Abstract
The envelope (E) protein is composed of three domains (ED1, ED2 and ED3) with ED3 targeted by the most potent neutralizing antibodies. DENV-2 strains can be divided into six genotypes. Comparison of ED3 of representative strains of the six genotypes revealed that there are nine variable residues that are specific to a given genotype. Recombinant ED3s (rED3s) of six different DENV-2 strains representing all nine variable residues were expressed, and their reactivity against a panel of two DENV-2 type-specific and three DENV complex-reactive monoclonal antibodies (mAbs) were compared. The differences in binding affinity to the rED3s representing different DENV-2 genotypes were relatively small, with the exception of type-specific-mAb 3H5 that showed up to 10-fold differences in binding between genotypes. Overall the binding differences did not lead to detectable differences in neutralization. Based on these results, DENV-2 ED3-specific neutralizing antibodies will likely be effective against DENV-2 strains from all six genotypes.
Keywords: antigenic sites, DENV-2 genotypes, antibody-mediated neutralization
Introduction
The disease dengue (DEN) is caused by four mosquito-borne, genetically and serologically related viruses, termed dengue virus 1 (DENV-1), DENV-2, DENV-3, and DENV-4. The DENVs are members of the genus Flavivirus, family Flaviviridae. There are approximately 100 million DENV infections annually, with 2.1 million cases of severe disease manifestations, including dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) (Kyle and Harris, 2008). The geographic range of the DENVs encompasses most of the tropical and subtropical regions of the world, putting approximately 3 billion people at risk for infection and this places a large burden on public health infrastructure (Kyle and Harris, 2008). DENVs are maintained in nature by two transmission cycles, the sylvatic cycle (occurring between forest dwelling mosquitoes and non-human primates) and the urban cycle (occurring between the primary vectors Aedes aegypti and Aedes albopictus and humans). Studies by Sabin have shown that human volunteers previously infected with DENV-1 or DENV-2 had short term herterotypic immunity to infection by both viruses, followed by long-term homotypic immunity to the original infecting DENV (Sabin, 1952).
Even though the four DENVs cause very similar disease manifestations, they are quite genetically diverse, having about 40% amino acid sequence divergence. Each DENV can be further subdivided into specific genotypes, which can have up to 6% genetic divergence (Twiddy et al., 2002). Specifically, six DENV-2 genotypes have been identified: American, American/Asian, Asian I, Asian II, cosmopolitan, and sylvatic (Twiddy et al., 2002). There is evidence to suggest that the DENV genotypes have different phenotypes and disease outcomes, which has become an area of great interest (Leitmeyer et al., 1999; Rico-Hesse et al., 1997). Limited studies suggest that the Asian genotypes are epidemiologically associated with greater disease outcomes (i.e. DHF/DSS), whereas the American genotype has been considered less virulent, since it is associated primarily with the less severe dengue fever (Rico-Hesse et al., 1997; Watts et al., 1999).
The DEN virion is enveloped, 50 nM in diameter, and contains a single-stranded, positive-sense RNA genome that is approximately 11kb in length (Chambers et al., 1990). The genome encodes one open reading frame that is translated into a single polyprotein that is co-and post-translationally processed by host and viral proteases to yield three structural proteins (capsid (C), pre-membrane/ membrane (prM/M), and envelope (E)) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Rice et al., 1985). Ninety, anti-parallel E protein homodimers are arranged on the surface of the viral envelope in a herringbone fashion that lacks traditional T=3 symmetry (Kuhn et al., 2002). The molecular arrangement of the E proteins on the viral surface leads to three chemically distinct environments at the two-, three-, and five-fold axes of symmetry that have been shown to be important for antibody binding (Kaufmann et al., 2006; Kuhn et al., 2002; Lok et al., 2008; Nybakken et al., 2005).
The E protein plays multiple roles in the virus life cycle, which include receptor binding, entry and fusion with the endosomal membrane. The E protein is composed of three ectodomains designated domain 1 (ED1), ED2 and ED3. ED1 is the central domain connecting ED2 and ED3, ED2 is the dimerization domain and also contains the highly conserved fusion loop (Allison et al., 2001; Rey et al., 1995), and ED3 is thought to be the receptor-binding domain. Evidence in support of ED3 as the receptor binding domain include the fact that ED3 protrudes farthest from the viral surface and soluble forms of ED3 block infection (by potentially competing for the receptor), as well as ED3-specific antibodies can neutralize virus infectivity (Chin, Chu, and Ng, 2007; Crill and Roehrig, 2001; Hung et al., 2004; Kuhn et al., 2002; Rey et al., 1995). At least twelve major antigenic sites have been identified on the DENV-2 E protein (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008; Roehrig, Bolin, and Kelly, 1998; Sukupolvi-Petty et al., 2007). There are two overlapping antigenic sites located on the surface of ED3; one is a DENV-2 type-specific antigenic site and the other is a DENV complex-reactive antigenic site (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008; Roehrig, Bolin, and Kelly, 1998; Sukupolvi-Petty et al., 2007). Evidence suggests that the majority of antibodies produced during infection target epitopes in proximity to the fusion loop at the distal end of ED2 (Crill and Roehrig, 2001; Throsby et al., 2006), however, those that bind ED3 are the most potent at neutralizing virus ((Roehrig, Bolin, and Kelly, 1998). The mechanism(s) by which flaviviruses are neutralized by antibody are not well understood, but it is thought that neutralization occurs by potentially blocking receptor binding, preventing endocytosis, inhibiting fusion with the endosome, or a combination of these mechanisms (Pierson et al., 2008).
The objective of the current study was to determine if amino acid sequence variation within ED3 among different DENV-2 genotypes has an effect on the affinity and neutralization efficacy of DENV-2 type-specific and DENV complex-reactive ED3-specific mAbs.
Results
Amino acid variation among the DENV-2 genotypes
DENV-2 amino acid diversity was determined by aligning the E protein sequences from 84 representative strains (downloaded from Genbank). Variable residues among genotypes were defined as amino acid substitutions that occurred in at least half of the representative strains composing a particular genotype. The alignment revealed that there are 31 variable residues in the E protein, nine of which were located in ED3: V308I, I322V, G330D, R345K, V365I, I378V, I379V, and N390D or N390S (Figure 1A). Six of these variable residues are surface exposed (V308I, G330D, R345K, H346Y, I379V, and N390D/S); and three variable residues have conservative substitutions (V308I, R345K, and I379V) (Figure 1B). The remaining three residues are non-conservative substitutions including G330D (found in the sylvatic genotype only), H346Y, (identified in half of the Asian I genotype isolates), and the N390S (cosmopolitan genotype) or N390D (American genotype). These variable residues were not critical for the binding of the DENV-2 type-specific or DENV complex-reactive antibodies in previous studies (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008; Sukupolvi-Petty et al., 2007).
Figure 1.
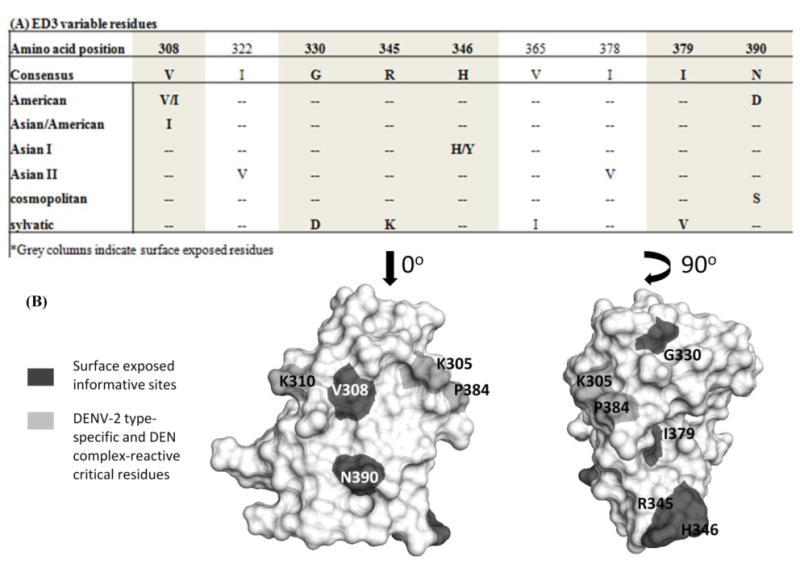
ED3 of different DENV-2 genotypes. Panel A: Alignment representing the ED3 informative sites, the residues that are bolded are surface exposed. Panel B: Structure of ED3 with the type-specific and complex-reactive critical residues highlighted light gray, and the surface exposed informative sites highlighted in dark gray.
Six DENV-2 strains from four different genotypes (Asian II, cosmopolitan, sylvatic and American) were chosen to evaluate if variable residues between genotypes and/or virus strain specific variation, within ED3, affected antibody binding affinity and virus neutralization (virus passage history shown in table 1). We did not investigate Asian I or Asian/American genotype viruses because most of these viruses had either identical ED3 sequences or contained conservative substitutions in ED3 and were represented by strains already being used. The DENV-2 Asian II genotype virus strain, New Guinea C (NGC), served as a reference as it has been used in our previous studies (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008). Strain H8-2027 was chosen as the representative of the cosmopolitan genotype. Two sylvatic genotype virus strains (DakHD10674 and DakAr578) were chosen because they contain the most variable residues among DENV-2 genotypes. Finally, two American genotype virus strains (PR159 and SML6420) were chosen. The specific amino acid sequences from these strains are shown in Figure 2.
Table 1. DENV-2 Passage history.
| Straina | Host | Year isolated | Country of origin | Passage historyb |
|---|---|---|---|---|
| New Guinea C | Human | 1944 | New Guinea | SM p26, C6/36 p17 |
| H8-2027 | Mosquito | 1969 | Indonesia | Vero p1, C6/36 p3 |
| DaKHD10674 | Human | 1970 | Senegal | Vero p1, C6/36 p10 |
| DaKaRA578 | Mosquito | 1980 | Ivory Coast | SM p8, C6/36 p7 |
| PR159 | Human | 1969 | Puerto Rico | PGMK p6, C6/36 p7 |
| SML6420 | Human | 1994 | United States of America, Texas | C6/36 p11 |
DENV-2 strains were obtained from the University of Texas Medical Branch World Reference Center of for Emerging Virus and Arboviruses
SM = suckling mouse; C6/36 = Aedes albopictus cell line; Vero = African green monkey kidney cell line; PGMK = primary green monkey kidney cells; pX = times passaged
Figure 2.

Alignment of the ED3 amino acid sequences (E residues 294-400) of six representative DENV-2 strains from four genotypes: NGC (Asian II), H8-2027 (cosmopolitan), SML6420 (American), PR159 (American), DaKaR578 (sylvatic), and DaKHD10674 (sylvatic).
Effect of amino acid substitutions on physical binding by mAbs
The DENV-2 type-specific and DENV complex-reactive antigenic sites have been shown to each consist of overlapping epitopes (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008), therefore, a panel of five mAbs (two DENV-2 type-specific and three DENV complex-reactive antibodies) were used to evaluate if the variable residues within ED3 among DENV-2 genotypes affected mAb affinity. The eptiopes and neutralization properties of three of these mAbs (3H5, GTX29202 and 20-783-74014) were previously characterized using the DENV-2 Asian genotype II strain NGC (Gromowski and Barrett, 2007).
The amino acid variation among the six viruses tested in this study had the largest effects on the binding of the DENV-2 type-specific mAb 3H5, followed by mAb ICL2 (Table 2). The largest difference in the affinities of mAb 3H5 for rED3 were between DakHD10674 (KD= 0.5 ± 0.04nM), H8-2027 (KD=3.2 ± 0.3nM) and SML6420 (KD= 4.9 ± 0.5nM), including a 10-fold difference between DakHD10674 and SML6420. The differences among these viruses were shown to be statistically significant by one-way ANOVA (P < 0.05). For mAb ICL2, the largest difference in affinity was between DakHD10674 (0.7 ± 0.1nM) and PR159 (2.4 ± 0.3nM), a 3.4 fold difference, which was statistically significant (p < 0.05). The binding of the two DENV-2 type-specific mAbs (3H5, and ICL2) to rED3 from the six DENV-2 representatives followed a particular pattern; the KD of the two mAbs increased (lower affinity) when assayed with rED3 containing substitutions of N390D/S (Table 2).
Table 2. mAb dissociation constants (KD) for representative DENV-2 rED3s.
| DENV-2 Genotype |
DENV-2 Strains | Type-specific mAbs | Complex-reactive mAbs | |||
|---|---|---|---|---|---|---|
|
| ||||||
| 3H5 (nM) | ICL2 (nM) | 20-783-74014 (nM) | GTX29202 (nM) | MD-05-0104 (nM) | ||
| Asian II | New Guinea C | 1.2 ± 0.1 | 1.3 ± 0.1 | 0.46 ± 0.04 | 0.25 ± 0.02 | 0.46 ± 0.03 |
| cosmopolitan | H8-2027 | 3.2 ± 0.3 a | 2.1 ± 0.2 a | 0.30 ± 0.06a | 0.23 ± 0.02 | 0.52 ± 0.5 |
| sylvatic | DaKHD10674 | 0.5 ± 0.04 b | 0.67 ± 0.1a,b | 0.30 ± 0.05 | 0.20 ± 0.02 | 0.32 ± 0.03 |
| sylvatic | DaKaR578 | 1.2 ± 0.1 b | 1.0 ± 0.1 b | 0.28 ± 0.04 | 0.22 ± 0.02 | 0.34 ± 0.03 |
| American | SML6420 | 4.9 ± 0.5 a,b,c,d,f | 2.1 ± 0.3 a,c,d | 0.22 ± 0.03 a | 0.17 ± 0.02 | 0.21 ± 0.02 |
| American | PR159 | 2.6 ± 0.3 c | 2.4 ± 0.3 a,c,d | 0.35 ± 0.07 | 0.20 ± 0.03 | 0.52 ± 0.06 |
(significantly different from NGC)
(different from H8-2027)
(different from DaKHD10674)
(different from DAKAR578)
(different from SML6420)
(different from PR159)
The effects of the amino acid variation on the DENV complex-reactive antigenic site were also evaluated. Three DENV complex-reactive mAbs (20-783-74014, GTX29202 and MD-05-0104) were used to determine if the substitutions among the viruses altered the affinities of the mAbs for their epitopes. In comparison to the DENV-2 type-specific mAbs, the affinities of most of the DENV complex-reactive mAbs for the six rED3s were not significantly affected by the amino acid differences among the rED3s (Table 2). The affinities of mAbs GTX29202 and MD-05-0104 did not differ significantly for any of the rED3s. The mAb 20-783-74014 was the only DENV complex-reactive mAb whose affinities for the rED3s differed significantly; the largest difference occurred between NGC (KD = 0.46 ± 0.04 nM) and SML6420 (V308I and N390D, KD= 0.22 ± 0.03nM), a two-fold difference; as determined by one-way ANOVA (P <0.05), this did not occur with V308I alone (KD= 0.5 ± 0.05nM).
The effects of antibody affinity on neutralization
Since neutralization has been correlated with the affinity of an antibody for its epitope (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008), neutralization assays were used to determine if the differences in physical binding translated into biological differences in neutralization. Differences in neutralization between the genotype representatives were evaluated using plaque reduction neutralization tests (PRNT50) (Table 3). Analysis of mAb 3H5 neutralization showed that the PRNT50 concentrations for NGC, DakHD10674, H8-2027 and SML6420 viruses did not differ significantly. An example of a neutralization curve is shown in Figure 3A. Even though the physical binding (as determined by KD) of the mAb 3H5 for rED3 was different among the viruses tested, the biological affects as shown by PRNT50 concentrations were not statistically different. The Hill slopes for the PRNT50 with mAb 3H5 also did not differ significantly either, with 1.1 ± 0.1 for NGC, 2.0 ± 0.3 for DAKHD10674, 1.3 ± 0.2 for SML6420, and 1.1±0.2 for H8-2027. The similar shapes of the neutralization dose-response curves for these four viruses indicate that the efficacy of mAb 3H5 to neutralize these viruses is equivalent.
Table 3. Plaque reduction neutralization test (50%) values for representative DENV-2s.
| DENV-2 Genotype |
DENV-2 Strains | Type-specific mAbs | Complex-reactive mAbs | |||
|---|---|---|---|---|---|---|
|
| ||||||
| 3H5 (nM) |
ICL2 (nM) |
GTX29202 (nM) |
20-783-74014 (nM) |
MD-05-0104 (nM) |
||
| Asian II | New Guinea C | 1.1 ± 0.1 | 4.2 ± 0.6 | 3.3 ± 0.6 | 7.5 ± 1.8 | 10.5 ± 1.7 |
| cosmopolitan | H8-2027 | 2.7 ± 0.6 | 3.3 ± 0.8 | 8.0 ± 2.0 | 6.8 ± 1.1 | 6.9 ± 1.4 |
| sylvatic | DaKHD10674 | 1.7 ± 0.1 | 3.7 ± 0.8 | 4.3 ± 0.5 | 9.1 ± 1.3 | 11.7 ± 1.6 |
| American | SML6420 | 1.5 ± 0.2 | 3.5 ± 0.5 | 3.6 ± 1.2 | 3.1 ± 1.2 | 9.1 ± 2.6 |
Figure 3.
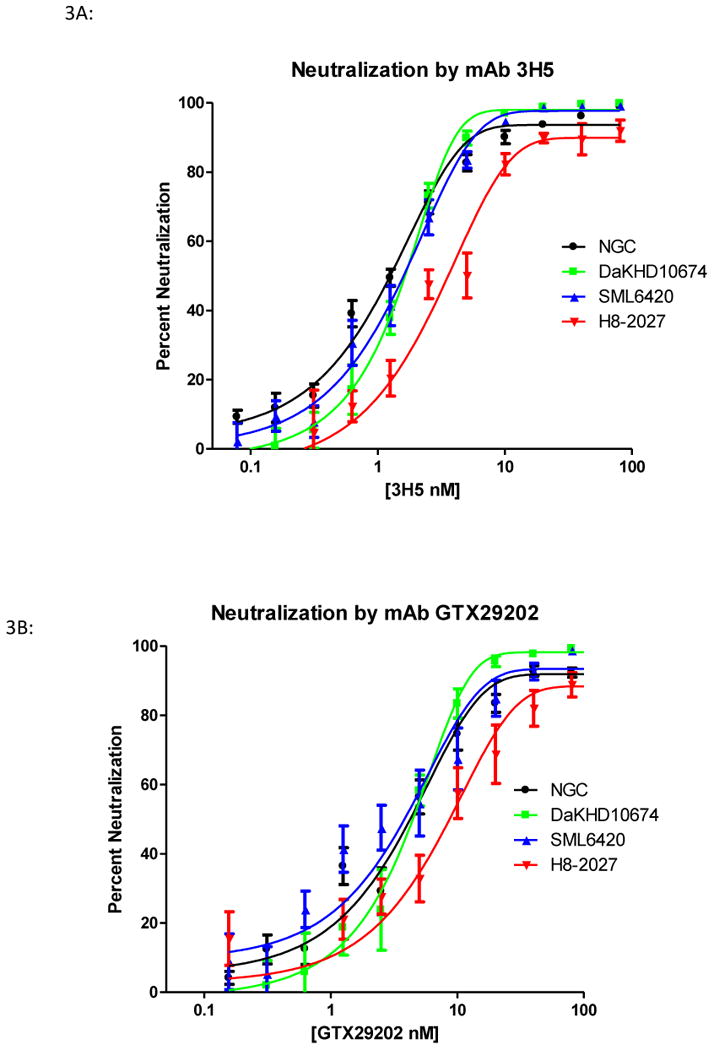
Neutralization of different DENV-2 genotype viruses by DENV-2 type specific and DENV complex reactive mAbs. Panel A: A representative neutralization curve of mAb 3H5 with NGC (● black), DakHD10674 (○ green), SML6420 (▲ blue), and H8-2027 (▼ red). Panel B: A representative neutralization curve with complex-reactive mAb GTX29202 with NGC (● black), DakHD10674 (○ green), SML6420 (▲ blue), H8-2027 (▼ red).
The neutralization data for the DENV complex-reactive mAbs yielded similar results to the DENV-2 type-specific mAbs (Table 3). All three DENV complex reactive mAbs had similar PRNT50 concentrations and Hill slopes. The neutralization curve for GTX29202 is shown in Figure 3B as an example. Even though the PRNT50 concentrations are not significantly different between the four viruses, the Hill-slopes for these viruses did differ significantly. The Hill-slopes for these viruses were 1.0 ± 0.6, 1.7 ± 0.3, 1.0 ± 0.2, and 0.7 ± 0.1 for mAb GTX29202 with NGC, DaKHD10674, and SML6420, respectively. The significant difference was between the Hill-slopes of strains DaKHD10674 and SML6420. As seen in Figure 3B, the neutralization curve for DaKHD10674 is steeper than the other two viruses, whereas both NGC and SML6420 have shallow neutralization curves indicating differences in mAb efficacy between DaKHD10674 and NGC and SML6420. The PRNT50 concentrations for each virus demonstrated an approximate 20-fold difference between the KD of the mAb for rED3 and the PRNT50 concentration, which was similar to that reported previously for DENV-2 strain NGC (Gromowski, Barrett, and Barrett, 2008).
Conservation of the DENV-2 type-specific and DENV complex-reactive critical residues in rED3
Since our panel of mAbs was able to bind and neutralize a diverse group of DENV-2 strains, we investigated whether or not the DENV-2 type-specific and DENV complex-reactive critical residues identified using strain NGC were also important for mAb binding to other DENV-2 viruses. We (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008) and others (Sukupolvi-Petty et al., 2007) have shown that a limited subset of residues are critically important for mAb binding to rED3. The critical residues identified were G304Y (Sukupolvi-Petty et al., 2007), K305A/E (Gromowski and Barrett, 2007; Sukupolvi-Petty et al., 2007), K307N/Q/I/E (Sukupolvi-Petty et al., 2007), K310E/A (Gromowski, Barrett, and Barrett, 2008; Sukupolvi-Petty et al., 2007), E383G (Gromowski and Barrett, 2007; Sukupolvi-Petty et al., 2007), P384A/N (Gromowski and Barrett, 2007; Sukupolvi-Petty et al., 2007). We chose to focus on the critical residues that are shared among mAbs studied thus far; K305A and P384A for the DENV-2 type-specific antigenic site and K310A for the DENV complex-reactive mAbs. We engineered each DENV rED3 used in this study with either K305A and P384A substitutions (DENV-2 type-specific antigenic site) or K310A substitution (DENV complex-reactive antigenic site). These proteins were used in ELISAs to determine if these critical residues are conserved among the four DENV-2 genotypes tested.
For the DENV-2 type-specific mAbs 3H5 and ICL2, regardless of the virus strain, residues K305 and P384 were critical for antibody binding (Table 4). The affinity of these mAbs for rED3 containing these substitutions was greatly reduced; the rED3s from NGC, DakHD10674, PR159, and SML6420 that contained K305A and P384A all resulted in affinity changes of greater than 50-fold, and for mAb ICL2 many of these substitutions resulted in essentially complete loss of binding to rED3. As expected, both of the DENV-2 type-specific mAbs were unaffected by the DENV complex-reactive epitope substitution of K310A in any of the different virus strains.
Table 4. relative dissociation constants of mutant rED3 as compared to the wild type rED3.
| rED3 | Type-specific mAbs | Complex-reactive mAbs | |||
|---|---|---|---|---|---|
|
| |||||
| 3H5 | ICL2 | 20-783-74014 | GTX29202 | MD-05-0104 | |
| NGC | 1 | 1 | 1 | 1 | 1 |
| NGC K305A + P384A | >50.0 | >50.0 | 3.5 | 4.8 | 5.2 |
| NGC K310A | 4.4 | 1.9 | 39 | >50.0 | >50.0 |
| DaKHD10674 | 1 | 1 | 1 | 1 | 1 |
| DaKHD10674 K305A + P384A | >50.0 | 46.9 | 1 | 1 | 0.8 |
| DaKHD10674 K310A | 1.4 | 1.9 | 9.7 | >50.0 | >50.0 |
| PR159 | 1 | 1 | 1 | 1 | 1 |
| PR159 K305A + P384A | >50.0 | >50.0 | 1 | 1.4 | 1.3 |
| PR159 K310A | 1.6 | 1.7 | >50.0 | >50.0 | >50.0 |
| SML6420 | 1 | 1 | 1 | 1 | 1 |
| SML6420 K305A + P384A | >50.0 | >50.0 | 1.7 | 1.3 | 1.5 |
| SML6420 K310A | 3.2 | 2.8 | >50.0 | >50.0 | >50.0 |
The binding of the DENV complex-reactive mAbs to the DENV-2 rED3 mutants followed a similar pattern with binding of mAbs greatly reduced for all of the rED3s containing the K310A substitution (Table 4). All three mAbs bound well to the wild-type rED3 proteins, and as expected binding were unaffected by the DENV-2 type-specific substitutions of K305A and P384A for all of the virus backgrounds. Only the DENV-2 NGC rED3 with the K305A and P384A substitutions showed a small change in affinity of 3.5- (mAb 20-783-74014) to 5.2- (mAb MD-05-0104) fold.
Discussion
The objective of this study was to assess the impact of the naturally occurring amino acid diversity in ED3 among DENV-2 genotypes on the physical binding of antibodies to ED3, and their subsequent biological activity of neutralization. In addition, another goal of this study was to determine if the DENV-2 type-specific and DEN complex-reactive antigenic sites were conserved among diverse DENV-2 genotypes. These questions were investigated using a panel of five mAbs (two DENV-2 type-specific and three DENV complex-reactive mAbs) that recognize two major overlapping antigenic sites on ED3, namely DENV-2 type-specific and DEN virus complex-reactive. In these assays we utilized representatives from four DENV-2 genotypes: Asian II (NGC), Cosmopolitan (H8-2027), sylvatic (DakHD10674 and DakAr578), and American (SML6420 and PR159). These six viruses represent most of the amino acid variability among the DENV-2 genotypes in ED3.
The impact of the amino acid variability among the six DENV-2s on the physical binding was evaluated by ELISA. These assays demonstrated that each mAb was able to bind rED3 regardless of the differing residue substitutions among the viruses. However, the amino acid substitutions among the viruses did alter the affinities of particular mAbs for rED3. Specifically, both DENV-2 type-specific mAbs were affected by the differing residue combinations more than the three DEN complex-reactive mAbs. The affinities of mAb 3H5 for almost all of the rED3s were different, the largest differences in KD were approximately 10-fold between DaKHD10674 and SML6420; the ED3 sequences of DaKHD10674 and SML6420 differ by twelve amino acids (Figure 2). DaKHD10674 has five surface exposed residue changes (D329E, G330D, R345K, T359I, and E360G) compared to the two surface exposed residue changes in SML6420 (V308I and N390D). Residues that are surface exposed may be more important for ligand interactions because they can directly contribute to the binding interaction and their side-chains are not buried inside the protein. Interestingly, when mAb 3H5 was used in a neutralization assay (PRNT50), there was no significant difference in the neutralization for any of the four viruses tested.
Similar patterns were seen with the other DENV-2 type-specific mAb and with DENV complex-reactive mAb 20-783-74014. Taken together, these data show that mAb affinity for rED3 can differ 10-fold without having a direct affect on the biological activity of neutralization. Clearly, neutralization is determined by factors other than antibody affinity. A similar phenomenon has been reported for DENV-1 strains 16007 and West Pac-74; where the affinity of mAb for ED3 did not differ between the viruses, but the PRNT50 values for these viruses were strikingly different (Shrestha et al., 2010).
Our results complement a recent study by Sukupolvi-Petty et al (Sukupolvi-Petty et al., 2010), where the effects of DENV-2 amino acid diversity in the E protein on antibody-mediated neutralization with mAbs generated in alpha/beta interferon receptor-deficient C57BL/6 mice were analyzed. As with our study, Sukupolvi-Petty et al (Sukupolvi-Petty et al., 2010) also showed that all of the ED3 mAbs tested could effectively neutralize DENV-2s from diverse genotypes. The major impact of genotypic variation was seen with the sylvatic virus PM33974 (three amino acid substitutions, G330D, R345K, V365I), where the substitution G330D reduced binding of two mAbs to rED3, and increased the quantity of these mAbs needed to neutralize the virus (Sukupolvi-Petty et al., 2010). In comparison, none of the mAbs used in this study were directly affected by the G330D substitution. Two of the rED3s used in this study contained the G330D substitution (DaKHD10674 and DakAr578), and the substitutions in these rED3s either increased the affinity (DakHD10674) or had no effect (DakAr578) as compared to the binding of mAbs to rED3 of NGC. Both DENV-2 type-specific and DEN complex-reactive mAbs neutralized DakHD10674 at the same concentration for all viruses studied. One major difference between our study and that of Sukupolvi-Petty et al is the source of the mAbs used. The mAbs used in this study were generated in a BALB/C background immunized with whole virus; whereas the mAbs used in (Sukupolvi-Petty et al., 2010) were generated in a C57BL/6 background following immunization with whole virus and then boosted with rED3. Even though many of the critical residues identified by Sukupolvi-Petty et al are shared by the mAbs used in this study, some of their mAbs were sensitive to residue changes in other regions of ED3 not shared by our mAbs, indicating that antibodies produced in the C57BL/6 background may not be identical to those produced in BALB/c background, which may be due to different MHC haplotypes of the mice. In contrast to recent studies on DENV-3 ((Brien et al., 2010; Wahala et al., 2010) ), DENV-2 type-specific and DEN complex-reactive mAbs targeting ED3 are able to effectively bind and neutralize diverse DENV-2 strains from most of the genotypes; whereas mAbs that recognize DENV-3 ED3 cannot bind and neutralize viruses from all genotypes. DENV-3 has a greater number of variable residues in ED3 compared to DENV-2 and more of these variable residues are critical for mAb binding (Brien et al., 2010; Wahala et al., 2010). DENV-3 also has more critical residues for the DENV-3 type-specific and DEN complex-reactive antigenic sites than DENV-2 (Brien et al., 2010; Matsui et al., ; Matsui et al., 2009; Wahala et al., 2010). Thus, amino acid differences in DENV-3 ED3 are more likely to disrupt antibody binding and neutralization than amino acid variation in DENV-2.
It was important to determine if the DENV-2 type-specific and DEN complex-reactive critical residues were shared across all of the DENVs tested. This was investigated by expressing rED3s with alanine substitutions at the positions of the previously identified critical residues K305A and P384A (DENV-2 type-specific) or K310A (DEN complex-reactive)(Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008; Sukupolvi-Petty et al., 2007) in the background of NGC, DaKHD10674, PR159, and SML6420 rED3. The two type-specific mAbs 3H5 and ICL2 were both affected by the substitutions K305A and P384A regardless of the virus background. This indicates that the DENV-2 type-specific antigenic site has the same critical residues for different genotypes. The complex-reactive mAbs were all sensitive to the substitution at K310A, regardless of the virus genotype indicating that the critical residue for the DEN complex reactive antigenic site is conserved between DENV-2 genotypes. Overall, these data demonstrate that the most of the DENV-2 type-specific and DEN complex-reactive mAbs bind the same region of ED3 regardless of the DENV-2 genotype and that substitutions affecting one antigenic site are specific and do not broadly affect the other antigenic site on the surface of ED3. This was expected since in our previous study (Gromowski et al., 2010) we demonstrated that substitutions of the critical residues K305A, P384A, or K310A do not dramatically alter the structure of DENV-2 NGC ED3.
The results of this study have shown that the amino acid variation in ED3 can affect the binding of the DENV-2 type-specific and one of the DEN complex-reactive mAbs for ED3, but that these changes in mAb affinity for ED3 do not affect the concentration at which neutralization takes place. Furthermore, we have shown in this study that the critical residues that make up the DENV-2 type-specific and DEN complex-reactive antigenic sites are essentially conserved among four of the DENV-2 genotypes and that substitution of these residues ablates antibody binding. Taken together the results indicate that the amino acid variation among in ED3 among the genotypes does not greatly affect antibodies that bind these antigenic sites and that ED3 targeting antibodies will be protective against diverse DENV-2 strains. This is important for a potential DENV-2 vaccine since it will be based upon a single representative DENV-2 strain.
Materials and Methods
Cells and viruses
Vero cells (African green monkey kidney) were maintained at 37°C in a 5% CO2 incubator in minimum essential media (Gibco) supplemented with 10% bovine growth serum, 1% L-glutamine, 1% non-essential amino acids, and 1% penicillin/streptomycin. C6/36 mosquito cells were maintained at 28°C in minimum essential media supplemented with 10% fetal bovine serum, 5% tryptose phosphate buffer, 1% L-glutamine, 1% non-essential amino acids, and 1% penicillin/streptomycin. The DENV-2 strains used in this study, New Guinea C (NGC) (Asian II), H8-2027 (cosmopolitan), DakHD10674 (sylvatic), and SML6420 (American) were amplified in C6/36 cells and then titrated by plaque assay in Vero cells.
Monoclonal antibodies (mAbs)
Five commercially available mAbs were used in this study: 3H5 (Millipore, immunogen DENV-2 New Guinea C (Gentry et al., 1982), ICL2 (Santa Cruz Biotechnologies, DENV-2 immunogen), 20-783-74014 (Gen Way Bio, immunogens: mixture of DENV-1, -2, -3, and -4), GTX29202 (GeneTex, immunogens: mixture of DENV-1, -2, -3, and -4), and MD-05-0104 (Ray Biotech, immunogens: DENV-1 Hawaii, DENV-2 New Guinea C, DENV-3 H87, and DENV-4 H-241). All five antibodies were affinity purified. MAbs 3H5, and ICL2 are mouse IgG1 DENV-2 type-specific while the mAbs 20-783-74014, GTX29202, and MD-05-0104 are mouse IgG2a DENV sub-complex-reactive. The mAb 3H5 (Millipore) was used as a control in all enzyme linked immunoabsorpbent assays (ELISAs), and 50% plaque reduction neutralization tests (PRNT50) to ensure consistency among the replicates and assays.
Site-directed mutagenesis
Site-directed mutagenesis of the rED3 MBP fusion construct contained in the pMAL-C2x vector was performed using the QuikChange XL 2 kit (Stratagene) according the manufacturer's directions. The engineered substitutions were confirmed by sequencing.
Expression and purification of recombinant DENV-2 ED3
The DENV-2 rED3s were cloned and expressed as described previously (Gromowski and Barrett, 2007). Briefly the bacterial lysates containing the expressed proteins were applied to 2ml centrifuge columns (Pierce) packed with amylose resin (New England Biolabs), and eluted using column buffer (10mM Tris pH 7.2, 100mM NaCl, 50mM EDTA) containing 10mM maltose. The concentration of the purified protein was quantified using the BCA kit (Pierce) as directed by the manufacturer.
Affinity measurements by indirect ELISA with rED3
The ELISAs with the rED3 were preformed as described previously (Gromowski and Barrett, 2007; Gromowski, Barrett, and Barrett, 2008) In the ELISAs the type-specific mAbs were diluted in blocking buffer to 80 nM (3H5, and ICL2). ICL2 was diluted to 160nM for wells containing rED3s for DENV-2 strains H8-2027, SML6420, and PR159, and 80nM for the rED3 of DENV-2 strains NGC, DakHD10674, and DakAr578. The complex-reactive mAbs 20-783-74014, GTX29202, and MD-05-0104 were diluted in blocking buffer to 10 nM (MD-05-0104 was diluted to 20 nM for the wells containing the rED3s H8-2027, SML6420, and PR159, and 10 nM for NGC, DakHD10674, and DakAr578). The mAbs were serially diluted two-fold and were incubated overnight at room temperature. The following day the mAbs were removed, the plates were washed, and incubated with goat anti-mouse IgG conjugated with horseradish peroxidase (IgG-HRP) diluted 1/1000. Antibody binding was visualized using 3, 3′, 5, 5′-tetramethylbenzidene substrate (Sigma) and read at 655nm on a UV-plate reader (BioRad). The KD values for each MAb calculated using non-linear regression analysis (single site binding saturation) SigmaPlot as described previously (Gromowski and Barrett, 2007). Each mAb was tested in duplicate per assay and results are an average of 5 experiments.
Plaque assays and Plaque reduction neutralization tests (PRNT50) were performed as described previously (Gromowski and Barrett, 2007).
Highlights.
We compared ED3 of representative strains of the six DENV-2 genotypes.
Differences in Kd of 5 MAbs to the rED3s were relatively small.
Physical binding differences did not lead to detectable differences in neutralization.
ED3-specific neutralizing antibodies will likely be effective against all DENV-2 genotypes.
Acknowledgments
This work was funded in part through a grant to ADTB from the NIAID through the Western Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (NIH grant U54 AI057156); G.D.G. was supported by NIAID T32 predoctoral fellowship AI060549.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75(9):4268–75. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien JD, Austin SK, Sukupolvi-Petty S, O'Brien KM, Johnson S, Fremont DH, Diamond MS. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol. 2010;84(20):10630–43. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–88. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chin JF, Chu JJ, Ng ML. The envelope glycoprotein domain III of dengue virus serotypes 1 and 2 inhibit virus entry. Microbes Infect. 2007;9(1):1–6. doi: 10.1016/j.micinf.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Crill WD, Roehrig JT. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J Virol. 2001;75(16):7769–73. doi: 10.1128/JVI.75.16.7769-7773.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry MK, Henchal EA, McCown JM, Brandt WE, Dalrymple JM. Identification of distinct antigenic determinants on dengue-2 virus using monoclonal antibodies. Am J Trop Med Hyg. 1982;31(3 Pt 1):548–55. doi: 10.4269/ajtmh.1982.31.548. [DOI] [PubMed] [Google Scholar]
- Gromowski GD, Barrett AD. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology. 2007;366(2):349–60. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- Gromowski GD, Barrett ND, Barrett AD. Characterization of dengue virus complex-specific neutralizing epitopes on envelope protein domain III of dengue 2 virus. J Virol. 2008;82(17):8828–37. doi: 10.1128/JVI.00606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromowski GD, Roehrig JT, Diamond MS, Lee JC, Pitcher TJ, Barrett AD. Mutations of an antibody binding energy hot spot on domain III of the dengue 2 envelope glycoprotein exploited for neutralization escape. Virology. 2010;407(2):237–46. doi: 10.1016/j.virol.2010.06.044. [DOI] [PubMed] [Google Scholar]
- Hung JJ, Hsieh MT, Young MJ, Kao CL, King CC, Chang W. An external loop region of domain III of dengue virus type 2 envelope protein is involved in serotype-specific binding to mosquito but not mammalian cells. J Virol. 2004;78(1):378–88. doi: 10.1128/JVI.78.1.378-388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc Natl Acad Sci U S A. 2006;103(33):12400–4. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–25. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle JL, Harris E. Global spread and persistence of dengue. Annu Rev Microbiol. 2008;62:71–92. doi: 10.1146/annurev.micro.62.081307.163005. [DOI] [PubMed] [Google Scholar]
- Leitmeyer KC, Vaughn DW, Watts DM, Salas R, Villalobos I, de C, Ramos C, Rico-Hesse R. Dengue virus structural differences that correlate with pathogenesis. J Virol. 1999;73(6):4738–47. doi: 10.1128/jvi.73.6.4738-4747.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, Diamond MS, Kuhn RJ, Rossmann MG. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15(3):312–7. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Matsui K, Gromowski GD, Li L, Barrett AD. Characterization of a dengue type-specific epitope on dengue 3 virus envelope protein domain III. J Gen Virol. 91(Pt 9):2249–53. doi: 10.1099/vir.0.021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Gromowski GD, Li L, Schuh AJ, Lee JC, Barrett AD. Characterization of dengue complex-reactive epitopes on dengue 3 virus envelope protein domain III. Virology. 2009;384(1):16–20. doi: 10.1016/j.virol.2008.11.013. [DOI] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437(7059):764–9. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe. 2008;4(3):229–38. doi: 10.1016/j.chom.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375(6529):291–8. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229(4715):726–33. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R, Harrison LM, Salas RA, Tovar D, Nisalak A, Ramos C, Boshell J, de Mesa MT, Nogueira RM, da Rosa AT. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 1997;230(2):244–51. doi: 10.1006/viro.1997.8504. [DOI] [PubMed] [Google Scholar]
- Roehrig JT, Bolin RA, Kelly RG. Monoclonal antibody mapping of the envelope glycoprotein of the dengue 2 virus, Jamaica. Virology. 1998;246(2):317–28. doi: 10.1006/viro.1998.9200. [DOI] [PubMed] [Google Scholar]
- Sabin AB. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1(1):30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Brien JD, Sukupolvi-Petty S, Austin SK, Edeling MA, Kim T, O'Brien KM, Nelson CA, Johnson S, Fremont DH, Diamond MS. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 2010;6(4):e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010;84(18):9227–39. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukupolvi-Petty S, Austin SK, Purtha WE, Oliphant T, Nybakken GE, Schlesinger JJ, Roehrig JT, Gromowski GD, Barrett AD, Fremont DH, Diamond MS. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol. 2007;81(23):12816–26. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, Geuijen C, Goudsmit J, Bakker AQ, Korimbocus J, Kramer RA, Clijsters-van der Horst M, de Jong M, Jongeneelen M, Thijsse S, Smit R, Visser TJ, Bijl N, Marissen WE, Loeb M, Kelvin DJ, Preiser W, ter Meulen J, de Kruif J. Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile Virus. J Virol. 2006;80(14):6982–92. doi: 10.1128/JVI.00551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298(1):63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, de Silva AM. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog. 2010;6(3):e1000821. doi: 10.1371/journal.ppat.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DM, Porter KR, Putvatana P, Vasquez B, Calampa C, Hayes CG, Halstead SB. Failure of secondary infection with American genotype dengue 2 to cause dengue haemorrhagic fever. Lancet. 1999;354(9188):1431–4. doi: 10.1016/S0140-6736(99)04015-5. [DOI] [PubMed] [Google Scholar]


