A novel evolutionarily conserved motif termed ROCC (regulation of cohesion and condensation) is identified in the regulatory subunit of cohesin. This study offers important insights into the regulation of the maintenance of cohesion and condensation and the molecular basis for cohesin's chromatin-tethering activity.
Abstract
Cohesin helps orchestrate higher-order chromosome structure, thereby promoting sister chromatid cohesion, chromosome condensation, DNA repair, and transcriptional regulation. To elucidate how cohesin facilitates these diverse processes, we mutagenized Mcd1p, the kleisin regulatory subunit of budding yeast cohesin. In the linker region of Mcd1p, we identified a novel evolutionarily conserved 10–amino acid cluster, termed the regulation of cohesion and condensation (ROCC) box. We show that ROCC promotes cohesion maintenance by protecting a second activity of cohesin that is distinct from its stable binding to chromosomes. The existence of this second activity is incompatible with the simple embrace mechanism of cohesion. In addition, we show that the ROCC box is required for the establishment of condensation. We provide evidence that ROCC controls cohesion maintenance and condensation establishment through differential functional interactions with Pds5p and Wpl1p.
INTRODUCTION
Cohesin, a founding member of the structural maintenance of chromosomes (SMC) family of proteins, was originally identified for its critical role in chromosome segregation (Strunnikov et al., 1993). Cohesin tethers sister chromatids in order to generate the cohesion necessary for proper chromosome segregation. However, additional studies revealed that cohesin is also important for condensation, regulation of gene expression, and DNA repair (Guacci et al., 1997; Guacci and Koshland, 2012; Donze et al., 1999; Rollins et al., 1999; Unal et al., 2004; Yeh et al., 2008). Because these different chromosomal processes have distinct spatial and temporal requirements, cohesin's activity (activities) must be strictly parsed and regulated. Many aspects of its precise regulation remain to be elucidated. For example, what are the activities of cohesin needed to maintain sister chromatid cohesion from S phase until anaphase onset? What prompts cohesin to tether chromosomes rather than condense them? Are these cues governed by distinct modes of regulation through distinct regulatory factors? In this study, we address these fundamental questions by examining the regulation of cohesin in both sister chromatid cohesion and condensation using the budding yeast Saccharomyces cerevisiae.
The persistence of cohesion in G2 and M phase is an active process that involves two auxiliary cohesin factors, Pds5 and Wpl1. Whereas Pds5p promotes cohesion maintenance, Wpl1p/Rad61p (Wapl in other organisms) inhibits it (Hartman et al., 2000; Kueng et al., 2006; Zhang et al., 2009; Peters and Nishiyama, 2012). Wpl1p's function can be explained in the context of the simple embrace model for cohesion. In the embrace model the cohesion between sister chromatids is generated when they are topologically entrapped by a single cohesin ring during S phase. So long as the cohesin ring remains intact and closed, cohesin will continue to entrap the two sister chromatids, maintaining both cohesion and its stable binding to chromosomes. In this simple model for cohesin function (the embrace model), the only way to dissolve cohesion is to disrupt the integrity of the cohesin ring, causing not only the escape of the sister chromatids but also the dissociation of cohesin from the chromosome. Consistent with this model, recent studies suggest that Wpl1p regulates cohesin binding to chromosomes, resulting in dissociation of cohesin from the chromosomes and dissolution of cohesion (Sutani et al., 2009). In this context, a potential positive activity for Pds5p in cohesion maintenance could be the protection of cohesin from the putative destabilizing activity of Wpl1p. However, in budding yeast, deletion of WPL1 (wpl1∆) does not rescue the cohesion maintenance defect of a pds5 mutant (Chan et al., 2013). Thus Pds5p must preserve cohesion by antagonizing a Wpl1p-independent mechanism of cohesion dissolution (Noble et al., 2006; Baldwin et al., 2009; Chan et al., 2013; D'Ambrosio and Lavoie, 2014). Does this alternative dissolution mechanism also destabilize cohesin binding to chromosomes or inhibit another activity of cohesin not predicted by the embrace model?
In budding yeast, cohesin and cohesin auxiliary factors also function in mitotic chromosome condensation. Inactivation of any cohesin complex subunit, as well as cohesin accessory factors Eco1p/Ctf7 and Pds5p, perturbed condensation at euchromatic single-gene-copy regions and at the heterochromatic ribosomal DNA (rDNA) repeats of the RDN locus (Guacci et al., 1997; Guacci and Koshland, 2012; Skibbens et al., 1999; Hartman et al., 2000). Thus factors that promote sister chromatid cohesion such as cohesin, Pds5p, and Eco1p also serve to promote cohesin-mediated condensation. In contrast, Wpl1p likely inhibits cohesin-mediated condensation, as well as cohesion. Indeed, the deletion of WPL1 (wpl1∆) suppresses the condensation defect of cells lacking Eco1p, and overexpression of Wpl1p in wild-type cells induces hypercondensation (Guacci and Koshland, 2012; Lopez-Serra et al., 2013). Together these results suggest a strong connection between factors that control the maintenance of cohesion and condensation. How do factors like Pds5p and Wpl1p communicate with cohesin to regulate condensation and cohesion?
Here we investigate how cohesin is regulated to ensure that cohesion is maintained in G2 and M phase yet still enables the establishment and maintenance of condensation. We chose to mutate the Mcd1p/Rad21p/Scc1p regulatory subunit of cohesin due to its regulatory roles in cohesion establishment, DNA repair, and cohesion dissolution at anaphase. We identified a cluster of conserved residues in the linker domain of Mcd1p, which we define as the regulation of cohesion and condensation (ROCC) box because they are required for the maintenance of cohesion and the establishment of condensation. The ROCC box together with Pds5p prevents cohesion dissolution by a mechanism independent of Wpl1p. Disruption of this ROCC-mediated mechanism does not destabilize cohesin binding to chromosomes, yet abrogates cohesion maintenance. Therefore, whereas stable binding of cohesin to chromosomes is necessary for cohesion maintenance, it is not sufficient. The ROCC box also promotes the establishment and maintenance of condensation by antagonizing Wpl1p's anticondensation activity. Taken together, our results suggest that ROCC controls cohesion maintenance and condensation establishment through differential functional interactions with Pds5p and Wpl1p, respectively.
RESULTS
Q266 region of Mcd1p is required for the maintenance of sister chromatid cohesion
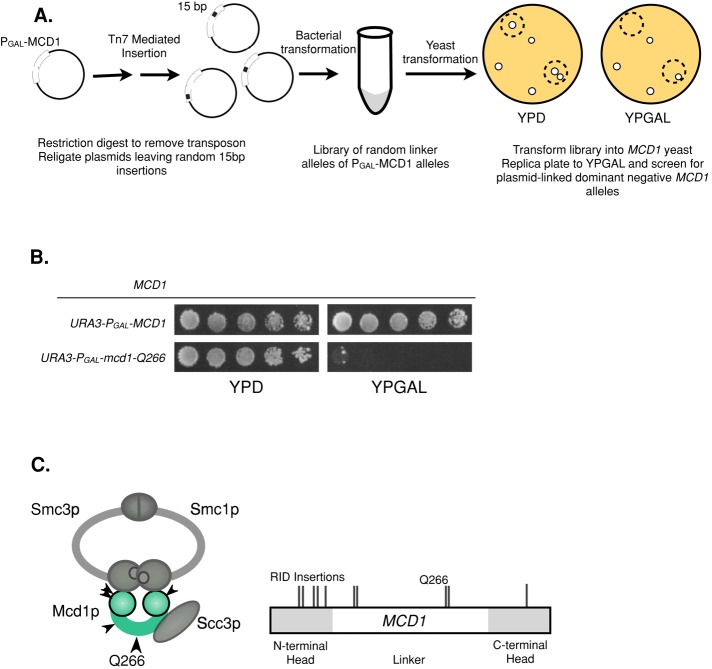
We sought to understand how cohesin function is coordinated in G2 and M phase to maintain sister chromatid cohesion while promoting chromosome condensation. We suspected that this coordination might be mediated by Mcd1p, the key cohesin regulatory subunit. Consequently we sought to identify MCD1 alleles for which the maintenance of cohesion was uncoupled from the establishment of condensation. Such partially functional, separation-of-function alleles were likely to be rare. We previously showed that partially functional alleles of Smc1p, another cohesin subunit, were highly enriched in a particular mutagenic strategy called random insertion dominant negative (RID; Milutinovich et al., 2007). Therefore we subjected MCD1 to RID mutagenesis to generate a DNA library of minichromosome-borne mcd1 mutants, each containing a single 15–base pair (five-residue) in-frame insertion (Figure 1A and Materials and Methods). The complexity of the library was calculated to span the MCD1 open reading frame (ORF) with one insertion every ∼2.5 base pairs of MCD1. This insertion library was transformed into wild-type yeast. The RID-mutagenized MCD1 was under control of the pGAL promoter. Therefore transformants were assayed for toxicity in the presence of galactose, which induces overexpression of the RID mutant proteins but not under noninducing conditions on dextrose.
FIGURE 1:
Isolation of RID mutants in MCD1. (A) Schematic of RID mutagenesis. Plasmid pVG385 containing MCD1 driven by the GAL1 promoter (pGAL-MCD1) was subjected to Tn7 transposition in vitro. The inserted transposon was removed by PmeI digestion, leaving random 15–base pair insertions. The library was transformed into either haploid WT (VG3349-1B) or mcd1-1 (VG3312-7A) strains, and transformants were screened for inviability or slow growth on galactose. (B) Haploid WT (VG3349-1B) yeast bearing pGAL-MCD1 or pGAL-mcd1-Q266 were grown at 23°C to saturation and then plated in 10-fold serial dilutions onto medium containing galactose (YPGAL) or dextrose (YPD) and incubated at 23°C for 3 d. pGAL-Mcd1-Q266 is semidominant over MCD1 and more penetrant at 16°C (unpublished data). (C) Schematic of cohesin structure and map of RID mutation positions in MCD1. The four subunits of the cohesin complex are represented. The head domains of Smc1p and Smc3p are shown as large gray balls at the base of these proteins. The amino and carboxy globular domains of Mcd1p are shown as green balls to the left and right, with the intervening green bar representing the linker region. Map of Rid mutations is shown to the right. Gray areas indicate globular domains as determined by sequence conservation.
We identified 32 RID-induced mutations that led to a dominant phenotype of slow growth or inviability (Figure 1B and Supplemental Table 2). Sequencing of these dominant-negative RID mutations identified 10 unique alleles of MCD1 (Figure 1C and Supplemental Table 2). Most of these RID alleles were in the amino- and carboxy-terminal globular domains, likely inhibiting the interaction of these domains with the head domains of the Smc3p and Smc1p, respectively (Figure 1C). The dominant phenotypes of these RID proteins were easily explained. We posited that by virtue of binding to only one of the two Smc head domains, these mutant proteins could assemble into a nonfunctional complex for cohesion, preventing the assembly of a functional cohesin with wild-type Mcd1p. In contrast, one RID allele arose from an insertion immediately following residue Q266 (mcd1-Q266) in the linker region of Mcd1p (Figure 1C). How this allele might generate a dominant phenotype was not clear. The unusual position of this insertion allele prompted us to pursue it further as a candidate for identifying a potentially novel regulatory domain for cohesin function in M phase.
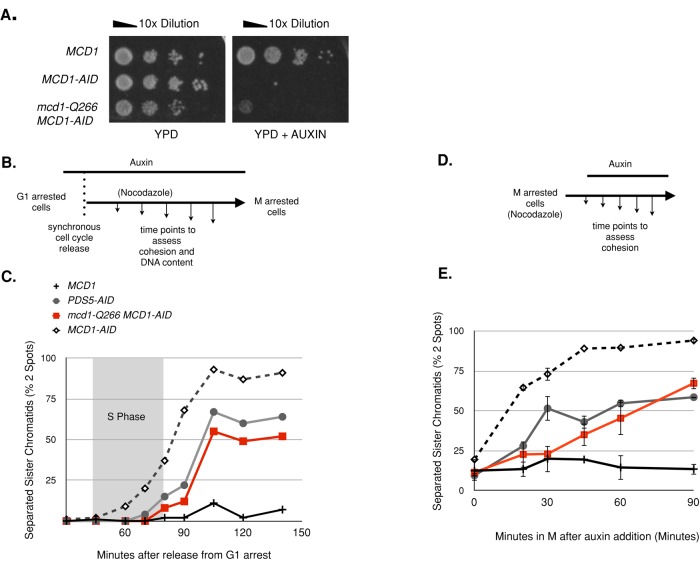
As a first step in characterizing this allele, we wanted to study its phenotypes under normal levels of expression and in the absence of Mcd1p. To eliminate Mcd1p, we generated a strain that contained MCD1-AID (at the MCD1 locus) and TIR1 (see Materials and Methods). AID (Auxin-Inducible Degron) encodes a small domain that binds the plant hormone auxin (Nishimura et al., 2009). TIR1 encodes Tir1p, an alternative F box required for SCF-mediated degradation of AID-tagged proteins (Gray et al., 1999). Because TIR1 was introduced into all strains in this study bearing an AID-tagged protein, it is omitted from all genotypes in the text for simplicity. In the presence of auxin, proteins coupled to AID are ubiquitinated and targeted for degradation. The effectiveness of auxin-induced degradation of MCD1-AIDp was evident by the inability of the MCD1-AID strain to grow in the presence of auxin and the dramatic reduction of Mcd1-AIDp within 30 min of auxin addition (Figure 2A and Supplemental Figures S1 and S2). Into this MCD1-AID strain we integrated an mcd1-Q266 allele under control of the MCD1 promoter at the URA3 locus. This mcd1-Q266 MCD1-AID strain grew normally in the absence of auxin but failed to grow in the presence of auxin (Figure 2A). Because MCD1-AID is degraded in response to auxin treatment, this result demonstrates that mcd1-Q266 expressed at wild-type levels is unable to support viability on its own and is recessive to wild-type MCD1. Thus Mcd1-Q266p is defective for one or more Mcd1p activities.
FIGURE 2:
mcd1-Q266 is recessive to MCD1 but exhibits a defect in cohesion maintenance after Mcd1p is removed via the auxin degron system. (A) Haploid yeast strains MCD1 (DK5531), MCD1-AID (DK5501), and mcd1-Q266 MCD1-AID (DK5535) were grown at 23°C to saturation, plated in 10-fold serial dilutions onto rich medium (YPD) or rich medium plus 500 μM auxin (YPD + auxin), and then incubated for 3 d at 23°C. (B) Schematic to measure cohesion establishment and maintenance as cells progress from G1 to M. Early-log-phase cells growing at 23°C in YPD were arrested in G1 using αF, and then auxin (500 μM) was added and cells incubated for an additional 1 h in G1. Cells were then released from G1 arrest into YPD medium containing auxin (500 μM) and nocodazole (15 μg/ml) to allow cell-cycle progression until arrest in M phase. Cell aliquots were fixed and processed every 15 min to assess cohesion and DNA content (see Supplemental Figure S3 and Material and Methods). (C) Analysis of cohesion in mcd1-Q266 cells during progression from G1 to M. Haploid MCD1 (DK5531, solid black crosses), PDS5-AID (DK5540, gray circles), MCD1-AID (DK5542, open diamonds), and mcd1-Q266 MCD1-AID (DK5535, red squares) were assessed for cohesion as described in B. The percentage of cells with two GFP foci at a CEN-distal locus (LYS4) is plotted. From 200 to 300 cells were counted at each time point. S phase is marked with a gray box (Supplemental Figure S3). The experiment was repeated three times, and representative time course is shown. (D) Schematic to measure cohesion maintenance in M phase–arrested cells. Early-log-phase cells growing at 23°C in YPD were arrested by addition of nocodazole (15 μg/ml) and incubation for 2.5 h. Auxin (500 μM) was added and cells incubated an additional 90 min. Cell aliquots were processed for the GFP cohesion assay before auxin addition (T = 0) and at 15-min intervals after auxin addition. (E) Analysis of cohesion in mcd1-Q266 strains in M-phase arrest. Haploid MCD1 (DK5531, solid black crosses), PDS5-AID (DK5540, gray circles), MCD1-AID (DK5542, open diamonds), and mcd1-Q266 MCD1-AID (DK5535, red squares) were treated and assessed for cohesion as described in D. Percentage of cells with two GFP foci at a CEN-distal locus LYS4. From 100 to 300 cells were counted at each time point. Data were generated from two independent experiments; error bars show SD.
We then examined cohesion in the MCD1 strain, MCD1-AID strain, and mcd1-Q266 MCD1-AID strain as cells progressed from G1 to M phase. In this experiment, G1-arrested cultures were treated with auxin and then released from their pheromone-induced arrest into media containing auxin and nocodazole (Figure 2B). This regimen allowed cells to progress synchronously through the cell cycle until they rearrested in M phase due to the presence of nocodazole. Of importance, auxin was present in the media from G1 to M to destroy Mcd1-AIDp and prevent its accumulation over the entire cell-cycle window. During cell cycle progression, aliquots of cells were fixed and assayed for cohesion using LacI–green fluorescent protein (GFP) tagged loci at LYS4 and DNA content by flow cytometry.
We first compared cohesion in the MCD1 strain and MCD1-AID strain to establish a basis for the kinetics of sister chromatid separation in the absence of cohesin. As expected for the MCD1 strain, very few separated sister chromatids appeared during the course of the experiment, as wild-type Mcd1p is not affected by auxin (Figure 2C). In contrast, under conditions in which the MCD1-AID strain lacked cohesin and any cohesion establishment, the strain showed a dramatic increase in sister chromatid separation during DNA replication (Figure 2C). The final fraction of cells with separated sister chromatids in the MCD1-AID strain (∼90%) was much greater than reported previously with conditional temperature-sensitive alleles in MCD1, SMC1, and SMC3 (∼65–70%; Guacci et al., 1997; Michaelis et al., 1997). The significant residual cohesion seen in the temperature-sensitive mutants has been interpreted to mean that cohesin-independent pathways for cohesion exist (Shimada and Gasser, 2007). The existence of these cohesin-independent pathways is challenged by the near-complete loss of cohesion in MCD1-AID strain at the genetic loci tested. Instead, the residual cohesion in the temperature-sensitive alleles likely resulted from incomplete inactivation of the mutant proteins.
Using these results as a foundation, we examined cohesion in the mcd1-Q266 MCD1-AID strain. The addition of auxin caused a dramatic increase in sister chromatid separation (Figure 2C). However, in contrast to the separation in MCD1-AID cells, precocious sister separation in the mcd1-Q266 MCD1-AID strain was not detected until ∼20 min after the completion of S phase (see flow cytometry, Supplemental Figure S3). Identical behavior was observed at a centromere- proximal locus, TRP1 (Supplemental Figure S4). This delay indicates that mcd1-Q266p establishes cohesion during S phase and maintains it for at least 20 min before losing it in G2 and M phase.
To corroborate our conclusion that mcd1-Q266p has a cohesion maintenance defect, we compared the kinetics of its cohesion loss to that seen in a PDS5-AID strain (Figure 2C). The kinetics and degree of cohesion loss in the PDS5-AID and mcd1-Q266 MCD1-AID strains were indistinguishable. Thus mcd1-Q266 causes a cohesion maintenance defect very similar that seen upon inactivation of Pds5p, a protein whose characterization set the paradigm for cohesion maintenance (Noble et al., 2006).
We next assessed cohesion maintenance of mcd1-Q266 by a second assay in which cells were arrested in M phase and then AID-tagged proteins were destroyed. Cultures of four strains (MCD1, mcd1-Q266 MCD1-AID, MCD1-AID, and PDS5-AID) were first arrested in M phase using nocodazole and then analyzed for cohesion (Figure 2D). All strains had robust cohesion (Figure 2E, 0 min). Auxin was added to medium that still contained nocodazole to deplete Mcd1-AIDp while cells remained arrested in M phase (Figure 2D). As expected, MCD1 cells retained cohesion, whereas the MCD1-AID cells rapidly lost cohesion (Figure 2E). Auxin addition also induced cohesion loss in the mcd1-Q266 MCD1-AID and PDS5-AID strains to similar levels (Figure 2E). This result further corroborates the cohesion maintenance defect of mcd1-Q266, as well as its similarity to the cohesion defect of PDS5-AID. Their cohesion loss was delayed relative to the MCD1-AID strain (Figure 2E). This delay suggests that the mechanism of cohesion dissolution in mcd1-Q266 MCD1-AID and PDS5-AID strains is distinct from what occurs when the sole Mcd1p reservoir in cells is rapidly degraded.
mcd1-Q266 identifies a mechanism for cohesion dissolution without destabilization of cohesin binding to chromosomes
A number of possible explanations existed for the defective cohesion maintenance of the mcd1-Q266 allele. The proximity of Q266 insertion to the Esp1 (separase) cleavage site suggested that the Q266 residue might be part of a motif required to prevent precocious cleavage of Mcd1p by Esp1p before anaphase (Uhlmann et al., 1999). This seemed unlikely, given that cells were arrested in M phase using nocodazole, a time in the cell cycle when Pds1p is present (Yamamoto et al., 1996a, b), so it should inhibit Esp1p. Indeed, we did not detect cleavage or any other degradation of mcd1-Q266p in nocodazole-treated cells (Supplemental Figure S5). Moreover, the overall Mcd1p levels were the same for MCD1 and mcd1-Q266 alleles (Supplemental Figure S5). These results ruled out Mcd1p degradation as a cause for the cohesion maintenance defect.
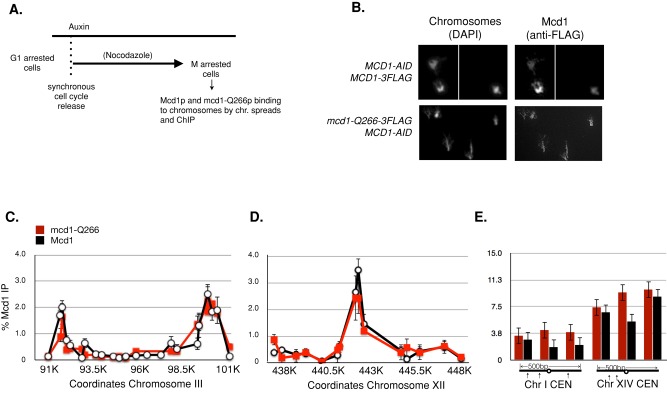
We then turned to possibility that mcd1-Q266p caused a defect in cohesion maintenance because it disrupted the binding of cohesin to chromosomes. G1-arrested cultures of MCD1-3FLAG MCD1-AID strain and mcd1-Q266-3FLAG MCD1-AID strain were treated with auxin and then released from their pheromone-induced arrest into media containing auxin and nocodazole to allow synchronous progression through the cell cycle until M-phase arrest (Figure 3A). Auxin was present in the media from G1 to M to destroy Mcd1-AIDp and prevent its accumulation, leaving only the epitope-tagged versions of either Mcd1p or mcd1-Q266p in cells. After reaching M phase, when the maximal cohesion defect of mcd1-Q266 strains is manifested (Figure 2C), both cultures were fixed. The fixed cultures were processed for chromosome spreads and chromatin immunoprecipitation (ChIP) to assess Mcd1p and mcd1-Q266p binding to chromosomes. Note that the chromosomal binding of these two Mcd1p variants is a surrogate measure of cohesin's binding to chromosomes because in yeast, Mcd1p and the other three subunits of cohesin are all completely interdependent for chromosomal binding (Tóth et al., 1999; Unal et al., 2008; Heidinger-Pauli et al., 2010b).
FIGURE 3:
mcd1-Q266p and Mcd1p exhibit similar and robust binding to chromosomes. (A) Schematic for analysis of cohesin binding as determined by chromatin immunoprecipitation. Early-log-phase cells were arrested in G1, treated with auxin to deplete MCD1-AID, and released from G1 and rearrested in M phase using nocodazole at 23°C as described in Figure 2, A and B. Cells were processed for chromosome spreads (B) and for chromatin immunoprecipitation (C–E). For ChIP experiments, the average cohesin binding from two biological replicates is plotted. Error bars represent SD between both experiments. (B) Chromosome spreads. Mcd1-3FLAG MCD1-AID (DK5536; top) and mcd1-Q266-3FLAG MCD1-AID (DK5535; bottom) cells were processed for chromosome spreads (see Materials and Methods). MCD1-Q266-3FLAGp or MCD1-3FLAGp was visualized using mouse anti-FLAG antibody (anti-FLAG) and DNA (DAPI). Data are from one of three independent experiments. (C–E) ChIP. Strains in B were processed for ChIP to assess FLAG tagged Mcd1p binding using rabbit anti-Mcd1p antibodies (Materials and Methods). Mcd1p (black lines or black bars) or mcd1-Q266p (red lines or red bars). (C) Chromosome III pericentric domain ChIP. Mcd1p binding was assessed by quantitative PCR (qPCR) using primers spanning a 10-kb region. Primer pairs were spaced every ∼500 base pairs. (D) Chromosome XII ChIP at a single-copy domain near the rDNA. Mcd1p binding over a 10-kb region was assessed by qPCR using primer pairs spaced every ∼500 base pairs. (E) CEN1 and CEN14 ChIP. Mcd1p binding at three loci immediately flanking CEN1 and CEN14. All primer sets examine DNA sequences within 500 base pairs of either centromere.
By chromosome spreads, we observed robust staining of Mcd1-3FLAGp and mcd1-Q266-3FLAGp on chromosomes. Thus the general binding of cohesin to chromatin was unaffected by mcd1-Q266p (Figure 3B). The more extended nature of the spread chromosomes of mcd1-Q266 cells compared with MCD1 cells made quantification of the staining difficult. However, we were able to use ChIP to quantify the binding of the Mcd1-3FLAGp and mcd1-Q266p at specific loci. The pattern and amount of their binding were indistinguishable at the centromere-proximal CARC1, the centromere-distal CARL1, and the centromeres of chromosomes 1 and 14 (Figure 3, C–E). Our results from both chromosome spreads and ChIP indicate that mcd1-Q266p does not compromise the total amount or the locus-specific targeting of cohesin to chromosomes.
The fact that steady-state levels of chromosomal binding for Mcd1-Q266p and Mcd1p were identical did not eliminate the possibility that mcd1-Q266p increased the dissociation of cohesin from chromosomes. Cohesin containing mcd1-Q266p might have been undergoing rapid cycles of disassociation and reassociation with chromosomes. To assess this possibility, we decided to inactivate the cohesin loader after cells were arrested in M phase. This strategy would allow cohesin to assemble onto chromosomes normally during S phase, but then, after the loader depletion in M, any cohesin that fell off chromosomes could no longer be reloaded onto chromosomes. Any instability in cohesin binding on chromosomes would be detected as a decrease in the amount remaining bound on chromosomes with time. The use of ChIP would enable us to monitor the stability of cohesin binding at small and specific chromosomal loci at any part of a chromosome. In previous studies, the stability of cohesin binding to chromosomes was measured by fluorescence recovery after photobleaching. However, the bleaching method in yeast is limited to measuring the stability of cohesin bound over a cohesin barrel, which forms over a 10- to 15-kb region flanking each of the centromeres (Yeh et al., 2008).
To execute this loader-inactivation strategy, we replaced the chromosomal copy of SCC2, the best-characterized subunit of the cohesin loader complex, with an SCC2-AID allele. As expected, the SCC2-AID strain was inviable on plates containing auxin (Supplemental Figure S2). We then assessed how effective depletion of the Scc2-AIDp was blocking cohesion generation. The SCC2-AID strain was staged in G1 and then depleted of Scc2-AIDp by the addition of auxin. Cells were synchronously released from G1 and in the presence of both nocodazole and auxin to continually destroy the loader and prevent its accumulation during cell cycle progression. Western blot analysis revealed that Scc2-AIDp was reduced below the level of detection (Supplemental Figure S6). We also assessed sister chromatid cohesion and cohesin binding to chromosomes (Supplemental Figure S6). Sister chromatid cohesion was reduced to levels comparable to that seen after inactivation of a cohesin subunit. In addition, cohesin binding at CARC1 was reduced to background levels. These results indicated that Scc2-AIDp function was severely compromised in the presence of auxin, making it an effective tool with which to study the stability of cohesin binding to chromosomes.
To ask whether inactivation of Scc2-AIDp can efficiently block cohesin loading in M phase, we generated a strain that contained the SCC2-AID, MCD1, and MCD1-6HA tagged under control of the pGAL promoter (pGAL-MCD1-6HA). We arrested a culture of this strain in M phase using nocodazole. The culture was split, and auxin was added to one-half to deplete Scc2-AIDp. Western blot analysis indicated that Scc2-AIDp was reduced to undetectable levels by 15 min after auxin addition (Supplemental Figure S7). Thirty minutes later, galactose was added to both cultures and allowed to incubate for an additional 60 min. Mcd1-6HAp was induced to high levels as assayed by Western blot in both cultures (Supplemental Figure S7). In the auxin-free culture, Mcd1-6HAp exhibited robust chromosomal binding, as assayed by chromosome spreads (Supplemental Figure S7). This result is consistent with published results that the cohesin loader is active and competent to load cohesin in M phase (Ström et al., 2004). In contrast, in the auxin-treated (Scc2-AIDp depleted) culture, Mcd1-6HAp failed to load onto chromosomes. Thus inactivation of the loader in M phase successfully prevented nucleoplasmic cohesin from binding chromosomes during M.
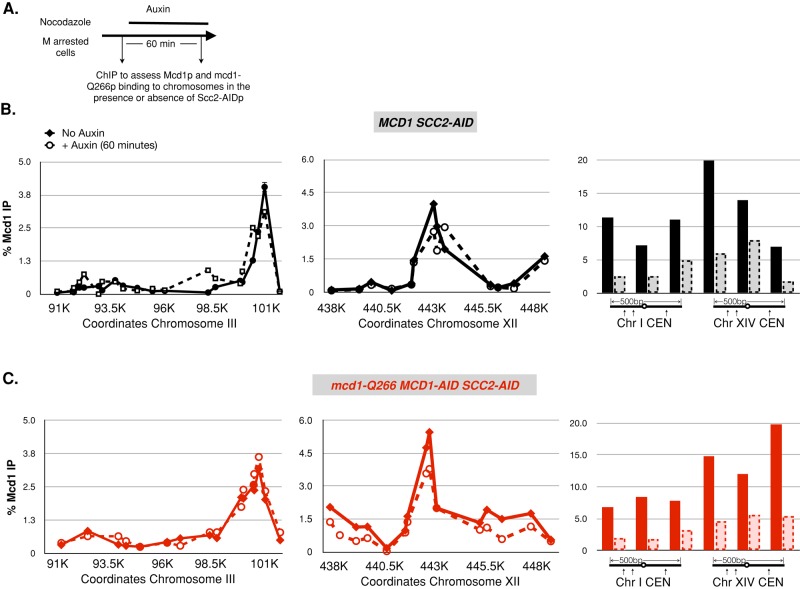
With the conditional SCC2-AID in hand, we analyzed the dissociation of both mcd1-Q266p and Mcd1p from chromosomes. We first generated a strain bearing the SCC2-AID and the MCD1-AID as the sole SCC2 and MCD1 alleles in cells. We then integrated a second copy of either MCD1-3FLAG or an mcd1-Q266-3FLAG allele at the URA3. We refer to these strains as the MCD1 and mcd1-Q266 stability strains, respectively. We arrested these two strains in M phase using nocodazole to allow normal loading of Mcd1-AIDp and MCD1-3FLAGp or mcd1-Q266-3FLAGp (Figure 4A). Cultures were split, and auxin was added to one-half. Auxin addition rapidly depleted the Scc2-AIDp loader within 15 min and the MCD1-AID at least within 30 min, leaving only the MCD1-3FLAGp or mcd1-Q266-3FLAGp on chromosomes (Supplemental Figures S1 and S6). After an additional 45 min in auxin, the two M-phase cultures were processed for ChIP to assess the binding of Mcd1-3FLAGp and mcd1-Q266-3FLAGp to chromosomes. Because Scc2-AIDp was fully depleted by 15 min after auxin addition, any ChIP signal that persisted at 60 min reflected Mcd1-3FLAGp or mcd-Q266-3FLAGp that remained bound to their chromosomal sites for at least 45 min (Supplemental Figure S7). Of importance, the percentage separated sister chromatids in MCD1 (15%) and mcd1-Q266 (50%) stability strains were the same as we observed previously in MCD1 SCC2 and mcd1-Q266 SCC2 strains (Figure 2E). Thus the depletion of Scc2-AIDp did not suppress or enhance the cohesion maintenance defect of mcd1-Q266.
FIGURE 4:
mcd1-Q266 and wild-type cohesin are stably bound on chromosomes at CAR sites but equally unstable at CENs. (A) Schematic to assess stability of the cohesin complex on chromosomes in M-phase cells. Cells were arrested in M phase as described in the Figure 2D legend. The culture was split in half, and auxin (500 μM) was added to one half; each half was incubated for 1 h in M phase, and then samples were processed for ChIP using rabbit anti-Mcd1 antibody (see Materials and Methods). (B, C) Analysis of cohesin binding to chromosomes in M phase after Scc2p loader depletion (stability strains). We used two haploid MCD1 stability strains, which contained (SCC2-AID MCD1-AID) and either MCD1-3FLAG (DK5560) or mcd1-Q266-3FLAG (DK5539). Strains were grown and processed as in A. FLAG-tagged Mcd1p binding at specific chromosomal sites was assessed by ChIP using qPCR. Two independent experiments were performed and gave equivalent results, one of which is shown here. (B) Effect of loader depletion on Mcd1-3FLAGp cohesin binding to chromosomes. ChIP of Mcd1-3FLAGp stability strain DK5560 when Scc2p was active (No Auxin; solid line) or after Scc2p loader was depleted (+ Auxin, dotted black line). Chip analysis at chromosome III pericentric region (left), chromosome XII single-copy arm region (middle). and within 500 base pairs of CEN3 and CEN14 (right). Data are the average of two qPCR replicates for each primer pair. (C) Effect of loader depletion on mcd1-Q266p cohesin binding to chromosomes. ChIP of Mcd1-Q266-3FLAG stability strain DK5539 when Scc2p was active (no auxin, solid red line) or after Scc2p loader was depleted (+ Auxin, dotted red line). ChIP analysis was performed at same loci as in B.
In the MCD1 stability strain, the amount of Mcd1-3FLAGp binding at CARC1 was unaffected by Scc2-AIDp depletion (Figure 4B, left), suggesting that cohesin binds very stably to this site. The proximity of CARC1 to the centromere places it likely very near or within the centromere barrel. Previous studies using fluorescence recovery after photobleaching of the centromere barrel also indicated stable cohesin binding in this region (Yeh et al., 2008; Mishra et al., 2010). This similarity validates our Scc2-AIDp degradation method as a metric for assessing stably bound cohesin in M phase.
The SCC2-AID tool also allowed us to measure the stability of cohesin binding to chromosomal sites outside the centromere barrel and at much finer resolution. The amount of Mcd1-3FLAGp binding at CARL1, a cohesin-binding site in the middle of the arm of chromosome XII, was also unaffected by Scc2-AIDp inactivation in our MCD1 stability strain (Figure 4B, middle). Thus cohesin also bound very stably to a representative arm CAR site. In contrast, Mcd1-3FLAGp binding at two different centromeres decreased nearly threefold to fivefold in the absence of the loader (Figure 4B, right). Thus most of the cohesin in immediate proximity of centromeres is only transiently bound, implicating the cohesin barrel for chromosome-remodeling activity.
In the mcd1-Q266 stability strain, the mcd1-Q266-3FLAGp binding at both the arm CARL1 and the pericentric CARC1 remained at the same high levels in the presence or absence of the loader (Figure 4C, left and middle). Furthermore, binding at the two centromeres was reduced in the absence of loader to the same low levels as seen in the MCD1 stability strain (Figure 4C, right). Thus the binding of mcd1-Q266-3FLAGp to these three chromosome regions was indistinguishable from that of Mcd1p. Of importance, despite the stable binding of mcd1-Q266-3FLAGp at noncentromeric CARs, mcd1-Q266 cells failed to maintain cohesion. Taken together, these results suggest that the disruption of cohesion maintenance in mcd1-Q266 occurs by a mechanism distinct from cohesin dissociation from chromosomes. Moreover, it suggests that the wild-type (WT) residues in the Q266 linker region of Mcd1p are required to suppress this mechanism.
A recent study analyzed another allele of MCD1, scc1-V137K, which henceforth will be referred to as mcd1-V137K (Chan et al., 2013). This allele had hyperstable binding to chromosomes as assayed by fluorescent recovery after photobleaching of the cohesin barrel. This study concluded that the mcd1-V137K allele caused a defect in cohesion establishment based on an endpoint assay that revealed increased spacing of the centromeric clusters that generate the cohesin barrel (Chan et al., 2013). However, an endpoint assay is not suitable for distinguishing between a defect in cohesion establishment or maintenance. Therefore we made an mcd1-V137K MCD1-AID strain with a GFP-LacI/LacO reporter at LYS4 to follow cohesion through a time-course experiment. For this purpose, we used our assay of MCD1-AID depletion in G1 arrest and release into nocodazole plus auxin to assess timing of cohesion loss as cells progressed from G1 to M (Supplemental Figure S8). In the mcd1-V137K strain, sister chromatids separated ∼20 min after MCD1-AID strain and ∼15 min after the completion of S phase. This delay in sister chromatid separation was indistinguishable from the cohesion maintenance defect observed in PDS5-AID and mcd1-Q266 strains (Supplemental Figure S8 and Figure 2). Therefore, by our direct measurements, the mcd1-V137K allele does indeed have a cohesion defect, but its defect is in cohesion maintenance. This result also means that the maintenance defect in mcd1-V137K, like that in mcd1-Q266, occurs without destabilizing cohesin binding to chromosomes. Residue V137, like the Q266 region of Mcd1p, must protect against a mechanism of cohesion dissolution that disrupts a cohesin activity other than stable chromosome binding. Furthermore, stable binding of cohesin to chromosomes is insufficient to maintain cohesion.
Q266 inhibits a Wpl1-independent mechanism for cohesion dissolution
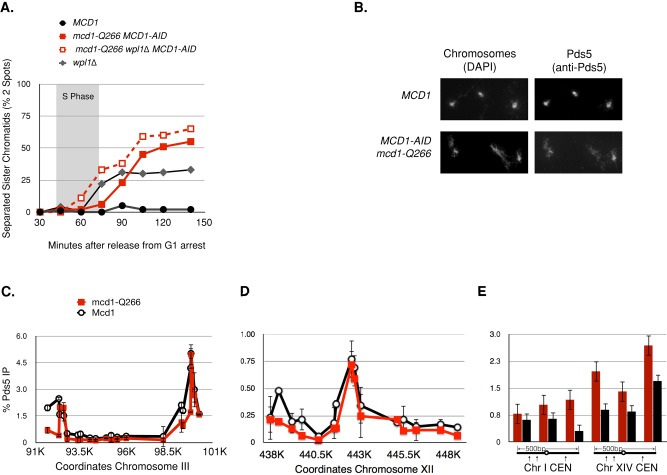
The phenotypic similarities of mcd1-Q266, and PDS5-AID prompted us to test whether the cohesion maintenance defect in these two mutants resulted from a failure to suppress a common process of cohesion dissolution. An obvious choice for this process was one mediated by Wpl1p, a cohesin inhibitor known to be active before anaphase (Uhlmann et al., 1999; Heidinger-Pauli et al., 2010b). If so, a deletion of WPL1 (wpl1∆) should suppress the precocious cohesion dissolution found in both PDS5-AID and mcd1-Q266 strains. However, a previous study revealed that cohesion dissolution in pds5 mutants was not suppressed by wpl1∆ (Chan et al., 2013). To test whether cohesion dissolution in mcd1-Q266 cells also occurs by a WPL1-independent mechanism, we deleted WPL1 in our mcd1-Q266 MCD1-AID strain. We assayed cohesion in these cells as they progressed from G1 to M to test cohesion establishment and maintenance (Figure 5).
FIGURE 5:
Mechanism of cohesion maintenance. (A) Effect of wpl1∆ on the cohesion maintenance defect of mcd1-Q266. Four haploid strains were subjected to auxin depletion in G1 phase then allowed to progress into M phase arrest under auxin depletion as described in the Figure 2B legend to assess whether WPL1 affects the mcd1-Q266 cohesion defect. Cohesion loss in MCD1 (DK5531, solid black circles), wpl1∆ (DK5561, gray diamonds), mcd1-Q266 MCD1-AID (DK5530, solid red squares), and wpl1∆ mcd1-Q266 MCD1-AID (DK5535, dotted red squares) was assessed by plotting the percentage of cells with two GFP foci at a CEN-distal locus (LYS4). From 200 to 300 cells were counted at each time point. Data are from two independent experiments. S phase is marked with a gray box. (B–E). Effect of mcd1-Q266 on Pds5p binding to chromosomes. Haploids MCD1 (DK5531) and mcd1-Q266 MCD1-AID (DK5501; bottom) were grown as described in A and then processed for chromosome spreads (B) and ChIP (C–E). (B) Chromosome spreads to detect Pds5p. The Pds5p staining in MCD1 cells (top) and mcd1-Q266 cells (bottom). Pds5p was detected using rabbit anti-Pds5p antibodies (right) and DNA detected using DAPI (left). Data were generated from two independent experiments, which gave similar results. A representative field is shown. (C–E) Pds5p ChIP. Strains in B were processed for ChIP to assess Pds5p binding using rabbit anti-Pds5p antibodies (Materials and Methods). Strains containing WT Mcd1p cohesin (DK5531, black lines) or mcd1-Q266 cohesin (DK5535, red lines) as shown. (C) Chip analysis at chromosome III pericentric region. (D) ChIP at chromosome XII single-copy arm region (middle) and (E) ChIP within 500 base pairs of CEN3 and CEN14 (right). Data from average cohesin binding from two biological replicates. Error bars represent SD between both experiments.
The wpl1∆ mcd1-Q266 MCD1-AID mutant exhibited the partial defect in cohesion establishment characteristic of wpl1∆ alone (Rowland et al., 2009; Sutani et al., 2009; Guacci and Koshland, 2012; Figure 5A, 75-min time point). Of importance, the cohesion maintenance defect in the wpl1∆ mcd1-Q266 strain was as severe as, or possibly exacerbated compared with, the mcd1-Q266 strain (Figure 5A, time points 90–150 min; see Figure 2C, MCD1-AID, for a true cohesion establishment defect). These results indicated that the precocious cohesion dissolution in mcd1-Q266, like that in pds5 mutants, occurred by a Wpl1p-independent mechanism. This independence fit with our observations that the mechanism of cohesion dissolution in mcd1-Q266 and pds5 compromised cells was inconsistent with Wpl1p's known inhibitory activity, promoting cohesin dissociation from chromosomes (Sutani et al., 2009). Taken together, our results are consistent with Pds5p and the Q266 region inhibiting a common but novel Wpl1p-independent process of cohesion dissolution.
The next question we addressed was whether the mcd1-Q266, pds5, and mcd1-V137K mutants failed to suppress this novel process because they shared a common molecular defect. In a previous study, mcd1-V137K strains were shown to be defective in the binding of Mcd1p to Pds5p by their failure to coimmunoprecipitate from soluble extracts (Chan et al., 2013). To characterize this defect further, we took advantage of the fact that Pds5p is recruited to chromosomes by cohesin and this binding is dependent upon Mcd1p (Hartman et al., 2000; Panizza et al., 2000). We assayed the ability of mcd1-V137K to impair binding of Pds5p to chromosomes by chromosome spreads (Supplemental Figure S8). The staining of Pds5p to chromosomes was dramatically reduced, indicating that mcd1-V137K blocked Pds5p binding to chromosome-bound cohesin, as well as to soluble cohesin. Thus mcd1-V137K and Pds5 mutants both affect Pds5 activity.
We then asked whether mcd1-Q266 also compromised Pds5p recruitment to chromosomes. We allowed mcd1-Q266 MCD1-AID and MCD1 strains to progress from G1 to M in the presence of auxin to generate chromosomes bound only with mcd1-Q266p or Mcd1p. We assessed Pds5p recruitment to chromosomes by chromosome spreads and ChIP. By chromosome spreads, Pds5p was bound to chromosomes containing only mcd1-Q226p, and its staining was similar to that of chromosomes containing wild-type Mcd1p (Figure 5B). Furthermore, by ChIP, the amount and position of Pds5p binding were indistinguishable at CARC1 and CARL1 in wild-type MCD1 and mcd1-Q266 cells (Figure 5, C and D). At two centromeres, there appears to be a moderate increase in Pds5p binding in mcd1-Q266 cells (Figure 5E). Thus the dramatic difference in Pds5p recruitment to chromosomes and CARs in mcd1-V137K and mcd1-Q266 suggests that these mutants have different molecular defects that lead to precocious sister chromatid separation.
The Q266 region promotes the establishment of chromosome condensation by antagonizing Wpl1p
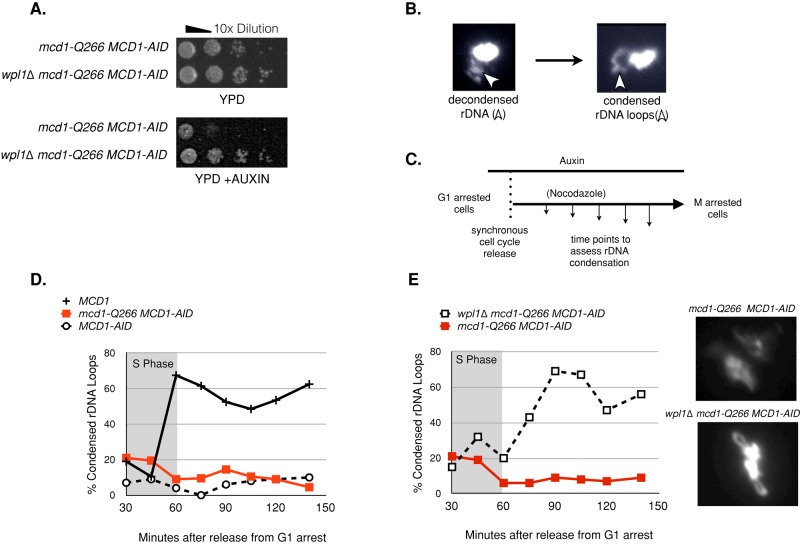
A deletion of WPL1 suppresses the auxin-induced lethality of the mcd1-Q266 MCD1-AID strain (Figure 6A). However, this suppression of inviability could not have been caused by suppression of the cohesion defect, since wpl1∆ exacerbated rather than suppressed the cohesion defect of mcd1-Q266 MCD1-AID (Figure 5A). Previously it was shown that wpl1∆ could suppress the lethality of eco1∆ (Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Sutani et al., 2009; Guacci and Koshland, 2012). Of importance, whereas wpl1∆ suppresses the lethality of an eco1∆ (wpl1∆ eco1∆), it fails to suppress the cohesion establishment defect; instead, it correlates with suppression of condensation defects characteristic of eco1 mutants (Guacci and Koshland, 2012). The surprising viability in the absence of robust cohesion establishment is believed to result from a surrogate pathway for bipolar attachment that results from the unusual assembly of the spindle during S phase in budding yeast (Guacci and Koshland, 2012). The correlation between the restoration of condensation and viability in eco1∆ wpl1∆ double mutants suggested that the condensation function of cohesin was essential for viability. By analogy, we wondered whether mcd1-Q266 caused a defect in condensation as well as cohesion maintenance, and, if so, whether wpl1∆ restored viability of the mcd1-Q266 MCD1-AID strain by suppressing its condensation defect.
FIGURE 6:
WPL1 deletion suppresses both the inviability and condensation defects of mcd1-Q266. (A) Haploid yeast strains mcd1-Q266 MCD1-AID (DK5530) and wpl1∆ mcd1-Q266 MCD1-AID (DK5550) were grown at 23°C and plated in 10-fold serial dilutions onto rich media (YPD) or rich media with 500 μM auxin (YPD + auxin). (B) Effect of wpl1∆ on the cohesion maintenance defect of mcd1-Q266. Four haploid strains were subjected to auxin depletion in G1 phase and then allowed to progress into M-phase arrest under auxin depletion as described in the Figure 2B legend. Cohesion loss in MCD1 (DK5531, solid black circles), wpl1∆ (DK5561, gray diamonds), mcd1-Q266 MCD1-AID (DK5530, solid red squares), and wpl1∆ mcd1-Q266 MCD1-AID (DK5535, dotted red squares) was assessed by plotting the percentage of cells with two GFP foci at a CEN-distal locus (LYS4). S phase is marked with a gray box. From 200 to 300 cells were counted at each time point. A representative time course is shown from two independent biological replicates. (B) Representative cytological morphology of the rDNA locus, as prepared by chromosome spreads and stained with DAPI (see Materials and Methods; Lavoie et al., 2000). Decondensed rDNA (left), condensed rDNA (right). Arrow indicates the location of rDNA. (C–F) Assessment of chromosome condensation in budding yeast. Samples were collected at 15-min intervals and processed to examine rDNA morphology by chromosome spreads stained with DAPI. At least 100 chromosome masses were scored per time point. (C) Schematic of auxin depletion in G1 and subsequent release to M-phase arrest. (D) Effect of mcd1-Q266 on rDNA condensation as described in C. Haploids MCD1 (DK5531, solid black crosses), MCD1-AID (DK5542, dashed black circles), and mcd1-Q266 MCD1-AID (DK5535, red squares) were subjected to auxin depletion in G1 phase and then allowed to progress into M-phase arrest under auxin depletion as described in the Figure 2B legend. rDNA condensation was assessed at various times points after G1-phase release. Percentage of cells where the rDNA “loop” was well formed is plotted. S phase as determined by flow cytometry is marked with a gray box. A representative biological replicate is shown from two biological replicates. (E) Effect of wpl1∆ on the rDNA condensation defect of mcd1-Q266 during progression from G1 to M. Haploids mcd1-Q266 (DK5530) and wpl1∆ mcd1-Q266 (DK5550) strains were grown and treated and then the rDNA scored for condensation as described in D and E. Right, representative plot from two biological replicates with similar results. Left, representative DAPI-stained chromosome masses from mcd1-Q266 (DK5530) (top) and wpl1∆ mcd1-Q266 (DK5550) (bottom) from the 150-min postrelease time point.
Budding yeast chromosomes condense ∼1.7-fold between interphase and mitosis (Guacci et al., 1994). Hence the chromosomes are indistinguishable in interphase and mitosis, exhibiting the same tight circular mass in 4′,6-diamidino-2-phenylindole (DAPI)–stained chromosome spreads. However, the 500-kb rDNA locus undergoes a dramatic shift in morphology from a diffuse puff in interphase to a short, line-like loop in mitosis (Figure 6B). We showed that this cell cycle–dependent change in morphology depends on cohesin and condensin (Guacci et al., 1997; Lavoie et al., 2000).
To analyze the effect of mcd1-Q266p on condensation, we compared rDNA morphology in mcd1-Q266 MCD1-AID, MCD1-AID, and MCD1 strains. We followed rDNA morphology by chromosome spreads as cells progressed from G1 to M in the presence of auxin (Figure 6C). In the MCD1 strain, the compacted loops of the rDNA appeared at the exit from S and was maintained through M (Figure 6D). In the MCD1-AID strain, the rDNA in most cells never formed short, line-like loops but instead remained as nondescript puffs (Figure 6D). These results corroborate previous studies linking cohesin function with proper condensation (Guacci et al., 1997; Guacci and Koshland, 2012; Heidinger-Pauli et al., 2010a). In the mcd1-Q266 MCD1-AID strain, the rDNA never condensed (Figure 6D). In addition, the morphology of the entire chromosome mass, as well as the rDNA, became distended as cells progressed longer into M (Figures 3B and 5B). Thus the establishment of rDNA condensation requires the Q266 region of Mcd1p.
Having established a condensation defect for mcd1-Q266 strains, we asked whether condensation was restored in the absence of Wpl1p. We examined rDNA morphology in the wpl1∆ mcd1-Q266 MCD1-AID strain by chromosome spreads (Figure 6E). We observed that rDNA condensation was restored to near-wild-type levels, albeit with delayed kinetics (Figure 6E). This result leads to three important conclusions. First, the Q266 region of Mcd1p promotes the establishment of chromosome condensation by antagonizing Wpl1p. Second, it corroborates previous observations that Wpl1p is an inhibitor of chromosome condensation in yeast. Finally, it further correlates an essential function of cohesin with chromosome condensation.
Partially conserved residues adjacent to Mcd1-Q266 define a Q266-inclusive region termed ROCC
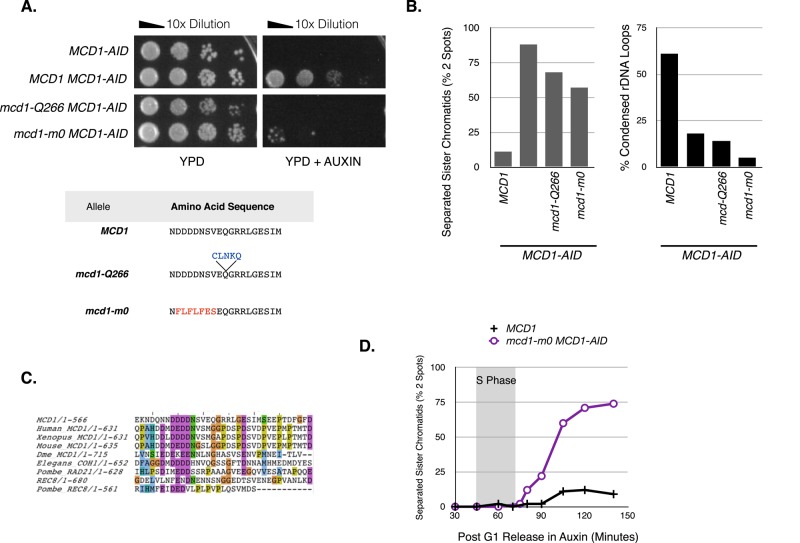
The mcd1-Q266 allele had a unique combination of phenotypes, including defects in condensation establishment and cohesion maintenance but not cohesion establishment. To assess whether these phenotypes were a peculiarity of this specific allele or representative of alleles in this region of Mcd1p, we subjected 14 residues flanking mcd1-Q266 to oligo-based mutagenesis (Materials and Methods). Mutants were screened for the inability to support viability in the absence of MCD1 and for defects in cohesion and condensation.
One allele, mcd1-m0, caused inviability and defects in cohesion and condensation (Figure 7, A and B). This allele changed seven residues (258–264) amino terminal to Q266 (Figure 7A). To help identify particularly important residues within the m0 allele for its phenotype, we asked whether any of these residues were conserved among Mcd1p orthologues. Alignments of yeast Mcd1p orthologues showed that the linker region including the Mcd1-Q266 residue has overall very poor conservation. However, a five–amino acid sequence, DDDDN (258–262), mutated in the m0 allele has an identical or near-identical match at very similar positions from the amino termini in mitotic Mcd1p orthologues in most species, including humans (Figure 7C). Using this sequence to align the adjacent sequences revealed potentially additional conserved residues. This conservation in the midst of an otherwise highly divergent linker sequence suggested that Q266 might alter an evolutionarily conserved region that participates in the regulation of cohesin function.
FIGURE 7:
Mutagenesis of residues adjacent to q266 identify an evolutionarily conserved region termed ROCC. (A) Residues in the region adjacent to Q266 were mutagenized and subjected to screening to identify mutant alleles that cannot support viability on their own but form a full-length Mcd1p (see Materials and Methods). One such allele, termed mcd1-m0, had a seven–amino acid substitution. (B) Assessment of cohesion and condensation of mcd1-m0 allele. Plasmids bearing mutant alleles generated in A were transformed into an MCD1-AID background (mcd1-m0 MCD1-AID: DK5543). Cells were depleted for Mcd1-AIDp in G1 phase and then released into M-phase arrest as described in Figure 2D. Cohesion (at LYS4) and condensation (at rDNA) were scored in M phase as described in Figure 6, B and D respectively. (C) Sequences of Mcd1p homologues were imported from GENBank into ClustalW and N-terminus aligned. No major homology was predicted by ClustalW. We manually aligned the homologues by shifting amino acids from Mcd1p homologues by several bases to highlight the presence of a poly–aspartic acid patch located similar distances from the N-terminus. (D) Analysis of cohesion in mcd1-m0 strain during progression from G1 to M phase. Haploid MCD1 (DK5542) and mcd1-m0 mcd1-AID (DK5543) strains during progression from G1 to M were subjected to auxin depletion in G1 phase and then allowed to progress into M-phase arrest under auxin depletion as described in Figure 2B. The percentage of cells with two GFP foci at a CEN-distal locus LYS4 was plotted to assess cohesion loss. S phase is marked with a gray box. A representative plot is shown from two biological replicates.
To assess the cohesion and condensation phenotypes further, we assessed cohesion and condensation in mcd1-m0 MCD1-AID cells after they had progressed from G1 to M in the presence of auxin. Both mutants exhibited defects in condensation and cohesion (Figure 7B). We then assayed cohesion in the mcd1-m0 MCD1-AID cells. Like mcd1-Q266, the m0 allele failed to maintain cohesion (Figure 7D). Moreover, the timing of cohesion loss was indistinguishable from that for the Q266 allele (Figures 2C and 7D). Thus the unique combination of defects in condensation and cohesion maintenance are shared between the Q266 allele and at least m0. Thus multiple residues proximal to Q266 contribute to the maintenance of cohesion and the establishment of condensation in budding yeast. We propose that this region comprises a new region, which we term the ROCC box, for “regulation of cohesion and condensation.”
DISCUSSION
For cohesin to mediate its diverse biological functions, its activity (or activities) must be parsed and coordinated through complex regulation. To begin to elucidate the mechanism(s) of this complex regulation, we focused on how cohesin is regulated in budding yeast during M phase to ensure the maintenance of sister chromatid cohesion and the establishment of condensation. Here we identified and characterized the ROCC box, a 10-residue region within the linker region of the Mcd1p subunit of cohesin. Our data indicate that the ROCC domain regulates both the maintenance of cohesion and the establishment of condensation.
The study of the ROCC box revealed that rocc mutants establish but cannot maintain cohesion. The kinetics of loss of cohesion in the rocc mutants is very similar to that seen in cells defective for Pds5p function, either pds5 mutants or mcd1/scc1 (mcd1-V137K; this study). In all these mutants, sister chromatids start separating ∼20 min after a cohesin-null mutant and 15–20 min after the completion of S phase. This temporal similarity of the cohesion maintenance defect caused by rocc and pds5 mutants suggests that their dysfunction leads to a failure to inhibit a common cohesion dissolution process in G2. One candidate for a dissolution inducer is Wpl1p (Wapl in vertebrates), as its overexpression in mammalian cells induces precocious sister separation (Gandhi et al., 2006). However, deletion of WPL1 does not suppress the cohesion maintenance defects of either rocc or pds5 mutants (this study; Chan et al., 2013). The Wpl1p-independent cohesion defects in these two mutants not only further tie together Pds5p and ROCC function, but they also further hint at the existence of a novel mechanism of cohesion regulation yet to be appreciated. We suggest that Pds5p bound to cohesin protects the core complex against this novel cohesion dissolution process. This is akin to the idea of Pds5p as a molecular shield to protect cohesin and cohesion maintenance (Stead et al., 2003; D'Ambrosio and Lavoie, 2014). Abrogation of Pds5p function makes cohesin susceptible to this dissolution process. We suggest that mcd1-V137K blocks Pds5p binding to cohesin, whereas mcd1-Q266 perturbs a Pds5p-associated activity (either Pds5p itself or another protein) that acts after Pds5p binds to cohesin.
Here we began to elucidate the mechanism of this novel Wpl1p-independent process for cohesion dissolution. Wpl1p disrupts the integrity of the cohesin complex, which in turn increases cohesin dissociation from chromosomes and dissolution of cohesion (Sutani et al., 2009). In contrast, we show that the novel Wpl1p-independent mechanism disrupts cohesion in rocc and mcd1-V137K mutants without altering the stable binding of cohesin to chromosomes (this study; Chan et al., 2013). Indeed, we observed almost no cohesin dissociated from chromosomes during a 30-min window after inhibition of the cohesin loader. Of importance, this low rate of cohesin dissociation from chromosomes in rocc mutants was measured at many chromosomal loci/regions. Furthermore, the persistent binding of cohesin to chromosomes in the scc1-V137K mutant was demonstrated by completely different method of fluorescence recovery after photobleaching (Chan et al., 2013). Finally, in these studies of the rocc and scc1-V137K mutants, the stable cohesin binding to chromosomes was monitored using different cohesin subunits as reporters, making it highly unlikely that a lone cohesin subunit could be functioning independent of a functional complex (Chan et al., 2013). Taken together, we showed that cohesion dissolution can occur without increasing cohesin dissociation from chromosomes through the study of two different mutants, two independent methods, and two different reporters of the cohesin complex.
The observation that cohesion can be dissolved without dissociating cohesin from chromosomes provides a key mechanistic insight into understanding how sister chromatids are tethered. In the simple embrace model, the only mechanism capable of dissolving cohesion is the disruption of the ring's integrity by either opening or cleaving it. This disruption of ring's integrity not only would allow the entrapped sister chromatids to escape, but would also cause cohesin dissociation from chromosomes. Clearly, this obligatory coupling of cohesion dissolution with cohesin dissociation from chromosomes does not fit the properties of dissolution observed in the rocc and mcd1-V137K mutants (this study). Instead, the cohesion dissolution in these mutants demands a second function of cohesin that can be disrupted besides its chromosome-binding activity. One of several possibilities for this second activity is cohesin oligomerization, as suggested by the handcuff model (Petrushenko et al., 2010; for review, see Onn et al., 2008). In this model, cohesion arises from the oligomerization of two cohesins, each of which topologically entraps a single sister chromatid. Additional models of tethering involving oligomerization have been proposed based on studies of bacterial SMC complexes (Gloyd et al., 2011). Further analysis of cohesion dissolution in rocc and pds5 mutants provides a potentially powerful tool to elucidate the molecular nature of this second step.
Our analyses of rocc mutants revealed a second important phenotype: an ability to establish cohesion but not condensation of the rDNA. This result demonstrated a role for ROCC in the regulation of the establishment of condensation. This rocc phenotype suggests that while cohesin is required for both cohesion and condensation, its activity (activities) in the establishment of cohesion and condensation can be uncoupled. Previously we reported evidence for this uncoupling, as the introduction of wpl1∆ into eco1∆ suppressed its condensation but not its cohesion defect (Guacci and Koshland, 2012). Of interest, a wpl1∆ also suppressed the condensation defect of rocc strains but not the cohesion-maintenance defect. Thus ROCC and Eco1p appear to antagonize an anticondensation activity of Wpl1p. Because rocc mutants do not destabilize cohesin binding to chromosomes, this anticondensation activity of Wpl1p must be distinct from its ability to dissociate cohesin from chromosomes. We suggest that ROCC with Eco1p modulates the multiple activities of Pds5p, Wpl1p, or other, yet-to-be-defined factors as part of a key regulatory network that controls cohesin's function in cohesion and condensation.
The role of cohesin and Pds5p in condensation is evolutionarily conserved, as they modulate meiotic chromosome condensation in budding yeast, fission yeast, and vertebrates (Ding et al., 2006; Novak et al., 2008; Jin et al., 2009). The role of Wpl1p in regulating condensation may also be conserved in vertebrates. A recent study showed that depletion of the WPL1 homologue (WAPL) in vertebrate cells led to cohesin-dependent chromosome compaction in serum-starved G0 cells (Tedeschi et al., 2013). As we observed in yeast, the presence or absence of WAPL in these G0 cells did not change the binding of cohesin to chromosomes, implying that the anticondensation function of Wapl in vertebrates is also distinct from its ability to dissociate cohesin from chromosomes. In this context, the apparent conservation of the ROCC box in mammalian orthologues of Mcd1p is very intriguing (this study). One possibility is that ROCC, Eco1p, and Wpl1p regulate a signal cascade, independent of any cohesin-tethering activity, which triggers condensation, such as by activating condensin. Alternatively, cohesin may promote condensation by its proposed ability to generate intrastrand tethers as well as the interstrand tethers needed for cohesion (Kagey et al., 2010; Tedeschi et al., 2013). In this scenario, ROCC, Eco1p, and Wpl1p may be part of a second pathway that regulates intrastrand tethers. Distinguishing between these models by further characterizing ROCC should provide important new insights into the fascinating complexity of cohesin function and regulation.
MATERIALS AND METHODS
Yeast strains, media, and reagents
Yeast strains used in this study are the A346a background, and their genotypes are listed in Supplemental Table 1. Synthetic dropout and yeast extract/peptone/dextrose (YPD) media were prepared as previously described (Guacci et al., 1997). For experiments using the AID system, a 1 M stock of 3-indoleacetic acid (Sigma-Aldrich, St. Louis, MO) was made in dimethyl sulfoxide and added to plates or liquid cultures at a final concentration of 500 μM, with cooling agar used in plates to ∼55°C before addition of auxin to each batch.
Dilution plating assays
Cells were grown to saturation in YPD medium at 23°C (or 30°C when listed), diluted to OD660 1.0 using YPD, and then plated in 10-fold serial dilutions. Cells were incubated on plates at relevant temperatures as described.
Cohesion assays
To monitor cohesion from G1 to M, haploid yeast cells were initially grown to mid log phase at 23°C in YPD culture. Cells were staged to G1 by addition of α factor (Sigma-Aldrich) to 10−8 M final concentration and incubation for 3 h. Cells were examined by light microscopy to confirm the presence of the schmoo morphology in 95% of the cells, at which point auxin was added (500 μM final) to induce degradation of AID-tagged proteins and cells incubated 1 h more. Cells were released from G1 arrest by washing six times in YPD containing auxin and 0.1 mg/ml Pronase E (Sigma-Aldrich), resuspended in fresh YPD containing auxin and nocodazole (Sigma-Aldrich) at 15 μg/ml final, and incubated at 23˚C to allow cell-cycle progression until arrest in mid M phase. To monitor cohesion and cell cycle progression and cohesion, cell aliquots were fixed every 15 min for DNA content by fluorescence-activated cell sorting and for cohesion via the LacI-GFP cohesion assay.
To monitor cohesion in M phase–arrested cells, haploid yeast cells were grown at 23˚C to mid log phase, and then nocodazole (15μg/ml) was added and cells incubated 3 h at 23°C to arrest at mid M phase. Arrests were confirmed by light microscopy for the classical large-budded cellular phenotype (∼95% of the population). Auxin (500 μM final) was added to cultures, and cells were incubated at 23°C. Aliquots were fixed and processed at various time points after auxin addition to assess cohesion via the GFP-LacI system.
Microscopy
Images were acquired with an Axioplan2 microscope (100× objective, numerical aperture [NA] 1.40; Zeiss, Thornwood, NY) equipped with a Quantix charge-coupled device camera (Photometrics, Tucson, AZ).
Chromosome spreads and microscopy
Chromosome spreads were performed as previously described (Wahba et al., 2013). Slides were incubated with a mouse anti-FLAG monoclonal (Sigma-Aldrich) at 1:2000, mouse anti-V5 (Life Technologies, Carlsbad, CA) at 1:2000, polyclonal rabbit anti-Pds5 at 1:2000, or polyclonal rabbit anti-Mcd1 at 1:2000 dilutions. The primary antibody was diluted in blocking buffer (5% bovine serum albumin, 0.2% milk, 1× phosphate-buffered saline, and 0.2% Triton X-100). The secondary Cy3-conjugated goat anti-mouse antibody (115-165-003) was obtained from Jackson ImmunoResearch (West Grove, PA) and diluted 1:2000 in blocking buffer. Indirect immunofluorescence was observed using an Olympus IX-70 microscope with a 100×/NA 1.4 objective and Orca II camera (Hamamatsu, Bridgewater, NJ).
Chromatin immunoprecipitation
Cells used for ChIP experiments were processed in the same manner as cells examined for cohesion assays as described in Wahba et al. (2013). Briefly, after synchronous release from G1 into mitotic arrest, 5 × 108 large-budded cells were fixed for 2 h with 1% formaldehyde. After cell lysis, chromatin was sheared 20 times for 45 s each (settings at duty cycle, 20%; intensity, 10; cycles/burst, 200; 30 s of rest between cycles) using a Covaris S2 (Covaris, Woburn, MA). Immunoprecipitation of epitope-tagged proteins were isolated using anti-Pds5 polyclonal antibody (Covance Biosciences, Princeton, NJ). Mcd1p was alternatively immunoprecipitated with a polyclonal rabbit anti-Mcd1 antibody (Covance Biosciences). A no-primary-antibody control was also run to ensure specificity. Appropriate dilutions of input and immunoprecipitated DNA samples were used for PCR analysis to ensure linearity of the PCR signal. PCR and data analysis was carried out as described previously (Wahba et al., 2013). All experiments were done at least twice, and a representative data set is shown. ChIP primers are available upon request.
Flow cytometry
Flow cytometry was conducted as described previously (Yamamoto et al., 1996a), with slight modifications. SYBR Green DNA I dye was used to examine DNA content (Life Technologies) at 1:10,000 dilution in sodium citrate buffer (pH 7.4, 50 mM). Twenty thousand events were collected for each time point.
Generation of auxin degron cassettes
The original auxin degron cassettes from Nishimura et al. (2009) were modified as followed. A 3V5 epitope tag and five–amino acid polylinker were added at the N-terminus of the AID ORF by standard cloning techniques.
Random insertion screen for dominant-negative alleles (RID screening)
Briefly, linker scanning mutagenesis was performed essentially as described in Milutinovich et al. (2007). Plasmid pVG385, a CEN URA3 plasmid that contains MCD1 under control of the pGAL promoter, was mutagenized in vitro using the TnABC transposase following the manufacturer's protocol (GPS-LS Linker Scanning System; New England BioLabs, Ipswich, MA). The in vitro–mutagenized pVG385 was transformed into bacteria, and a Tn7 random insertion DNA library (from ∼4000 bacterial colonies) was generated. Tn7 inserts at only one site in each plasmid and generates a PmeI site at each end of the Tn7. This Tn7 library DNA was digested with PmeI to excise Tn7, and the linearized plasmid was gel purified and recircularized (ligated at low density). This recircularized DNA has a 15–base pair insertion at the site of Tn7 excision and was transformed into Escherichia coli. DNA from ∼12,000 individual bacterial colonies was harvested to form the bacterial MCD1-RID library. This pVG385-RID library was then transformed into either MCD1 or mcd1-1 haploid yeast strains and plated onto URA- dextrose medium to select URA+ transformants. Approximately 6000 transformants from wild-type yeast and 2300 transformants from mcd1-1 yeast were screened by replica plating to both URA–dextrose and URA–galactose media. Transformants that were inviable or slow growing on galactose but grew well on dextrose were selected for further analysis. These candidate clones were patched onto URA–dextrose plates and then replica plated to URA–galactose and 5-fluoroorotic acid (5′FOA) galactose media to confirm linkage of the inviable phenotype on galactose media without the candidate plasmid. Plasmids from these candidates were isolated and retransformed into yeast to confirm pVG385-MCD1-RID lethality. Thus confirmed, plasmids were sequenced using standard methods to identify the site of insertion.
Oligo-based mutagenesis of the ROCC domain
Briefly, we generated mutagenized PCR fragments of the MCD1 reading frame at codons specifying amino acids 258–265 and, separately, 265–272. These mutated MCD1 fragments were cotransformed with a gapped MCD1 plasmid (pVG164 CEN TRP MCD1) into DK5521 (mcd1∆ CEN URA mcd1-1) such that resulting TRP+ colonies repaired the CEN TRP1 plasmid using the mutagenized MCD1 fragment as template. About 4000 TRP+ independent transformants were recovered, and, of these, 50 candidate transformants were inviable on 5′FOA. Sequenced plasmids were rebuilt and retested in an MCD1-AID background for cohesion and condensation analysis.
Supplementary Material
Acknowledgments
We thank Koshland lab members for helpful discussions, reagents, and constructive feedback on the manuscript. Katie Hogan and Carol Davenport conducted the initial RID overexpression screen. This work was supported by Grant R01 GM092813 from the National Institute of General Medical Sciences/National Institutes of Health to D.E.K. and a National Science Foundation Graduate Research Fellowship to T.E.
Abbreviations used:
- AID
auxin-induced degron
- CEN
centromere
- DAPI
4′,6-diamidino-2-phenylindole
- rDNA
ribosomal DNA
- SCF
Skp, Cullin, F-box Complex
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-04-0929) on June 25, 2014.
REFERENCES
- Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–3419. doi: 10.4161/cc.8.20.9911. [DOI] [PubMed] [Google Scholar]
- Chan K-L, Gligoris T, Upcher W, Kato Y, Shirahige K, Nasmyth K, Beckouët F. Pds5 promotes and protects cohesin acetylation. Proc Natl Acad Sci USA. 2013;110:13020–13025. doi: 10.1073/pnas.1306900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio LM, Lavoie BD. Pds5 Prevents the polySUMO-dependent separation of sister chromatids. Curr Biol. 2014;24:361–371. doi: 10.1016/j.cub.2013.12.038. [DOI] [PubMed] [Google Scholar]
- Ding D-Q., Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloyd M, Ghirlando R, Guarné A. The role of MukE in assembling a functional MukBEF complex. J Mol Biol. 2011;412:578–590. doi: 10.1016/j.jmb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13:1678–1691. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D. Cohesin-independent segregation of sister chromatids in budding yeast. Mol Biol Cell. 2012;23:729–739. doi: 10.1091/mbc.E11-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Mert O, Davenport C, Guacci V, Koshland D. Systematic reduction of cohesin differentially affects chromosome segregation, condensation, and DNA repair. Curr Biol. 2010a;20:957–963. doi: 10.1016/j.cub.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Onn I, Koshland D. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics. 2010b;185:1249–1256. doi: 10.1534/genetics.110.116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Guacci V, Yu H-G. Pds5 is required for homologue pairing and inhibits synapsis of sister chromatids during yeast meiosis. J Cell Biol. 2009;186:713–725. doi: 10.1083/jcb.200810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters J-M. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Lavoie BD, Tuffo KM, Oh S, Koshland D, Holm C. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol Biol Cell. 2000;11:1293–1304. doi: 10.1091/mbc.11.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F. Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol. 2013;23:64–69. doi: 10.1016/j.cub.2012.11.030. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Milutinovich M, Unal E, Ward C, Skibbens RV, Koshland D. A multi-step pathway for the establishment of sister chromatid cohesion. PLoS Genet. 2007;3:e12. doi: 10.1371/journal.pgen.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Hu B, Kurze A, Beckouët F, Farcas A-M, Dixon SE, Katou Y, Khalid S, Shirahige K, Nasmyth K. Both interaction surfaces within cohesin's hinge domain are essential for its stable chromosomal association. Curr Biol. 2010;20:279–289. doi: 10.1016/j.cub.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Noble D, Kenna MA, Dix M, Skibbens RV, Unal E, Guacci V. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism. Cell Cycle. 2006;5:2528–2536. doi: 10.4161/cc.5.21.3405. [DOI] [PubMed] [Google Scholar]
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Hoog C. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol. 2008;180:83–90. doi: 10.1083/jcb.200706136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Peters J-M, Nishiyama T. Sister chromatid cohesion. Cold Spring Harb Perspect Biol. 2012;4:a011130. doi: 10.1101/cshperspect.a011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrushenko ZM, Cui Y, She W, Rybenkov VV. Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. EMBO J. 2010;29:1126–1135. doi: 10.1038/emboj.2009.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Shimada K, Gasser SM. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell. 2007;128:85–99. doi: 10.1016/j.cell.2006.11.045. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Larionov VL, Koshland D. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- Tedeschi A, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–568. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J Cell Biol. 1996a;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996b;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, et al. Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS One. 2009;4:e5232. doi: 10.1371/journal.pone.0005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.