Abstract
Activation-induced cytidine deaminase (AID) is essential for class switch recombination (CSR) and somatic hypermutation (SHM) of Ig genes. The AID C terminus is required for CSR but not for S region DNA DSBs during CSR, and it is not required for SHM. AID lacking the C terminus (ΔAID) is a dominant negative (DN) mutant, as human patients heterozygous for this mutant fail to undergo CSR. In agreement, we show that ΔAID is a DN mutant when expressed in AID-sufficient mouse splenic B cells. In order to have DN function,ΔAID must have deaminase activity, suggesting that its ability to induce DSBs is important for the DN function. Supporting this hypothesis, Msh2-Msh6 have previously been shown to contribute to DSB formation in S regions, and here we find that Msh2 is required for the DN activity, as ΔAID is not a DN mutant in msh2−/− cells. Our results suggest that the DNA DSBs induced by ΔAID are unable to participate in CSR, and might interfere with the ability of full-length AID to participate in CSR. We propose thatΔAID is impaired in its ability to recruit non-homologous end joining (NHEJ) repair factors, resulting in accumulation of DSBs that undergo aberrant resection. Supporting this hypothesis, we find that the S-S junctions induced by ΔAID have longer microhomologies than those induced by full-length AID. In addition, our data suggest that AID binds Sµ regions in vivo as a monomer.
Introduction
AID initiates antibody gene diversification after immunization or infection by deamination of cytosines in Ig S regions, leading to DSBs and CSR, and also in recombined V(D)J gene segments, leading to SHM (1–4). It has been known for several years that the C terminus of AID is required for CSR, although it does not appear to have a role during SHM of antibody genes (5–7). Some Hyper-IgM (HIGM) human patients, who cannot undergo CSR, are heterozygous for mutant AID proteins lacking 8 or more amino acids at the C terminus of AID (ΔAID), indicating that ΔAID is a dominant negative (DN) mutant (5, 8). Although it is unknown why ΔAID is a DN mutant, it has been suggested that AID functions as a dimer or tetramer (5, 9–12), and perhaps the full-length AID and ΔAID heterodimerize and fail to perform a function, or fail to interact with a protein essential for CSR (5). However, dimerization of full-length AID and ΔAID would not explain the very low level of CSR in human HIGM patients, since both proteins are likely to be simultaneously induced, and therefore some dimers of full-length AID should be present.
Mouse and human AID have a deaminase domain between amino acids (aa) 56 and 94, a C terminal domain reported to be required for CSR but not for SHM between aa 182 and 198(2, 5–7), and a nuclear export signal also located at the C-terminus between aa 190 and 198 (13, 14). AID predominantly resides in the cytoplasm, and nuclear AID undergoes rapid ubiquitin-mediated proteasomal degradation, causing the half-life of nuclear AID to be 3 times shorter than that of cytoplasmic AID (15). However, nuclear export is not required for CSR, as mouse AIDF198A is not exported from nuclei, and yet CSR and SHM are only modestly reduced in cells expressing this mutant (14). Also, some C terminus substitution mutants that retain a functional nuclear export signal cannot potentiate CSR (16).
A relevant difference between CSR and SHM is that the latter does not require DSBs or recombination. During CSR, the dUs in S regions generated by AID are converted into DSBs in the donor (Sµ) and acceptor Sx regions (17). The dU bases are excised by uracil-N-glycosylase (UNG) and UNG deficiency causes a dramatic reduction in S region DSBs and CSR (18–20). Apurinic/apyrimidinic endonuclease 1 and 2 (APE1/2) can nick the abasic sites generated by UNG and are important for creating S region DSBs during CSR (21). When the single-strand breaks (SSBs) created by APE activity on opposite DNA strands within Sµ are sufficiently near, they can form a DSB, which can recombine with a DSB in a downstream S region. When the SSBs created by AID-UNG-APE activities are too far apart to spontaneously form a DSB, mismatch repair (MMR) proteins are thought to convert these distal SSBs into DSBs during CSR, and in MMR-deficient B cells S region DSBs are decreased by 50–80% (17, 22–24). The DSBs in two different S regions are recombined by non-homologous end joining (NHEJ), resulting in CSR.
Retrovirally transduced AID tagged at the C terminus with the estrogen receptor (ER) binds to S regions, as assayed by chromatin immunoprecipitation (ChIP) in aid−/− splenic B cells (25). In that study (25), we reported that ΔAID associated poorly with S region DNA. However, our current ChIP results, now obtained many times, show that ΔAIDassociates with Sµ as well as full-length AID. We cannot repeat the previous result and do not understand the basis of this discrepancy. We have published a correction (26). All other results reported previously are reproducible. The new finding that ΔAID binds Sµis in better agreement with the fact that ΔAID is competent for introducing mutations and DSBs at the Sµ and Sγ3 regions (7, 25, 27, 28).
ΔAID-ER introduces as many mutations in unrearranged (germline) Sµ in B cells induced to undergo CSR as does AID-ER (7); thus, ΔAID can produce the DNA substrate for UNG and Msh2-Msh6 (MutSα). However, these repair proteins bind poorly to Sµ in cells expressing ΔAID, indicating the importance of the C terminus for their binding (25, 29). Consistent with the dependence on the C terminus for the binding of UNG, there is an increase in the proportion of transition mutations at G:C bp in the Sµ region in cells expressing ΔAID, as expected if UNG does not readily access the mutations (7). CSR is reduced 2–3 fold in Msh2- or Msh6-deficient B cells relative to MutSα-sufficient cells (30–33), and this effect is reproduced in our retroviral system when full-length RV-AID is expressed in Msh2- or Msh6-deficient aid−/− B cells (25). However, when ΔAID is expressed, the small amount of CSR induced (~10% of full-length AID) is not further reduced in MutSα-deficient cells. This indicates that MMR does not contribute to CSR efficiency in cells expressing ΔAID, consistent with the poor binding of Msh2 and Msh6 to Sµ in cells expressing ΔAID (25).
It is not understood why the AID C terminus is required for CSR, nor is it understood how its deletion creates a DN mutation. Aid−/− B cells transduced with ΔAID-ER have S region DSBs (25, 27), indicating that the AID C terminus is important for events after DSB formation. However, it is possible that S region DSBs are aberrantly processed and/or inefficiently introduced in cells expressing ΔAID, and that they accumulate due to lack of repair and/or inefficient S-S recombination. Thus, the C terminus might be required for recruiting proteins involved in producing non-resected DSBs that are appropriate for NHEJ and/or for recruiting proteins involved in the end-joining process itself, as previously suggested (29, 34, 35). If AID functions as a dimer, it is possible that a heterodimer of AID and ΔAID cannot recruit these proteins. Numerous proteins have been demonstrated to associate with AID (36), and several require the AID C terminus for their association (14, 37, 38). These proteins have been shown to help export AID from the nucleus (14, 39), or help maintain it in the cytoplasm (38), or help recruit AID to S regions (37, 40).
In this report, we show that the DN function of ΔAID observed in humans is also found in mouse B cells, and we also show thatΔAID lacking deaminase activity does not have DN function and does not associate stably with Sµin aid+/+ cells. We also find that the DN phenotype of ΔAID depends upon the MMR protein Msh2. Our results suggest that the DN function depends upon the ability of ΔAID to bind Sµ and to induce DSBs in S regions, and also indicate that ΔAID-induced DSBs are not recombined by NHEJ. It is possible that the inability of these DSBs to recombine by NHEJ results in inefficient S-S recombination and generation of aberrantly resected DSBs that interfere with the ability of full-length AID to induce normal DSBs that can be recombined properly during CSR.
Materials and Methods
Mice
Mice were extensively (≥8 generations) backcrossed to C75BL/6. AID-deficient mice were obtained from T. Honjo (Kyoto University, Kyoto, Japan) (1). Msh2-deficient mice were obtained from T. Mak, Univ. of Toronto CA (41). Mlh1-deficient mice were obtained from R.M. Liskay, Oregon Health Sciences University, Portland OR (42). UNG-deficient mice were obtained from D. Barnes and T. Lindahl, London Research Institute, London UK (43). Msh6-deficient mice were obtained from W. Edelmann, Albert Einstein Medical College, NY (44). For each experiment, splenic B cells were isolated from littermates. Mice were housed in the IACUC-approved specific pathogen-free facility at the University of Massachusetts Medical School; mice were used according to the guidelines from University of Massachusetts Medical School IACUC.
Antibodies
Antibodies to ER (sc-8002X), GAPDH (sc-25778), growth factor receptor-bound protein 2 (Grb2) (sc–255), and Msh6 (sc-10798) were purchased from Santa Cruz, and antibody for Lamin A/C was from Cell Signaling (#2032). Rabbit antibodies to mouse AID (20) and UNG (23) were previously described. For ChIP experiments using untagged AID, we used rabbit AID antibody provided by J. Chaudhuri (10).
Production of retroviruses in Phoenix-E cells
pMX-PIE-AID-FLAG-ER-IRES-GFP-puro and pMX-PIE-ΔAID-FLAG-ER-IRES-GFP-puro(7) were received from Drs V. Barretto and M. Nussenzweig (The Rockefeller University, NY). The control retrovirus, pMX-PIE-ER-IRES-GFP, was constructed and viruses were prepared as previously described (25). To create the ΔAIDH56R/E58Q and AIDH56R/E58Q mutants, the AID-ER gene was subcloned into Bluescript (Stratagene), mutated using Quik-Change (Stratagene), sequenced, and then reinserted into pMX-PIE. pMIG-AID and pMIG (45) were received from J. Chaudhuri, Rockefeller University. pMIG-ΔAIDwas created by converting amino acid position 189 to a nonsense codon.
Chromatin immunoprecipitation (ChIP)
Live cells were isolated by flotation on Lympholyte M (Cedar Lane, Ontario Canada) 24 hrs after retroviral transduction. After recovery and washing twice, ~2 × 107 live cells were resuspended in SBSS, and were cross-linked with formaldehyde at a final concentration of 1% for 5 min at 37°C. Cross-linking was stopped by adjustment to 125 mM glycine, and incubation for 5 min at room temp. Cross-linked cells were intermittently sonicated at 4°C for 60 min total. Samples were filtered through glass wool. 2 × 106 cell equivalents were incubated overnight with antibody at 4°C. On the following day, Protein G or Protein A Dynabeads (Invitrogen) were added to the samples and incubated for 2 h at 4°C. The beads were then washed 5 times for 10 min and reverse crosslinking was performed at 65°C in the presence of RNAse A. The samples were treated with Proteinase K at 55°C and DNA was recovered using phenol-chloroform extraction followed by ethanol precipitation. ChIP results were assayed by real time PCR using Sybr Green. Significance was calculated by a paired two-tailed T test. Primers for Sµ were forward primer DK99: AACTAGGCTGGCTTAACCGAGATG and reverse primer DK100: GTCCAGTGTAGGCAGTAGAGTTTA. Primers for mb-1/CD79a were: mb-1FW: CCACGCACTAGAGAGAGACTCAA and mb-1REV: CCGCCTCACTTCCTGTTCAGCCG. Primers for Cµ were: forward primer CuPG-F: TCTGACAGGAGGCAAGAAGACAGATTCTTA and reverse primer: CuPG-R: GCCACCAGATTCTTATCAGACAGGGG (46), except for the experiment shown in Fig 5C, in which Cµ primers previously described were used (25).
B cell purification and cultures
Mouse splenic B cells were isolated by T cell depletion with antibody and complement (21). B cells were cultured at 105 cells/ml. LPS (25 µg/ml), anti-IgD dextran (0.3 ng/ml)(Fina Biosolutions) +/− IL-4 (20 ng/ml) were used to induce class switch recombination to IgG1 and IgG3, respectively. Human BLyS/BAFF (50 ng/ml)(Human Genome Sciences) was included in all cultures. Retroviral infection and assay of CSR by FACS was performed as previously described (25). Briefly, cells were activated for 2 days, and then infected. One day later they were harvested for all experiments. The one exception is noted in the Results section. Statistical difference in CSR between different cultures was determined by a 2-tailed T test.
Western blotting
Preparation of extracts and western blots were previously described (21).
RT-PCR
RNA was isolated using Trizol (Ambion) from splenic B cells after 2 days of activation under IgG3 switching conditions. The RNA was treated with DNase I twice (DNA-free Kit, Ambion), and cDNA synthesis was performed using oligop (dT)10 and Super Script II reverse transcriptase (Invitrogen). Primers to amplify endogenous AID mRNA were located in the 4th exon (forward primer: AACTTCGGCGCATCCTTTTG) and in the 3’ untranslated region (reverse primer: CGTGTGACATTCCAGGAGGT). Primers to amplify RV-AID-ER were the same forward primer as for endogenous mRNA and the reverse primer was located in the ER tag (GGTTGGCAGCTCTCATGTCT).
Ligation-mediated PCR (LM-PCR)
After culture for 2 days, viable cells were isolated by flotation on Ficoll/Hypaque gradients (=1.09), or Lympholyte (Cedar Lane, Ontario, Canada); cells were imbedded in low melt agarose plugs, and DNA isolated as described (20). For linker ligation, 50 µL 1× ligase buffer was added to the plugs which were then heated to 68°C to melt the agarose. 20 µL DNA (about 10,000 cell equivalents) was added to 2 µL T4 DNA Ligase (2 Weiss units, MBI Fermentas, Hanover, MD), 10 µL ds annealed linker in 1× ligase buffer, 3 µL 10× Ligase buffer, 5µL 50% PEG 4000 and 25 µL dH20 and incubated overnight at 16°C. Linker was prepared by annealing 5 nmoles each of LMPCR. 1 (5’-GCGGTGACCCGGGAGATCTGAATTC-3’) and LMPCR.2 (5’-GAATTCAGATC-3’) in 300 µL 1× ligase buffer, which results in a dsoligo with a 14 ntss overhang that can only ligate unidirectionally. Ligated DNA samples were heat inactivated at 70°C for 10 min, diluted 3× in dH20, and then heated to 70° C for an additional 20 min. This sample was then assayed for mb1 DNA (primer sequences same as used for ChIP) by PCR to adjust DNA input prior to LM-PCR. The primer 5’Sμ (5’-GCAGAAAATTTAGATAAAATGGATACCTCAGTGG-3’) was used in conjunction with linker primer (LMPCR.1) to amplify DNA breaks. Three-fold dilutions of input DNA (0.5, 1.5 and 4.5 µL) were amplified by HotStarTaq (Qiagen) using a touchdown PCR program. PCR products were electrophoresed on 1.25% agarose gels and blotted onto nylon membranes. Blots were hybridized with an Sµ-specific oligonucleotide probe (mu probe5’: AGGGACCCAGGCTAAGAAGGCAAT for 5’ Sµ LM-PCR; end-labeled with [γ32P]-ATP at 37°C overnight and washed at 50°C with 2× SSC/0.1% SDS.
Amplification and sequencing of Sµ-Sα junctions
Genomic DNA (100ng) from RV-transduced cells induced to switch to IgA (47) is amplified (in 12 reactions per genotype) using 2 nested PCRs (Expand Long Template system, Roche), using the same program for each PCR. Primers for the first round were: 5u3 – AATGGATACCTCAGTGGTTTTTAATGGTGGGTTTA and SaR3 - CCCATCCCATCCCATCCCATC and for the second round were: u3H3 – AACAAGCTTGGCTTAACCGAGATGAGCC and SaR2 – CCAGCCCAGCTCAGGCCATTT. The products were cloned using the Topo TA cloning kit (Invitrogen) and sequenced by Macrogen.
Results
ΔAID-ER is a DN mutant in aid+/+ cells
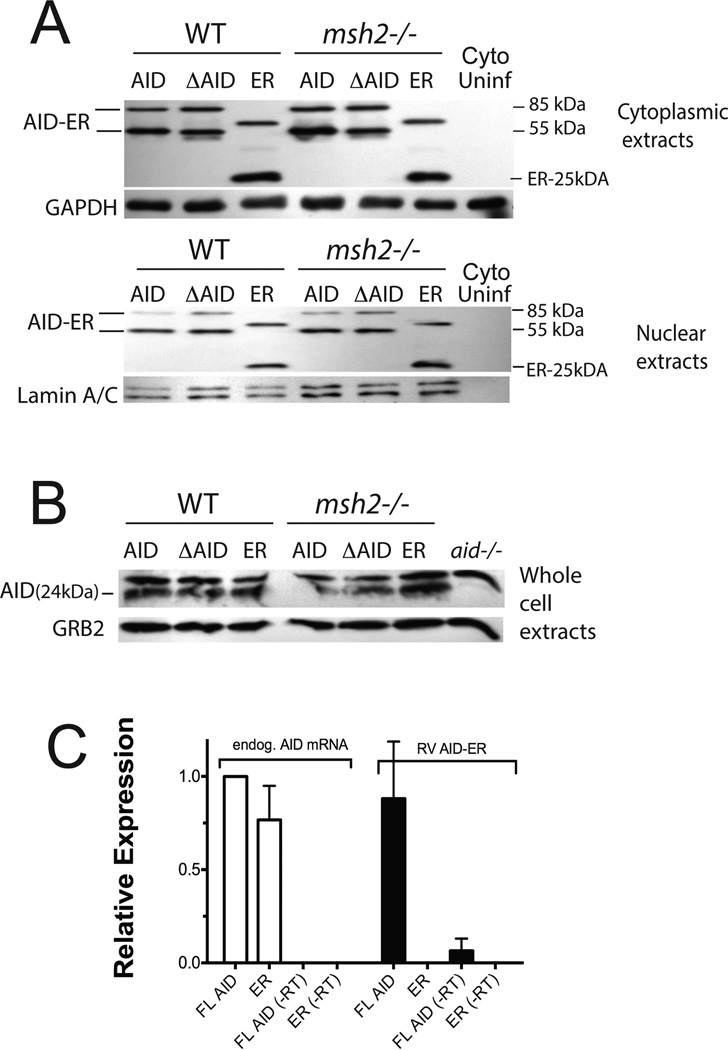
To study the DN effect of ΔAID, we transduced RV constructs encoding full-length AID-ER, ΔAID-ER, or the ER tag alone into WT mouse splenic B cells two days after activation to induce CSR. The cultures were treated with tamoxifen (OHT) at the time of retroviral transduction to induce nuclear localization of ER-tagged proteins, allowing AID to reach its target. We harvested the cells one day after transduction, because we found this was optimal for their viability (25). We detected no consistent difference in cell viability and recovery from cultures expressing full-length AID, ΔAID or the ER tag alone at this time point; it averaged ~90%.Two days after transduction, cell viability was reduced to ~70% among all cultures. To compare expression of the AID constructs, we prepared extracts of transduced WT cells for western blot analysis of AID and ΔAID expression. Full-length AID-ER and ΔAID-ER are expressed at similar levels in cytoplasmic and nuclear extracts from WT cells with similar GFP expression (Fig 1A: 3 left-most lanes). Note that the ER-tagged constructs have two forms of protein, which probably are monomers and dimers, although their apparent molecular weights do not have a clear 2-fold relationship on these gradient polyacrylamide gels. To determine if the transduced AID-ER or ΔAID-ER affects endogenous AID levels, we also assayed endogenous AID in total cell extracts, and did not find a difference between cells transduced with the three retroviruses (Fig 1B). This indicates that expression of ΔAID does not cause degradation of endogenous AID, ruling out one possible explanation for the mechanism of its DN effect. As our AID antibody is specific for the C terminus of AID, which is where the ER tag is located, we cannot detect transduced ΔAID with this antibody. To compare the levels of transduced AID-ER and endogenous AID, we compared their relative mRNA levels by qRT-PCR. As shown in Fig 1C, the levels of mRNAs for endogenous and transduced AID are similar. Taken together, the data in Fig 1 indicate that endogenous AID, AID-ER and ΔAID-ER are expressed at similar levels in aid+/+ cells one day after transduction, and that ΔAID-ER does not cause degradation of endogenous AID.
Figure 1. Quantitation of transduced AID-ER and endogenous AID levels in splenic B cells induced to switch to IgG3.
(A) Western blots of cytoplasmic and nuclear extracts (20 µg) from RV-infected splenic B cells one day after viral transduction were probed with anti-ER antibody, and with antibodies to GAPDH or Lamin A/C for loading controls. (B) Western blot of total cell extracts (75 µg) from the RV-infected or uninfected splenic B cells (from the same experiment shown in A) probed with antibody to mouse AID to detect endogenous AID. (C) qRT-PCR analysis shows that endogenous AID mRNA and AID mRNA transcribed from RV-AID-ER are present at similar levels in B cells activated to switch, one day after transduction with RV-AID-ER or RV-ER. Results of 2 independent experiments (2 mice) are normalized to endogenous AID mRNA in RV-AID-ER transduced cells. (−RT) indicates samples to which reverse transcriptase was not added.
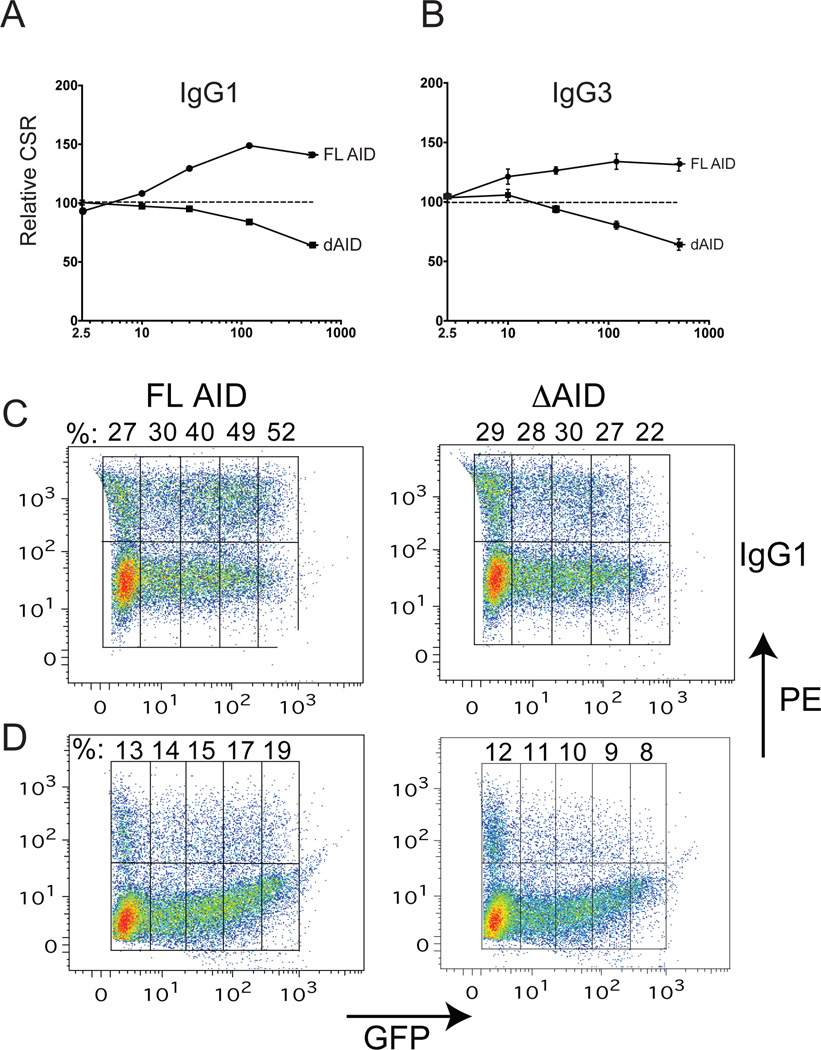
To determine whether ΔAID-ER has a DN effect on CSR when expressed in WT mouse splenic B cells, we performed a dose-response assay to compare IgG1 and IgG3 CSR as a function of GFP mean fluorescence intensity (MFI), which is an indicator of AID and ΔAID expression levels. Increasing expression of full-length AID-ER results in increased IgG1 CSR (Fig 2A, C), as previously reported (48), whereas increasing doses of ΔAID results in decreasing CSR to IgG1. CSR to IgG3 increases with increasing expression of AID in WT B cells, but decreases with increasing expression of ΔAID (Fig 2B, D). These data demonstrate that ΔAID has a DN effect on CSR in WT mouse B cells. The weaker DN effect we observe relative to the nearly complete inhibition of CSR found in most HIGM patients with the heterozygous C terminal AID deletion (5, 8) is likely due to the fact that in our experiments endogenous full-length AID is expressed prior to retroviral transduction. Endogenous AID is already highly expressed in mouse splenic B cells two days after treatment with switch inducers (20), which is the day of retroviral transduction and one day prior to harvesting.
Figure 2. Dose-response assays of IgG1 (A, C) and IgG3 (B, D) switching in aid+/+ mouse splenic B cells expressing increasing levels of AID-ER or ΔAID-ER, as assayed by GFP expression (MFI).
Each data point on the curves in A and B represent the average%IgG1 or %IgG3 (±SEM) at the indicated MFI (binned as shown in C and D) of 3 cultures from 1 mouse transduced with the indicated retrovirus relative to %CSR in cells transduced with the empty ER retrovirus. Data points on the Y-axis (MFI=2.5) in A and B represent CSR in GFP-negative cells, expressing only endogenous AID. CSR in the GFP-negative cells are shown in the left most boxes of the FACS dot plots in C and D. The procedure for gating is illustrated in Fig 7A.
Untagged ΔAIDhas DN activity
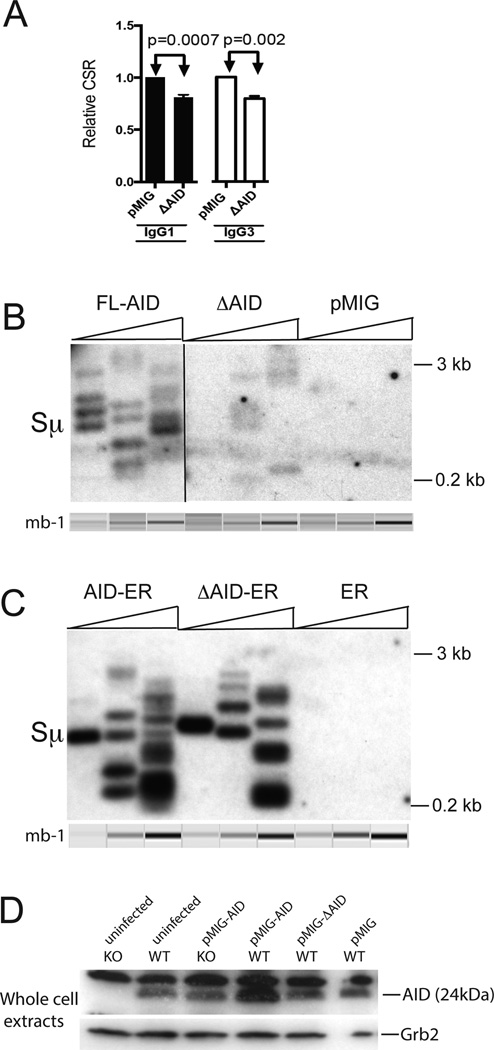
To better compare with endogenousΔAID in HIGM patients, we tested whether an untagged ΔAID expressed in the pMIG retrovirus would show DN function in RV transduction experiments. Fig 3A shows that CSR to IgG3 and to IgG1 is significantly lower in WT cells expressing ”AID than in cells expressing the empty pMIG, demonstrating the DN effect is not dependent upon the ER tag. In these experiments we did not add tamoxifen to the cultures, and we infected the cells one day after activation and harvested two days later. The increased duration of infection used in experiments with untagged AID (2 days instead of 1 day) was required to obtain the DN effect with these constructs, probably because unlike ER-tagged ΔAID, the untagged ΔAID appears to be poorly expressed, as reported previously for human ΔAID (29, 39). This is consistent with the facts that ΔAID lacks a nuclear export signal, and AID is degraded more rapidly in nuclei than in cytoplasm (15). We could not examine the expression of ΔAID directly, as our AID antibody is directed against the C terminus. We tested the two commercially available antibodies that were reported to be against different epitopes in mouse AID, but, unlike our AID antibody, they did not detect mouse AID in our western blots. Again, we did not detect consistent differences in cell viability or recovery after infection with the different viruses, although the cells were less viable when harvested two days after infection (~70%) than when harvested after one day (~90%), independent of the particular AID expression construct. Although it has been speculated that the reason untagged ΔAID poorly induces CSR is due to its low expression, the finding that both ΔAID-ER and untagged ΔAID poorly induce CSR, and both have DN effects on CSR in WT cells indicate that the phenotype of ΔAID is not solely due to its low expression.
Figure 3. Untagged ΔAID has DN activity, induces very few Sµ DSBs, and does not cause degradation of endogenous AID.
(A) Compilation of IgG1 and IgG3 CSR results (+SEM) for the untagged DAID in WT cells relative to CSR in cells expressing pMIG. The control retrovirus pMIG does not affect CSR significantly, as GFP-negative cells switched similarly to cells transduced with pMIG (not depicted). In these experiments, cells were activated to switch for 24 hrs, transduced and harvested 2 days later. The procedure for gating is illustrated in Fig 7A. In addition, we gated on the brightest GFP+ cells (~50% of the cells). Results from two independent experiments (2 mice) with two cultures each are shown. (B) LM-PCR assay of Sµ DSBs in aid−/− cells induced to switch to IgG3 transduced with untagged AID, ΔAID or the empty retrovirus pMIG. Transduction was performed on day 2 after activation, and cells harvested on day 3. The line indicates where the image was cut to remove irrelevant lanes. Three fold titrations of input DNA were performed, and the mb-1 gene was amplified as an internal control for template input. The mb-1 PCR bands shown were obtained by electrophoretic analysis on a QIAxcel Advanced instrument, which subjects each sample to electrophoresis in a capillary, and provides an image and quantitation of each lane. (C) LM-PCR assay of Sµ DSBs in aid−/− cells induced to switch to IgG3 transduced with AID-ER, ΔAID-ER or the control retrovirus ER. Methods similar to B. (D) Westerns blot of 80 µg of total cell extracts of aid−/− and aid+/+ cells transduced with untagged AID and ΔAIDexpressed in the retrovirus pMIG, and then probed with antibody to AID and to Grb2 for loading controls. Cells were cultured and transduced as in A, but similar results were observed if cells were cultured for 2 days prior to transduction, and harvested one day later.
As untagged ΔAID might be less stable than ΔAID-ER, we asked if untagged ΔAID might destabilize endogenous AID. As shown in Fig 3D, the levels of endogenous AID in WT cells expressing ΔAID are not reduced relative to untransduced WT cells or cells transduced with pMIG, similar to the results obtained with ΔAID-ER, and confirming that ΔAID does not exert a DN effect by causing degradation of endogenous AID.
We used LM-PCR to determine if untagged ΔAID induces DSBs in the Sµ region in aid−/− cells, and found that although ΔAID induces Sµ DSBs, there are many fewer than in cells expressing full-length AID (Fig 3B). This result differs from what we obtain in cells transduced with ΔAID-ER, in which the DSB frequency is similar to that observed in cells transduced with AID-ER (Fig 3C) and (25). This difference is consistent with the likely reduced expression of untagged ΔAID relative to that of ΔAID-ER. Note that LM-PCR only detects DSBs that are blunt, orif T4 DNA Polymerase is added prior to ligation, staggered DSBs can be detected. However, addition of T4 Pol does not change the relative frequency of DSBs induced by AID-ER or ΔAID-ER (25). If ΔAID induces extensively end-resected DSBs, as suggested by the results of Zahn et al (29), these DSBs would not be detected by this assay.
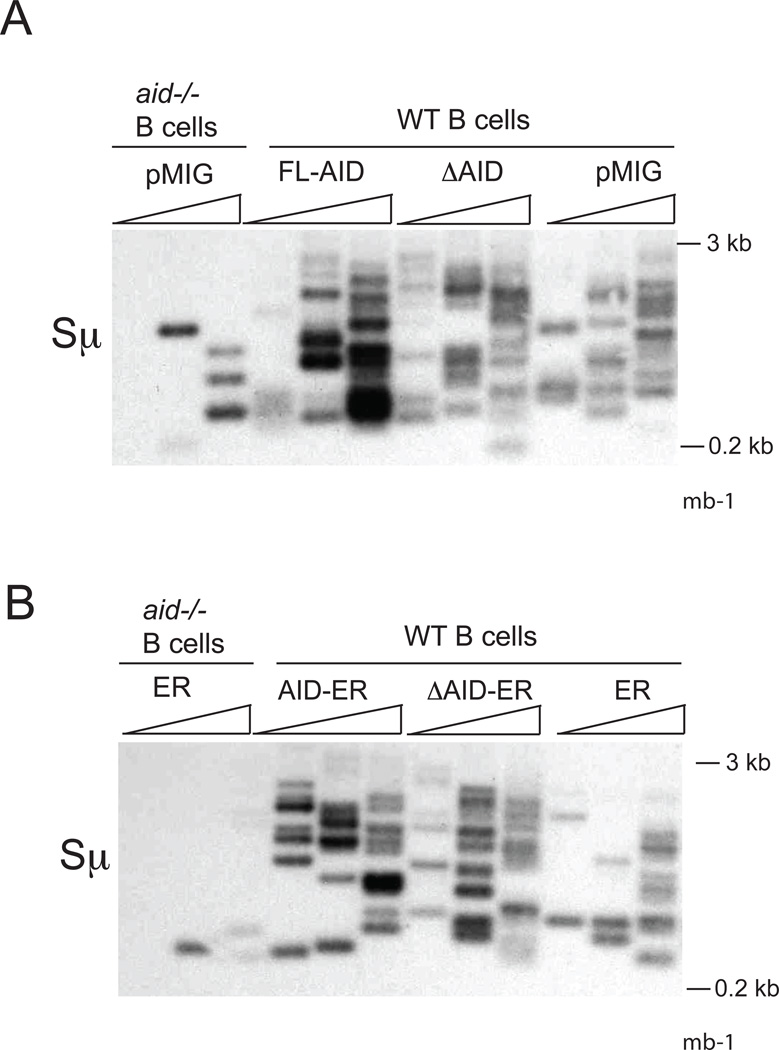
To determine if ΔAID might affect Sµ DSBs detected in WT cells, we performed LM-PCR experiments in WT cells transduced with untagged and tagged AID viruses, using T4 Pol to fill in staggered ends. In WT cells, we detect similar numbers of DSBs in cells expressing ΔAID as in cells expressing full-length AID, whether ΔAID is untagged (Fig 4A) or tagged (Fig 4B). As the intensity of the signals is slightly greater in cells expressing full-length AID, it is possible that these DSBs are slightly more numerous or more efficiently amplified. Similar results were obtained in the absence of T4 Pol (not depicted). Again, extensively end-resected DSBs would not be detected. As pointed out previously (25, 27), the role of the C terminus of AID appears to manifest itself subsequent to DSB formation.
Figure 4. LM-PCR assays of Sµ DSBs in WT (aid+/+) or aid−/− cells induced to switch to IgG3 transduced with pMIG expressing untagged AID constructs (A) or pMXpie expressing ER-tagged AID constructs (B).
Methods are described in Fig 3.
ChIP assays show that ΔAID binds Sµ in WT cells
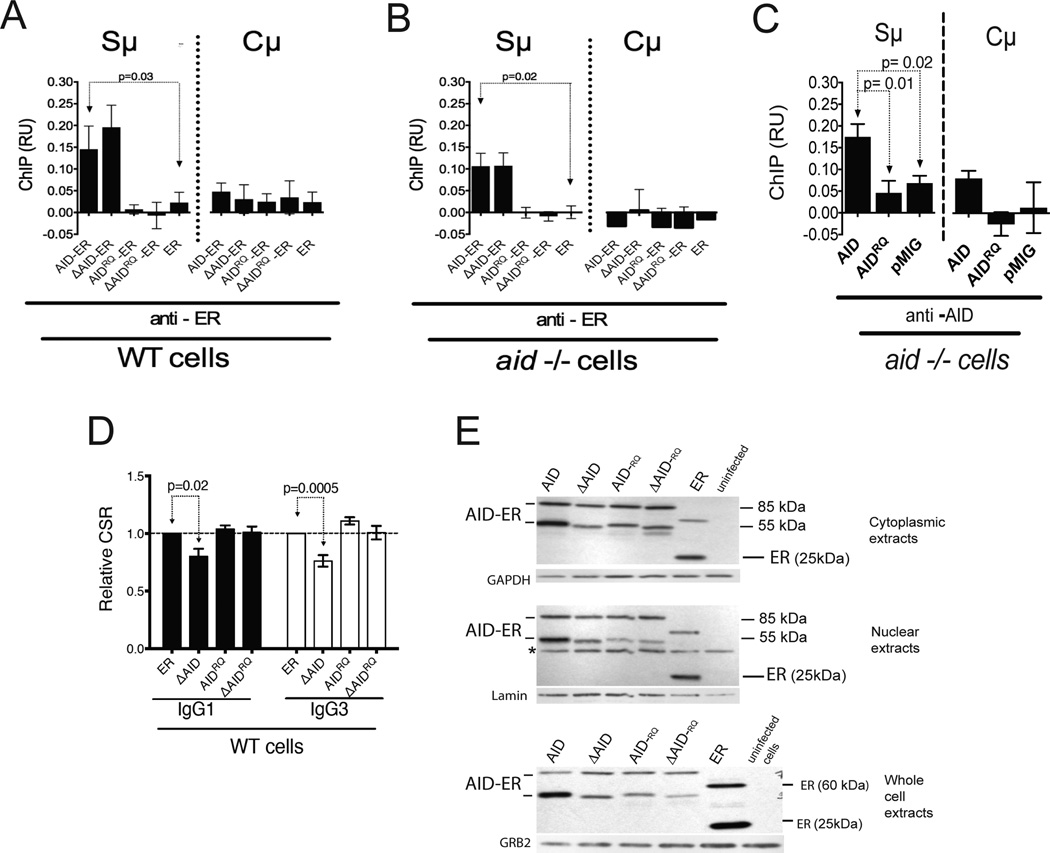
To further explore the mechanism of the DN effect, we asked whether ΔAID-ER binds Sµ in WT cells expressing endogenous AID, or whether endogenous AID might compete and thereby inhibit its binding. WT B cells transduced with ER were used as negative controls. As shown in Fig 5A, ”AID-ER is recruited at least as well as AID-ER to the S¼ region in AID-sufficient cells, similar to results inaid−/− cells (Fig 5B). Binding of AID and ΔAID was not detected at the Cµ gene in WT oraid−/− cells.
Figure 5. Both the DN effect and association of ΔAID-ER with Sµ depend upon deaminase activity of ΔAID-ER.
(A) ChIP of the AID-ER proteins at Sµ and Cµ in WT cells relative to input DNA, using anti-ER antibody. Error bars indicate SEM. 5 ChIPs (2 independent experiments, 2 mice) were performed, except for the no antibody control for FL-AID, where 4 were performed. ChIPs were analyzed by qPCR; % input was calculated, and % input in absence of antibody was subtracted. (B) ChIP of the AID-ER proteins at Sµ and Cµ in aid−/− cells relative to input DNA. 6 ChIPs for AID, ΔAID, and ER and 3 ChIPs for AIDRQ and ΔAIDRQ were performed (2 mice). Analysis as in A. (C) ChIP experiments performed with aid−/− cells transduced with untagged AID pMIG constructs, immunoprecipitated with anti-AID antibody (10). Error bars indicate SEM. 5–6 ChIPs (2 independent experiments, 2 mice) were performed. ChIPs were analyzed by qPCR; % input was calculated, and % input in absence of antibody was subtracted. P values determined by the two-tailed T test. (D) Compilation of IgG1 and IgG3 CSR results (+SEM) for the indicated RV-AID constructs in GFP-hi WT splenic B cells. Results are normalized to AID-ER results in one of the cultures each for IgG1 and IgG3. Six cultures (3 mice, 2 cultures each) were performed. CSR is plotted for GFP-hi cells, which are ~50% of the total GFP+ cells. Cells transduced with ER switch similarly to GFP-negative cells in the same cultures (not depicted). (E) Western blots of AID-ER in nuclear, cytoplasmic, and whole cell extracts from transduced WT splenic B cells. 20 µg protein was loaded in each lane. *Unknown irrelevant protein.
ΔAID lacking deaminase activity is not a DN mutant and does not bind Sµ in WT cells
We previously showed that AID lacking deaminase activity due to mutations in the catalytic domain, AIDH56R/E58Q (AIDRQ) (10, 49), does not bind to Sµ and Sγ3 in our ChIP assays in aid−/− cells (25) (see also Fig 5B). This result differs from that reported by Vuong et al (50) who found that AIDRQ expressed without a tag binds similarly as WT AID to Sµ, so we asked whether the difference was due to the ER tag. However, we found that untagged AIDRQis also unable to bind Sµ under our ChIP conditions (Fig 5C). There are several differences between the ChIP conditions in our lab compared to Vuong et al (50), and we do not know which ones are important. Taken together, the data from our lab and the Chaudhuri lab (50) suggest that AIDRQ binds Sµ but less stably than WT AID.
We now asked whether ΔAIDRQ-ER would have DN function by analyzing CSR in WT cells transduced with ΔAIDRQ in comparison with ER and full-length AIDRQ, and ΔAID. Fig 5D shows the combined CSR data, normalized to CSR in cells expressing ER alone. Again, CSR in cells expressing ΔAID is significantly lower than in cells expressing ER. However, there is no DN effect observed in cells expressing ΔAIDRQ or AIDRQ. This is consistent with our finding that ΔAIDRQ or AIDRQ do not stably associate with Sµ in WT cells (Fig 5A). As shown in the western blot in Fig 5E, the AIDRQ-ER proteins are expressed at similar levels toΔAID-ER in nuclei, cytoplasm and in whole cell extracts, ruling out the possibility that ΔAIDRQ does not have a DN effect due to lack of expression. These data suggest that the DN effect depends upon the ability of ΔAID to bind Sµ, and/or the ability of ΔAID to deaminate dC and to induce Sµ DSBs.
If AIDRQ and ΔAIDRQheterodimerize with endogenous AID, they might be expected to bind Sµ in WT cells. Although AID and ΔAID are detected at Sµ in WT cells, neither AIDRQ nor ΔAIDRQ bind (Fig 5A), suggesting that a functional deaminase domain must be present in both partners of the heterodimer for detectable binding to Sµ,or that endogenous WT AID does not heterodimerize with catalytically inactive AID, or that AID functions as a monomer.
Sµ-Sα junctions show increased microhomology in cells expressing ΔAID
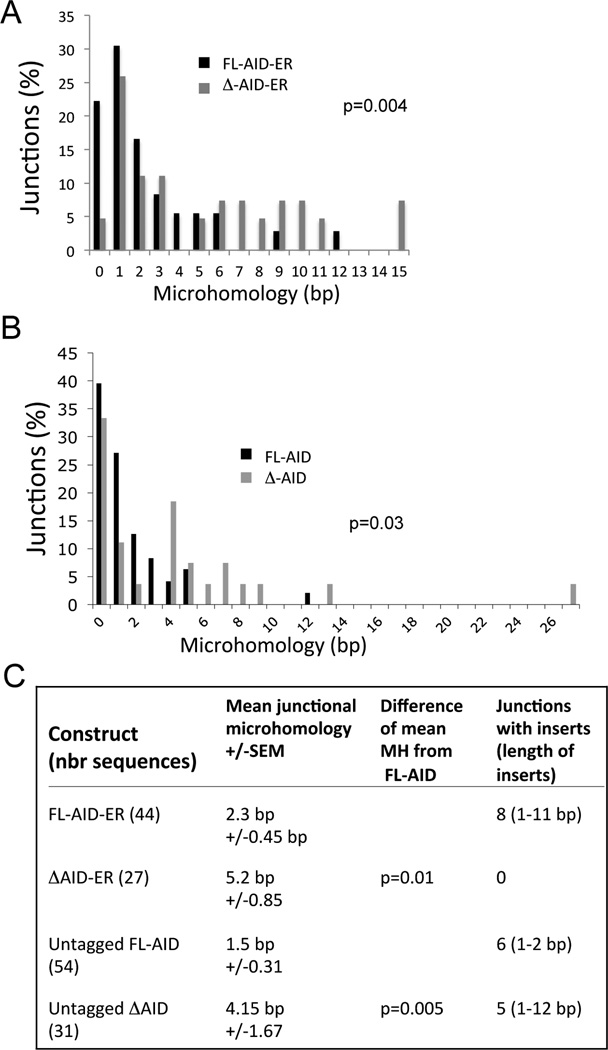
Because the deaminase activity of AID is essential for S regions DNA breaks, we hypothesize that the DN function of ΔAID depends upon its ability to induce S region breaks. Perhaps the breaks are aberrantly processed, and cannot participate in S-S recombination by NHEJ. They might interfere with normal S-S recombination induced by full-length AID. To begin to address this possibility, we examined the S-S junctions in aid−/− cells expressing ΔAID to determine if they showed increased amounts of microhomologyas would be expected if they were not recombined by NHEJ, which is the predominant mechanism for recombining S-S junctions. We examined Sµ-Sα junctions because Sα has the greatest amount of homology to Sµ of any S region, thus increasing the sensitivity of the assay (51). Indeed, we found that Sµ-Sα junctions showed an average of 2.3-fold greater lengths of microhomology in aid−/− cells expressing ΔAID-ER compared to cells expressing AID-ER (Fig 6C), similar to observations in human HIGM patients expressing both C terminally deleted AID and full-length AID (35), and recent reports studying a human AID with a 17 amino acid C terminal deletion in mouse aid−/− B cells (29, 52). We also examined junctions in cells expressing untagged ΔAID, and found a 2.8-fold increase in microhomology lengths in comparison to cells expressing full-length untagged AID (Fig 6C). There were also significant differences in the distribution of microhomology lengths between cells expressing AID and ΔAID, whether it was tagged or untagged (Fig 6A and B, respectively). Note that in cells expressing ΔAID, we observed junctions with up to 15 and 27 bp of microhomology. These data suggest that the AID C terminus is involved in directing S region DSBs toward recombination by NHEJ, as suggested previously (29, 35, 52).
Figure 6. Microhomology (MH) at Sµ-Sα junctions in aid−/− cells transduced with AID-ER, ΔAID-ER, and untagged AID and ΔAID.
(A) Graph indicates the percent of junctions with the indicated lengths of microhomology cloned from aid−/− cells transduced with AID-ER and ΔAID-ER retroviruses. Junctions with inserts are not included. (B) Same as A except junctions from cells transduced with untagged AID and ΔAID. The indicated p values show differences in distributions of MH between AID and ΔAID, determined by a Mann-Whitney 2-tailed test. (C) Compiled mean junctional MH +/− SEM for the ER tagged and untagged AID and ΔAID, as indicated. Sequences with inserts are not included in the calculations of MH.
The DN effect of ΔAID depends on the presence of Msh2
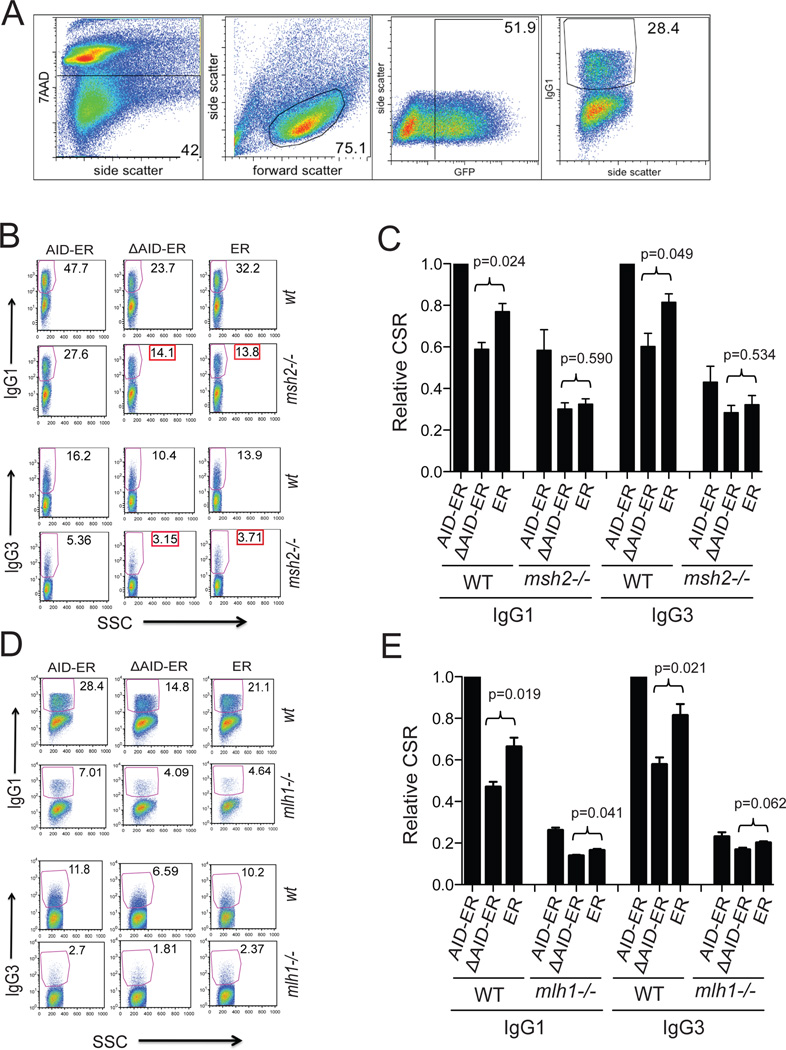
Msh2 and Msh6 are required for optimal CSR in B cells expressing full-length AID-ER but do not contribute to CSR in B cells expressing ΔAID-ER (25). It is clear that at least one of the roles of MMR in CSR is to increase S region DSBs, most likely by converting distal SSBs to DSBs (17, 22–24). Furthermore, in the absence of the MMR proteins Mlh1 or Pms2, S-S junctions show increased microhomology (53–55), similar to the junctions in cells expressing ΔAID. To determine if the DN effect depends on MMR, we expressed AID-ER and ”AID-ER in AID-sufficient msh2−/− or mlh1−/− B cells and analyzed CSR by flow cytometry. Fig 7A shows the gating strategy used in these experiments. Figs 7B,D provide examples of the switching results in the GFP+ transduced cells, and Figs 7C,E present the compiled results. Demonstrating the DN effect, CSR is significantly lower in WT cells expressing ΔAID than in cells expressing ER (Fig 7C,E). However, ΔAID does not have a DN effect in msh2−/− cells, as CSR is not significantly lower in ΔAID-expressing msh2−/− cells than in ER-expressing cells (Figs 7B,C). The levels of RV-AID proteins or endogenous AID in the transduced cells are not affected by lack of the MMR protein Msh2, as shown in Figs 1A, B. In mlh1−/− cells the DN effect borders on significance (Fig 7E), perhaps due to the presence of Msh2-Msh6 in these cells. MMR is partially functional in the absence of Mlh1-Pms2 (17, 56). These results suggest that the DN effect of ΔAID in AID-sufficient cells depends upon a function that Msh2 provides. At first glance, this seems surprising because we have previously shown that MMR does not contribute to CSR induced by ΔAID, and because the AID C terminus is important for recruiting MMR proteins to S regions (25). However, endogenous AID can recruit MMR proteins in cells expressing ΔAID, and thus it is possible that the ability of MMR to increase S region DSBs is important for the DN effect.
Figure 7. FACS analyses of CSR in WT, msh2−/−, and mlh1−/− cells demonstrates that the DN effect of ΔAID-ER requires the presence of Msh2.
(A) Gating strategy used for all FACS experiments in this manuscript, proceeding from left to right. Example is from panel D: WT cells transduced with AID-ER transduced and induced to switch to IgG1. Retrovirally-infected cultured cells were stained with 7-AAD to detect dying/dead cells, and with antibody (Fab’2) to IgG1 or IgG3 conjugated to PE. The percent of viable (7AAD-neg), retrovirally-infected (GFP+) cells that were IgG1+ of IgG3+ was determined as shown. Compensation and gating were performed using FlowJo software (Treestar). (B) FACS results for one representative CSR experiment comparing the DN effect of ΔAID-ER in WT and msh2−/− B cells (expressing endogenous AID). The FACS plots show only viable and GFP+ cells; the gates represent PE-IgG1/PE-IgG3 positive cells within the GFP+ populations as indicated. The entire GFP+ population was analyzed in these experiments. Side scatter (SSC) is plotted on the X-axis. (C) Compilation of IgG1 and IgG3 CSR results (+SEM) for the indicated constructs in WT and msh2−/− cells relative to CSR in WT cells expressing AID-ER. Two independent experiments (2 mice, 3 cultures each) were performed. IgG1 and IgG3 CSR are significantly increased in WT cells expressing AID-ER relative to ER (p=0.004 and 0.011, respectively). IgG1 and IgG3 CSR in msh2−/− cells expressing AID-ER relative to ER were not significantly increased (p=0.065 and 0.284, respectively). (D) FACS results for one experiment comparing the DN effect of ΔAID-ER in WT and mlh1−/− B cells. (E) Compilation of IgG1 and IgG3 CSR results (+SEM) for the indicated constructs in WT and mlh1−/− cells relative to CSR in WT cells expressing AID-ER). Three independent experiments (3 mice, 3 cultures each) were performed. IgG1 and IgG3 CSR were significantly increased in WT cells expressing AID-ER relative to ER (p=0.002 and 0.027, respectively). IgG1 CSR in mlh1−/− cells expressing AID-ER relative to ER was significantly increased (p=0.004) but not IgG3 CSR (p=0.256).
Discussion
ΔAID has been found to function as a DN mutant in HIGM patients that retain one allele encoding WT AID (5), and here we address the mechanism of this DN effect. We show that the DN effect of ΔAID requires Msh2, and that ΔAID must have deaminase activity. The deaminase activity is required for binding of ΔAID-ER, Msh2-Msh6, and UNG to Sµin our ChIP assays (25)(and data herein). However, deaminase activity is also essential for AID to induce DNA breaks, suggesting that the DN effect might require the induction of DNA breaks. Consistent with this hypothesis, MMR proteins are important for creating DSBs during CSR (23).
We have shown that Msh2 and Msh6 are recruited to Sµ, dependent upon both the C terminus of AID and deaminase activity (25). Thus, it seemed surprising that the DN effect of ΔAID depends upon Msh2, as ΔAID poorly recruits Msh2 to Sµ. However, full-length endogenous AID would recruit Msh2-Msh6 in cells expressing ΔAID, which could increase S region DSBs. Thus, the dependence of the DN effect upon Msh2 supports the hypothesis that DSBs are required for the DN effect. MMR is normally involved in post-replicative repair during S phase, whereas CSR occurs during G1 phase (23, 57), so it is possible that MMR must be specifically recruited in order to increase DSBs during CSR. As ΔAID does not recruit UNG to Sµ as well as does full-length AID (25, 29, 52), it is also likely that ΔAID inefficiently induces SSBs. Thus, it is possible that in cells expressing haploid amounts of both AID and ΔAID that DSBs might be even more dependent upon MMR than in cells expressing only full-length AID. MMR appears to help generate DSBs that can be recombined by NHEJ, or perhaps contributes to recruitment of NHEJ proteins, as S-S junctions in cells lacking Mlh1 or Pms2 show increased microhomology similar to those in cells lacking NHEJ proteins (53, 54, 58, 59).
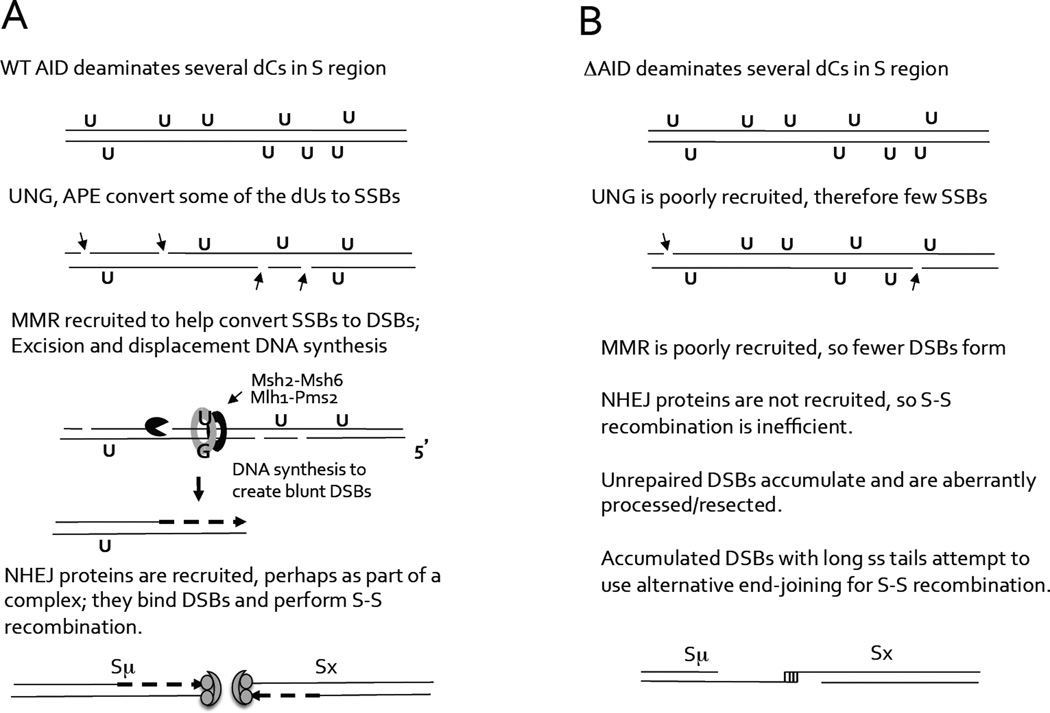
It is clear that the DSBs induced in aid−/− cells expressing ΔAID cannot undergo efficient S-S recombination by NHEJ, as CSR is 90% reduced compared to cells expressing FL-AID, and the S-S junctional microhomology is increased. It is possible that AID, but not ΔAID, recruits NHEJ proteins, which normally perform S-S recombination. Thus, in cells expressing ΔAID, DSBs might undergo extensive end-resection. We hypothesize that the reason the numbers of DSBs in S regions in bothaid−/− and WT cells expressing ΔAID-ER appear similar in LM-PCR experiments to those in cells expressing AID-ER, is that, while DSBs are created less efficiently due to poor recruitment of UNG and MMR, those that are made are also repaired less efficiently due to poor recruitment of NHEJ. Also, ΔAID has been reported to have higher deaminase activity than full-length AID (28, 29). To explain how the putative inability of ΔAID to recruit NHEJ proteins could have a DN effect, it is possible that aberrantly resected DSBs predominate over breaks induced by endogenous full-length AID, due to their inability to recombine efficiently, and these DSBs interfere with recombination by NHEJ. Figure 8 presents an outline model of this hypothesis.
Figure 8. Outline model for role of the AID C terminus in the introduction of S region DSBs and S-S recombination.
UNG and Msh2 do not appear to bind AID directly, and we only detect their interaction with AID by ChIP, dependent upon the C terminus of AID (25). The fact that deaminase activity is required for binding of Msh2 and UNG to Sµ in our ChIP experiments (25) suggests that UNG and MutSα require their DNA substrates, dU and U:G mismatches, respectively, for binding to DNA. It is possible that NHEJ proteins might also be part of a complex that is recruited by AID, dependent upon the C terminus and deaminase activity. DNA-PKcs, a DNA-dependent kinase, is very important for NHEJ (60). It has been reported to bind to the AID C terminus, but only in the presence of DNA (34). Consistent with this, DNA-PKcs has been shown to ChIP with Sµ in CH12 B lymphoma cells expressing full-length AID but not human ΔAID (52). Also, KU70 has recently been shown to ChIP at Sµ in mouse splenic B cells expressing human AID but not ΔAID (29). Recent evidence indicates that APE1 and AID interact directly, dependent upon phosphorylation of AID at S38 (50). Thus, NHEJ proteins could be recruited as part of a complex that converts AID-induced dUs to DSBs and then forms S-S junctions.
If AID functions as a dimer, then AID lacking deaminase activity should be able to heterodimerize with endogenous WT AID, as the catalytic site is not located in regions thought to be involved in AID dimerization (11, 12) or in DNA binding (61). However, as do not detect binding of AID lacking deaminase activity (AIDRQ) to S regions in WT cells, one possible explanation for our results is that both subunits of the putative AID dimer must have deaminase activity in order for AID to bind detectably to Sµ. If heterodimers of ΔAIDRQ and AID cannot bind DNA, they should not interfere with the function of WT AID, as transduced AID is not expressed in excess over endogenous AID in our experiments. Alternatively, our results could be interpreted to indicate that AID functions as a monomer. Although several reports suggest that AID functions as a dimer or tetramer (5, 9, 62), another report indicated that purified AID functions preferentially as a monomer in vitro acting on single strand DNA and also on a small loop in dsDNA (63). Thus, our data suggest either that an AID-ΔAIDRQ heterodimer does not form or if it forms, it cannot bind DNA, perhaps due to an inability to recruit UNG and/or MutSα, or alternatively, that AID functions as a monomer.
Acknowledgments
We thank J. Chaudhuri (Sloan-Kettering) for pMIG, pMIG-AID, and for anti-AID antibody, and for very helpful discussions. We thank V. Barreto and M. Nussenzweig (The Rockefeller University, NY) for the pMX-PIE-AID-FLAG-ER-IRES-GFP-puro and pMX-PIE-ΔAID-FLAG-ER-IRES-GFP-puro plasmids. We thank the UMMS Flow Cytometry Core Facility for excellent technical assistance.
The research was supported by grants from NIAID to JS: R01 AI023283 and R21 AI099988, and to CES: RO3 AI092528. J. Limauro and E. Xie were supported by the SURE fellowship program, funded by the Univ of Massachusetts Medical School. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Abbreviations
- AID
Activation-induced cytidine deaminase
- aa
Amino acid
- anti-δdex
anti-IgD dextran
- APE1/2
Apurinic/apyrimidinic endonuclease 1 and 2
- ChIP
chromatin immunoprecipitation
- ER
estrogen receptor
- HIGM
Hyper-IgM
- CSR
Class switch recombination
- AIDRQ
AIDH56R/E58Q (AID lacking deaminase activity)
- DN
Dominant negative
- Mismatch repair
MMR; proteins in this pathway are Msh2, Msh6, Mlh1, Pms2, Exonuclease I
- MH
Microhomology
- NHEJ
non-homologous end joining
- RV
Retroviral
- SHM
Somatic hypermutation
- UNG
Uracil-N-glycosylase
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 2.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 3.Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–104. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- 4.Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, Gearhart PJ. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, Kinoshita K, Muramatsu M, Honjo T. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 6.Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 7.Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 8.Imai K, Zhu Y, Revy P, Morio T, Mizutani S, Fischer A, Nonoyama S, Durandy A. Analysis of class switch recombination and somatic hypermutation in patients affected with autosomal dominant hyper-IgM syndrome type 2. Clin Immunol. 2005;115:277–285. doi: 10.1016/j.clim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 11.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 12.Jaszczur M, Bertram JG, Pham P, Scharff MD, Goodman MF. AID and Apobec3G haphazard deamination and mutational diversity. Cellular and molecular life sciences : CMLS. 2013;70:3089–3108. doi: 10.1007/s00018-012-1212-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride KM, Barreto V, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic Hypermutation Is Limited by CRM1-dependent Nuclear Export of Activation-induced Deaminase. J Exp Med. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoufouchi S, Faili A, Zober C, D'Orlando O, Weller S, Weill JC, Reynaud CA. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellyard JI, Benk AS, Taylor B, Rada C, Neuberger MS. The dependence of Ig class-switching on the nuclear export sequence of AID likely reflects interaction with factors additional to Crm1 exportin. Eur J Immunol. 2011;41:485–490. doi: 10.1002/eji.201041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Ann Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 19.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 20.Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent upon AID and UNG. J Exp Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min I, Schrader C, Vardo J, D'Avirro N, Luby T, Stavnezer J, Selsing E. The Sm tandem repeat region is critical for isotype switching in the absence of Msh2. Immunity. 2003;19:515–524. doi: 10.1016/s1074-7613(03)00262-0. [DOI] [PubMed] [Google Scholar]
- 23.Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 24.Cortizas EM, Zahn A, Hajjar ME, Patenaude AM, Di Noia JM, Verdun RE. Alternative End-Joining and Classical Nonhomologous End-Joining Pathways Repair Different Types of Double-Strand Breaks during Class-Switch Recombination. J Immunol. 2013;191:5751–5763. doi: 10.4049/jimmunol.1301300. [DOI] [PubMed] [Google Scholar]
- 25.Ranjit S, Khair L, Linehan EK, Ucher AJ, Chakrabarti M, Schrader CE, Stavnezer J. AID binds cooperatively with UNG and Msh2-Msh6 to Ig switch regions dependent upon the AID C terminus. J. Immunol. 2011;187:2464–2475. doi: 10.4049/jimmunol.1101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjit S, Khair L, Linehan EK, Ucher AJ, Chakrabarti M, Schrader CE, Stavnezer J. Correction: AID Binds Cooperatively With UNG and Msh2-Msh6 to Ig Switch Regions Dependent upon the AID C Terminus. J Immunol. 2014;192:4934. doi: 10.4049/jimmunol.1101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doi T, Kato L, Ito S, Shinkura R, Wei M, Nagaoka H, Wang J, Honjo T. The C-terminal region of activation-induced cytidine deaminase is responsible for a recombination function other than DNA cleavage in class switch recombination. Proc Natl Acad Sci U S A. 2009;106:2758–2763. doi: 10.1073/pnas.0813253106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A Portable Hot Spot Recognition Loop Transfers Sequence Preferences from APOBEC Family Members to Activation-induced Cytidine Deaminase. J Biol Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahn A, Eranki AK, Patenaude AM, Methot SP, Fifield H, Cortizas EM, Foster P, Imai K, Durandy A, Larijani M, Verdun RE, Di Noia JM. Activation induced deaminase C-terminal domain links DNA breaks to end protection and repair during class switch recombination. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1320486111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrader CE, Edelmann W, Kucherlapati R, Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrenstein MR, Neuberger MS. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. Embo J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Scherer SJ, Ronai D, Iglesias-Ussel MD, Peled JU, Bardwell PD, Zhuang M, Lee K, Martin A, Edelmann W, Scharff MD. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of MutS homologues in antibody diversification. J. Exp. Med. 2004;200:47–59. doi: 10.1084/jem.20040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Geraldes P, Platt JL, Cascalho M. The double-edged sword of activation-induced cytidine deaminase. J Immunol. 2005;174:934–941. doi: 10.4049/jimmunol.174.2.934. [DOI] [PubMed] [Google Scholar]
- 35.Kracker S, Imai K, Gardes P, Ochs HD, Fischer A, Durandy AH. Impaired induction of DNA lesions during immunoglobulin class-switch recombination in humans influences end-joining repair. Proc Natl Acad Sci U S A. 2010;107:22225–22230. doi: 10.1073/pnas.1012591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stavnezer J. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol. 2011;32:194–201. doi: 10.1016/j.it.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, Steinacker P, Li Z, Yates J, 3rd, Herron B, Otto M, Zan H, Fu H, Casali P. 14-3-3 adaptor proteins recruit AID to 5'-AGCT-3'-rich switch regions for class switch recombination. Nature structural & molecular biology. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasler J, Rada C, Neuberger MS. Cytoplasmic activation-induced cytidine deaminase (AID) exists in stoichiometric complex with translation elongation factor 1alpha (eEF1A) Proc Natl Acad Sci U S A. 2011;108:18366–18371. doi: 10.1073/pnas.1106729108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geisberger R, Rada C, Neuberger MS. The stability of AID and its function in class-switching are critically sensitive to the identity of its nuclear-export sequence. Proc Natl Acad Sci U S A. 2009;106:6736–6741. doi: 10.1073/pnas.0810808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reitmair AH, Redston M, Cai JC, Chuang TC, Bjerknes M, Cheng H, Hay K, Gallinger S, Bapat B, Mak TW. Spontaneous intestinal carcinomas and skin neoplasms in Msh2-deficient mice. Cancer Res. 1996;56:3842–3849. [PubMed] [Google Scholar]
- 42.Baker SM, Plug AW, Prolla RA, Bronner CE, Harris AC, Yao X, Christie D-M, Monell C, Arnheim N, Bradley A, Ashley T, Liskay RM. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nature Gen. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 43.Baar J, Pennell NM, Shulman MJ. Analysis of a hot spot for DNA insertion suggets a mechanism for Ig switch recombination. J. Immunol. 1996;157:3430–3435. [PubMed] [Google Scholar]
- 44.Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, Liedtke W, Cohen PE, Kane MF, Lipford JR, Yu N, Crouse GF, Pollard JW, Kunkel T, Lipkin M, Kolodner R, Kucherlapati R. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91:467–477. doi: 10.1016/s0092-8674(00)80433-x. [DOI] [PubMed] [Google Scholar]
- 45.Vuong BQ, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight GS, Chaudhuri J. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat Immunol. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ucher AJ, Linehan EK, Teebor GW, Schrader CE, Stavnezer J. The DNA glycosylases Ogg1 and Nth1 do not contribute to Ig class switching in activated mouse splenic B cells. PloS one. 2012;7:e36061. doi: 10.1371/journal.pone.0036061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sernandez IV, de Yebenes VG, Dorsett Y, Ramiro AR. Haploinsufficiency of activation-induced deaminase for antibody diversification and chromosome translocations both in vitro and in vivo. PloS one. 2008;3:e3927. doi: 10.1371/journal.pone.0003927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papavasiliou FN, Schatz DG. The activation-induced deaminase functions in a postcleavage step of the somatic hypermutation process. J Exp Med. 2002;195:1193–1198. doi: 10.1084/jem.20011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vuong BQ, Herrick-Reynolds K, Vaidyanathan B, Pucella JN, Ucher AJ, Donghia NM, Gu X, Nicolas L, Nowak U, Rahman N, Strout MP, Mills KD, Stavnezer J, Chaudhuri J. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol. 2013;14:1183–1189. doi: 10.1038/ni.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stavnezer J, Bjorkman A, Du L, Cagigi A, Pan-Hammarstrom Q. Mapping of switch recombination junctions, a tool for studying DNA repair pathways during immunoglobulin class switching. Adv. Immunol. 2010;108:45–109. doi: 10.1016/B978-0-12-380995-7.00003-3. [DOI] [PubMed] [Google Scholar]
- 52.Sabouri S, Kobayashi M, Begum NA, Xu J, Hirota K, Honjo T. C-terminal region of activation-induced cytidine deaminase (AID) is required for efficient class switch recombination and gene conversion. Proc Natl Acad Sci U S A. 2014;111:2253–2258. doi: 10.1073/pnas.1324057111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ehrenstein MR, Rada C, Jones AM, Milstein C, Neuberger MS. Switch junction sequences in PMS2-deficient mice reveal a microhomology- mediated mechanism of Ig class switch recombination. Proc Natl Acad Sci U S A. 2001;98:14553–14558. doi: 10.1073/pnas.241525998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schrader CE, Vardo J, Stavnezer J. Role for Mismatch Repair Proteins Msh2, Mlh1, and Pms2 in Immunoglobulin Class Switching Shown by Sequence Analysis of Recombination Junctions. J Exp Med. 2002;195:367–373. doi: 10.1084/jem.20011877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peron S, Metin A, Gardes P, Alyanakian MA, Sheridan E, Kratz CP, Fischer A, Durandy A. Human PMS2 deficiency is associated with impaired immunoglobulin class switch recombination. J Exp Med. 2008;205:2465–2472. doi: 10.1084/jem.20080789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5'-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 57.Sharbeen G, Yee CW, Smith AL, Jolly CJ. Ectopic restriction of DNA repair reveals that UNG2 excises AID-induced uracils predominantly or exclusively during G1 phase. J Exp Med. 2012;209:965–974. doi: 10.1084/jem.20112379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 59.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Advances in immunology. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 60.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchimura Y, Barton LF, Rada C, Neuberger MS. REG-gamma associates with and modulates the abundance of nuclear activation-induced deaminase. J Exp Med. 2011;208:2385–2391. doi: 10.1084/jem.20110856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larijani M, Petrov AP, Kolenchenko O, Berru M, Krylov SN, Martin A. AID associates with single-stranded DNA with high affinity and a long complex half-life in a sequence-independent manner. Mol Cell Biol. 2007;27:20–30. doi: 10.1128/MCB.00824-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brar SS, Sacho EJ, Tessmer I, Croteau DL, Erie DA, Diaz M. Activation-induced deaminase, AID, is catalytically active as a monomer on single-stranded DNA. DNA Repair (Amst) 2008;7:77–87. doi: 10.1016/j.dnarep.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]