far exceeding other OTU’s, suggesting these two OTUs are the most associated with the combined endpoint. We then fit the following Cox proportional hazards models additionally adjusted for GERD and Shannon Diversity Index (SDI) based on this dimensionally reduced data: (1) model with PC1 and PC2 ; (2) model with Streptococcus sp. OTU 1345 and Staphylococcus sp. OTU 1348 treated as continuous variables based on OTU relative abundance; and (3) model with Streptococcus sp. and Staphylococcus sp. dichotomized using a threshold identified using the function cox.main from the R AIM package (http://cran.r-project.org/web/packages/AIM) which simultaneously optimized the best OTU relative abundance thresholds for our combined clinical endpoint19. Cox models presented in the primary paper have the property of assuming proportional hazards in providing overall hazard ratios. As an average hazard ratio may be under- or overestimated by this method, a weighted Cox regression using the Rcoxphw package (http://cran.r-project.org/package=coxphw) is also presented the Supplemental Appendix. These models also assume independent censoring conditional on the covariates in the model, although with only 5 individuals subject to non-administrative censoring, this was of little concern.
Role of the Funding Source
The study sponsor had no role in the analysis or interpretation of the data. Nor was the study sponsor involved in the writing of the manuscript or decision to submit the paper for publication. The corresponding author had full access to the data and final responsibility for the decision to submit for publication.
Results
Baseline demographics are outlined in Table 1. Of 71 available COMET subjects, 55 subjects had complete data available for analysis. Supplemental Table 1 summarizes missing data that resulted in non-inclusion of 16 subjects in this analysis. A comparison of demographics for those subjects not included to those included is presented in Supplemental Table 2 which were not significantly different, although the SDI and Inverse Simpson were both slightly lower in the excluded group. Mean age for the analysis group was 64·3 years. A greater percentage of subjects were male consistent with the gender distribution of this disease20. Subjects on average had a 47·6 pack-year smoking history, FVC% of 70·1 % predicted and DLCO of 42·3 % predicted. Over half of subjects (56·4%) reported a history of reflux. Median length of time from diagnosis to enrollment was 119 days. With respect to IPF therapies, three individuals were on prednisone at the time of enrollment and two individuals were on a zathioprine. No individuals were on antibiotics at the time of enrollment. Mean follow-up was 355·8 days. In terms of general microbial descriptors, the Shannon Diversity Index for the group was 2·41 and Inverse Simpson Index 8·52. Supplemental Table 3 outlines the top 50 OTU’s in order of mean % relative abundance among subjects where the OTU was detected. OTUs classified as Prevotella sp., Veillonellasp. and Escherichia sp.21 were the OTUs with greatest % relative abundance and occurred in 37, 41 and 35 subjects respectively.
Table 1.
Patient population baseline characteristics.
| Demographics | N=55 |
|---|---|
| Age (years) | 64·3 (7·5) |
| Male gender | 37/55 (67·2%) |
| Past Smoker | 35/55 (63·6%) |
| Current Smoker | 2/55 (3·64%) |
| Pack-years | 47·6 (52·3) |
| FEV1% predicted | 73·2 (18·3) |
| FVC % predicted | 70·1 (17·0) |
| DLCO % predicted | 42·3 (14·0) |
| Desaturation <88% during 6MWT | 29/55 (52·7%) |
| History of Gastroesophageal Reflux Disease | 31/55 (56·4%) |
| Mean follow-up time (days) | 355·8 |
| Microbiome Indices | |
| Shannon Diversity Index | 2·41 (0·46) |
| Inverse Simpson Index | 8·52(3·86) |
In order to identify OTUs associated with disease progression, we first conducted Cox regression analyses adjusted for factors known to influence survival: age, gender, smoking status and desaturation during six minute walk testing. Forty-three OTUs met p<0·10 significance in a Cox proportional hazards analysis associated with the combined endpoint. These OTUs were then used to construct principal components. PC 1 was most strongly influenced by the presence of a single OTU classified as Streptococcus(OTU 1345, PC weight 62·13), distantly followed by Prevotella sp. (OTU 1196, PC weight 5·08). PC 2 was most strongly influenced by the presence of a Staphylococcus OTU (OTU 1348, PC weight 32·78) distantly followed by one classified as Sphingomona. (OTU 1058, PC weight 3·88).
In Table 2A, Principal Components 1 and 2 were associated with disease progression with HR 1·80 (95% CI 1·21, 2·68; p=0·003) and 1·72 (95% CI 1·16, 2·56, p=0·007), respectively. In this model, desaturation<88% on 6MWT (HR 4·18, 95% CI 1·40, 12·42; p=0·01) and gastroesophageal reflux (HR 3·05, 95% CI 1·19, 7·83; p=0·02) were also significant. As the Streptococcus-and Staphylococcus-classified OTUs were the strongest drivers of PC1 and PC2 respectively, we examined the relationship of these organisms alone as compared to the principal components to the combined endpoint. As can be seen in Table 2B, both OTUs were significantly associated with the outcome, with hazard ratios that were even greater than for the principal components, HR 1.11 (95% CI 1·04, 1·18, p=0·0009) for the Streptococcus-classified OTU 1345 and 1·16 (95% CI 1·03, 1·31; p=0·01) for the Staphylococcus-classified OTU 1348. The concordance index for the two models is nearly identical, 0.71 using the principal components and 0·72 using the organisms themselves, indicating equally good model fit. As another Streptococcus-classified OTU (1350) also had high % relative abundance, we also tested this OTU in the survival model along with OTU’s 1345 and 1348 as a continuous variable (data not shown), and this was not statistically significant (HR 0·98; 95% CI 0·87,1·11; p=0·77).
Table 2.
Adjusted Cox regression models to determine the association of pulmonary IPF microbiome to disease progression defined as death, acute exacerbation, lung transplant, or decline in FVC of 10% or DLCO of 15%. In model 2A, microbiome is represented in the model by Principal Components (PC) 1 and 2*. In model 2B, the strongest drivers of PC1 and PC2, Streptococcus OTU 1345 and Staphylococcus 1348 represent the microbiome component of the model. In model 2C, as opposed to using the relative abundance of Streptococcus and Staphylococcus as continuous covariates as was done in 2B, we determined thresholds for the relative abundance of Streptococcus OTU 1345 and Staphylococcus 1348 that simulataneously optimized the model. This approach best optimizes the model as is indicated by the index of concordance.
| Table 2A. | |||
|---|---|---|---|
| Variable | Relative Risk | 95% Confidence Interval | P-value |
| Age (per 10 years) | 0·83 | 0·.41,1·67 | 0·60 |
| Male | 0·70 | 0·30,1·65 | 0·42 |
| Ever Smoker | 2·20 | 0·89,5·40 | 0·09 |
| FVC (per 10%) | 1·26 | 0·92,1·73 | 0·14 |
| DLCO (per 10%) | 0·87 | 0·52,1·46 | 0·59 |
| Desaturation <88% | 4·18 | 1·4,12·42 | 0·01* |
| Gastroesophageal reflux | 3·05 | 1·19,7·83 | 0·02* |
| Principal Component 1 | 1·80 | 1·21,2·68 | 0·003* |
| Principal Component 2 | 1·72 | 1·16,2·56 | 0·007* |
| Shannon Diversity Index | 0·67 | 0·44,1·01 | 0·06 |
| Concordance Index | 0·71 | ||
| Table 2B. | |||
|---|---|---|---|
| Variable | Relative Risk | 95% Confidence Interval | P-value |
| Age (per 10 years) | 0·82 | 0·41,1·64 | 0·57 |
| Male | 0·72 | 0·31,1·69 | 0·45 |
| Ever Smoker | 2·17 | 0·89,5·32 | 0·09 |
| FVC (per 10%) | 1·27 | 0·93,1·74 | 0·13 |
| DLCO (per 10%) | 0·87 | 0·52,1·47 | 0·61 |
| Desaturation <88% | 4·33 | 1·44,12·95 | 0·009* |
| Gastroesophageal reflux | 3·05 | 1·19,7·82 | 0·02* |
| Streptococcus OTU 1345 % relative abundance | 1·11 | 1·04,1·18 | 0·0009* |
| Staphylococcus OTU 1348 % relative abundance | 1·16 | 1·03, 1·31 | 0·01* |
| Shannon Diversity Index | 0·68 | 0·45,1·02 | 0·06 |
| Concordance Index | 0·72 | ||
| Table 2C. | |||
|---|---|---|---|
| Variable | Relative Risk | 95% Confidence Interval | P-value |
| Age (per 10 years) | 0·71 | 0·35, 1·43 | 0·33 |
| Male | 0·87 | 0·36, 2·12 | 0·77 |
| Ever Smoker | 1·54 | 0.67, 3·52 | 0·31 |
| FVC (per 10%) | 1·30 | 0·94, 1·79 | 0·11 |
| DLCO (per 10%) | 0·99 | 0·60, 1·64 | 0·99 |
| Desaturation <88% | 7·13 | 2·25, 22·5 | 0·0008* |
| Gastroesophageal reflux | 3·41 | 1·28, 9·08 | 0·01* |
| Streptococcus OTU 1345 >threshold (3.9% relative abundance) | 10·19 | 2·94, 35·35 | 0·0002* |
| Staphylococcus OTU 1348 >threshold (1.8% relative abundance) | 5·06 | 1·71,14·93 | 0·003* |
| Shannon Diversity Index | 0·53 | 0·33, 0·84 | 0·007* |
| Concordance Index | 0·77 | ||
Prior to building these models, in order to identify OTUs associated with disease progression, we first conducted Cox regression analyses adjusted for factors known to influence survival: age, gender, smoking status and desaturation during six minute walk testing. Forty-three OTUs met p<0·10 significance in a Cox proportional hazards analysis associated with the combined endpoint. These OTUs were then used to construct principal components.
Finally we wanted to determine if a threshold for % relative abundance for either Streptococcus OTU 1345 or Staphylococcus OTU 1348 was associated with progression. In Table 2C, a threshold of greater than 3·9% relative abundance for Streptococcus OTU 1345 (HR 10·19, 95% CI 2·94, 35·35; p=0·0002) and 1·8% relative abundance for Staphylococcus OTU 1348 (HR 5·06, 95% CI 1·71, 14·93, p=0·003) optimized the model. These thresholds were chosen based on a function that simultaneously optimized the best relative abundance thresholds of our OTUs of interest for our combined clinical endpoint (see statistical methods). In fact, the concordance index for this model is 0·77 indicating even better fit than the models based on principal components or linear values for the quantities of the Streptococcus-and Staphylococcus-classified OTUs. If the microbiome data are removed from the Cox model, the index of concordance drops to 0·699. We also included immunosuppression at the time of enrollment as a covariate in the Cox model and it was non-significant (p=0.97) and did not meaningfully change the effect size or significance of the microbiome variables. Similarly, we also investigated any influence of clinical center of recruitment which yielded results consistent with those reported in Table 2 (see Supplemental Table 4).Clinical centers also differed in their bronchoscopic approach (nasal vs. oral). A Chi-square test found no evidence of an association between approach and presence of Streptococcus OTU 1345 or Staphylococcus OTU 1348 greater than our specified threshold (p=0·66 and p=0·19, respectively). The study also collected information on presence of emphysema on the CT as this can be present in IPF patients and could confound results. It was reported as present in 8. We included this as a covariate in the Cox model. It was not statistically significant (p=0.81) and the hazard ratios and significance for other covariates were not meaningfully changed. Finally, the Cox model used assumes proportional hazard rates. As using the average hazard ratio may be under- or overestimated using this methodology, we also performed a weighted Cox regression analysis which did not appreciably impact the results (see Supplemental Table 5).
Adjusted survival curves are displayed in Figure 1. Figure 1A demonstrates difference in survival for individuals above and below the 3·9% relative abundance threshold for Streptococcus OTU 1345; figure 1B demonstrates survival difference of individuals above and below the 1·8% relative abundance threshold for Staphylococcus OTU 1348. Both models are adjusted for the average profile patient. A total of 36 individuals met the combined endpoint. The number at risk at 100 day intervals is also evident in the figure. A breakdown of the event types experienced by each individual is in Supplemental Table 6.
Figure 1.
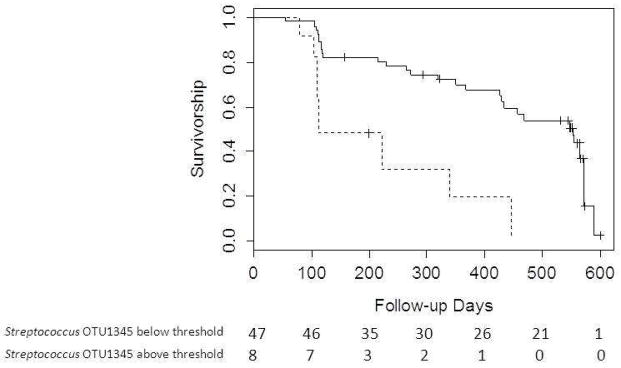
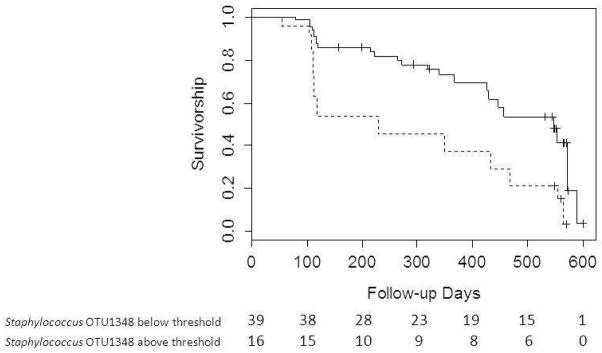
Figure 1A. Survival curves generated from Cox regression model stratified by threshold of 3·9% relative abundance for Streptococcus OTU 1345 with average patient profile values substituted for other covariates. Solid line indicates relative abundance below 3·9%; Dashed line indicates relative abundance above 3·9%. Number at risk at 100 day intervals displayed below the figure.
Comparison of survival between patients with Streptococcus OTU 1345 below 3·9% relative abundance and patients with Streptococcus OTU 1345 above 3·9% relative abundance, adjusted for the following mean values: age-64·3 years; probability of being male-67·2%; probability of smoking (current and past)-67·3%; FVC%-70·1; DLCO%-42·3; probability of desaturation<88%-52·7%; probability of history of reflux-56·4%; and probability of having Staphylococcus OTU 1348 above threshold relative abundance-29%
Figure 1B. Survival curves generated from Cox regression model stratified by threshold of 1·8% for Staphylococcus OTU 1348 with average patient profile values substituted for other covariates. Solid line indicates relative abundance below 1·8%; Dashed line indicates relative abundance above 1·8%. Number at risk at 100 day intervals displayed below the figure.
Comparison of survival between patients with Staphylococcus OTU 1345 below 1·8% relative abundance and patients with Staphylococcus OTU 1345 above 1·8% relative abundance, adjusted for the following mean values: age-64·3 years; probability of being male-67·2%; probability of smoking (current and past)-67·3%; FVC%-70·1; DLCO%-42·3; probability of desaturation<88%-52·7%; probability of history of reflux-56·4%; and probability of having Streptococcus OTU 1348 above threshold relative abundance-14·5%
A distribution of subjects based on the bacterial thresholds is displayed in Table 3. Here we can see that either Streptococcus OTU 1345 or Staphylococcus OTU 1348 was identified in 23/55 subjects (41·2%). More patients had a relative abundance of Staphylococcus OTU 1348 above the threshold than Streptococcus OTU 1345 above threshold. Only one subject had both organisms present above the identified threshold. Supplemental Table 7 shows the differences in demographics between subjects with and without the Streptococcus OTU 1345 and Staphylococcus OTU 1348 above and below threshold. No significant difference in any of the clinical metrics was identified. Individuals with Streptococcus did demonstrate greater microbial diversity with higher SDI scores, 2·62 vs. 2·33, p=0·03.
Table 3.
Frequency distribution of individuals where Streptococcus and Staphylococcus OTU s were identified above threshold (3.9% for Streptococcus and 1.8% for Staphylococcus).
| Staphylococcus OTU 1348 − | Staphylococcus OTU 1348+ | Total | |
|---|---|---|---|
| Streptococcus OTU 1345- | 32 | 15 | 47 |
| Streptococcus OTU 1345 + | 7 | 1 | 8 |
| Total | 39 | 16 | 55 |
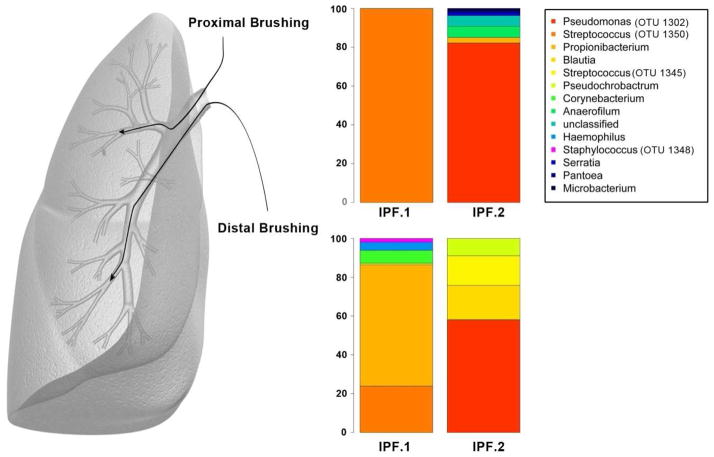
As bronchoscopies were performed through the upper airway, we sought additional confirmation that Streptococcus sp.OTU 1345 and Staphylococcus sp. OTU 1348 could be identified in the IPF lungin the absence of bronchoscopy. Data from two IPF lung explants are presented in Figure 2 with accompanying relative abundance data presented in Supplemental Table 8. Protected airway brushings were performed after the lungs were removed. Both Streptococcus sp. and Staphylococcus sp. were identified, with Lung 1 containing both Streptococcus OTU 1350 and OTU1345 as well as Staphylococcus OTU 1348. Lung 2 had one Streptococcus OTU (1345). Thus, Streptococcus OTU 1345 was present in both explants. Staphylococcus OTU 1348 was present in one of the explants.
Figure 2.
% Relative abundance of OTU’s from two IPF lung explants taken from proximal and distal airway brushings after the lung was removed. IPF Lung 1 demonstrated 100% relative abundance of Streptococcus OTU 1350 in the proximal brushing. In the distal brushing, two Streptococcus OTU’s were identified, OTU 1350 at 23.89% and OTU 1345 at 0.81% relative abundance. Staphylococcus OTU 1348 was also identified in the distal brush at 1.86% relative abundance. In IPF Lung 2, neither Streptococcus nor Staphylococcus was identified in the proximal brush, but the distal brush demonstrated 15.29% relative abundance for Streptococcus OTU 1345.
We previously used the COMET cohort as part of a larger meta-analysis to help identify genetic variants associated with IPF susceptibility. The 11p15.5/TOLLIP/rs5743890_G allele was identified to be associated with decreased susceptibility to IPF but when examined in additional analyses in other cohorts, was also demonstrated to be associated with increased mortality1, which was the converse relationship of 11p15.5/MUC5B/rs3570590_T. However, only the TOLLIP_G attained significance in the COMET cohort while MUC5B_T did not. Here using a Fisher’s exact test, we did not find an association between this TOLLIP allele with either Streptococcus OTU 1345 (p=0·66) or Staphylococcus OTU 1348 (p=0·75) above our defined thresholds. In our Cox regression model of disease progression that includes mortality as an endpoint, the hazard ratios for exceeding Streptococcus and Staphylococcus thresholds maintained statistical significance with similar magnitude of effect size when adjusted for this allele HR 9.32 (95% CI 2·53, 44·4; p<0·001) and HR 7·35 (95% CI 2·49, 21·7;p<0·001), respectively. In this model, the TOLLIP G allele maintained independent statistical significance, HR 4·70 (95% CI 1·79, 12·32;p=0·002) suggesting that the genetic factors are not required for the microbial associations with disease progression in IPF.
Discussion
In this report we demonstrate an association between IPF disease progression and the lung pulmonary microbial community assessed via BAL. Most importantly, we demonstrate that the presence of specific Streptococcus and Staphylococcus OTUs appear to be associated with poorer outcomes. To our knowledge, this is the first report of an association between the presence of specific microbes and disease progression in IPF. These preliminary data generate hypotheses regarding the potential role of the microbiome in the pathogenesis in IPF that could support further investigation of the role of antibiotics as a potential therapeutic approach in this recalcitrant disease.
The etiology of IPF is not well understood but thought to result from repeated alveolar injury and abnormal repair22. However, the exact triggers for injury are not known. While immunosuppressive agents that might target inflammation have been investigated in IPF, one of the largest and most rigorously conducted studies (PANTHER-IPF) comparing prednisone, azathioprine and N-acetylcysteine versus placebo in IPF demonstrated significantly greater mortality in those receiving active treatment.23 It has also been hypothesized that infectious agents could play a role8. Studies examining a potential pathogenic contribution of viruses including Hepatitis C, Human Herpes Virus and Transfusion Transmitted Virus demonstrate conflicting results8. Viruses in acute exacerbations of IPF have also been investigated. While animal models suggest that viral infection can exacerbate established fibrosis24, human studies utilizing PCR analysis of BAL fluid have failed to reliably identify viral DNA in the majority of acute exacerbation subjects studied25,26. Culture-independent techniques have identified bacterial communities within both the healthy lung and in association with other respiratory disorders including asthma and COPD27–29, but much less data is available in IPF (panel).
If, as our data suggests, there is an association between bacteria and disease progression, a potential link could be the TLR9 receptor which is present in higher concentrations in surgical lung biopsies from rapidly progressive patients33. Staphylococcus exerts some of its effects such as type I IFN signaling via the TLR9 pathway34. Interestingly, TOLLIP is involved in TLR signaling. The independence of this marker may be associated with other infectious agents too small to have been studied in this cohort and its independence from Staphylococcus and Streptococcus may speak to additive effects that would require additional investigation.
The Streptococcus and Staphylococcus OTU’s of interestin our study were not seen in all patients, and we did not see any statistically significant differences in the baseline clinical aspects of subjects with or without Streptococcus or Staphylococcus above our optimized threshold, although likely our power to make such comparisons was low. It is interesting, however, that subjects predominantly had one organism or the other and not both. We also demonstrated that only one Streptococcus OTU (1345), and not another Streptococcus-classified OTU (1350) was associated with poorer outcomes. The association of these organisms with disease progression despite similar baseline clinical characteristics provides additional support to the hypothesis that they at a minimum are biomarkers of disease progression and may be involved in disease pathogenesis. Alternate approaches to microbial identification, including culture and microbe-specific PCR, may further identify which specific species of the Streptococcus and Staphylococcus taxonomic groups are associated with disease progression.
We also noted that both GERD35 and the bacteria retained statistical significance in the Cox models. A high prevalence of GERD in IPF has previously been noted although whether GERD causes or worsens the disease and if so via what mechanism is not clear. Our data suggest however, that regardless of the relationship between GERD and the lung microbiome that both GERD and the Streptococcus and Staphylococcus bacteria have at least in part independent associations with disease progression in IPF.
There are clearly limitations to this analysis. The 454 pyrosequencing methodology used here has the advantage of being able to detect bacteria at low levels or that are difficult to grow in culture. Further study will be required to determine which specific species of the Streptococcus and Staphylococcus generaare associated with disease progression and to compare the clinical significance of microorganisms detected via culture independent methods versus traditional, culture-based methods. Our patient population is also small and recruited from tertiary medical centers and therefore may not be reflective of the general IPF patient population.
Bronchoscopic sampling was not performed throughout the lung and therefore the influence of regional variation in the IPF lung cannot be completely explored in this study. In addition, no oral decontamination mouthwash was performed, no attempt was made to reduce the possibility of oral contamination and no oral samples were collected. However, the additional data from our IPF explants suggest there is regional variation in the IPF lung and help to confirm the presence of Streptococcus OTU 1345 and Staphylococcus OTU 1348 in IPF lungs with severely progressed disease. There is clearly a relationship between the oral and lung microbiome. Data were recently published comparing the oral and lung microbiome in healthy individuals where it was shown that many of the bacteria identified in the lung also reside in the mouth.36 The differences in community structure seen are then likely to be the result of a selective pressure that the lung environment places on the bacterial community.
Regardless of whether the organisms originated in the mouth or the lung, the presence of certain microbesin BAL is clearly associated with disease progression suggesting that at a minimum, the presence of such organisms obtained in this fashion could be used as a biomarker of disease progression. These associations appear to be independent of the contribution of the previously documented 11p15.5/TOLLIP/rs5743890_G allele to increased mortality in IPF. More data in the form of a clinical intervention trial will be needed to determine whether these organisms, however, are causal in the pathway of disease progression or rather reflect the fibrotic milieu in the local pulmonary environment that are the true drivers of disease.
Finally, for purposes of this analysis, we have chosen to utilize a composite endpoint. It has been argued that composite endpoints can be problematic, particularly in the conduction of therapeutic clinical trials where discordant effects of a drug on various components of a composite endpoint can obscure efficacy information. Here much of the effect we report was driven by changes in lung function. Changes in FVC have been correlated with survival time in multiple large cohorts of patients with IPF which makes it a useful prognostic factor. However, change in lung function may be problematic as a surrogate for mortality.37 Our results suggest that the lung microbiome correlates with disease progression and is a biomarker for more severe disease. While certainly these results lead to hypotheses about potential treatments, these results should be viewed as preliminary. Data from another cohort where samples for microbiome analyses are collected prospectively would help to confirm our findings. If the specific species of Streptococcus and Staphylococcus can be identified in future analyses, such data would support a randomized, placebo controlled antibiotic trial in IPF stratified on the baseline presence of these particular Streptococcus sp. and/or Staphylococcus sp. Such data would be needed to support the routine use of antibiotics as treatment for IPF.
Supplementary Material
Research in Context Panel.
Systematic Review
A PubMed search utilizing the terms “bacteria” and “idiopathic pulmonary fibrosis” limited to humans revealed the following relevant literature. Richter, et al. used culture based techniques to evaluate BAL from 22 stable IPF patients and identified bacteria in eight subjects30. Pathogens in IPF identified included Haemophilus, Streptococcus, Moraxella and Pseudomonas. While no Staphylococcus sp. were identified in culture, the BAL fluid from IPF patients was noted to promote growth of Staphylococcus aureus as compared to BAL from control subjects. Another recent study performed 454 pyrosequencing on BAL fluid from 18 subjects with a variety of respiratory conditions including five with idiopathic interstitial pneumonia and noted that the bacterial families Staphylococcaceae and Streptococcaceae were among the top 20 bacterial families with respect to relative abundance.31
A small, pilot study of 20 patients with interstitial pneumonia revealed that treatment with co-trimoxazole improved forced vital capacity (FVC), dyspnea (MRC score) and shuttle walk distance over a three month period32. A follow-up larger, placebo-controlled clinical trial of co-trimoxazole in fibrotic interstitial pneumonia (IPF or fibrotic NSIP) was subsequently conducted demonstrating no impact on FVC, MRC score or 6MWT. However, a significant reduction in all-cause mortality was seen, HR 0·21 (95% CI 0·06, 0·78) as well as a reduction in the percentage of patients requiring an increase in oxygen therapy (OR l·05 (95% CI 0·00, 0·61).
Interpretation
Our analysis uses culture-independent techniques based on 454 pyrosequencing on BAL fluid to determine the relationship between bacterial signatures and disease progression in IPF defined as death, acute exacerbation, lung transplant, or decline in FVC of 10% or DLCO of 15%. We detected significant associations between increased % relative abundance of two OTUs, a Streptococcus OTU. (p<0·0009) and a Staphylococcus OTU (p=0·01) and disease progression. These preliminary data suggest IPF disease progression is associated with presence of specific members within the Staphylococcus and Streptococcus genera although additional research will be needed both to identify the specific species and to determine whether this is a causal relationship.
Acknowledgments
We acknowledge John Erb-Downward and Robert Dickson for their expertise and insights with respect to bioinformatics and the scientific discussion.
Funding: This work was funded by NIH/NHLBI RC2 HL101740
COMET investigators
Brown University, Providence, RI - Kevin Dushay (site PI), Jonathon Kurtis
Cleveland Clinic Foundation, Cleveland, OH – Jeffrey T Chapman (site PI)
Duke University – Kevin Anstrom (site PI)
National Jewish Health, Denver, CO - Kevin K Brown (site PI), Gregory Cosgrove, Joshua Solomon, Jeffrey Swigris, Evans Fernandez-Perez
Temple University, Philadelphia, PA – Gerard Criner (site PI), Francis Cordova, Namrata Patel, Thomas Rogers
University of California, Los Angeles, CA – John Belperio (site PI)
University of California, San Francisco, CA – Talmadge E King Jr. (site PI), Harold R. Collard
University of Chicago, Chicago, IL – Imre Noth (site PI), Cathy Brown, Joe GN Garcia, D Kyle Hogarth, Yong Huang, Yves Lussier, Shwu-Fan Ma, Michael Wade
University of Michigan, Ann Arbor, MI – Fernando J. Martinez, Kevin R Flaherty, Galen B Toews, Eric S. White (Principal Investigators), Cory Hogaboam, Vibha Lama, Bethany Moore, Thomas Moore, Susan Murray, Cathie Spino.
Vanderbilt University, Nashville, TN – James E Loyd (site PI)
Footnotes
Author Contributions:
MH, SM, IN, VL, BM, EW, KF, GH and FM contributed to conceptualization of the study. MH, IN, VL, KF and FJM were involved in data collection. MH, YZ, SM, NT, IN, KF, GH and FJM contributed to data analysis. MH, YZ, SM, NT, IN, VL, BM, EW, KF, GH and FM participated in manuscript writing and editing.
Conflicts of Interest:
MH reports grants from NHLBI during the conduct of the study; grants and personal fees from GSK, personal fees from Pfizer, personal fees from BoehringerIngelheim, personal fees from Forest, personal fees from Novartis, personal fees from Medimmune, from Ikaria, personal fees from Regeneron, personal fees from Grifols, personal fees from Uptodate, outside the submitted work.
YZ, SM, and NT report grants from NIH during the conduct of this study.
IN reports grants from NIH during the conduct of the study. In addition, Dr. Noth has a patent TOLLIP SNPs in IPF pending.
VL and BM report no conflict of interest.
EW reports grants from the NIH during the conduct of this study.
KF reports personal fees and non-financial support from BoehringerIngelheim, during the conduct of the study; personal fees from BoehringerIngelheim, personal fees from Fibrogen, personal fees from Genentech, personal fees from Gilead, personal fees from Ikaria, personal fees from ImmuneWorks, personal fees from MedImmune, personal fees from Novartis, personal fees from Takeda, personal fees from Vertex, personal fees from Veracyte, personal fees from Roche, personal fees from Pulmonary Fibrosis Foundation, grants from ImmuneWorks, grants and personal fees from Intermune, grants from Bristol-Myers Squibb, personal fees from Glaxo Smith Klein, personal fees from Forest, personal fees from Up To Date, personal fees from NACE, personal fees from Excel, personal fees from France Foundation outside the submitted work.
GH reports reports grants from The National Institutes of Health during the conduct of the study.
FM reports grants from NHBLI, during the conduct of the study; personal fees from Able, personal fees from American Institute for Research, personal fees from Axon, personal fees from Grey Healthcare, personal fees from Merion, personal fees from Sudler& Hennessey, personal fees from Actelion, from Centocor, from Gilead, personal fees from Amgen, personal fees from Astra Zenneca, personal fees from Forest, personal fees from GSK, personal fees from Ikaria, personal fees from Jannsens, personal fees from Merck, personal fees from Nycomed/Takeda, personal fees from Pearl, personal fees from Pfizer, personal fees from Forest, personal fees from GSK, personal fees from Nycomed/Takeda, personal fees from American College of Chest Physicians, personal fees from Center for Healthcare Education, personal fees from CME Incite, personal fees from Inova, personal fees from MedScape/Web MD, personal fees from National Association for Continuing Education, personal fees from NCME, personal fees from Peer Voice, personal fees from Projects in Knowledge, personal fees from St. John's Hospital, personal fees from St. Mary's Hospital, personal fees from University of Illinois, Chicago, personal fees from University of Texas, Southwestern, personal fees from University of Virginia, personal fees from UpToDate, personal fees from Wayne State University, personal fees from Carden Jennings, personal fees from Ikaria, personal fees from MedImmune, personal fees from Nycomed/Takeda, personal fees from Vertex, personal fees and other from BoehringerIngelheim, personal fees from Bayer, personal fees from Forest, personal fees from GSK, personal fees from Nycomed/Takeda, personal fees from Prescott, personal fees from Informa, from Stromedix, from Promedior, personal fees from CSA Medical, personal fees from Miller Medical Communications, personal fees from Veracyte, outside the submitted work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Noth I, Zhang Y, Swhu-Fan M, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. The Lancet Respiratory Medicine. 2013;1(4):309–17. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fingerlin TE, Murphy E, Zhang W, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45(11):1409. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Dwyer DN, Armstrong ME, Trujillo G, et al. The Toll-like Receptor 3 L412F Polymorphism and Disease Progression in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2013;188(12):1442–50. doi: 10.1164/rccm.201304-0760OC. [DOI] [PubMed] [Google Scholar]
- 4.Peljto AL, Zhang Y, Fingerlin TE, et al. Association between the MUC5B promoter polymorphism and survival in patients with idiopathic pulmonary fibrosis. JAMA. 2013;309(21):2232–9. doi: 10.1001/jama.2013.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herazo-Maya JD, Noth I, Duncan SR, et al. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Sci Transl Med. 2013;5(205):205 ra136. doi: 10.1126/scitranslmed.3005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilani SR, Vuga LJ, Lindell KO, et al. CD28 down-regulation on circulating CD4 T-cells is associated with poor prognoses of patients with idiopathic pulmonary fibrosis. PLoS One. 2010;5(1):e8959. doi: 10.1371/journal.pone.0008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosas IO, Richards TJ, Konishi K, et al. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med. 2008;5(4):e93. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molyneaux PL, Maher TM. The role of infection in the pathogenesis of idiopathic pulmonary fibrosis. Eur Respir Rev. 2013;22(129):376–81. doi: 10.1183/09059180.00000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shulgina L, Cahn AP, Chilvers ER, et al. Treating idiopathic pulmonary fibrosis with the addition of co-trimoxazole: a randomised controlled trial. Thorax. 2013;68(2):155–62. doi: 10.1136/thoraxjnl-2012-202403. [DOI] [PubMed] [Google Scholar]
- 10.Han MK, Huang YJ, Lipuma JJ, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67(5):456–63. doi: 10.1136/thoraxjnl-2011-201183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naik PK, Bozyk PD, Bentley JK, et al. Periostin promotes fibrosis and predicts progression in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;303(12):L1046–56. doi: 10.1152/ajplung.00139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghu G, Collard HR, Egan JJ, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183(6):788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaherty KR, Andrei AC, King TE, Jr, et al. Idiopathic Interstitial Pneumonia: Do Community and Academic Physicians Agree on Diagnosis? Am J Respir Crit Care Med. 2007 doi: 10.1164/rccm.200606-833OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flaherty KR, King TE, Jr, Raghu G, et al. Idiopathic interstitial pneumonia: what is the effect of a multidisciplinary approach to diagnosis? Am J Respir Crit Care Med. 2004;170(8):904–10. doi: 10.1164/rccm.200402-147OC. [DOI] [PubMed] [Google Scholar]
- 15.Collard HR, Moore BB, Flaherty KR, et al. Acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2007;176(7):636–43. doi: 10.1164/rccm.200703-463PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason KL, Erb Downward JR, Mason KD, et al. Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect Immun. 2012;80(10):3371–80. doi: 10.1128/IAI.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daigle D, Simen BB, Pochart P. High-throughput sequencing of PCR products tagged with universal primers using 454 life sciences systems. Curr Protoc Mol Biol. 2011;Chapter 7(Unit7):5. doi: 10.1002/0471142727.mb0705s96. [DOI] [PubMed] [Google Scholar]
- 18.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer - Verlag; 2009. [Google Scholar]
- 19.Tian L, Tibshirani R. Adaptive index models for marker-based risk stratification. Biostatistics. 2011;12(1):68–86. doi: 10.1093/biostatistics/kxq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han MK, Murray S, Fell CD, et al. Sex differences in physiological progression of idiopathic pulmonary fibrosis. Eur Respir J. 2008;31(6):1183–8. doi: 10.1183/09031936.00165207. [DOI] [PubMed] [Google Scholar]
- 21.Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7(6):e39315. doi: 10.1371/journal.pone.0039315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maher TM, Wells AU, Laurent GJ. Idiopathic pulmonary fibrosis: multiple causes and multiple mechanisms? Eur Respir J. 2007;30(5):835–9. doi: 10.1183/09031936.00069307. [DOI] [PubMed] [Google Scholar]
- 23.Idiopathic Pulmonary Fibrosis Clinical Research N. Raghu G, Anstrom KJ, King TE, Jr, Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366(21):1968–77. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMillan TR, Moore BB, Weinberg JB, et al. Exacerbation of established pulmonary fibrosis in a murine model by gammaherpesvirus. Am J Respir Crit Care Med. 2008;177(7):771–80. doi: 10.1164/rccm.200708-1184OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wootton SC, Kim DS, Kondoh Y, et al. Viral infection in acute exacerbation of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183(12):1698–702. doi: 10.1164/rccm.201010-1752OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konishi K, Gibson KF, Lindell KO, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;180(2):167–75. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the "healthy" smoker and in COPD. PLoS One. 2011;6(2):e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang YJ, Kim E, Cox MJ, et al. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS. 2010;14(1):9–59. doi: 10.1089/omi.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter AG, Stockley RA, Harper L, Thickett DR. Pulmonary infection in Wegener granulomatosis and idiopathic pulmonary fibrosis. Thorax. 2009;64(8):692–7. doi: 10.1136/thx.2008.110445. [DOI] [PubMed] [Google Scholar]
- 31.Garzoni C, Brugger SD, Qi W, et al. Microbial communities in the respiratory tract of patients with interstitial lung disease. Thorax. 2013;68(12):1150–6. doi: 10.1136/thoraxjnl-2012-202917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varney VA, Parnell HM, Salisbury DT, Ratnatheepan S, Tayar RB. A double blind randomised placebo controlled pilot study of oral co-trimoxazole in advanced fibrotic lung disease. Pulm Pharmacol Ther. 2008;21(1):178–87. doi: 10.1016/j.pupt.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Trujillo G, Meneghin A, Flaherty KR, et al. TLR9 differentiates rapidly from slowly progressing forms of idiopathic pulmonary fibrosis. Sci Transl Med. 2010;2(57):57ra82. doi: 10.1126/scitranslmed.3001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker D, Prince A. Staphylococcus aureus induces type I IFN signaling in dendritic cells via TLR9. J Immunol. 2012;189(8):4040–6. doi: 10.4049/jimmunol.1201055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raghu G, Freudenberger TD, Yang S, et al. High prevalence of abnormal acid gastro-oesophageal reflux in idiopathic pulmonary fibrosis. Eur Respir J. 2006;27(1):136–42. doi: 10.1183/09031936.06.00037005. [DOI] [PubMed] [Google Scholar]
- 36.Morris A, Beck JM, Schloss PD, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187(10):1067–75. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raghu G, Collard HR, Anstrom KJ, et al. Idiopathic pulmonary fibrosis: clinically meaningful primary endpoints in phase 3 clinical trials. Am J Respir Crit Care Med. 2012;185(10):1044–8. doi: 10.1164/rccm.201201-0006PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.