Abstract
DNA repair aberrations and associated chromosomal instability is a feature of chronic lymphocytic leukemia (CLL). To evaluate if DNA repair insufficiencies are related to methylation changes, we examined the methylation of nine promoter regions of DNA repair proteins by bisulfide sequencing in 26 CLL primary samples and performed quantitative PCR on a subset of samples to examine BRCA1 expression. We also investigated if changes in cytogenetic or expression level of DNA repair proteins led to changes in sensitivity to a novel PARP inhibitor, CEP-8983, alone and in combination with bendamustine. No changes in promoter methylation were identified in BRCA1, BRCA2, FANC-C, FANC-F, FANC-L, ATM, MGMT, hMLH1 and H2AX except for two cases of minor BRCA1 hypermethylation. CLL samples appeared to have reduced BRCA1 mRNA expression uniformly in comparison to non-malignant lymphocytes irrespective of promoter hypermethylation. CEP-8983 displayed single agent cytotoxicity and the combination with bendamustine demonstrated synergistic cytotoxicity in the majority of CLL samples. These results were consistent across cytogenetic subgroups, including 17p deleted and previously treated patients. Our results provide rationale for further exploration of the combination of a PARP inhibitor and DNA damaging agents as a novel therapeutic strategy in CLL.
Keywords: Chronic lymphocytic leukemia (CLL), poly (ADP-ribose) polymerase (PARP), CEP-8983, bendamustine
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is the most common leukemia among adults in Western countries [1]. The disease remains incurable with chemotherapy-based approaches[2]. Traditional therapies for CLL include the nucleoside analog fludarabine and the alkylating agents chlorambucil and cyclophosphamide, which damage cellular DNA [2]. In 2008, bendamustine (Supplemental Fig. 1A), a nitrogen mustard analog that shows significant activity against B cell neoplasms and has a unique cytotoxicity and DNA damage profile, was approved for treatment of CLL [3–7]. As with other malignancies, DNA repair defects are thought to be important to the pathogenesis of CLL and its resistance to therapy [8]. The majority of gross chromosomal lesions in CLL appear to often involve at least one gene important to proper DNA repair: 17p deletion (TP53), 13q deletion (BRCA2), 11q deletion (ATM), and trisomy 12 (MDM2; [9–12]). Interestingly, CLL cells appear to have significant defects in both major DNA double strand break (DSB) repair pathways: the error-free homologous recombination (HR) and the error-prone non-homologous end joining (NHEJ; [8, 13–17]. Lastly, upregulation of NHEJ pathway members in CLL is associated with resistance to alkylating agents [18, 19].
The enzymes PARP1 and PARP2 are involved in a wide variety of nuclear processes, notably DNA damage sensing and repair through the base excision repair (BER), single strand break (SSB) repair, and double strand break (DSB) repair pathways [20, 21]. Working models suggest that inhibition of PARP may lead to an increase in SSBs, which can form DSBs upon encountering a replication fork (Fig. 1B). Regardless of the specific mechanism of action, PARP inhibition can be synthetically lethal in a defective DSB repair background, as seen in patients with defective BRCA, Ataxia Telengietasia Mutated (ATM), or Fanconi Anemia (FA) proteins [22–25]. This approach has validated clinically in breast cancer type 1/2 susceptibility protein (BRCA1/2) mutated/deficient breast and ovarian cancers using poly (ADP ribose) polymerase (PARP) inhibitors [26–28]. CEP-8983 (Supplementary Fig. 1A) is a potent and selective 4-methoxy-carbazole inhibitor of PARP1/2, with low nanomolar enzyme half maximal inhibitory concentration (IC50) values reported [29, 30].
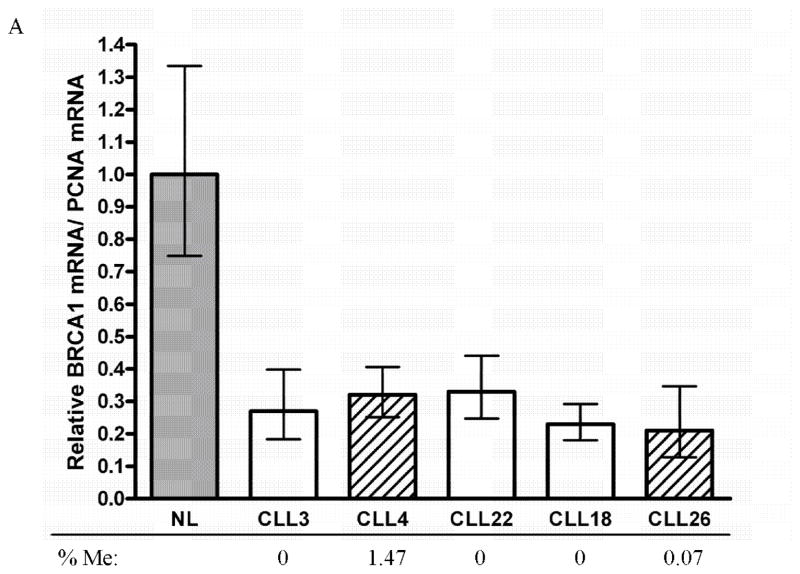
Figure 1. BRCA1 expression in CLL.
BRCA1 mRNA expression levels were examined in CLL patient samples using RT-qPCR. Values normalized to PCNA expression and relative to normal lymphocytes (NL).
The goal of this study was to investigate if DNA repair deficiencies in CLL are due to changes in promoter hypermethylation and examine if agents targeting DNA repair defective cells are synergistic in CLL primary samples. Given that CLL cells may have defects in one or more DSB repair pathways (BRCA, ATM, FA, etc.), we hypothesized inhibition of BER/SSB repair (i.e. PARP inhibition) could lead to enhanced cytotoxicity through combination therapy with DNA damaging agents leading to increased double strand breaks and cell death(Supplementary Fig. 1B). DNA repair genes have been reported to be hypermethylated in human leukemias, including BRCA1 in AML, and hMLH1 in Richter’s transformation of CLL [31, 32]. Furthermore, BRCA1 promoter hypermethylation has been shown to predict response to PARP inhibitors in other malignancies [33–35]. To explore if changes in DNA repair function in CLL are due to methylation changes, we examined DNA repair pathway proteins BRCA1, BRCA2, FANC-C, FANC-F, FANC-L, ATM, MGMT, hMLH1, H2AX.
MATERIALS AND METHODS
Drugs
CEP-8983 (CEP-9722 metabolite) and bendamustine were obtained from Cephalon Inc. Upon arrival, powder forms of the drugs were dissolved in dimethyl sulfoxide (DMSO; American Type Culture Collection [ATCC]) at stock concentrations of 10 mM. Stocks were aliquoted into 25 μl volumes and stored at −80°C and thawed immediately before use. All samples in the described experiments contained identical concentrations of DMSO. The upper dose range analyzed in assays(CEP-8983 50uM, bendamustine 50uM) were chosen through prior reports of plasma in vivo concentrations [36, 37].
Patient samples
CLL patient samples were provided by the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins Tumor and Cell Procurement Bank, supported by the Regional Oncology Center Grant # 2 P30 CA 006973-44. All patients gave informed consent according to the Declaration of Helsinki under a protocol approved by the Johns Hopkins Institutional Review Board. FISH cytogenetic data and immunoglobulin heavy chain variable region (IgVH) mutation status were gathered from patient records for each sample. Most patient samples were processed via Ficoll-Hypaque (GE Healthcare) density gradient purified mononuclear cells, which were then sorted via magnetic bead conjugated CD34 antibody to give CD34 negative lymphoid cells. These were aliquoted and frozen in 10% DMSO/90% fetal bovine serum (FBS; Gemini Bioproducts) at −80°C until use. Before use, the samples were thawed rapidly at 37°C, washed twice with RPMI, and resuspended in an appropriate volume of culture medium. Post thaw viability was assessed by trypan blue exclusion. Three patient samples were obtained as fresh whole blood in heparin tubes (BD), processed and used in experiments without freezing or CD34 sorting.
Bisulfite Conversion
Genomic DNA (gDNA) extracted using the Wizard Genomic Purification Kit (Promega) was bisulfite treated with the EZ DNA methylation kit (Zymo Research) for 16 cycles of 95°C for 10 minutes, 50°C for 60 minutes.
Quantitative Methylation-specific PCR (qMSP), MSP
For qMSP, bisulfite-converted gDNA was added to QuantiTect SYBR Green mix (Qiagen) containing either the unmethylated (U) or methylated (M) primer pairs for analysis on the iCycler iQ real-time PCR detection system (Bio-Rad) according to manufacturer’s recommendations. Primer sequences are listed in supplementary table S1. To quantify the qMSP products, bisulfite-converted gDNA mixtures as indicated below were used to generate standard curves for the unmethylated (U) and methylated (M) qMSP reactions using gDNA from normal peripheral lymphocytes (NL) and in vitro CpG methylated (IVD) Jurkat gDNA (N4002S, New England BioLabs). Genomic sequences were obtained from the Ensembl Genome Browser (www.ensembl.org). The percent methylation of each sample was calculated by the ratio of the M reaction quantity to the sum quantity of the U and M products. Each sample was performed in duplicate and called positive for methylation when its M amplicon matched the melting temperature of IVD product and had the same product size when visualized on a 2.5% agarose gel. MSP analysis of BRCA2 and Fanconi Anemia genes (FANC-A, FANC-C, FANC-F, FANC-L, ATM, MGMT, MLH1, H2AX) was performed as described previously [38]. Primers were designed using MSPPrimer [39].
| Unmethylated Standard | NL gDNA | IVD gDNA |
|---|---|---|
| 1x | 1ng | 999ng |
| 5x | 5ng | 995ng |
| 10x | 10ng | 990ng |
| 50x | 50ng | 950ng |
| 100x | 100ng | 900ng |
| 200x | 200ng | 800ng |
| 500x | 500ng | 500ng |
| 1000x | 1000ng |
| Methylated Standard | NL gDNA | IVD gDNA |
|---|---|---|
| 1x | 999ng | 1ng |
| 5x | 995ng | 5ng |
| 10x | 990ng | 10ng |
| 50x | 950ng | 50ng |
| 100x | 900ng | 100ng |
| 200x | 800ng | 200ng |
| 500x | 500ng | 500ng |
| 1000x | 1000ng |
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
1 μg total RNA extracted using the Trizol reagent (Invitrogen Life Technologies) was reverse transcribed into cDNA using the iScript supermix kit (Bio-Rad). Real-time PCR was performed in triplicates on an iCycler iQ real-time PCR detection system (Bio-Rad) using the SsoAdvanced SYBR green supermix (Bio-Rad) according to manufacturer’s instructions. Primer sequences are listed in supplementary table S2. BRCA1 gene expression levels were calculated using the 2−ΔΔCt method after normalization with cell cycle marker PCNA.
Cell lines and culture methods
All cell lines and CLL primary patient samples were cultured in RPMI 1640+ (Gibco), supplemented with 10% FBS (Gemini Bioproducts), 1% L-Glutamine (Gibco), and 1% Penicillin Streptomycin (Gibco), at 37°C in 5% CO2. SEM cells were obtained from Deutsche Sammlung von Mikrooganismen und Zellkulturen (DSMZ). HL-60 and TF-1 cells were obtained from ATCC. NCI-H929 and U-266 cell lines were provided by Ivan Borrello (Johns Hopkins University). Dami and HEL cell lines were provided by Michael McDevitt (Johns Hopkins University).
Cytotoxicity Assays
Cytotoxicity was assessed using an MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) assay (Roche). Cells were incubated in 96 well plates with increasing doses of drug (0, 1, 5, 10, 20, 50 μM) for 72 hours at 37°C in 5% CO2. Each condition was performed in quadruplicate (4 wells of plate). For drug combinations, a 1:1 ratio of drug concentration was used, with identical incubation times. Untreated cells were used as negative control. At the end of the 72-hour incubation, the MTT reagents were added as per manufacturer’s protocol, and the colorimetric output was read using a Bio-Rad microplate reader.
Western blotting
Western blotting was performed as previously described [40]. Primary antibodies (anti-PAR rabbit polyclonal [Trevigen]; anti-caspase 3 rabbit polyclonal [Cell Signaling]; anti-β actin rabbit polyclonal [Cell Signaling]; anti-γH2AX mouse monoclonal [Upstate]) were diluted at 1:1000 in blocking buffer and incubated with membranes overnight at 4°C while rocking. Horse radish peroxidase conjugated secondary antibodies (sheep-anti-mouse IgG and donkey-anti-rabbit IgG [GE Healthcare]) were diluted at 1:5000 in blocking buffer and incubated with membranes for 1 hour at room temperature while rocking. Proteins were visualized using Amersham enhanced chemiluminescence (ECL; GE Healthcare), exposed on BioMax XAR Film (Kodak), developed, and scanned using a Bio-Rad GS800 densitometer.
Statistical Analyses
MTT assays were analyzed using Microplate Manager version 5.2.1 (Bio-Rad). Western blot films were analyzed using Quantity One version 4.5.0 (Bio-Rad). Effective dose curves, isobolograms, IC50 values, and combinational indices were determined using CalcuSyn version 2.1 (Biosoft). Figures and statistical analyses were generated in CalcuSyn and GraphPad Prism 4. The in vitro data were analyzed using two-tailed t tests.
RESULTS
Promoter methylation analysis of DNA repair genes and reduced BRCA1 mRNA expression in CLL
To determine if epigenetic silencing of genes involved in DNA repair can underlie sensitivity to DNA damaging therapies, we investigated the promoter CpG island methylation status of BRCA1 using qMSP and BRCA2, FANC-C, FANC-L, FANC-F, ATM, MGMT, MLH1 and H2AX using MSP in a series of CLL patient samples. We observed no significant frequency of methylation of most of the DNA repair genes (Table 1). However, two CLL samples had detectable BRCA1 hypermethylation, but at levels lower than previously observed in breast and ovarian cancer (0.07, 1.47%) as determined by qMSP (Figure 1). BRCA1 expression was normalized to PCNA, a cell cycle marker, as the expression of BRCA1 is cell cycle dependent. While the expression of BRCA1 was reduced in both samples with DNA methylation, we found that all samples in this small cohort of CLL samples investigated for BRCA1 mRNA showed significantly reduced expression (60–80% reduction) compared to normal peripheral blood mononuclear cells (Figure 1). The consistency across the limited samples we analyzed likely indicates that reduced BRCA1 expression is not a rare occurrence in CLL.
Table 1.
Methylation analysis of CLL patient samples.
| Gene | BRCA1 | BRCA2 | FANC-C | FANC-F | FANC-L | ATM | MGMT | hMLH1 | H2AX | |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Methylated Samples | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Total Samples | 26 | 16 | 20 | 26 | 21 | 22 | 24 | 26 | 26 | |
|
| ||||||||||
| Freq. of Methylation | 7.69% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
The combination of CEP-8983 and bendamustine results in synergistic cytotoxicity in a significant proportion of CLL primary patient samples in vitro
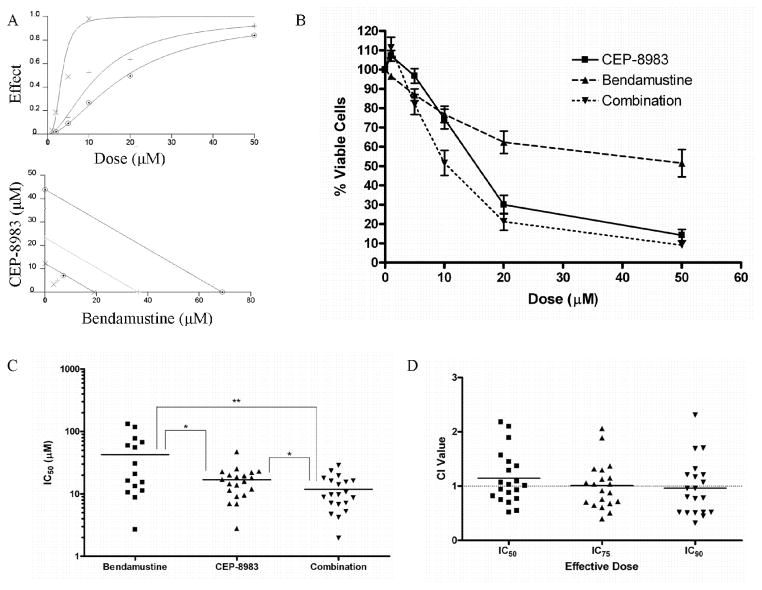
In order to explore the in vitro activity of PARP inhibitor in CLL cells, we investigated sensitivity to the PARP inhibitor CEP-8983 alone and in combination with the alkylating agent bendamustine in primary patient CLL samples. A total of 26 CLL samples were used for this study (Supplementary Table S1). Twenty-two of the samples (20 unique patients) were incubated for 72 hours with increasing doses of CEP-8983, bendamustine, and a 1:1 combination of CEP-8983 and bendamustine, and then analyzed for cytotoxic effect using an MTT assay. Dose response curves for each treatment were generated for each of the samples (Figure 2A, B). The mean IC50 values were 16.84, 42.53, and 11.85 μM for CEP-8983, bendamustine, and the combination, respectively (Figure 2C). CEP-8983 and bendamustine each sensitized the CLL cells to the other’s cytotoxic effects in vitro, as the mean IC50 of the combination was significantly lower than the mean IC50 values for each monotherapy (p<0.05; Figure 2C). The most sensitive samples for each treatment had IC50 values in the low single-digit micromolar range. Five samples, including one previously treated with bendamustine, one trisomy 12, one 11q deleted, and two 17p deleted, were resistant to bendamustine in our assay, as defined by clearly unachievable IC50 values (e.g. in the 1000s of μM). These values were excluded from statistical analyses. However, none of the samples were resistant (as defined by ability to calculate combined IC50 below 50uM) to the combination suggesting the possibility of this drug combination may overcome therapeutic bendamustine resistance of CLL cells.
Figure 2. Cytotoxicity analysis of CLL primary patient samples.
(A) Dose-effect curve (⊙=bendamustine, +=CEP-8983, ×=combination at 1:1 ratio). Bottom graph: Isobologram of combination (lower line=90% inhibitory concentration (IC90), middle line=IC75, upper line=IC50). The markings ×, +, and ⊙, represent a measure of the drug interactions at the IC50, IC75, and IC90, respectively. (B) Composite dose curve depicting % viable cells (relative to untreated) after treatment with CEP-8983, bendamustine, and combination at a 1:1 ratio. (C) Plot depicting the IC50 values of CLL primary patient samples treated with CEP-8983, bendamustine, and combination. Mean IC50 values were 16.84, 42.53, and 11.85 μM for CEP-8983, bendamustine, and the combination, respectively (*p<0.05, **p<0.01). (D) Plot depicting the combinational index (CI) of CLL samples at the IC50, IC75, and IC90. Values below 1 are synergistic. Mean CI values are 1.146, 1.010, and 0.9650 at the IC50, IC75, and IC90, respectively.
To further quantify drug interactions in these samples we used the Chou-Talalay method to calculate a combinational index (CI) value indicative of synergistic (0<CI<1), additive (CI=1–1.1), or antagonistic (CI>1.5) cytotoxic effects at the IC50, IC75, and IC90 [41]. A qualitative isobologram of drug interactions (Figure 2A), as well as quantitative CI values, were generated for each sample. The mean CI values were close to 1 at all effective doses (1.146, 1.010, and 0.9650 at the IC50, IC75, and IC90, respectively; Figure 2D). Importantly, 45%, 50%, and 60% of the patient samples analyzed displayed synergistic interactions between the drugs at the IC50, IC75, and IC90, respectively (Figure 2D). Samples that did not display synergy using this calculation generally had CI values below 1.5, indicating additive effects or only minor antagonism, if any. The combination of CEP-8983 and bendamustine resulted in synergistic cytotoxicity in a significant proportion of CLL primary patient samples in vitro. However, it is also evident that there is a range of sensitivity and synergistic interactions across the general CLL patient population.
Sensitivity to CEP-8983, alone and in combination with bendamustine, and synergistic interactions are consistent across genetic and cytogenetic subtypes of CLL, including previously treated patients
In order to understand the differential sensitivity to CEP-8983, alone and in combination with bendamustine, and their synergistic effects, we stratified the 20 unique patients who were analyzed by the MTT assays described above into previously established genetic and cytogenetic subgroups. We defined groups based on IgVH mutation status and cytogenetic abnormalities identified by FISH (e.g. trisomy 12, deletion 11q, deletion 13q, deletion 17p). 39%, 25%, 20%, 40%, and 35% of patient samples were positive for IgVH mutation, trisomy 12, deletion 11q, deletion 13q, and deletion 17p, respectively (Supplementary Table S1). Comparison of the IC50 and CI values for CEP-8983 alone and in combination with bendamustine across these subtypes revealed no significant differences (Supplementary Fig. 2A, 2B). Further analysis of the number of FISH abnormalities (data not shown) and prior treatment (Supplementary Figure 3A, B) revealed no significant differences between groups. Patients who had been treated with bendamustine prior to this analysis had significantly higher bendamustine IC50 values in vitro compared to bendamustine naïve patient samples (p<0.05; Supplemental Figure 3C). Notably, this combination has in vitro activity in 17p deleted CLL patients (Supplemental Figure 2A, B), who are known to have worse prognoses and therapeutic resistance, as well as previously treated patients (Supplemental Figure 3A, B).
Investigation into the mechanism of action of CEP-8983 and bendamustine in B cell malignancies
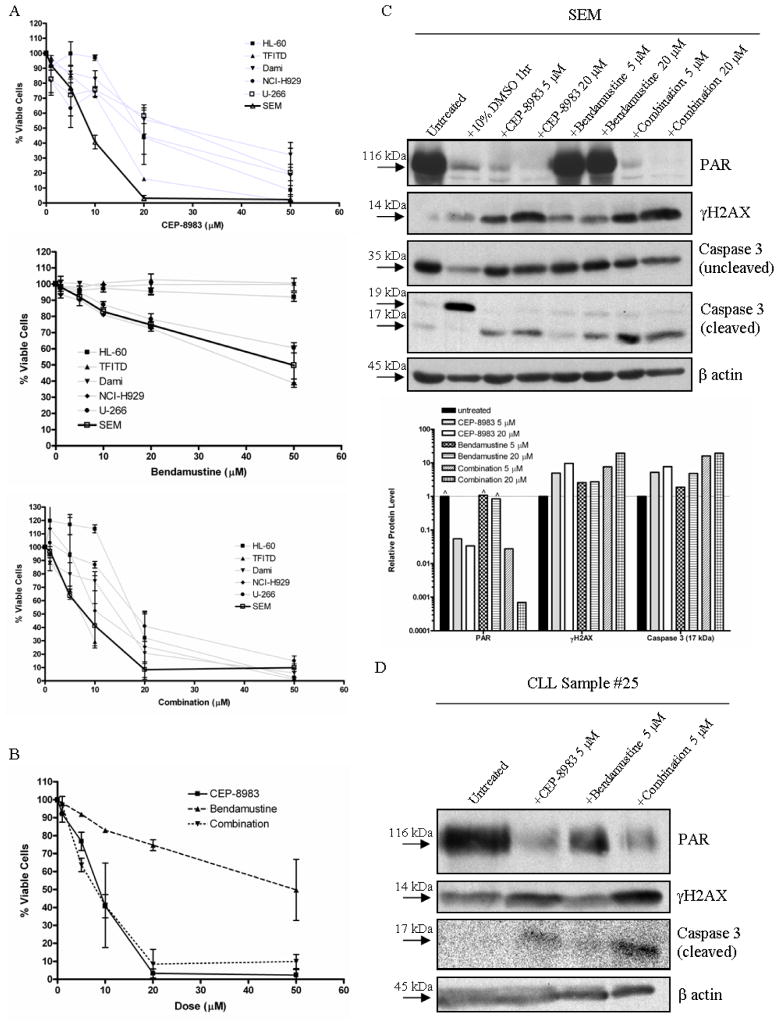
Due to the lack of a standard model cell line for CLL, we investigated the mechanism of action of CEP-8983 alone and in combination with bendamustine in other lymphoid and myeloid cell lines. We examined general cytotoxicity due to CEP-8983 in 7 cell lines with or without known p53 mutations, which revealed moderate IC50 values in the mid- double digit micromolar range for most cell lines examined (Figure 3A). We examined the mechanisms of action of CEP-8983 and bendamustine in the p53 WT established B cell precursor acute leukemia cell line SEM since it was the most sensitive cell line we tested (Figure 3A).
Figure 3. Responses of cell lines and CLL samples to CEP-8983, bendamustine and combination of both drugs.
(A) The acute myeloid leukemia (AML) cell lines with p53 mutations HL-60, TFITD, and Dami, the multiple myeloma cell lines U-266(p53 mut) and NCI-H929(p53 WT), and the B cell precursor leukemia cell line SEM(p53 WT) were treated with increasing doses (0–50 μM) of CEP-8983 (top graph), bendamustine (middle graph), and a 1:1 combination of the two drugs (bottom graph) for 72 hours. The cytotoxicity/% viability (relative to untreated) was analyzed using the MTT assay. SEMs were the most sensitive to CEP-8983 and the combination. The dose curves for the three B cell cancer cell lines treated with bendamustine are similar and significantly different from the three AML cell lines (p<0.05). (B) Composite of SEM dose curves. (C) Western blot analysis of SEM cells treated with CEP-8983, bendamustine, and the combination, all at two different dose levels (5 and 20 μM) for 72 hours. The 10% DMSO treatment represents a positive control for apoptosis. The graph below the blots is a log scale representation of the relative protein levels, determined by their optical density in the blot, and normalized to β actin levels. Quantification allows the categorization of synergistic interactions, including synergistic induction of DSBs, as shown by γH2AX levels, and synergistic induction of apoptosis, as shown by cleaved caspase 3 levels (^ indicates saturation of densitometer). (D) Western blot analysis of a chronic lymphocytic leukemia (CLL) primary patient sample treated with CEP-8983, bendamustine, and the combination, all at 5 μM for 72 hours. The results mimic those seen in SEM.
MTT assays of SEM revealed a dose-dependent decrease in cell viability, with the majority of the cytotoxic effect caused by CEP-8983 and the combination with bendamustine occurring between 0–20 μM in vitro (Figure 3B). Notably, dose-dependent bendamustine cytotoxicities in SEM and the two multiple myeloma cell lines NCI-H929(p53WT) and U-266 (p53 mut) were significantly higher than the acute myeloid leukemia (AML) cell lines HL-60, TF-ITD (TF-1 with FLT3 ITD), and Dami (p<0.05; Figure 3A) which all have a p53 mutation.
To determine in more detail the cellular and molecular mechanisms involved in CEP-8983-mediated cytotoxity in SEM, we examined target proteins via western blotting after in vitro exposure to drug. As expected, CEP-8983 treatment resulted in a dose-dependent inhibition of PAR levels (Figure 3C), consistent with its proposed mechanism of action. The reduction in PAR correlated well with cytotoxicity in the MTT assay, with >90% inhibition achieved at the IC50 (Figure 3B, C). CEP-8983 treatment also resulted in a dose-dependent increase in DNA damage (i.e. toxic DSBs), as shown by increased γH2AX protein levels, as well as apoptosis, as shown by increased cleaved caspase 3 protein levels (Figure 3C). Bendamustine treatment also resulted in increased DSBs and apoptosis, but not to the same extent as equimolar CEP-8983 (Figure 3C). The combination of CEP-8983 and bendamustine resulted in reduction in PAR levels similar to CEP-8983 monotherapy, even in the presence of the DNA damaging activity of bendamustine (Figure 3C). Importantly, the combination resulted in synergistic induction of DSBs and apoptosis as shown by nonlinear increases in γH2AX and cleaved caspase 3 protein levels, respectively (Figure 3C). Together, these results indicate the expected mechanism of action of CEP-8983 and reveal toxic synergistic interactions with bendamustine at the molecular level in a B cell malignancy.
We next performed a similar protein target assessment in two primary CLL samples to corroborate the results obtained in SEM cells. We found similar but less drastic reductions in PAR levels with CEP-8983 and the combination with bendamustine in both samples. We further assessed one sample for DNA damage and apoptosis, which revealed synergistic induction of DSBs and apoptosis upon treatment with the drug combination (Figure 3D). In summary, our primary CLL samples responded in a manner similar to SEM, suggesting a common mechanism of action.
DISCUSSION
We have reported several novel and important findings from our studies: 1) Promoter hypermethylation is not a common cause of DNA repair deficiencies in CLL 2)Reduced expression of BRCA1 mRNA appears to be a common occurrence in CLL; 3) the combination of CEP-8983 and bendamustine induces synergistic cytotoxicity in a significant portion of CLL patient samples, with a range of sensitivity and synergy.
Unfortunately, promoter hypermethylation is not a common finding in the nine genes we examined but two cases were found to have BRCA1 promoter hypermethylation. The lower level of hypermethylation seen in our two CLL samples as compared to solid tumor models may indicate only the most extreme form of BRCA1 inactivation, which may involve chromatin modifications which repress BRCA1 expression in CLL as we have seen for CTNNA1 in AML [42]. Despite infrequent promoter methylation changes, reduced expression of BRCA1 at the level of mRNA appears more common in CLL and is due to unknown causes. Future chromatin immunoprecipitation studies could provide greater insight into this form of epigenetic silencing, which in viable samples could be linked to defects in homologous recombination using Rad51/γH2AX foci assays. Nonetheless, the identification of this aberration, in conjunction with previous reports of DNA repair defects in CLL, strengthens the rationale for using PARP inhibitors and DNA damaging agents as targeted therapies for CLL.
The only previous study looking at PARP inhibition in CLL identified ATM deficiency as a determinant of sensitivity to the PARP inhibitor olaparib in CLL cells [43]. Additionally, the authors showed that synergy between olaparib and bendamustine could be observed in the ATM mutant mantle cell lymphoma cell line Granta-519 [43]. Our study extends these observations and is the most extensive study of a PARP inhibitor in combination with a DNA damaging agent in CLL primary patient samples to date. The IC50 values and cytotoxic responses to bendamustine in the MTT assay were similar to those expected for CLL samples[44]. Moreover, our results indicate that sensitivity to CEP-8983 and synergy with bendamustine are likely independent of ATM/11q status, and in contrast, span all genetic and cytogenetic subtypes. Exploiting this synergistic sensitization may increase the therapeutic index of these drugs by allowing lower doses of each to be given compared to monotherapy.
In conclusion, our data provide support for the study of the combination of a PARP inhibitor and bendamustine to the clinical setting, for use in CLL and possibly other B cell neoplasms. Early phase clinical trials will help determine the in vivo activity and interactions of the drugs, giving us a better indication of their utility for altering disease course of patients with CLL. Additionally, correlative studies, such as evaluation of DNA repair genes and their functionality, as well as PAR levels, of patients treated with this combination will help facilitate the discovery of suitable biomarkers of response.
Supplementary Material
Acknowledgments
The authors would like to thank Mark Levis for providing reagents and laboratory space and Ivan Borrello for providing U-266 and NCI-H929 cell lines.
Funding Support
D.E.G. and K.W.P. received research funding from Cephalon Inc.
J.E.K, J.E.H & K.W.P are members of the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins which is supported from the NIH core grant P30 CA006973
Footnotes
Authors Contributions
R.E.D, W.P, J.E.H, M.A.M, K.W.P, participated in design of the study, acquisition of the data, analysis and interpretation of the data, and writing and revising the manuscript. J.E.K, M.M.S, D.E.G. participated in acquisition of biospecimens, design of the study, and revision of the manuscript.
Potiential Conflicts of Interest
D.E.G. and K.W.P received research funding from Cephalon Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rozman C, Montserrat E. Chronic lymphocytic leukemia. The New England journal of medicine. 1995;333:1052–7. doi: 10.1056/NEJM199510193331606. [DOI] [PubMed] [Google Scholar]
- 2.Gribben JG, O’Brien S. Update on therapy of chronic lymphocytic leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:544–50. doi: 10.1200/JCO.2010.32.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rummel MJ, Gregory SA. Bendamustine’s emerging role in the management of lymphoid malignancies. Semin Hematol. 2011;48 (Suppl 1):S24–36. doi: 10.1053/j.seminhematol.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Leoni LM, Bailey B, Reifert J, Bendall HH, Zeller RW, Corbeil J, et al. Bendamustine (Treanda) displays a distinct pattern of cytotoxicity and unique mechanistic features compared with other alkylating agents. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:309–17. doi: 10.1158/1078-0432.CCR-07-1061. [DOI] [PubMed] [Google Scholar]
- 5.Leoni LM, Hartley JA. Mechanism of action: the unique pattern of bendamustine-induced cytotoxicity. Semin Hematol. 2011;48 (Suppl 1):S12–23. doi: 10.1053/j.seminhematol.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Bergmann MA, Goebeler ME, Herold M, Emmerich B, Wilhelm M, Ruelfs C, et al. Efficacy of bendamustine in patients with relapsed or refractory chronic lymphocytic leukemia: results of a phase I/II study of the German CLL Study Group. Haematologica. 2005;90:1357–64. [PubMed] [Google Scholar]
- 7.Knauf WU, Lissichkov T, Aldaoud A, Liberati A, Loscertales J, Herbrecht R, et al. Phase III randomized study of bendamustine compared with chlorambucil in previously untreated patients with chronic lymphocytic leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:4378–84. doi: 10.1200/JCO.2008.20.8389. [DOI] [PubMed] [Google Scholar]
- 8.Sampath D, Plunkett W. The role of DNA repair in chronic lymphocytic leukemia pathogenesis and chemotherapy resistance. Current oncology reports. 2007;9:361–7. doi: 10.1007/s11912-007-0048-6. [DOI] [PubMed] [Google Scholar]
- 9.Fenaux P, Preudhomme C, Lai JL, Quiquandon I, Jonveaux P, Vanrumbeke M, et al. Mutations of the p53 gene in B-cell chronic lymphocytic leukemia: a report on 39 cases with cytogenetic analysis. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 1992;6:246–50. [PubMed] [Google Scholar]
- 10.Garcia-Marco JA, Caldas C, Price CM, Wiedemann LM, Ashworth A, Catovsky D. Frequent somatic deletion of the 13q12.3 locus encompassing BRCA2 in chronic lymphocytic leukemia. Blood. 1996;88:1568–75. [PubMed] [Google Scholar]
- 11.Watanabe T, Hotta T, Ichikawa A, Kinoshita T, Nagai H, Uchida T, et al. The MDM2 oncogene overexpression in chronic lymphocytic leukemia and low-grade lymphoma of B-cell origin. Blood. 1994;84:3158–65. [PubMed] [Google Scholar]
- 12.Schaffner C, Stilgenbauer S, Rappold GA, Dohner H, Lichter P. Somatic ATM mutations indicate a pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood. 1999;94:748–53. [PubMed] [Google Scholar]
- 13.Christodoulopoulos G, Malapetsa A, Schipper H, Golub E, Radding C, Panasci LC. Chlorambucil induction of HsRad51 in B-cell chronic lymphocytic leukemia. Clinical cancer research: an official journal of the American Association for Cancer Research. 1999;5:2178–84. [PubMed] [Google Scholar]
- 14.Bello VE, Aloyz RS, Christodoulopoulos G, Panasci LC. Homologous recombinational repair vis-a-vis chlorambucil resistance in chronic lymphocytic leukemia. Biochemical pharmacology. 2002;63:1585–8. doi: 10.1016/s0006-2952(02)00954-1. [DOI] [PubMed] [Google Scholar]
- 15.Jones GG, Reaper PM, Pettitt AR, Sherrington PD. The ATR-p53 pathway is suppressed in noncycling normal and malignant lymphocytes. Oncogene. 2004;23:1911–21. doi: 10.1038/sj.onc.1207318. [DOI] [PubMed] [Google Scholar]
- 16.Stankovic T, Stewart GS, Fegan C, Biggs P, Last J, Byrd PJ, et al. Ataxia telangiectasia mutated-deficient B-cell chronic lymphocytic leukemia occurs in pregerminal center cells and results in defective damage response and unrepaired chromosome damage. Blood. 2002;99:300–9. doi: 10.1182/blood.v99.1.300. [DOI] [PubMed] [Google Scholar]
- 17.Xu ZY, Loignon M, Han FY, Panasci L, Aloyz R. Xrcc3 induces cisplatin resistance by stimulation of Rad51-related recombinational repair, S-phase checkpoint activation, and reduced apoptosis. J Pharmacol Exp Ther. 2005;314:495–505. doi: 10.1124/jpet.105.084053. [DOI] [PubMed] [Google Scholar]
- 18.Muller C, Christodoulopoulos G, Salles B, Panasci L. DNA-Dependent protein kinase activity correlates with clinical and in vitro sensitivity of chronic lymphocytic leukemia lymphocytes to nitrogen mustards. Blood. 1998;92:2213–9. [PubMed] [Google Scholar]
- 19.Deriano L, Guipaud O, Merle-Beral H, Binet JL, Ricoul M, Potocki-Veronese G, et al. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood. 2005;105:4776–83. doi: 10.1182/blood-2004-07-2888. [DOI] [PubMed] [Google Scholar]
- 20.Dantzer F, Schreiber V, Niedergang C, Trucco C, Flatter E, De La Rubia G, et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- 21.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 23.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–7. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 24.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dedes KJ, Wetterskog D, Ashworth A, Kaye SB, Reis-Filho JS. Emerging therapeutic targets in endometrial cancer. Nat Rev Clin Oncol. 2011;8:261–71. doi: 10.1038/nrclinonc.2010.216. [DOI] [PubMed] [Google Scholar]
- 26.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. The New England journal of medicine. 2009;361:123–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 27.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–51. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 28.Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 29.Miknyoczki S, Chang H, Grobelny J, Pritchard S, Worrell C, McGann N, et al. The selective poly(ADP-ribose) polymerase-1(2) inhibitor, CEP-8983, increases the sensitivity of chemoresistant tumor cells to temozolomide and irinotecan but does not potentiate myelotoxicity. Molecular cancer therapeutics. 2007;6:2290–302. doi: 10.1158/1535-7163.MCT-07-0062. [DOI] [PubMed] [Google Scholar]
- 30.Ator MBR, Chatterjee S, Dunn D, Hudkins R. Novel multicyclic compounds and their use thereof. 2001 [Google Scholar]
- 31.Scardocci A, Guidi F, D’Alo F, Gumiero D, Fabiani E, Diruscio A, et al. Reduced BRCA1 expression due to promoter hypermethylation in therapy-related acute myeloid leukaemia. British journal of cancer. 2006;95:1108–13. doi: 10.1038/sj.bjc.6603392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fulop Z, Csernus B, Timar B, Szepesi A, Matolcsy A. Microsatellite instability and hMLH1 promoter hypermethylation in Richter’s transformation of chronic lymphocytic leukemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2003;17:411–5. doi: 10.1038/sj.leu.2402792. [DOI] [PubMed] [Google Scholar]
- 33.Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10:1192–9. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drew Y, Mulligan EA, Vong WT, Thomas HD, Kahn S, Kyle S, et al. Therapeutic potential of poly(ADP-ribose) polymerase inhibitor AG014699 in human cancers with mutated or methylated BRCA1 or BRCA2. Journal of the National Cancer Institute. 2011;103:334–46. doi: 10.1093/jnci/djq509. [DOI] [PubMed] [Google Scholar]
- 35.Veeck J, Ropero S, Setien F, Gonzalez-Suarez E, Osorio A, Benitez J, et al. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:e563–4. doi: 10.1200/JCO.2010.30.1010. author reply e5–6. [DOI] [PubMed] [Google Scholar]
- 36.Miknyoczki SJ, Pritchard S, McGann N, Grobelny J, Burns C, Diebold J, et al. The selective PARP inhibitor, CEP-8983 exhibits significant chemosensitization in combination with TMZ and SN38 against chemoresistant tumor cell lines and xenografts. AACR Meeting Abstracts. 2005;2005:1206-c-7. [Google Scholar]
- 37.Ogura M, Uchida T, Taniwaki M, Ando K, Watanabe T, Kasai M, et al. Phase I and pharmacokinetic study of bendamustine hydrochloride in relapsed or refractory indolent B-cell non-Hodgkin lymphoma and mantle cell lymphoma. Cancer Science. 2010;101:2054–8. doi: 10.1111/j.1349-7006.2010.01633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brandes JC, Carraway H, Herman JG. Optimal primer design using the novel primer design program: MSPprimer provides accurate methylation analysis of the ATM promoter. Oncogene. 2007;26:6229–37. doi: 10.1038/sj.onc.1210433. [DOI] [PubMed] [Google Scholar]
- 40.Pratz KW, Cortes J, Roboz GJ, Rao N, Arowojolu O, Stine A, et al. A pharmacodynamic study of the FLT3 inhibitor KW-2449 yields insight into the basis for clinical response. Blood. 2009;113:3938–46. doi: 10.1182/blood-2008-09-177030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer research. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 42.Ye Y, McDevitt MA, Guo M, Zhang W, Galm O, Gore SD, et al. Progressive chromatin repression and promoter methylation of CTNNA1 associated with advanced myeloid malignancies. Cancer research. 2009;69:8482–90. doi: 10.1158/0008-5472.CAN-09-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–87. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 44.Schwanen C, Hecker T, Hubinger G, Wolfle M, Rittgen W, Bergmann L, et al. In vitro evaluation of bendamustine induced apoptosis in B-chronic lymphocytic leukemia. Leukemia. 2002;16:2096–105. doi: 10.1038/sj.leu.2402651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.