Gut microbiota regulation by mucosal immunity
Keywords: commensal, microbiota, mucosal immunity
Abstract
The benefits of commensal bacteria to the health of the host have been well documented, such as providing stimulation to potentiate host immune responses, generation of useful metabolites, and direct competition with pathogens. However, the ability of the host immune system to control the microbiota remains less well understood. Recent microbiota analyses in mouse models have revealed detailed structures and diversities of microbiota at different sites of the digestive tract in mouse populations. The contradictory findings of previous studies on the role of host immune responses in overall microbiota composition are likely attributable to the high β-diversity in mouse populations as well as technical limitations of the methods to analyze microbiota. The host employs multiple systems to strictly regulate their interactions with the microbiota. A spatial segregation between the host and microbiota is achieved with the mucosal epithelium, which is further fortified with a mucus layer on the luminal side and Paneth cells that produce antimicrobial peptides. When commensal bacteria or pathogens breach the epithelial barrier and translocate to peripheral tissues, the host immune system is activated to eliminate them. Defective segregation and tissue elimination of commensals result in exaggerated inflammatory responses and possibly death of the host. In this review, we discuss the current understanding of mouse microbiota, its common features with human microbiota, the technologies utilized to analyze microbiota, and finally the challenges faced to delineate the role of host immune responses in the composition of the luminal microbiota.
Introduction
The mammalian body harbors trillions of microbes, including eubacteria, archaea, fungi and protozoa; these groups are composed of thousands of species (1). The majority of these microbes reside in the digestive tract, where rich nutrients foster the formation of well-organized microbial communities through interactions among microbes and host factors.
By coexisting as either symbionts or pathobionts within the host, these microbes bring about beneficial or detrimental impacts on the health of the host, respectively. However, because of the lack of knowledge on most of these microbes, many of them are simply regarded as commensals, while their effects on the health of the host remain largely unknown (1, 2). One of the most appreciated benefits to the host of the gut microbiota, which refers to the microbial community in the gastrointestinal tract, comes from metabolites of bacteria such as vitamins and short-chain fatty acids (SCFA), which are important for both systemic and intestinal development of the host (3). SCFA-producing colonic bacteria and certain types of colonic Clostridia, as well as ileal segmented filamentous bacteria (SFB), are critical in shaping the host immune balance in mice (4–7).
Given the significant impacts of commensals on host health, our next question is whether the host, in a reciprocal fashion, regulates the commensal microbes living inside its body. Understanding and identifying the host factors that control the populations and localization of symbionts and pathobionts is important for developing therapeutic treatments for human diseases that are affected by these microbes. To tackle this question experimentally, the host factors that affect microbial ecology can be manipulated. Murine models have great advantages for this type of study, due to the feasibility to genetically modify immune or metabolic components and the availability of well-established genetic knock-in and knock-out models. In addition, the microbiota in humans and mice share many common features. Here, we review the current understanding of how the host immune system regulates control of the microbiota; we base the article mainly on studies that have utilized genetically manipulated mouse models.
The mouse microbiota shares common features with the human microbiota but also has unique commensals that affect host immune responses and disease
We would like to first provide an overview of the mouse microbiota in the digestive tract to better understand its regulation. Post-weaning mice harbor 108, 109–10, and 1010–11 commensal bacteria in the oral cavity, small intestine, and large intestine, respectively, whereas pre-weaning mice harbor <108 bacteria in whole digestive tract (8). Whereas the microbiota in these areas of the digestive tract in pre-weaning mice are relatively simple and uniform, the composition of microbiota in intestine of adult mice is complex and unique (8). After weaning, the microbiota in the upper digestive tract, namely the oral cavity and small intestine, continues to maintain its simple form as it is dominated by Lactobacillales, whereas the large intestine begins to harbor mostly Bacteroidales and Clostridiales that are able to digest more-complex carbohydrates (8, 9).
The oral microbiota in mice is characterized by a low α-diversity (i.e. the composition in each individual host) and a high abundance of proteobacteria and Lactobacillales, especially γ-proteobacteria and streptococci (9, 10). These features are common with human microbiota (11). Several commensal groups have identical species that colonize the human and mouse digestive tracts (e.g. Enterobacteriaceae species), but many commensals in humans and mice are not identical, even though they are similar. For example, although NI1060, the murine commensal that accumulates and induces periodontitis at the ligature-damaged gingival site, cannot be found in humans, a phylogenetically related bacterium is associated with human aggressive periodontitis (10).
Despite these similarities, the mouse microbiota in the digestive tract has several unique features, including a low abundance of oral obligate anaerobes associated with major dental diseases, a high abundance of ileal SFB that induces Th17-oriented immune responses, a low abundance of bifidobacteria which affects the susceptibility to infection of pathogens such as Escherichia coli O157:H7 (12), and a different abundance of Lachnospiraceae species (from the Clostridia class) which can control Treg cells (4, 5, 8). The complexity in murine colonic microbiota is mainly associated with phylotypic α-diversity in mouse-specific Porphyromonaceae of Bacteroidales and Lachnospiraceae of Clostridiales, which represent about half of the murine colonic bacteria, and can be detected by denaturing gradient gel electrophoresis (DGGE) analysis despite being indistinguishable in 16S rRNA phylotype analysis of operational taxonomic unit (OTU) clustered at 97% nucleotide identity (8). Therefore, some immunological effects of microbiota in the digestive tract might be species-specific.
Difficulties that can complicate the assessment of the role of the host immune system in microbiota composition
The roles of the host immune system in the regulation of microbiota in the lumen of the digestive tract are still under debate. Contradictory results have been reported for the roles of individual host factors in the control of microbiota, many of which were based on comparisons between wild-type control mice and mice deficient in specific host factors (Table 1). For example, previous studies on the role of the innate immune receptors such as TLR5 and Nod2 in the regulation of intestinal microbiota composition have shown contradictory conclusions.
Table 1.
Microbiota composition in genetically modified mice
| Mice | Microbiota difference (⇧, increased; ⇩,decreased) | Samples | Methods | Co-housing | Reference |
|---|---|---|---|---|---|
| MyD88−/−/TRIF−/− | No difference | Skin, oral, fecal | Pyrosequencing | Littermates | (13) |
| MyD88−/− | No difference | Ileal, cecal | Pyrosequencing | Littermates | (14, 15) |
| MyD88−/−NOD | Porphyromonadaceae ⇧, Lactobacillaceae ⇧, Rikenellaceae ⇧ | Cecal | PCR | Littermates | (16) |
| MyD88ΔIECa | Bacteroidetes ⇩, Proteobacteria ⇧, some Firmicutes ⇧ | Fecal | Pyrosequencing | Littermates | (17) |
| TLR2−/− | No difference | Cecal | Pyrosequencing | Littermates | (15) |
| TLR4−/− | No difference | Cecal | Pyrosequencing | Littermates | (15) |
| TLR5−/− | No difference | Cecal | Pyrosequencing | Littermates | (15) |
| Species-level difference, phylum-level no difference | Cecal | Pyrosequencing | Littermates | (18) | |
| Enterobacteria ⇧ in colitic but not non-colitic mice | Fecal | Pyrosequencing | Littermates | (19) | |
| TLR9−/− | No difference | Cecal | Pyrosequencing | Littermates | (15) |
| Rip2−/− | No difference | Ileal, cecal, fecal | Pyrosequencing | Littermates | (20) |
| Nod1−/− | No difference | Ileal, cecal, fecal | DGGE, pyrosequencing | Littermates | (20, 21,22) |
| Nod2−/− | Bacteroidetes ⇧, Firmicutes ⇧ (terminal ileum), no difference (feces) | Ileal, fecal | PCR | Littermates | (23) |
| Bacteroidetes ⇧, Firmicutes ⇧ (both ileum and feces) | Ileal, fecal | Pyrosequencing | Littermates | (24) | |
| Bacteroidaceae ⇧, Rikenellaceae ⇧, Prevotellaceae ⇧ | Fecal | Pyrosequencing | Unspecified | (25) | |
| No difference | Ileal, cecal, fecal | DGGE, pyrosequencing | Littermates | (20,22) | |
| ASC−/− | No difference | Fecal | DGGE, pyrosequencing | Yes | (26, 27, 28) |
| TM7 ⇧, Prevotellaceae ⇧, Lactobacillus ⇩ | Fecal | Pyrosequencing | No | (26) | |
| NLRP3−/− | Citrobacter ⇧, Proteus ⇧, Shigella ⇧, Mycobacterium ⇧ | Fecal | T-RFLP | Littermates | (29) |
| NLRP6−/− | TM7 ⇧, Prevotellaceae ⇧, Lactobacillus ⇩ | Fecal | Pyrosequencing | No | (26) |
| No difference | Fecal | Pyrosequencing | Yes | (26) | |
| IL-18−/− | TM7 ⇧, Prevotellaceae ⇧, Lactobacillus ⇩ | Fecal | Pyrosequencing | No | (26) |
| No difference | Fecal | Pyrosequencing | Yes | (26) | |
| Rag1−/− | Lachinospiraceae ⇧, Porphyromonadaceae ⇩ (feces) | Skin, oral, fecal | Pyrosequencing | Littermates | (13) |
| Neisseriaceae ⇧, Streptococcaceae ⇩ (oral), no difference (skin) | |||||
| Ig μ−/− | No difference | Oral, ileal, fecal | Culture-based | Littermate | (30) |
| AID−/− | Bacteroidaceae ⇧, Peptostreptococcus ⇧, Bifidobacterium | Ileal | Culture-based | No | (31) |
| SFB ⇧ | Ileal | PCR | Littermate | (32) | |
| pIgR−/− | Pasteurellaceae ⇧, Lachnospiraceae ⇧ | Fecal | Pyrosequencing | Littermate | (33) |
| PDCD1−/− | Erysipelotrichaceae ⇧, Prevotellaceae ⇧, Alcaligenaceae ⇧, TM7 ⇧ | Cecal | Pyrosequencing | unspecified | (34) |
| IL-22−/− | Bacteroides ⇩, Porphyromonadaceae ⇧, Prevotellaceae ⇧ | Fecal | Pyrosequencing | No | (35) |
| Clostridiaceae ⇩, Lactobacillus ⇩, Rikenellaceae ⇧ | |||||
| No difference | Fecal | Pyrosequencing | Yes | (35) | |
| RegIIIγ−/− | No difference in lumen | Ileal | Pyrosequencing | Littermates | (14) |
| Eubacterium rectal ⇧, SFB ⇧ groups in mucosa | Ileal | PCR | Littermates | (14) | |
| STAT3−/− | No difference | Fecal | DGGE | Yes | (36) |
| IRF9−/− | Variation ⇧ | Fecal | DGGE | Yes | (36) |
| PPARγ−/− | No difference | Fecal | PCR | Littermates | (37) |
| DEFA5 Tgb | Bacteroides ⇧, MIB groups ⇧ | Fecal | PCR | Yes | (38) |
| MMP7−/− | Firmicutes ⇧ | Fecal | PCR | Yes | (38) |
| C1galt1ΔIECa | Lactobacillus ⇧, Clostridium ⇧, Lachnospiraceae ⇧, Ruminococcus ⇧ | Fecal, cecal | Pyrosequencing | Unspecified | (39) |
| Bacteroidetes ⇧, Firmicutes ⇩ | Fecal | Pyrosequencing | Littermates | (40) | |
| B4galnt2−/− | Helicobacter spp ⇩ | Ileal, cecal, fecal | Pyrosequencing | Littermates | (41) |
| βGalT1 Tgb | Bacteroidetes ⇩, Firmicutes ⇧ | Fecal | PCR | Yes | (42) |
| TMF−/− | No difference | Fecal | Pyrosequencing | Yes | (43) |
| Ruminococcaceae , Roseburia , Lactobacillus | Fecal | Pyrosequencing | No | (43) | |
| Fut2−/− | Bacteroides ⇧, Parabacteroides ⇧, Parasutterella ⇧, | Fecal | Pyrosequencing | Littermates | (44) |
| Eubacterium ⇧, Clostridiales ⇩ |
aΔIEC, specifically deleted in intestinal epithelial cells. bTg, transgenic.
Comprehensive review of these studies underscores two main issues likely accountable for these discrepancies: (i) technical challenges in the determination of the microbiota composition and (ii) the limited knowledge of the high diversities and the dramatic changes in microbiota among individuals. For the former, the main problem is that many commensals are currently uncultivable. Non-biased and non-culture-based analytic techniques are essential to accurately assess the microbiota composition. Up to now, non-culture-based analyses commonly practiced include genomic hybridization, quantitative PCR (qPCR) using bacterial group-specific primers, DGGE, terminal restriction fragment length polymorphism (T-RFLP), species-specific microarray, random sequencing of amplified 16S rRNA gene libraries and meta-genomic pyrosequencing (45) (Table 1). In particular, the accuracy of microbiota analysis has been greatly improved by recent development of cost-effective next-generation sequencing techniques (46), although technical issues such as cross-hybridization and chimeric sequences may still potentially undermine the accuracy of 16S ribosomal RNA-based and meta-genomics-based analyses.
The high diversity of the microbiota composition in each individual host (α-diversity) and among individual hosts (β-diversity) also presents great challenges to deciphering the importance of specific host factors in regulating the microbiota. With great advances in analyzing the microbiota composition led by the Human Microbiome Project and other groups, studies have shown that microbiota compositions among human individuals are highly diverse (11). Moreover, the microbiota composition in murine intestine changes drastically with different diets, aging, and inflammatory states (8, 11, 47, 48).
Because of these high diversities and variations of the microbiota among individuals, even mice of the same genotype show different microbiota compositions if housed in separate cages within the same facility (49). In addition, different animal facilities and providers have reported unique profiles of microbiota compositions (4). Another issue is the presence of complexity among cryptic species in genomic DNA-based analysis. For example, the majority of the colonic bacterial population is composed of Bacteroidales and Clostridiales species that possess identical or very similar 16S rRNA phylotypes but have distinct metabolic profiles (8, 50). Therefore, abundance of these species is greatly affected by diet ingredients, but alteration in their abundance might not be reflected by 16S ribosomal RNA-based analyses (47, 50). Post-weaning mice possess extremely high diversities in these bacterial groups (8). Therefore, appropriate experimental controls must be incorporated and environmental contributions should be taken into account in studies to address the roles of host factors in regulating the microbiota.
One way to circumvent the obstacles to comparing the composition of the microbiota is co-housing the wild-type and genetically deficient mice, since murine commensals are transferable to mice of different genotypes during co-housing (26, 35, 50). Up to now, no particular bacterium is known to be absolutely untransferable among cohabiting mice. The microbiota in mice of different genotypes eventually equilibrates after several weeks of co-housing unless deficiency in the host factor of interest intrinsically alters the microbiota composition (4, 26, 35).
Location determines whether the host targets pathogens and commensals for elimination
While much more work is still required to reach a consensus on the regulation of overall lumenal microbiota by the host immune system, there has been increasing understanding of the importance of host factors in the controlling of particular bacteria, including pathogenic bacteria, in the mucosa. Pathogenic bacteria possess unique ways to colonize in the host and induce host complications. Therefore, colonization by pathogenic bacteria in many ways is subject to natural selection during evolution and the host must acquire defense strategies to eliminate pathogens that do not naturally inhabit the host.
Many pathogenic bacteria cannot colonize in the presence of commensals, although the precise mechanisms in which commensals prevent colonization of pathogenic bacteria are not still well understood. For example, overgrowth of Salmonella enterica species, Enterococcus faecalis and Clostridium difficile in intestine requires dysbiosis caused by antibiotics (51–53). However, some pathogens including enteropathogenic E. coli (EPEC) and a related rodent pathogen, Citrobacter rodentium, have systems to attach to the host epithelium and obtain nutrients from the epithelium even in the presence of colonic commensals (54) (Fig. 1). Importantly, elimination of C. rodentium is mediated by CD4+ T cells and IgG (55–57). However, germ-free mice that are monocolonized with C. rodentium do not eliminate C. rodentium that have turned off virulence genes responsible for attachment to the host (58). These facts suggest that the host immune system targets pathogenic bacteria only when they locate near the epithelium and thereby pose threats to the host.
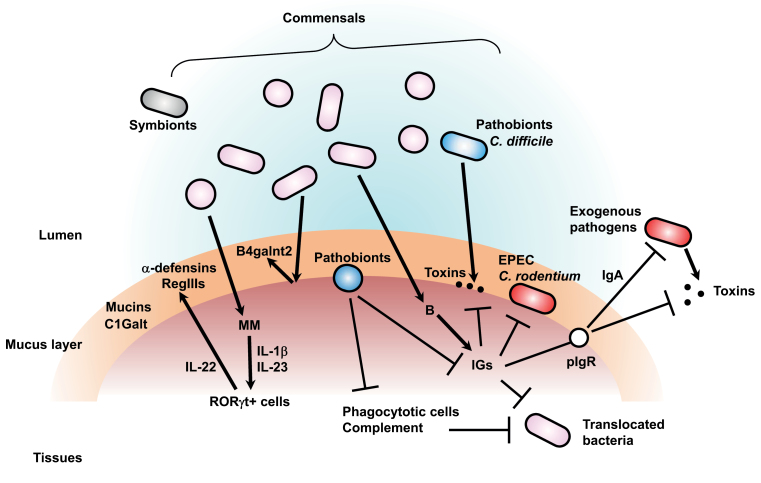
Fig. 1.
Regulation of microbiota by the host immune system. Exogenous enteric pathogens and commensals including symbionts and pathobionts can colonize the lumen of the digestive tract. Host epithelium is protected by a mucus layer containing mucins and their glycosylating enzymes such as C1Galt. Commensals are segregated by the mucus layer and antimicrobial proteins (e.g. α-defensins and RegIII proteins) from epithelium. Commensals augment expression of immunoglobulins (IGs), of mucosal glycan-modifying enzymes such as B4galnt2 and of antimicrobial proteins. Toxin-secreting and non-toxin-secreting pathogens are eventually eliminated by immunoglobulins including IgA. Secretory IgA, which pIgR transfers to the lumen, is important to eliminate pathogens and their toxins. Immunoglobulins are also important for elimination of pathogens that either attach to the epithelium (i.e. Citrobacter rodentium and EPEC) or invade into host tissue. Furthermore, immunoglobulins neutralize toxins secreted from pathobionts such as Clostridium difficile. Translocated pathogens and commensals are also eliminated by multiple host defense mechanisms, including phagocytotic cells and complement components. However, some pathobionts, including periodontal pathobionts, subvert the host attack and induce diseases. Commensals also enhance their segregation from the host by stimulating a particular subset of myelomonocytic cells (MM) that induce IL-22 secretion from RORγt+ lymphoid cells through IL-1β and IL-23. IL-22 induces expression of antimicrobial RegIII proteins for bacterial segregation.
In support of this hypothesis, T cell-independent IgA against toxin A of C. difficile was found to be protective against C. difficile infection independently of the polymeric immunoglobulin receptor (pIgR), which is required for translocation of secretory IgA to the lumen (59). In this case the immunoglobulins produced by the host likely neutralizes toxin A once C. difficile is detected in host tissues. In contrast to infections by pathobionts such as C. difficile, elimination of certain pathogens including Vibrio cholerae requires the pIgR (60), suggesting that secretory IgA controls only pathogens but not commensals. Of note, the host immune system removes the commensals only when they are translocated into tissues, which can be caused by loss of epithelial barrier. The translocated commensals are eliminated by complement components in a sepsis model (61) and phagocytotic cells, which are recruited upon Nod1- and IL-1β-mediated signaling in the C. difficile infection model (21, 27). Other immune responses mediated by multiple inflammatory signaling as well as adaptive immune responses also contribute to elimination of commensals in tissues, as extensively described by previous studies. Therefore, the host immune defense is activated against commensals only when they have escaped from where they are supposed to be (the lumen) and disseminate to host tissues.
The mucosal epithelium segregates commensals from the host in bacterial stimulation-dependent and stimulation-independent ways
Spatial segregation of commensals and the host is essential for their mutually beneficial relationship as well as for the maintenance of a balanced homeostatic state in the host. The mucosal lumen is a non-protected area within the host where commensals are licensed to exist freely without interference from the host. The lumenal area close to the intestinal epithelium is protected from microbes by mucus and antimicrobial factors secreted from epithelial cells and particular specialized cells including Paneth cells in the small intestine (62) (Fig. 1).
Deficiencies of transglycosylases, C1Galt and B4galnt2, result in alterations of the microbiota composition (Table 1) (39–41). Antimicrobial peptides, including individual RegIII proteins and defensins, specifically regulate the abundance of certain bacterial types (63, 64). RegIIIγ is important in segregating bacteria such as Eubacterium rectale and SFB so that they are ≈50 μm away from intestinal epithelium, and the loss of RegIIIγ results in colonization of Gram-positive bacteria on the epithelium, although RegIIIγ deficiency is not associated with alteration in overall microbiota composition (14). Importantly, the expression of RegIII proteins and α-defensins, but not mucins, is enhanced in the presence of commensals [(62, 65), also see GEO GDS2968, GDS4319 and GDS640 for the global gene expression profiles in germ-free and conventional mice] (66–68).
The expression of all RegIII proteins is dependent on IL-22 (14), suggesting that the microbiota composition is not influenced by differential expression of RegIII proteins. However, the lack of B4galnt2, which is also induced by commensals and modifies mucosal glycans, alters the microbiota composition (40). Therefore, commensals indeed promote the spatial segregation of themselves from the host by providing stimulation to the host. From an evolutionary point of view, this may represent a strategy of immune evasion by commensals to favor their residence in the host.
Production of IL-22 in RORγt+ innate lymphoid cells is dependent on IL-23 and IL-1β, which are produced by bacteria-sensing resident macrophages and a subset of dendritic cells (69–73). Since normal gut microbiota is found mice lacking ASC, an essential component of the IL-1β-producing inflammasome (74), segregation of commensals from epithelium likely does not affect the whole microbial population. However, particular commensals, including Bacteroides thetaiotaomicron and Akkermansia muciniphila, have the ability to interact with mucus components and utilize the intestinal mucus as an energy source (75). Thus, given the importance of the mucus layer and antimicrobial peptides in the spatial segregation between the host and gut microbiota, many ongoing studies are directed to delineating how alterations in these two key components of host defense contribute to dysbiosis and disease development.
Conclusion
In summary, our current understanding is that the host eliminates commensals and pathogens when they translocate into tissues or invade the lumenal site proximal to the epithelium, but at the distal site in the lumen the host exerts very little control over the bacteria. As discussed above, due to the high diversities and changes within and among individual hosts, rigorous and careful experimental controls must be included in studies to determine the ability of the host immune system to control the lumenal microbiota composition. For the same reason, discrepancies have arisen among previous studies that addressed the roles of various host factors in shaping the microbiota.
However, with the recent advances in technologies, more information and conclusive results are anticipated from future studies of the microbiota composition, commensal localization, and metabolomes. For example, whole-genome sequencing of many standard commensal strains is under way as a part of the Human Microbiome Project and other programs. This will greatly enhance the detection of more specific species related to the standard commensals by qPCR. Application of a long-through-type next-generation sequencing technique (45) to meta-genomic analysis will accelerate the assembly of more accurate contigs. On the other hand, there are currently no comprehensive searchable databases like NCBI GEO (76) that are available for microbiota composition, and the deposition of microbiota composition data to a public database is currently not mandatory for publication of the data. Such a public database, if available, will tremendously advance our understanding of host–commensal interactions and related diseases.
Finally, the ultimate goal is to translate the knowledge obtained from mouse-model studies to humans in order to understand dysbiosis-related diseases. Although the human microbiota has been extensively characterized and diet-based prebiotic/probiotic approaches are popular, the human microbiota is much more complex and diverse, and manipulations of the microbiota in human studies are subject to many ethical considerations. Despite the caveats, active research in recent years on the role of the host immune system in mouse microbiota has provided valuable insights into the regulation of human microbiota and the pathogenesis of intestinal disorders.
Funding
National Institutes of Health (R01 DE018503).
Acknowledgements
We are grateful to Melody Zeng and Nobuhiko Kamada (University of Michigan) for their critical review of the manuscript and stimulating discussions.
References
- 1. Kamada N., Chen G. Y., Inohara N., Núñez G. 2013. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14:685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chow J., Tang H., Mazmanian S.K. 2011. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr. Opin. Immunol. 23:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramakrishna B. S. 2013. Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 4:9. [DOI] [PubMed] [Google Scholar]
- 4. Ivanov I. I., Atarashi K., Manel N., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atarashi K., Tanoue T., Oshima K., et al. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500:232. [DOI] [PubMed] [Google Scholar]
- 6. Furusawa Y., Obata Y., Fukuda S., et al. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446. [DOI] [PubMed] [Google Scholar]
- 7. Arpaia N., Campbell C., Fan X., et al. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hasegawa M., Osaka T., Tawaratsumida K., et al. 2010. Transitions in oral and intestinal microflora composition and innate immune receptor-dependent stimulation during mouse development. Infect. Immun. 78:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gillilland M. G., 3rd, Erb-Downward J. R., Bassis C. M., et al. 2012. Ecological succession of bacterial communities during conventionalization of germ-free mice. Appl. Environ. Microbiol. 78:2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiao Y., Darzi Y., Tawaratsumida K., et al. 2013. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell Host Microbe 13:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson C. J., Bohannan B. J., Young V. B. 2010. From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukuda S., Toh H., Hase K., et al. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543. [DOI] [PubMed] [Google Scholar]
- 13. Scholz F., Badgley B. D., Sadowsky M. J., Kaplan D. H. 2014. Immune mediated shaping of microflora community composition depends on barrier site. PLoS One 9:e84019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vaishnava S., Yamamoto M., Severson K. M., et al. 2011. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ubeda C., Lipuma L., Gobourne A., et al. 2012. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209:1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wen L., Ley R. E., Volchkov P. Y., et al. 2008. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455:1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frantz A. L., Rogier E. W., Weber C. R., et al. 2012. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 5:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vijay-Kumar M., Aitken J. D., Carvalho F. A., et al. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carvalho F. A., Koren O., Goodrich J.K., et al. 2012. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe 12:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robertson S. J., Zhou J. Y., Geddes K., Rubino S. J., Cho J. H., Girardin S. E., Philpott D. J. 2013. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes 4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hasegawa M., Yamazaki T., Kamada N., et al. 2011. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J. Immunol. 186:4872. [DOI] [PubMed] [Google Scholar]
- 22. Natividad J. M., Petit V., Huang X., et al. 2012. Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in Nod1-/-; Nod2-/- mice. Inflamm. Bowel Dis. 18:1434. [DOI] [PubMed] [Google Scholar]
- 23. Petnicki-Ocwieja T., Hrncir T., Liu Y. J., et al. 2009. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl Acad. Sci. USA 106:15813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rehman A., Sina C., Gavrilova O., et al. 2011. Nod2 is essential for temporal development of intestinal microbial communities. Gut 60:1354. [DOI] [PubMed] [Google Scholar]
- 25. Mondot S., Barreau F., Al Nabhani Z., et al. 2012. Altered gut microbiota composition in immune-impaired Nod2-/- mice. Gut 61:634. [DOI] [PubMed] [Google Scholar]
- 26. Elinav E., Strowig T., Kau A. L., et al. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hasegawa M., Kamada N., Jiao Y., Liu M. Z., Núñez G., Inohara N. 2012. Protective role of commensals against Clostridium difficile infection via an IL-1β-mediated positive-feedback loop. J. Immunol. 189:3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Henao-Mejia J., Elinav E., Jin C., et al. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirota S. A., Ng J., Lueng A., et al. 2011. NLRP3 inflammasome plays a key role in the regulation of intestinal homeostasis. Inflamm. Bowel Dis. 17:1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marcotte H., Lavoie M.C. 1996. No apparent influence of immunoglobulins on indigenous oral and intestinal microbiota of mice. Infect. Immun. 64:4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fagarasan S., Muramatsu M., Suzuki K., Nagaoka H., Hiai H., Honjo T. 2002. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 298:1424. [DOI] [PubMed] [Google Scholar]
- 32. Suzuki K., Meek B., Doi Y., et al. 2004. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc. Natl Acad. Sci. USA 101:1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. PrgRogier E. W., Frantz A. L., Bruno M. E., et al. 2014. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc. Natl Acad. Sci. USA 111:3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawamoto S., Tran T. H., Maruya M., et al. 2012. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science 336:485. [DOI] [PubMed] [Google Scholar]
- 35. Zenewicz L. A., Yin X., Wang G., et al. 2013. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 190:5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson C. L., Hofer M. J., Campbell I. L., Holmes A. J. 2010. Community dynamics in the mouse gut microbiota: a possible role for IRF9-regulated genes in community homeostasis. PLoS One 5:e10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peyrin-Biroulet L., Beisner J., Wang G., et al. 2010. Peroxisome proliferator-activated receptor gamma activation is required for maintenance of innate antimicrobial immunity in the colon. Proc. Natl Acad. Sci. USA 107:8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salzman N. H., Hung K., Haribhai D., Chu H., et al. 2010. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perez-Munoz M., Bergstrom K., Peng V., et al. 2014. Discordance between changes in the gut microbiota and pathogenicity in a mouse model of spontaneous colitis. Gut. Microbes, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sommer F., Adam N., Johansson M. E., Xia L., Hansson G. C., Bäckhed F. 2014. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLoS One 9:e85254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Staubach F., Künzel S., Baines A. C., et al. 2012. Expression of the blood-group-related glycosyltransferase B4galnt2 influences the intestinal microbiota in mice. ISME J. 6:1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vanhooren V., Vandenbroucke R. E., Dewaele S., et al. 2013. Mice overexpressing β-1,4-Galactosyltransferase I are resistant to TNF-induced inflammation and DSS-induced colitis. PLoS One 8:e79883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bel S., Elkis Y., Elifantz H., et al. 2014. Reprogrammed and transmissible intestinal microbiota confer diminished susceptibility to induced colitis in TMF-/- mice. Proc. Natl Acad. Sci. USA, 111:4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kashyap P. C., Marcobal A., Ursell L. K., et al. 2013. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc. Natl Acad. Sci. USA 110:17059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Inglis G. D., Thomas M. C., Thomas D. K., Kalmokoff M. L., Brooks S. P., Selinger L. B. 2012. Molecular methods to measure intestinal bacteria: a review. J. AOAC Int. 95:5. [DOI] [PubMed] [Google Scholar]
- 46. Weinstock G. M. 2012. Genomic approaches to studying the human microbiota. Nature 489:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Salonen A, de Vos W. M. 2014. Impact of diet on human intestinal microbiota and health. Annu Rev Food Sci Technol. 5:239. [DOI] [PubMed] [Google Scholar]
- 48. Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl Acad. Sci. USA 104:13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Antonopoulos D. A., Huse S. M., Morrison H. G., Schmidt T. M., Sogin M. L., Young V. B. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koropatkin N. M., Cameron E. A., Martens E. C. 2012. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 10:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kaiser P., Diard M., Stecher B., Hardt W. D. 2012. The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen’s virulence factors, and the host’s mucosal immune response. Immunol. Rev. 245:56. [DOI] [PubMed] [Google Scholar]
- 52. Rice L. B. Antimicrobial resistance in Gram-positive bacteria. 2006. Am. J. Med. 119(Suppl. 1):S11. [DOI] [PubMed] [Google Scholar]
- 53. Chen X., Katchar K., Goldsmith J. D., et al. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135:1984. [DOI] [PubMed] [Google Scholar]
- 54. Mundy R., MacDonald T. T., Dougan G., Frankel G., Wiles S. 2005. Citrobacter rodentium of mice and man. Cell. Microbiol. 7:1697. [DOI] [PubMed] [Google Scholar]
- 55. Simmons C. P., Clare S., Ghaem-Maghami M., et al. 2003. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium . Infect. Immun. 71:5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bry L., Brigl M., Brenner M. B. 2006. CD4+-T-cell effector functions and costimulatory requirements essential for surviving mucosal infection with Citrobacter rodentium . Infect. Immun. 74:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Masuda A., Yoshida M., Shiomi H., et al. 2008. Fcγ receptor regulation of Citrobacter rodentium infection. Infect. Immun. 76:1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kamada N., Kim Y. G., Sham H. P., et al. 2012. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336:1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Johnston P. F., Gerding D. N., Knight K. L. 2014. Protection from Clostridium difficile infection in CD4 T cell- and polymeric immunoglobulin receptor-deficient mice. Infect. Immun. 82:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Uren T. K., Wijburg O. L., Simmons C., Johansen F. E., Brandtzaeg P., Strugnell R. A. 2005. Vaccine-induced protection against gastrointestinal bacterial infections in the absence of secretory antibodies. Eur. J. Immunol. 35:180. [DOI] [PubMed] [Google Scholar]
- 61. Dahlke K., Wrann C. D., Sommerfeld O., et al. 2011. Distinct different contributions of the alternative and classical complement activation pathway for the innate host response during sepsis. J. Immunol. 186:3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sommer F., Bäckhed F. 2013. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 11:227. [DOI] [PubMed] [Google Scholar]
- 63. Stelter C., Käppeli R., König C., et al. 2011. Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLoS One 6:e20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Selsted M. E., Ouellette A. J. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551. [DOI] [PubMed] [Google Scholar]
- 65. Zheng Y., Valdez P. A., Danilenko D. M., et al. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 14:282. [DOI] [PubMed] [Google Scholar]
- 66. Munakata K., Yamamoto M., Anjiki N., et al. 2008. Importance of the interferon-alpha system in murine large intestine indicated by microarray analysis of commensal bacteria-induced immunological changes. BMC Genomics 9:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. El Aidy S., van Baarlen P., Derrien M., et al. 2012. Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 5:567. [DOI] [PubMed] [Google Scholar]
- 68. Mutch D. M., Simmering R., Donnicola D., et al. 2004. Impact of commensal microbiota on murine gastrointestinal tract gene ontologies. Physiol Genomics 19:22. [DOI] [PubMed] [Google Scholar]
- 69. Sutton C. E., Lalor S. J., Sweeney C. M., Brereton C. F., Lavelle E. C., Mills K. H. 2009. Interleukin-1 and IL-23 induce innate IL-17 production from γδ T cells, amplifying Th17 responses and autoimmunity. Immunity 31:331. [DOI] [PubMed] [Google Scholar]
- 70. Reynders A., Yessaad N., Vu, Manh T. P., et al. 2011. Identity, regulation and in vivo function of gut NKp46+RORγt+ and NKp46+RORγt- lymphoid cells. EMBO J. 30:2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Verreck F. A., de Boer T., Langenberg D. M., et al. 2004 Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc. Natl Acad. Sci. USA 101:4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shaw M. H., Kamada N., Kim Y. G., Núñez G. 2012. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med. 209:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Melissa A. K., Charlie G. B., Gretchen E. D., et al. 2012. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G. 2009. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 10:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ouwerkerk J. P., de Vos W. M., Belzer C. 2013. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract. Res. Clin. Gastroenterol. 27:25. [DOI] [PubMed] [Google Scholar]
- 76. Barrett T., Troup D. B., Wilhite S. E., et al. 2011. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res. 39:D1005. [DOI] [PMC free article] [PubMed] [Google Scholar]