Abstract
The function of scaffolding proteins is to bring together two or more proteins in a relatively stable configuration, hence their name. Numerous scaffolding proteins are found in nature, many having multiple protein–protein interaction modules. Over the past decade, examples of scaffolding complexes long thought to be stable have instead been found to be surprisingly dynamic. These studies are scattered among different biological systems, and so the concept that scaffolding complexes might not always represent stable entities and that their dynamics can be regulated has not garnered general attention. We became aware of this issue in our studies of a scaffolding protein in microvilli, which forced us to reevaluate its contribution to their structure. The purpose of this Perspective is to draw attention to this phenomenon and discuss why complexes might show regulated dynamics. We also wish to encourage more studies on the dynamics of “stable” complexes and to provide a word of caution about how functionally important dynamic associations may be missed in biochemical and proteomic studies.
INTRODUCTION
Scaffolding proteins have critical roles in cellular signaling pathways in which they bring multiple binding partners together to facilitate their concerted interactions and functions. They achieve this by being composed of several protein–protein interaction modules, most notably PDZ (postsynaptic density 95/discs large/zona occludens-1) and SH3 (Src homology 3) domains (Pawson and Nash, 2003; Good et al., 2011). Additionally, scaffolding proteins and their partners generally show highly specific subcellular localizations. Some well-studied examples include MAPK signaling during mating in the budding yeast using the scaffold Ste5p (sterile 5; Printen and Sprague, 1994), neuronal synaptic signaling exploiting PSD-95 (postsynaptic density 95; Sampedro et al., 1981), and photosensory reception in Drosophila signaling using InaD (inactivation no after-potential D; Shieh and Zhu, 1996). Other scaffolds, such as members of the NHERF (Na+-H+ exchanger regulatory factor) family and SNX27 (sorting nexin family member 27), are involved in the stabilization, sorting, recycling, and localization of cell surface receptors (Shenolikar and Weinman, 2001; Lauffer et al., 2010; Ardura and Friedman, 2011; Romero et al., 2011).
Scaffolds also perform critical roles in cell polarity (Thompson, 2013). The scaffold Bem1 coordinates a feedback loop to generate localized activation of Cdc42 to ensure that budding yeast assembles a single bud (Johnson et al., 2011). The PDZ scaffolds par-3 and par-6 are essential for establishment of asymmetry and proper cleavage in the early embryo of Caenorhabditis elegans (Kemphues et al., 1988; Watts et al., 1996). In Drosophila, Scrib (scribble), Dlg (discs large), Baz (Bazooka), and Sdt (stardust) are all PDZ scaffolds that regulate epithelial polarity (Woods and Bryant, 1991; Bilder et al., 2003). Another PDZ scaffold, ZO-1 (zona occludens-1) is involved in the stabilization and barrier function of tight junctions (Stevenson et al., 1986). Additionally, the linking proteins α- and β-catenin play vital roles in cadherin-based cell–cell adhesion, which helps give rise to the functional organization of cells into tissues (Ozawa et al., 1989; Gumbiner, 2000). The overwhelming majority of these scaffolds involved in polarity are highly conserved across species, further highlighting their importance.
The name “scaffold” implies the formation of a stable complex, a notion further reinforced by their highly specific localizations. However, over the past decade, there have been examples of scaffolding protein complexes long thought to provide stable linkages but subsequently found to be surprisingly dynamic. These advances have been driven by the increased accessibility of techniques such as FRAP (fluorescence recovery after photobleaching) and photoactivation to examine the dynamics of components in vivo. Despite these advances, the in vivo dynamics of many scaffold complexes are often not considered. In this Perspective, we aim to draw attention to this phenomenon by discussing some examples of unexpectedly dynamic scaffold complexes and to discuss how this may relate to their physiological roles. Further, we wish to encourage more analyses of in vivo dynamics of cellular components, as unexpected insights can emerge. Finally, we explore the issue that dynamic protein complexes are likely systematically underrepresented in current proteomic data.
In the examples discussed below, the terms dynamic and stable serve as qualitative descriptors of the dynamics of components in the context of the stability of the structures in which they participate. The affinity of protein–protein interactions is a function of their on and off rates (Pollard, 2010). On rates are largely limited by diffusion (on the order of 106 to 107 M−1s−1), so the off rate is often the determining factor of binding affinity. Techniques such as FRAP measure the off rate of proteins based on their fluorescence recovery rates. A relatively low-affinity first-order interaction of 1 μM would have an off rate of 1 s−1 with a thalf of 0.7 s and might typically be considered dynamic, while a high-affinity interaction of 1 nM would have an off rate of 0.001 s−1 and a thalf of ∼12 min and, depending on the biological context, could be considered stable. Of course, the complexity of the intracellular environment often means that few proteins show dynamics fit by simple first-order reactions, but these rates nonetheless help put things in perspective.
THE MAPK SCAFFOLD Ste5p
The scaffold Ste5p is a major regulatory component of the MAPK (mitogen-activated protein kinase) signal cascade involved in budding yeast mating (Printen and Sprague, 1994). Ste5p was originally thought to stabilize the complex of Fus3p (a MAPK), Ste7p (a MAPKK), and Ste11p (a MAPKKK) and increase their local concentration to facilitate the phosphorylation cascade (Choi et al., 1994). In an early example examining dynamics, FRAP was used to investigate the dynamics of the individual components of this complex in vivo (van Drogen et al., 2001). In the presence of pheromone during mating, Ste5p, Fus3p, and Ste7p are all highly localized to the tips of mating projections. Remarkably, Ste11p is not detectably enriched at mating projections, although its interaction with Ste5p is required, which suggests its association with this complex is very transient. FRAP of Ste5p and Fus3p revealed that these components have very different dynamics. Interestingly, Fus3p shuttles rapidly in and out of the nucleus irrespective of its phosphorylation status and has a very dynamic association with mating tips (recovers at mating tips with thalf = 0.3 s), while Ste5p remains more stably bound to the membrane (thalf = 8 s). After its activation, Fus3p can phosphorylate Ste5p to negatively regulate its activity and thereby provide feedback regulation to the mating MAPK cascade (Bhattacharyya et al., 2006). Interestingly, Ste5p undergoes an auto-inhibitory intramolecular interaction that is released in the presence of mating factor, allowing Ste5p to then interact with Fus3p (Zalatan et al., 2012). The dynamic nature of the Ste5p-Fus3p complex in vivo was a pioneering discovery and led to two important concepts. First, the notion that scaffolding protein interactions can be highly dynamic to allow reequilibration for rapid changes in signaling, and second, the concept that the very transient nature of Fus3p association with the Ste5p-MAPK cascade may permit signal amplification by activation of multiple Fus3p molecules (Figure 1A). Thus an analysis of dynamics provided important insights into this well-studied pathway.
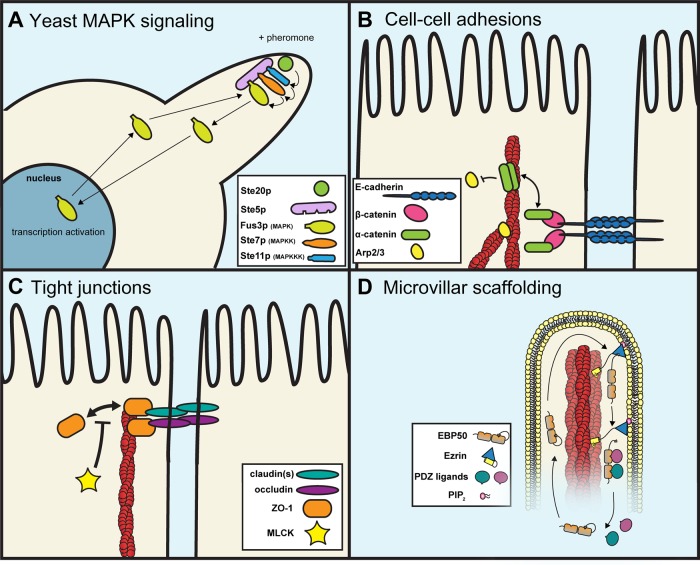
FIGURE 1:
Examples of dynamic scaffolding protein complexes once thought to be stable. (A) Ste5p, a scaffold for the MAPK cascade during mating in budding yeast, brings together Ste11p, Ste7p, and Fus3p at mating projection tips. Fus3p can rapidly shuttle between the cytoplasm and nucleus to activate transcription and also provides negative feedback via phospho-mediated inhibition of Ste5p activity. (B) The linking protein α-catenin is a critical component of cell–cell adhesions. As a monomer, it binds to the E-cadherin–β-catenin complex. As a homo-dimer, α-catenin can bind to F-actin (red) and blocks Arp2/3 binding, thereby preventing local actin polymerization. (C) The MAGUK family member ZO-1 plays an essential role in tight junction barrier function. It binds to the C-terminal tails of claudins and occludins and links them to the underlying F-actin. The linkage provided by ZO-1 is transient, as it freely exchanges with the cytoplasm. This dynamic exchange is suppressed by MLCK activity. (D) The PDZ scaffolding protein EBP50 provides a critical linkage between PDZ ligands and ezrin in microvilli on the apical surface of epithelial cells. The EBP50 tail has a high-affinity association with ezrin and is stable when not bound to PDZ ligands. On PDZ ligand binding, the EBP50 tail–ezrin interaction becomes dynamic, and EBP50 rapidly exchanges with the cytoplasm.
THE LINKING PROTEIN α-CATENIN
Cadherin-based cell–cell adhesion is critical in development and is disrupted in cancer metastasis. β-Catenin binds to the cytoplasmic tail of E-cadherin and to the linking protein α-catenin, which interacts with the underlying actin cytoskeleton and associated actin-binding proteins (Jamora and Fuchs, 2002). The stoichiometric complex of E-cadherin, β-catenin, and α-catenin was thought to provide a stable and persistent linkage between cadherin-mediated cell–cell adhesions and the actin cytoskeleton. However, the crucial experiment to show that α-catenin can bind simultaneously to both actin and the E-cadherin–β-catenin complex had not been performed. The nature of this adhesion complex was dissected using a combination of elegant biochemistry, FRAP, and photoactivation (Drees et al., 2005; Yamada et al., 2005). FRAP and photoactivation of fluorescently tagged adhesion complex components at cell–cell junctions indicated that actin was found to recover much faster (thalf = 0.16 min) at epithelial cell junctions than E-cadherin, β-catenin, and α-catenin, which all showed similar dynamics (thalf ≈ 0.5 min), thereby calling into question the existence of a stable link between the adhesion complex and actin cytoskeleton. Part of the explanation comes from the finding that monomeric α-catenin cannot bind to F-actin and β-catenin simultaneously in vitro and that binding to F-actin actually decreases α-catenin's affinity for β-catenin (Yamada et al., 2005). While this provides a likely explanation for the different dynamics seen by FRAP, it raised the question of how the adhesion complex connects to the underlying actin cytoskeleton if they are not stably connected as originally thought. Moreover, α-catenin can form a homo-dimer that is unable to bind β-catenin but can bundle F-actin and compete with Arp2/3 (actin-related protein 2/3) for actin filaments, thereby suppressing Arp2/3’s activity (Drees et al., 2005). Thus a combination of in vivo dynamics and biochemistry has suggested that α-catenin can switch between monomeric and dimeric forms, neither of which alone links the E-cadherin–β-catenin complex to F-actin (Figure 1B). These results suggest that α-catenin instead provides a regulatory role controlling the underlying actin dynamics, thereby casting doubt on a well-studied model. Despite these intriguing results, studies with chimeras and homologues from simpler organisms have not fully resolved whether α-catenin can provide a direct or indirect linkage between β-catenin and F-actin, and a number of additional roles for α-catenin have been suggested (Maiden and Hardin, 2011).
PDZ SCAFFOLDING PROTEINS
There are more than a hundred PDZ domain–containing proteins in the human proteome. PDZ domains bind to ligands through a very short region generally located at their C-terminus and are often found in scaffolding proteins with other protein–protein interaction domains. Over the past few years, several examples of the involvement of these proteins in dynamic associations have emerged.
ZO-1 is a tight junction scaffolding protein critical for the barrier function of epithelial cells. It is a member of the MAGUK (membrane-associated guanylate kinase) family and contains three PDZ domains and one SH3 domain. It localizes precisely to tight junctions, where it interacts with membrane-bound claudins and occludin to link them to actin filaments (Fanning and Anderson, 2009). The tight junction was considered to be a static structure composed of many stable protein–protein interactions. However, when Shen and colleagues examined the dynamics of different tight junction proteins using FRAP, FLIP (fluorescence loss in photobleaching), and photoactivation, they found that ZO-1 is much more dynamic than claudin-1 (ZO-1 showed 70% mobility with a thalf of ∼100 s; claudin-2 showed only 20% mobility and a thalf of ∼200 s) and much less dynamic than actin (99% mobility, thalf of ∼15 s), which is surprising if it links them together stably (Shen et al., 2008). Interestingly, inhibition or interference with MLCK (myosin light chain kinase) enhances transepithelial resistance and selectively stabilizes ZO-1 at tight junctions without affecting the dynamics of both occludin and claudin-1 (Figure 1C). Thus the dynamics of the scaffolding protein ZO-1 can be regulated in vivo, although the mechanism linking MLCK activity and change in dynamics is not yet clear (Yu et al., 2010). The dynamic nature of this complex revised the prevailing model that the tight junction was a static structure and gave new insights into its regulation.
Septate junctions in Drosophila epithelial cells perform a barrier function similar to that of vertebrate tight junctions and are also critical for development of the epithelium. Several claudin homologues localize to septate junctions together with the basolateral membrane–determinant PDZ-containing scaffolding proteins Dlg and Scrib (Wu and Beitel, 2004). To examine the nature of the interactions in the septate junction complex, Oshima and Fehon used FRAP and FLIP of individual septate junction proteins and found many of the core components to be very stably associated (thalf ≈ 30 min; Oshima and Fehon, 2011). Interestingly, Dlg, another MAGUK with three PDZ domains and an SH3 domain, is much more dynamic (thalf ≈ 1.6 min), although it is widely regarded as an authentic septate junction protein. The recovery rate of Dlg is unaffected in various septate junction binding-partner mutants, so the authors concluded that, while Dlg localizes to septate junctions and is required for their formation, it is not a core component of them. Because Dlg is a polarity determinant of the basolateral membrane, it makes sense that it is more dynamic than the other core structural junction proteins.
Scaffolding is also critical for microvilli on the apical domain of epithelial cells, which have a specialized protein composition and enhance apical signal reception. EBP50 (ERM-binding phosphoprotein of 50 kDa), a member of the NHERF family of scaffolding proteins, has two PDZ domains, localizes to microvilli, and is required for their formation (Morales et al., 2004; Garbett et al., 2010). It provides a linkage between PDZ ligands via its PDZ domains and active ezrin through its C-terminal tail (Reczek et al., 1997). Because it binds active ezrin in vitro with single nanomolar affinity (Terawaki et al., 2006) and a thalf of 21 min (Garbett and Bretscher, 2012), it was thought to stably link various membrane-associated proteins to the underlying actin cytoskeleton via ezrin (Fehon et al., 2010). However, examination of microvillar protein dynamics in vivo using FRAP and photoactivation revealed that EBP50 is remarkably dynamic (thalf ≈ 5 s) compared with ezrin and its PDZ ligand podocalyxin, which were both relatively stable (thalf ≈ 30–50 s; Garbett and Bretscher, 2012). Interestingly, the C-terminal region of EBP50 that binds ezrin is intrinsically highly dynamic in vivo, and the unoccupied PDZ domains negatively regulate its dynamics (Garbett et al., 2013). On ligand binding to either PDZ domain, the negative regulation is additively relieved to yield a highly dynamic protein—an unexpected scenario in which a scaffolding protein becomes more dynamic upon ligand binding. Biochemical data further support this model: because of the high dynamics, active ezrin immunoprecipitates very poorly with ligand-bound EBP50, yet a robust interaction can be seen with the ligand-free protein. These results imply that the linkage EBP50 provides between its PDZ ligands and ezrin is tuned to maintain a specific level of association (Figure 1D). One possibility is that its rapid dynamics might serve as a regulated linkage between the plasma membrane and actin (via interactions with transmembrane PDZ ligands and ezrin, respectively) to adapt to changing forces generated by high local membrane tension and actin treadmilling (Viswantha et al., 2014). The mechanism of this tuning has not yet been elucidated.
CONCLUSIONS AND FUTURE IMPLICATIONS
The examples discussed above have all revealed an unexpected degree of dynamics of linking and scaffolding proteins compared with their binding partners. Although the scales of these examples range from seconds to minutes, the underlying theme is the same—what we think of as stable complexes might in fact be intricately regulated by their dynamics. Because many scaffolding proteins participate in cell polarity, which often involves mutually exclusive dynamic protein complexes, as seen in the nematode early embryo or fly epithelium, the dynamics in these cases is not entirely unexpected. As in cell polarity, one clear benefit is the ability to rapidly adapt to changing environmental requirements or to diverse signaling cues. An additional benefit is that feedback can mediate the duration of specific interactions within a scaffolding complex to avoid otherwise adverse consequences, which is especially important in signaling pathways. For example, photoreception in Drosophila appears to require a refractory period to diminish the response after exposure to bright light. A key component of this pathway is the scaffolding protein InaD, which contains five PDZ domains. On acute light exposure, PDZ5 of InaD undergoes a conformational change stabilized by disulfide-bond formation to preclude ligand binding. On returning to the dark, the disulfide is reduced and ligand binding restored (Mishra et al., 2007). Very recently, we have found that the closest NHERF family member of EBP50, E3KARP (NHE3 kinase A regulatory protein), is normally stably associated with microvilli and ezrin but can become highly dynamic with either a single amino acid modification or in response to specific cellular signaling events (Cécile Sauvanet, unpublished data).
The importance of dynamics in the systems discussed above is clear, but the study of such systems at the biochemical level can be challenging. We were fortunate to discover EBP50 by its very high affinity for ezrin in vitro, yet were perplexed for years by our inability to efficiently coprecipitate it with ezrin from cell lysate (Reczek et al., 1997). We now know this was due to EBP50’s unexpectedly high dynamics in vivo, which represents a ∼250-fold increase from its in vitro off rate. Using in vitro reconstitution studies, we concluded that some factor(s) in cell lysate enhance the off rate of EBP50 from ezrin by regulating the intrinsic dynamic nature of the EBP50 tail (Garbett and Bretscher, 2012; Garbett et al., 2013). Similar regulation could be true of other scaffolds whose dynamics and biochemical nature have not yet been carefully dissected.
Similarly, it is noticeable in many of the systems discussed above that coprecipitation of ligands with scaffolding proteins has often been challenging. If we extrapolate this experience to systematic proteomic studies, important dynamic protein complexes are likely to be vastly underrepresented, although many may readily be demonstrated when tested with pure components in vitro. One way to account for this is to combine reversible cross-linking techniques to catch dynamic interactions coupled with the high sensitivity of modern mass spectrometry (Viswanatha et al., 2013). Alternatively, new techniques have been developed to label and identify proteins in close vicinity to a reporter, which should greatly aid in the discovery of dynamic interactions that would otherwise not be detected by standard analysis of stable protein complexes (Rhee et al., 2013).
Overall, scaffolding proteins and their binding partners are emerging as important regulators of signaling and other pathways through dynamic associations. If such interactions are difficult to capture by analysis of cell extracts using traditional approaches, we encourage researchers to investigate the in vivo dynamics of individual components of the system as well as employ newer and alternative biochemical approaches. Doing so may reveal interesting and unexpected mechanisms of regulation such as those described here.
Acknowledgments
We thank members of the Bretscher lab for helpful comments and advice. This work was supported by National Institutes of Health grant GM-036652.
Abbreviations used:
- Dlg
discs large
- EBP50
ERM-binding phosphoprotein of 50 kDa
- FLIP
fluorescence loss in photobleaching
- FRAP
fluorescence recovery after photobleaching
- InaD
inactivation no after-potential D
- MAGUK
membrane-associated guanylate kinase
- MAPK
mitogen-activated protein kinase
- MLCK
myosin light chain kinase
- NHERF
Na+-H+ exchanger regulatory factor
- PDZ
postsynaptic density 95/discs large/zona occludens-1
- Scrib
scribble
- SH3
Src homology 3
- Ste5p
sterile 5
- ZO-1
zona occludens-1
Footnotes
REFERENCES
- Ardura JA, Friedman PA. Regulation of G protein-coupled receptor function by Na+/H+ exchange regulatory factors. Pharmacol Rev. 2011;63:882–900. doi: 10.1124/pr.110.004176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya RP, Reményi A, Good MC, Bashor CJ, Falick AM, Lim WA. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- Bilder D, Schober M, Perrimon N. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat Cell Biol. 2003;5:53–58. doi: 10.1038/ncb897. [DOI] [PubMed] [Google Scholar]
- Choi K-Y, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning AS, Anderson JM. Zonula occludens-1 and -2 are cytosolic scaffolds that regulate the assembly of cellular junctions. Ann NY Acad Sci. 2009;1165:113–120. doi: 10.1111/j.1749-6632.2009.04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Bretscher A. PDZ interactions regulate rapid turnover of the scaffolding protein EBP50 in microvilli. J Cell Biol. 2012;198:195–203. doi: 10.1083/jcb.201204008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, LaLonde DP, Bretscher A. The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J Cell Biol. 2010;191:397–413. doi: 10.1083/jcb.201004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Sauvanet C, Viswanatha R, Bretscher A. The tails of apical scaffolding proteins EBP50 and E3KARP regulate their localization and dynamics. Mol Biol Cell. 2013;24:3381–3392. doi: 10.1091/mbc.E13-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Regulation of cadherin adhesive activity. J Cell Biol. 2000;148:399–404. doi: 10.1083/jcb.148.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–E108. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Jin M, Lew DJ. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr Opin Genet Dev. 2011;21:740–746. doi: 10.1016/j.gde.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Lauffer BE, Melero C, Temkin P, Lei C, Hong W, Kortemme T, von Zastrow M. SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J Cell Biol. 2010;190:565–574. doi: 10.1083/jcb.201004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden SL, Hardin J. The secret life of α-catenin: moonlighting in morphogenesis. J Cell Biol. 2011;195:543–552. doi: 10.1083/jcb.201103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Socolich M, Wall MA, Graves J, Wang Z, Ranganathan R. Dynamic scaffolding in a G protein-coupled signaling system. Cell. 2007;131:80–92. doi: 10.1016/j.cell.2007.07.037. [DOI] [PubMed] [Google Scholar]
- Morales FC, Takahashi Y, Kreimann EL, Georgescu MM. Ezrin-radixin-moesin (ERM)-binding phosphoprotein 50 organizes ERM proteins at the apical membrane of polarized epithelia. Proc Natl Acad Sci USA. 2004;101:17705–17710. doi: 10.1073/pnas.0407974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Fehon RG. Analysis of protein dynamics within the septate junction reveals a highly stable core protein complex that does not include the basolateral polarity protein Discs large. J Cell Sci. 2011;124:2861–2871. doi: 10.1242/jcs.087700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Baribault H, Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21:4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Printen JA, Sprague GF. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Berryman M, Bretscher a. Identification of EBP50: a PDZ-containing phosphoprotein that associates with members of the ezrin-radixin-moesin family. J Cell Biol. 1997;139:169–179. doi: 10.1083/jcb.139.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H-W, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero G, von Zastrow M, Friedman PA. Role of PDZ proteins in regulating trafficking, signaling, and function of GPCRs: means, motif, and opportunity. Adv Pharmacol. 2011;62:279–314. doi: 10.1016/B978-0-12-385952-5.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro MN, Bussineau CM, Cotman CW. Postsynaptic density antigens: preparation and characterization of an antiserum against postsynaptic densities. J Cell Biol. 1981;90:675–686. doi: 10.1083/jcb.90.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenolikar S, Weinman EJ. NHERF: targeting and trafficking membrane proteins. Am J Physiol Renal Physiol. 2001;280:F389–95. doi: 10.1152/ajprenal.2001.280.3.F389. [DOI] [PubMed] [Google Scholar]
- Shen L, Weber CR, Turner JR. The tight junction protein complex undergoes rapid and continuous molecular remodeling at steady state. J Cell Biol. 2008;181:683–695. doi: 10.1083/jcb.200711165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh B, Zhu M. Regulation of the TRP Ca channel by INAD in Drosophila photoreceptors. Neuron. 1996;16:991–998. doi: 10.1016/s0896-6273(00)80122-1. [DOI] [PubMed] [Google Scholar]
- Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terawaki S, Maesaki R, Hakoshima T. Structural basis for NHERF recognition by ERM proteins. Structure. 2006;14:777–789. doi: 10.1016/j.str.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Thompson BJ. Cell polarity: models and mechanisms from yeast, worms and flies. Development. 2013;140:13–21. doi: 10.1242/dev.083634. [DOI] [PubMed] [Google Scholar]
- van Drogen F, Stucke VM, Jorritsma G, Peter M. MAP kinase dynamics in response to pheromones in budding yeast. Nat Cell Biol. 2001;3:1051–1059. doi: 10.1038/ncb1201-1051. [DOI] [PubMed] [Google Scholar]
- Viswanatha R, Bretscher A, Garbett D. Dynamics of ezrin and EBP50 in regulating microvilli on the apical aspect of epithelial cells. Biochem Soc Trans. 2014;42:189–194. doi: 10.1042/BST20130263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R, Wayt J, Ohouo PY, Smolka MB, Bretscher A. Interactome analysis reveals ezrin can adopt multiple conformational states. J Biol Chem. 2013;288:35437–35451. doi: 10.1074/jbc.M113.505669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- Wu VM, Beitel GJ. A junctional problem of apical proportions: epithelial tube-size control by septate junctions in the Drosophila tracheal system. Curr Opin Cell Biol. 2004;16:493–499. doi: 10.1016/j.ceb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Marchiando AM, Weber CR, Raleigh DR, Wang Y, Shen L, Turner JR. MLCK-dependent exchange and actin binding region-dependent anchoring of ZO-1 regulate tight junction barrier function. Proc Natl Acad Sci USA. 2010;107:8237–8241. doi: 10.1073/pnas.0908869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science. 2012;337:1218–1222. doi: 10.1126/science.1220683. [DOI] [PMC free article] [PubMed] [Google Scholar]