Nup50 is a mobile nucleoporin with a pronounced presence both at the nuclear pore complex and in the nucleoplasm that can move between these different localizations. The dynamic behavior of Nup50 in both locations is dependent on active transcription by RNA polymerase II.
Abstract
The nuclear pore complex (NPC) plays a critical role in gene expression by mediating import of transcription regulators into the nucleus and export of RNA transcripts to the cytoplasm. Emerging evidence suggests that in addition to mediating transport, a subset of nucleoporins (Nups) engage in transcriptional activation and elongation at genomic loci that are not associated with NPCs. The underlying mechanism and regulation of Nup mobility on and off nuclear pores remain unclear. Here we show that Nup50 is a mobile Nup with a pronounced presence both at the NPC and in the nucleoplasm that can move between these different localizations. Strikingly, the dynamic behavior of Nup50 in both locations is dependent on active transcription by RNA polymerase II and requires the N-terminal half of the protein, which contains importin α– and Nup153-binding domains. However, Nup50 dynamics are independent of importin α, Nup153, and Nup98, even though the latter two proteins also exhibit transcription-dependent mobility. Of interest, depletion of Nup50 from C2C12 myoblasts does not affect cell proliferation but inhibits differentiation into myotubes. Taken together, our results suggest a transport-independent role for Nup50 in chromatin biology that occurs away from the NPC.
INTRODUCTION
Nuclear pore complexes (NPCs) have long been known to function as gates that control transport of protein and RNA between the nucleus and the cytoplasm (Wente and Rout, 2010). However, studies in yeast, fly, and mammalian systems have implicated various nucleoporins in transcriptional activation, transcriptional elongation, RNA processing, RNA stabilization, gene silencing, and heterochromatin formation (Pascual-Garcia and Capelson, 2014). These reports indicate that nucleoporins fulfill diverse roles in gene regulation and chromatin biology in addition to their canonical function in nucleocytoplasmic transport.
An early hint that NPCs might play a role in chromatin organization came from electron microscopic analysis of the nuclear envelope (NE), which noted that NPCs are interspersed between regions of densely packed heterochromatin (Blobel, 1985). Since this early observation, NPCs have been shown to moonlight as regulators of chromatin organization by tethering DNA “zip codes” (Light et al., 2010; Brickner et al., 2012) or looping 5′ and 3′ ends of genes (Tan-Wong et al., 2009), as well as scaffolding either gene activation (Taddei et al., 2006) or repression (Van de Vosse et al., 2013) in what appears to be a context-dependent manner. Further, NPCs appear to coordinate transcription with mRNA export by interacting both with the RNA processing and mRNA export machineries (Rougemaille et al., 2008).
A spate of recent studies demonstrated that chromatin-associated functions of individual nucleoporins extend beyond the NPC structure to chromatin sites that do not interact with the nuclear periphery (Capelson et al., 2010; Kalverda et al., 2010; Liang et al., 2013). This phenomenon may be explained by the finding that nucleoporins exhibit a range of NPC residence times that spans several orders of magnitude (Rabut et al., 2004a). For example, components of the central NPC scaffold are extremely stable at the NPC and exchange only when the nucleus disassembles in mitosis. Any role of these stable scaffold nucleoporins in gene regulation is thus likely to occur at the NPC structure. In contrast, components of the nuclear basket, such as Nup153 and Nup50, have residence times of seconds at the NPC and are thus capable of exploring the nucleoplasm between NPC-binding events. Nup153 has a Zn finger domain and interacts with tracts of transcriptionally active chromatin in Drosophila melanogaster (Vaquerizas et al., 2010). Similar behavior has been documented for the nuclear pool of Nup98, which accumulates in dynamic intranuclear foci termed GLFG bodies (Griffis et al., 2004) separately from the more stable NPC pool (Rabut et al., 2004a). This nucleoplasmic pool interacts with chromatin loci and regulates gene expression (Capelson et al., 2010; Kalverda et al., 2010; Liang et al., 2013). Finally, both Nup153 and Nup98 are sensitive to transcriptional inhibition in mammalian cells (Griffis et al., 2002, 2004), further implicating a link between the NPC and transcription.
Among mobile nucleoporins, Nup50 has the shortest residence time at the NPC, and only nuclear transport receptors such as importin β are more dynamic (Rabut et al., 2004a). It is unknown how Nup50 dynamics relate to that of Nup98 and Nup153 and whether it has other functions beyond its known role in nuclear transport. Nup50 interacts directly with transport receptors of the importin α subclass (Lindsay and Macara, 2002; Pumroy et al., 2012) and can be thought of as at the interface between the NPC structure and the transport machinery moving through it. This interaction potentiates protein transport under certain conditions (Lindsay and Macara, 2002), possibly by clipping importin α in a closed conformation to promote recycling (Pumroy et al., 2012).
Genetic data indicate that Nup50 is not required for nucleocytoplasmic transport, as fibroblasts lacking Nup50 proliferate normally in culture. However, the Nup50−/− mouse dies late in gestation due to massive defects in neural tube formation, indicating that Nup50 does perform an essential role in particular cell types during development (Smitherman et al., 2000). Nup50 is tethered to the nuclear basket via its interaction with Nup153 (Hase and Cordes, 2003; Makise et al., 2012), but whether it functions with Nup153 in any of its chromatin-related functions is unknown. Its presumptive yeast homologue, Nup2, has the ability to limit the spread of repressive regions on chromatin (so-called “boundary activity”; Dilworth, 2005), and Nup50 has been shown to bind chromatin loci in D. melanogaster (Kalverda et al., 2010). Further complicating this picture is the fact that components of the nuclear transport machinery also interact with chromatin loci (Casolari et al., 2004).
In this study, we investigate the nuclear functions of Nup50 and report the surprising finding that Nup50 requires neither its interaction with importin α nor Nup153 in order to become immobilized on chromatin by transcriptional inhibition. We test the functional importance of Nup50 and find that while dispensable for myoblast proliferation, it is essential for myogenic cell differentiation, suggesting a role for Nup50 in mediating normal responses to differentiation stimuli via its position on chromatin.
RESULTS
Nup50 is present in nucleoplasmic and NPC-associated pools
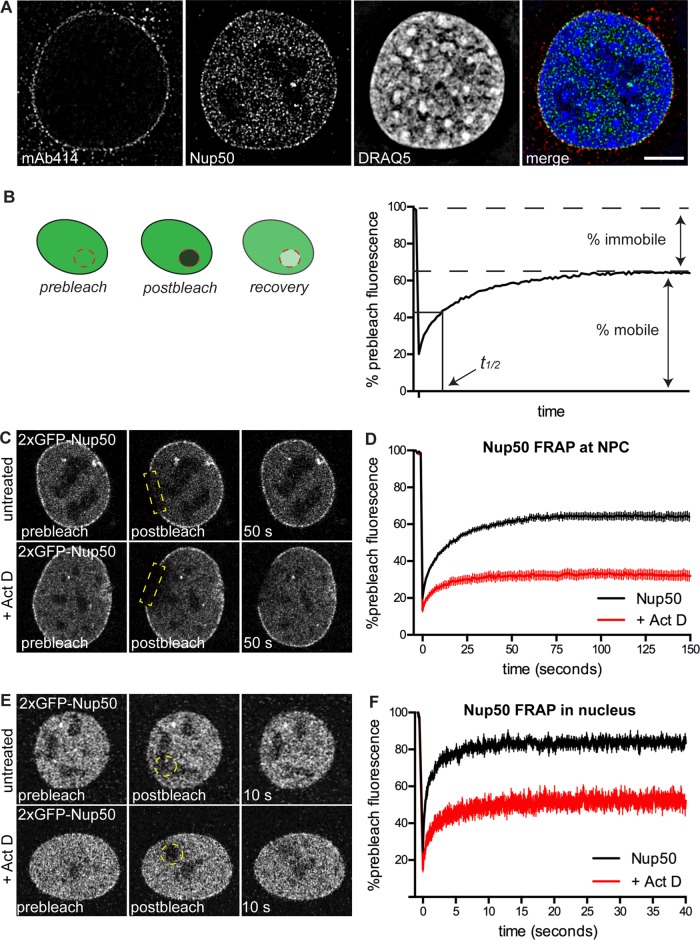
Nup50 is tethered to the NPC by interaction with Nup153. Unlike Nup153, however, Nup50 is present in a sizable nucleoplasmic pool (Figure 1A and Supplemental Figure S2; Guan et al., 2000). We analyzed mouse C2C12 myoblast cells by structured illumination microscopy (SIM) and compared Nup50 localization in the nucleus to the FG-repeat Nups stained by mAb414 (including Nup62 and Nup153). Whereas FG-repeat Nups are detectable only around the periphery of the nucleus in a central z-section, endogenous Nup50 is detectable both at nuclear pores (resolved at individual-NPC resolution by SIM) and throughout the nucleoplasm (Figure 1A). This nucleoplasmic pool is more easily extracted by detergent than the NPC-bound pool (Guan et al., 2000) and disappears along with the NPC-localized signal when cells are treated with Nup50 RNA interference (RNAi; Supplemental Figure S1). This bona fide localization of a nucleoporin to the nucleoplasm suggests that Nup50 may have important function(s) away from the NE.
FIGURE 1:
NPC-bound and nucleoplasmic pools of Nup50 are immobilized by transcription inhibition. (A) SIM of C2C12 cells costained for FG-repeat Nups (mAb414), Nup50, and DNA (DRAQ5). (B) Schematic of FRAP image acquisition and analysis; see Materials and Methods for detailed protocol. (C, D) FRAP of NPC-bound 2xGFP-Nup50 in the absence or presence of 1 μg/ml Act D. N > 15 cells/condition. (E, F) FRAP of nucleoplasmic 2xGFP-Nup50 in the absence or presence of 1 μg/ml Act D. N > 15 cells/condition. Points indicate averaged values, and error bars indicate SEM.
We next compared Nup50’s localization to Nup98, which also has an intranuclear fraction. As previously reported (Griffis et al., 2004), green fluorescent protein (GFP)–Nup98 can be found discretely accumulated in GLFG body puncta (Supplemental Figure S2B, top, arrowhead) or diffusely enriched within nucleoli (Supplemental Figure S2B, bottom, asterisk). Nup50 does not follow Nup98 into nucleoli (Supplemental Figure S2B, bottom) but will occasionally accumulate with Nup98 in GLFG foci (Supplemental Figure S2B, top). Overall, Nup50 is distinguished from Nup98 by its more uniform distribution through the nucleoplasm and general exclusion from nucleoli.
Both NPC-bound and free nucleoplasmic Nup50 are immobilized by transcription inhibition
Nup153 interacts with large tracts of transcriptionally active chromatin in D. melanogaster (Vaquerizas et al., 2010), and its dynamics at the NPC are sensitive to transcriptional inhibition (Griffis et al., 2004). These observations implicate Nup153 in transcription-related processes. We sought to confirm GFP-Nup153’s sensitivity to transcriptional inhibition by fluorescence recovery after photobleaching (FRAP) analysis (Figure 1B) in cells treated with actinomycin D (Act D). Act D functions by intercalating preferentially into regions of actively transcribing chromatin (Yu, 1983; Chen et al., 1990) and inhibits RNA Pol I, RNA Pol II, and RNA Pol III, roughly in that order of sensitivity (Bensaude, 2011). We tested the response of Nup153 to a range of Act D doses and found that within 30 min of incubation, immobilization of Nup153 at the NPC became apparent in the presence of at least 0.5 μg/ml Act D (Supplemental Figure S3). This was visible as a decreased plateau of fluorescence recovery in Act D–treated cells compared with controls (Figure 1B and Supplemental Figure S3B). The observed kinetics are consistent with that previously reported (Griffis et al., 2004). Of importance, neither nuclear transport receptors nor nucleocytoplasmic transport were significantly affected by transcriptional inhibition (Supplemental Figure S4).
Because Nup50 associates with Nup153, we tested whether Nup50 also responds to transcriptional inhibition. Similar to Nup153, we found that immobilization of Nup50 at the NPC became apparent within 30 min of treatment with 1 μg/ml Act D (Figure 1, C and D). We next tested the effect of Act D on Nup50’s dynamics in the nucleoplasm and found that the nucleoplasmic pool of Nup50 was immobilized when transcription was inhibited (Figure 1, E and F). We deduced that the nucleoplasmic pool of Nup50 appeared homogeneously sensitive to transcriptional inhibition, as equivalent FRAP dynamics were observed regardless of where the bleaching spot was positioned. This behavior is distinct from that observed for Nup153, which responds to transcriptional inhibition at the NPC but has little to no nuclear pool that can be detected by fluorescence microscopy (Supplemental Figure S3). It is also distinct from Nup98, which exchanges much more slowly at the NPC (Rabut et al., 2004a) and is sensitive to transcriptional inhibition only within intranuclear foci (Griffis et al., 2004). Transcriptional sensitivity of Nup50 throughout the nuclear compartment—both at the NPC and inside the nucleus—sets it apart from the previously identified transcriptionally sensitive Nups.
The Nup153-binding N-terminal domain of Nup50 responds to transcriptional inhibition
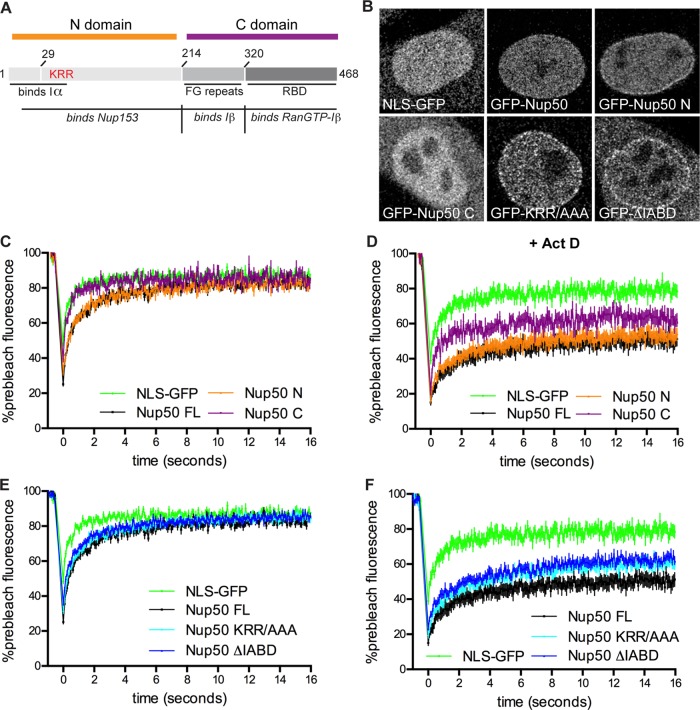
We next sought to define the regions of Nup50 that control sensitivity to transcriptional inhibition. To that end, we expressed either the N-terminal portion of Nup50, which contains importin α– and Nup153-binding domains, or the C-terminal portion of Nup50, which contains the small FG-repeat region (through which importin β associates) and a Ran-binding domain (Figure 2A; Lindsay and Macara, 2002; Makise et al., 2012). As would be expected based on Nup50’s reliance on Nup153 binding for NPC targeting (Hase and Cordes, 2003), Nup50’s N domain is found both at the NPC and in the nucleoplasm, whereas the C domain does not associate with the NPC (Figure 2B). We analyzed the dynamics of these protein domains in the nucleoplasm and found that the N domain of Nup50 phenocopies full-length Nup50. Both full-length Nup50 and the N domain exhibit slower diffusion kinetics through the nucleoplasm than does NLS-GFP (Figure 2C) and become immobilized in the nucleoplasm by transcriptional inhibition (Figure 2D). The former observation suggests that Nup50 is slowed by binding and release events as it diffuses through the nuclear volume and that these binding interactions occur through the N domain. Consistent with this, the C-terminal domain exhibits identical diffusive behavior to NLS-GFP (which interacts only with importin α in the nucleus; Figure 2C). The C-terminal domain responds intermediately to transcriptional inhibition (Figure 2D), possibly by dimerizing with endogenous Nup50. Overall, this domain analysis indicates that the N-terminal half of Nup50 is sufficient for dynamic interaction with unknown nucleoplasmic binding partners (Figure 2C) and for response to transcriptional inhibition (Figure 2D).
FIGURE 2:
The Nup153-binding N terminus of Nup50 mediates nuclear interactions and transcription sensitivity. (A) Diagram of Nup50 domain organization. (B) Localization of Nup50 and NLS-GFP constructs used in C–F. (C, E) FRAP of nucleoplasmic Nup50 and NLS-GFP constructs. N >18 cells/condition. (D, F) FRAP of nucleoplasmic Nup50 and NLS-GFP constructs in presence of 1 μg/ml Act D. N > 10 cells/condition. Points indicate averaged values, and error bars indicate SEM.
Transcription-dependent dynamics of Nup50 are independent of the NPC
Because the N-terminal portion of Nup50 interacts with Nup153 and importin α, it is possible that Nup50’s transcriptional response may be mediated by either of these proteins. To test for dependence on importin α, we mutated Nup50’s polybasic 44KRR46 importin α−binding motif or deleted the N-terminal importin α –binding domain (IABD; residues 1–46) entirely (Lindsay and Macara, 2002) and analyzed the dynamics of these mutants by FRAP. We found that Nup50’s nucleoplasmic dynamics (Figure 2E) and response to transcriptional inhibition (Figure 2F) were unchanged. In light of this result and our finding that neither NLS-GFP nor importin α is affected by Act D treatment (Supplemental Figure S4), we conclude that Nup50’s response to transcriptional inhibition is independent of the protein transport machinery.
Nup50 relies on Nup153 for targeting to the NPC (Hase and Cordes, 2003), and it is possible that it could rely on Nup153 for transcriptional sensing as well. We tested this possibility by stably knocking down Nup153 in 2xGFP-Nup50 C2C12 cells and then subjecting Nup50 to FRAP analysis in the absence or presence of Act D. Depleting Nup153 displaces Nup50 from the NPC (Hase and Cordes, 2003; Figure 3A), and Nup50 becomes more dynamic at the nuclear periphery without its NPC tether (Figure 3B, gray vs. black trace). We used these two criteria to identify Nup153-depleted cells. We found that transcriptional sensitivity was unchanged despite the change in Nup50’s localization (Figure 3B, orange vs. red trace). We performed a similar experiment in the presence of Nup98 RNAi, since Nup98 also responds to transcriptional inhibition (Griffis et al., 2002, 2004) and influences Nup50’s chromatin-binding loci in Drosophila (Kalverda et al., 2010). RNAi against Nup98 also knocks down Nup96, which is a product of the same transcript and is essential for NPC assembly (Doucet et al., 2010). Nup50 was displaced from the NPC and into the nucleoplasm by Nup98 knockdown (Figure 3A), possibly because depletion of Nup96 decreases NPC numbers (Doucet et al., 2010). Of importance, however, Nup50 retained the ability to respond to transcriptional inhibition in cells depleted of Nup98/96 (Figure 3C). Therefore Nup50’s ability to sense transcriptional status is not mediated by Nup98 or Nup153. These data collectively indicate that Nup50’s transcriptional response is independent of the NPC, as the phenotype persists when Nup50 is no longer targeted to the NPC (Figure 3) and cannot participate in nucleocytoplasmic transport (Figure 2). We therefore focused our further analysis on the free nucleoplasmic pool of Nup50.
FIGURE 3:
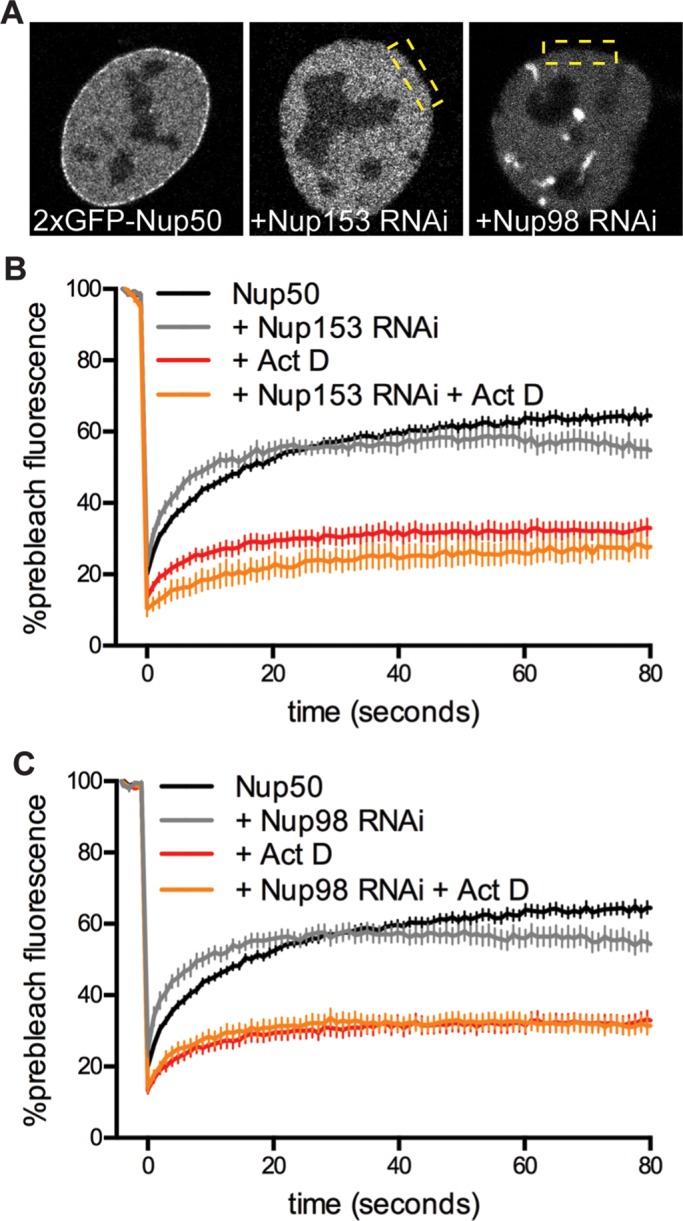
Nup50’s response to transcription inhibition persists when untethered from the NPC. (A) Localization of 2xGFP-Nup50 in cells stably transduced with control RNAi, Nup153 RNAi, or Nup98 RNAi. (B) FRAP of Nup50 in wild-type cells (black trace) or cells treated with Nup153 RNAi (gray trace) in the absence or presence of 1 μg/ml Act D. N > 6 cells/condition. (C) FRAP of Nup50 in wild-type cells (black trace) or cells treated with Nup98 RNAi (gray trace) in the absence or presence of 1 μg/ml Act D. N > 9 cells/condition. Data are pooled from two hairpins each for Nup153 and Nup98. Points indicate averaged values, and error bars indicate SEM.
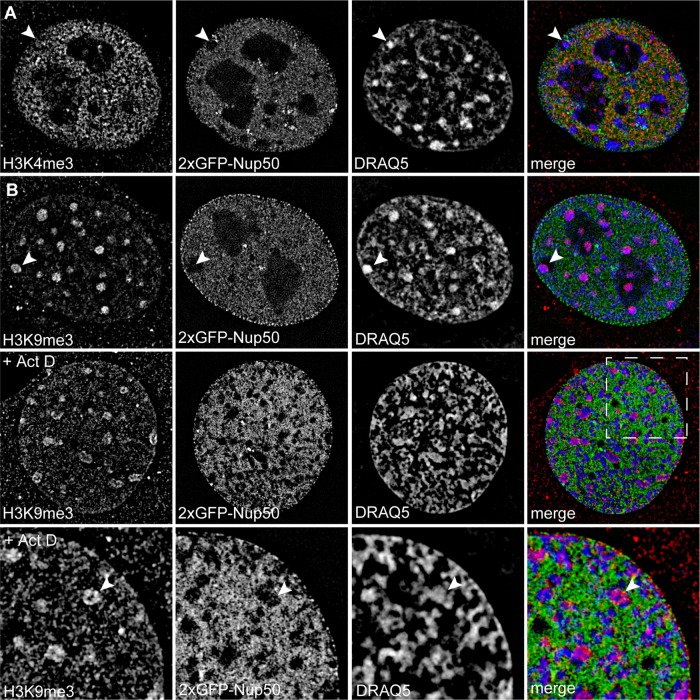
Within the nucleus, Nup50 segregates away from heterochromatic foci
To gain better resolution of any nuclear or chromatin structures with which Nup50 might be associating, we performed SIM on cells stably expressing 2xGFP-Nup50 (which exhibits similar localization to endogenous Nup50; see Figure 1, A and B) and costained for DNA and chromatin marks (Figure 4). Nup50 is broadly distributed through the nucleoplasm but is excluded from nucleoli; this pattern is generally similar to the distribution of the active, transcription-associated H3K4me3 chromatin mark (Figure 4A, top). Closer inspection of Nup50, DNA density, and the repressive chromatin mark H3K9me3 suggests that Nup50 is excluded from regions of highly condensed chromatin (Figure 4, A and B, arrowheads). This displacement becomes even more prominent in transcriptionally inhibited cells (Figure 4B, lower two panels, arrowhead).
FIGURE 4:
Within the nucleus, Nup50 overlaps with euchromatin and segregates from heterochromatin. (A) SIM of C2C12 cells stably expressing 2xGFP-Nup50 and costained for H3K4me3 and DNA (DRAQ5). (B) SIM of C2C12 cells stably expressing 2xGFP-Nup50 and costained for H3K9me3 and DNA (DRAQ5) in the absence (top) or presence (bottom two) of 5 μg/ml Act D. Arrowheads indicate heterochromatic foci.
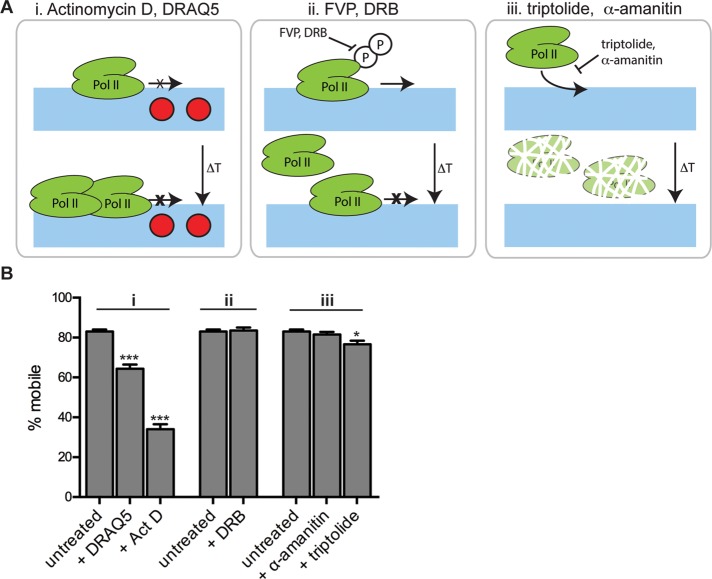
Nup50 is sensitive to transcription-targeting DNA intercalators
To better understand how Nup50 senses transcriptional inhibition, we analyzed Nup50 dynamics in the presence of a battery of drugs (Figure 5 and Supplemental Table S1). We tested transcription inhibitors of three main classes (Figure 5A): 1) DNA intercalators, 2) P-TEFb kinase inhibitors, and 3) inhibitors of transcription initiation. 1) The DNA intercalator Act D inhibits transcription by intercalating preferentially into regions of open chromatin (Yu, 1983; Chen et al., 1990), where costaining of Nup50 and chromatin marks suggests Nup50 may be present (Figure 4). Act D intercalation distorts DNA sufficiently to disrupt chromatin-associated processes, including transcription, without displacing the protein complement of chromatin from its nucleic acid docking sites (Chen et al., 1990). The result is that many chromatin-associated proteins, from RNA Pol II itself (Kimura, 2002) to transcription factors such as the estrogen receptor (Stenoien et al., 2001), become immobilized at the binding sites with which they normally interact dynamically. 2) The P-TEFb kinase inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) functions by preventing the regulatory phosphorylations of RNA Pol II that permit productive elongation of transcripts to begin (Bensaude, 2011). RNA Pol II's FRAP behavior reflects this, as it associates dynamically with transcription start sites and becomes stably engaged with chromatin only upon transition to productive transcript elongation. On treatment with DRB, the prominent fast-associating/dissociating phase remains, whereas the minor, slower-recovering phase disappears (Darzacq et al., 2007). Although RNA Pol II does not become kinetically trapped at transcription start sites in the presence of DRB, if one surveys the RNA Pol II population, it appears that more RNA Pol II is present in an initiating/paused/poised form. 3) α-Amanitin and triptolide prevent transcription initiation. α-Amanitin functions by binding competitively to the RNA Pol II subunit Rpb1, whereas triptolide binds and inhibits the ATPase subunit of TFIIH, thus preventing the opening of the transcription start site to accommodate RNA Pol II. In the short term, these drugs prevent RNA Pol II from engaging with transcription start sites; in the longer term, this inability to bind causes RNA Pol II to be targeted for proteolytic degradation (Bensaude, 2011).
FIGURE 5:
Nup50 dynamics are uniquely affected by transcription targeting intercalators. (A) Schematic of transcription inhibiting drugs used and their modes of action. i) Intercalators; ii) inhibitors of RNA Pol II regulatory phosphorylation; iii) competitive inhibitors of transcription initiation. (B) Summary of FRAP analysis of nucleoplasmic Nup50 in the presence of the indicated transcriptional inhibitors; see Material and Methods and Supplemental Table S1 for durations and concentrations of drugs used. N > 8 cells/condition. Bars, SEM. *p < 0.05 by t test.
We found that drugs from classes 2 and 3 had no effect on Nup50 mobility (Figure 5B and Supplemental Table S1). Instead, we found that Nup50 was uniquely sensitive to transcription-targeting DNA intercalators. Of interest, this was not the case for Nup98 and Nup153, which also became immobilized by DRB treatment (Griffis et al., 2004; our unpublished observations). In addition to Act D, we found that incubating cells for a short time with the DNA dye DRAQ5 had an intermediate but significant effect on Nup50 mobility (Figure 5B). Similar to Act D, DRAQ5 limits RNA Pol II progression through transcriptionally active chromatin (Richard et al., 2011). Of importance, the minor groove–binding dye Hoechst 33342 has little to no effect on transcription (White et al., 2000) and had no effect on Nup50 dynamics (Supplemental Table S1). Other intercalators, such as doxorubicin and cisplatin, also had no effect (Supplemental Table S1). Act D affects transcription by all three polymerases, with RNA Pol I being most sensitive (>0.05 μg/ml Act D), RNA Pol II intermediate (>0.5 μg/ml Act D), and RNA Pol III least sensitive (>5 μg/ml Act D; Bensaude, 2011). The range of Act D used here should affect both RNA Pol I and RNA Pol II (1 μg/ml); we found that Act D in the range that should affect only RNA Pol I (0.1 μg/ml) had a modest effect on Nup50 mobility (Supplemental Table S1). It is of note that only Act D and DRAQ5 are able to immobilize the transcription machinery on chromatin and that Nup50 appears uniquely responsive to these drugs.
Nup50 dynamics require ongoing RNA Pol II association with chromatin
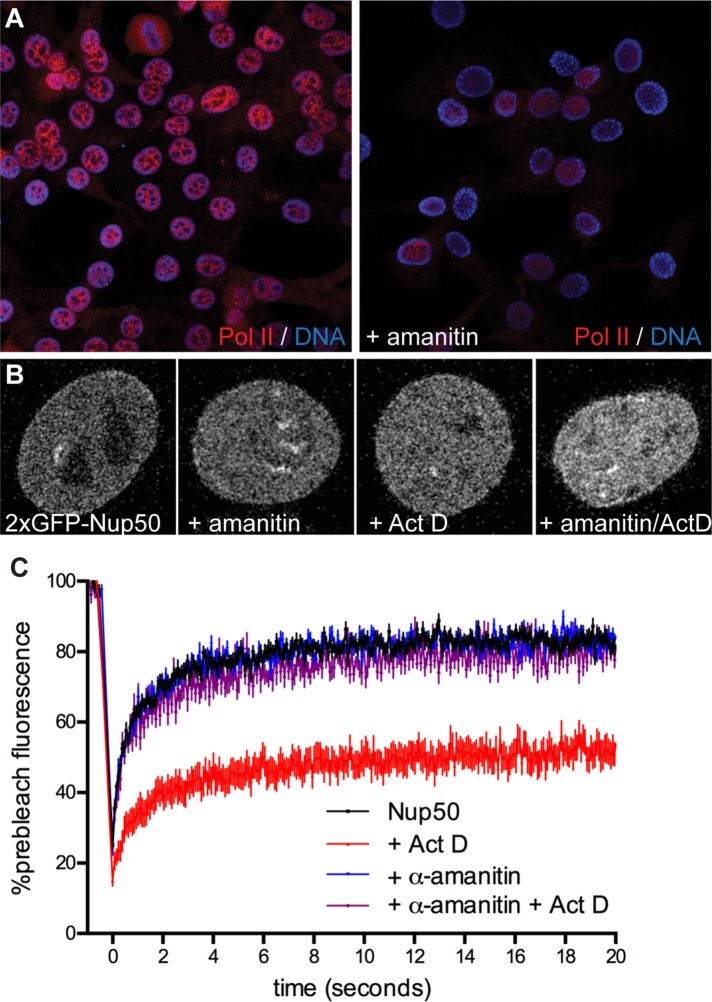
To probe more closely the dependence of Nup50 mobility on the transcription machinery, we sought a means to perturb RNA Pol II and assess the effects on Nup50. To achieve this, we preincubated cells with α-amanitin, which specifically destabilizes RNA Pol II and causes its eventual degradation (Bensaude, 2011) but on its own has no effect on Nup50 localization (Figure 6B) or dynamics (Figure 6C, blue trace). After 24 h of preincubation with 25 μg/ml α-amanitin, RNA Pol II is largely depleted (Figure 6A). We then tested the response of Nup50 to Act D and found that Nup50 no longer became immobilized under these conditions. This finding suggests that Nup50’s immobilization on chromatin requires that RNA Pol II is also engaged with chromatin.
FIGURE 6:
Nup50 dynamics depend on ongoing RNA Pol II transcription. (A) C2C12 cells stably expressing 2xGFP-Nup50 were preincubated with 25 μg/ml α-amanitin for 24 h before further treatment and analysis in C. (B) Representative images of 2xGFP-Nup50 under the indicated conditions. (C) FRAP of Nup50 in untreated cells (black trace), in cells treated with 1 μg/ml Act D (red trace), in cells treated with 25 μg/ml α-amanitin for 24 h (blue trace), or in cells first treated with α-amanitin followed by 1 μg/ml Act D (magenta trace). N > 17 cells/condition. Points indicate averaged values, and error bars indicate SEM.
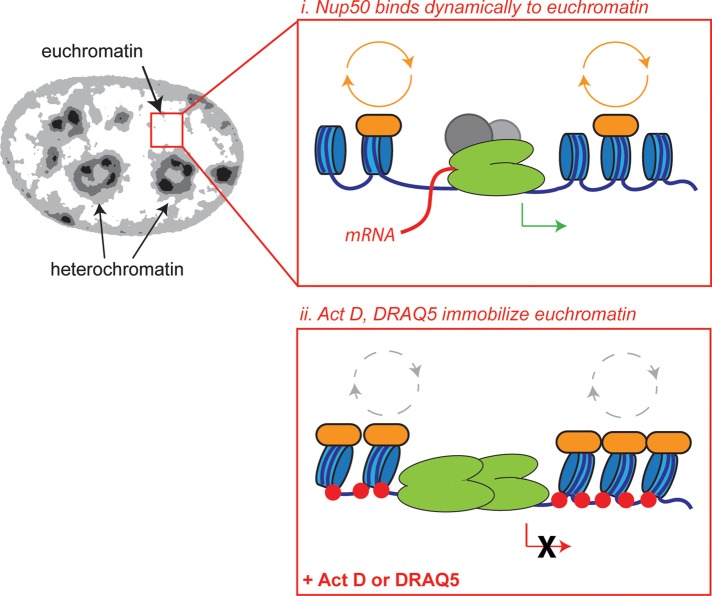
One interpretation of these data would be that Nup50 associates with RNA Pol II on chromatin. Confoundingly, however, only a small proportion of RNA Pol II itself becomes immobilized on chromatin when cells are treated with Act D (Supplemental Figure S6; Kimura, 2002). This is due to the fact that at any given time, only ∼20% of RNA Pol II molecules are engaged with and transcribing gene loci (Darzacq et al., 2007) and are thus capable of becoming trapped on chromatin by Act D intercalation. Nup50 is distributed broadly throughout the nucleoplasm, and there are no apparent nuclear subregions where Nup50 is more dramatically affected by changes in transcriptional status. We therefore consider it more likely that Nup50 interacts dynamically with an abundant component of actively transcribing euchromatin (see discussion of Figure 9) that relies on ongoing RNA Pol II activity for its deposition or stability.
FIGURE 9:
Model. Nup50 (orange) binds dynamically to an abundant factor on euchromatin. When transcription is inhibited, polymerases (green), nucleosomes (blue), and other associated proteins (not depicted), as well as Nup50, become immobilized on intercalated regions of euchromatin.
Nup50 dynamics correlate with global transcriptional activity levels
If the response of Nup50 dynamics to Act D is linked to ongoing transcription, we reasoned that Nup50 should also be sensitive to global changes in transcriptional output that occur in response to environmental stimuli. To address this possibility, we induced 2xGFP-Nup50–expressing C2C12 myoblasts to enter quiescence (G0) by incubating myoblasts in low-serum medium depleted of the essential amino acid methionine (Zhang et al., 2010). Quiescence is characterized by greatly reduced transcriptional output (Cheung and Rando, 2013). Cell cycle exit occurs after 24–48 h of methionine withdrawal (Supplemental Figure S5). On readdition of medium containing elevated serum and all essential amino acids, cells ramp up transcriptional activity within 1 h (Galbraith and Espinosa, 2011). After an ∼12-h lag phase, cells reenter G1 phase and resume cycling (Figure 7A and Supplemental Figure S5C). If Nup50 dynamics are indeed linked to transcriptional activity, clear differences should arise when comparing Nup50 dynamics in transcriptionally quiet G0 phase cells versus cells that have been exposed to a short phase of serum induction. We find that this is the case. The mobile fraction of Nup50 is significantly decreased in quiescent (G0) cells compared with cycling cells (Figure 7B, light gray trace vs. black trace), but addition of serum increases Nup50 mobility (Figure 7B, dark gray trace). We then treated either quiescent or cycling cells with Act D and found that in quiescent cells, Nup50 was even more dramatically immobilized (Figure 7B, orange trace vs. red trace). Strikingly, addition of full serum to quiescent cells quickly and completely reverted this phenotype, so that serum-stimulated cells again showed a response to Act D that was identical to that seen in cycling cells (Figure 7B, brown trace vs. red trace). We also visualized these differences in dynamics by fluorescence lifetime in photobleaching (FLIP; Figure 7C). Nup50 is rapidly bleached in the nuclei of cycling cells, as expected based on its high exchange rate on and off the NPC (Rabut et al., 2004a; Figure 7C, top). When cycling cells are treated with Act D, Nup50 slowly exchanges into the bleach region over time, resulting in a gradual broadening of the bleach region and eventual loss of nuclear fluorescence (Figure 7C, middle). When quiescent cells are exposed to Act D before FLIP, no detectable broadening of the bleach region is observed over several minutes of analysis (Figure 7C, bottom). Remarkably, when cells were fixed several hours later after completion of live imaging, the same cell was visible with a clearly defined square bleach region centered in the nucleoplasm (Figure 7C, bottom right). We conclude from these data that the dynamics of Nup50’s association with chromatin scale with the level of transcriptional output, ranging from one extreme in quiescent cells that have been further inhibited by pharmacologic intervention, to the most dynamic extreme of cycling, transcriptionally active cells. Of importance, even in these transcriptionally active cells, Nup50’s diffusion through the chromatin compartment is slowed by apparent binding and release events (Figure 2C).
FIGURE 7:
Changes in Nup50 dynamics correlate with changes in global transcriptional activity. (A) Schematic of growth conditions for inducing reversible quiescence in C2C12 myoblast cells. (B) FRAP of 2xGFP-Nup50 in cycling cells (black trace), cycling cells treated with 1 μg/ml Act D (red trace), quiescent (G0 phase) cells (light gray trace), quiescent cells treated with 1 μg/ml Act D (orange trace), serum-stimulated cells (medium gray trace), and serum-stimulated cells plus Act D (brown trace). N > 9 cells/condition. Points indicate averaged values, and error bars indicate SEM. (C) FLIP of 2xGFP-Nup50 in cycling cells (top), cycling cells treated with 1 μg/ml Act D (middle), or quiescent cells treated with 1 μg/ml Act D (bottom).
Nup50 is required for myoblast differentiation
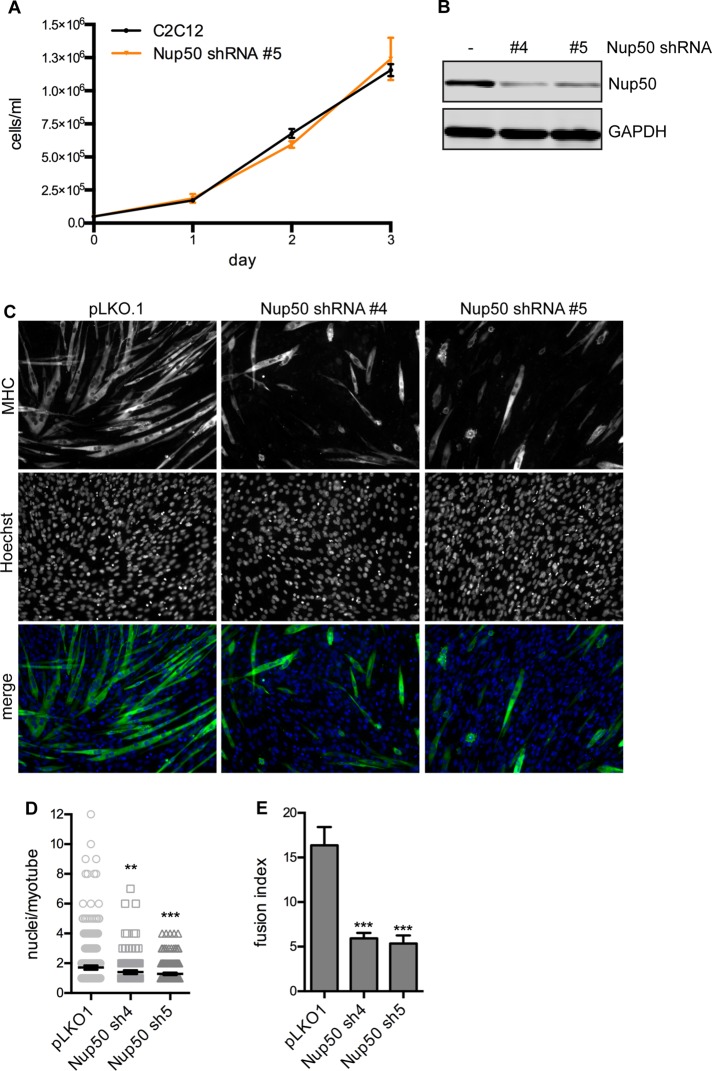
We depleted C2C12 myoblasts of Nup50 and found that they proliferated normally (Figure 8A). Consistently, mouse embryonic fibroblasts derived from a Nup50−/− mouse proliferate normally in culture, although the mouse is not viable (Smitherman et al., 2000). These data argue against an essential role for Nup50 in nucleocytoplasmic transport. The change in Nup50 dynamics in response to various means of perturbing transcriptional output suggests that Nup50 may have a role in some aspect of transcriptional regulation, and we reasoned that Nup50 may be required for execution of particular developmental and/or differentiation programs. We tested this possibility in vitro by testing the ability of Nup50-depleted myoblasts to differentiate into myotubes. Introduction of two independent short hairpin RNAs (shRNAs) targeted to Nup50 caused a significant decrease in the efficiency of myotube formation (Figure 8, C–E). Myotubes are marked by myosin heavy chain (MHC) expression and an elongated, multinucleate morphology (Figure 8C). Nup50 knockdown caused both a decrease in fusion index (the percentage of nuclei per field present in MHC+ cells containing more than two nuclei; Figure 8E) and in the number of nuclei per myotube (Figure 8D). These data suggest that Nup50 plays a yet-to-be-identified role in developmental gene regulation or chromatin biology.
FIGURE 8:
Depletion of Nup50 does not affect cell proliferation but limits differentiation. (A) Comparison of cell number over several days of growth of C2C12 cells (black trace) or C2C12 cells stably expressing a Nup50 shRNA (orange trace) that were initially seeded at equal density. (B) Western blot of Nup50 in C2C12 cells stably transduced with pLKO.1 empty vector or two different Nup50 shRNAs. (C) MHC staining of differentiated myotubes in C2C12 cells stably transduced with pLKO.1 empty vector or two different Nup50 shRNAs after 72 h in differentiation medium. (D) Quantification of number of nuclei per myotube observed in C2C12 cells expressing pLKO.1 empty vector or two different Nup50 shRNAs after 72 h in differentiation medium as shown in C. N > 189 cells/condition. *p <0.05 by t test with Welch's correction. (E) Fusion index (percentage of nuclei in myotubes containing n > 2 nuclei/total nuclei in field) for C2C12 cells expressing pLKO.1 empty vector or two different Nup50 shRNAs after 72 h in differentiation medium as shown in C. N > 2000 cells/condition. *p < 0.05 by t test.
DISCUSSION
We report that the dynamic nuclear basket nucleoporin Nup50 exhibits transcription-dependent dynamics both at the NPC and within the nucleoplasm. Of importance, Nup50’s response to transcription inhibition persists when Nup50 is unable to engage with the nuclear transport machinery (Figure 2) and occurs without any corresponding effect on nuclear transport (Supplemental Figure S4). We therefore conclude that Nup50 performs a function on chromatin that is independent of its known role in nuclear transport. We expect that chromatin-associated function of Nup50 is independent of the other known transcriptionally sensitive NPC proteins—Nup98 and Nup153—as its nuclear dynamics are not affected by either Nup98 or Nup153 knockdown (Figure 3). Because Nup50’s response to transcription-inhibiting drugs depends on RNA Pol II being engaged with chromatin and scales with global changes in transcriptional output, we expect that this represents a bona fide sensitivity to transcription-related processes on chromatin. Finally, we show that depletion of Nup50 from C2C12 myoblasts does not affect proliferation but inhibits differentiation into myotubes (Figure 8), suggesting a role for Nup50 in mediating normal responses to stimuli via its position on chromatin.
Because Nup50 is distributed broadly throughout the nucleoplasm and responds homogeneously to transcription inhibition, we propose that Nup50 interacts dynamically with an abundant component of euchromatin (Figure 9). When cells are exposed to DNA-intercalating drugs such as Act D or DRAQ5, these drugs incorporate into and halt processes occurring on active euchromatin, including polymerase progression and, presumably, consequent nucleosome turnover. Treatment with these drugs immobilizes Nup50, as well as elongating RNA Pol II (Kimura, 2002), histones (Catez et al., 2002), HP1a (Cheutin et al., 2003), certain transcription factors (Stenoien et al., 2001), and presumably many other proteins that are engaged with the affected stretches of DNA. It seems unlikely that Nup50 interacts directly with RNA Pol II because RNA Pol II itself responds less dramatically to Act D treatment than does Nup50 (compare Supplemental Figure S6 to Figure 1; Kimura, 2002). This is due to the fact that at any given time, only ∼20% of RNA Pol II molecules are engaged with and transcribing chromatin loci (Darzacq et al., 2007). Instead, a consistent explanation would be that Nup50 interacts dynamically with an abundant component of euchromatin that is deposited in a transcription-dependent manner, such as a subpopulation of histones or a histone-binding protein.
Key to a deeper understanding of the mechanism and importance of Nup50’s binding to chromatin is defining the chromatin loci and protein-binding partners with which it associates. We undertook experiments to answer both of these questions. Unfortunately, we were not able to identify a Nup50 antibody that performed well under chromatin-immunoprecipitating conditions. Recent DamID experiments in D. melanogaster–derived S2 cells identified a number of chromatin loci bound by Nup50 (Kalverda et al., 2010), which were enriched with actively transcribed genes having active chromatin modifications and developmentally significant roles. It is attractive to speculate that Nup50’s interaction with developmental genes may be functionally required for developmental and differentiation processes such as the formation of terminally differentiated myotubes (Figure 8).
We also performed immunoprecipitation followed by mass spectrometry of affinity-tagged Nup50 in an effort to identify protein-binding partners. We recovered known interactions at the NPC and in the nuclear transport machinery but did not identify any nuclear binding partners relevant to the phenotype under study (unpublished data). Future efforts focused on identifying interaction partners specifically in the nuclear compartment will be important to solving this mystery.
Nup50 has been termed a “tristable switch” because of its ability to interact with importin α, importin β, and Ran through distinct protein domains (Lindsay and Macara, 2002). This cycle of interactions potentiates protein transport under certain conditions (Lindsay and Macara, 2002). However, it is not required for nucleocytoplasmic transport, as Nup50-null or -knockdown cells proliferate normally (Smitherman et al., 2000; Figure 8). We find that Nup50 responds equivalently to transcriptional inhibition when its Ran-binding C-terminus is deleted or when its importin α–binding motif is mutated (Figure 2). That is, Nup50 responds equivalently to transcriptional inhibition when parts of the “switch” are deleted. It is an intriguing possibility, however, that Nup50 could bring importin α, importin β, Ran, or perhaps even transport cargoes along as passengers to its binding sites on chromatin.
Overall, our data suggest that Nup50 performs additional functions related to chromatin biology within the nucleoplasm that are independent of its roles in protein transport. How might Nup50 be apportioned between these two roles? We find that Nup50 responds to transcriptional inhibition even when not tethered to the NPC by Nup153. It is possible that tethering to the NPC via Nup153 may expose domains of Nup50 necessary for potentiating protein transport. Perhaps relevant to this is work from the Ullman laboratory showing that Nup153 binds to two regions of Nup50 and that one of these binding modes is mediated by importin α (Makise et al., 2012). Nup50 may then exist in a distinct conformation with distinct roles when unbound from Nup153 in the nucleoplasm.
It is becoming increasingly clear that a subset of nuclear pore proteins have functions that are not strictly related to nucleocytoplasmic transport. The cast of components of the mammalian NPC has been defined by proteomic analysis (Cronshaw, 2002), but recent studies have revealed that some nucleoporins, such as gp210, are present only in the NPCs of selected tissues (Olsson et al., 2004; Raices and D'Angelo, 2012). Up-regulation of gp210 is required for myogenesis and appears to regulate transcriptional programs without having any appreciable effect on nucleocytoplasmic transport (D'Angelo et al., 2012). Other nucleoporins appear to be multifunctional, having established roles in both nuclear transport and other processes. For instance, Nup98 has clearly defined roles in nucleocytoplasmic transport of proteins (Radu et al., 1995) and RNAs (Blevins et al., 2003) but also functions in gene regulation (Capelson et al., 2010; Kalverda et al., 2010; Liang et al., 2013) and has recently been implicated in RNA stabilization (Singer et al., 2012). Separately, recent work has demonstrated that the NPC functions as a tether for protein complexes that perform roles unrelated to nucleocytoplasmic transport, such as the TREX RNA export complex (Umlauf et al., 2013) and the Mad1/2 spindle checkpoint complex (Rodriguez-Bravo et al., 2014). Surprisingly, FRAP experiments indicate that both the TREX complex (Umlauf et al., 2013) and the Mad1/2 complex (Rodriguez-Bravo et al., 2014) are more stably associated with the NPC than many mobile nucleoporins. These findings suggest that NPCs should not strictly be considered as transport channels but instead as multifunctional hubs that integrate a variety of nuclear processes, many of which are critical for cell differentiation.
Our study adds Nup50 to the group of transcriptionally sensitive nucleoporins along with Nup153 and Nup98. These Nups have been shown to function in gene regulation, and we anticipate that Nup50 will have some role in gene regulation as well. Nup153 exchanges dynamically on and off the NPC structure (Rabut et al., 2004b) but is not clearly detected in a soluble nuclear pool (Supplemental Figure S2). It is thus unclear whether some or all of Nup153’s interactions with the genome occur on the NPC itself. Nup98 is very stable when incorporated into the NPC (Rabut et al., 2004b) but also exists in a dynamic intranuclear pool (Supplemental Figure S2). Work in Drosophila (Kalverda et al., 2010) and mammalian cells (Liang et al., 2013) demonstrated distinct roles of free versus tethered Nup98 in interacting with the genome. Soluble Nup98 interacts robustly with highly expressed developmental genes, whereas chromatin association with NPC-tethered Nup98 occurs at distinct loci that are not highly expressed. Compared to Nup98 and Nup153, Nup50 has a much more abundant nucleoplasmic pool, which seems to respond homogeneously to transcriptional inhibition (Figure 1), whereas Nup98 and Nup153 do so only in defined subregions of the nucleus. We therefore speculate that Nup50 associates with a very abundant component of chromatin to promote differentiation.
MATERIALS AND METHODS
Reagents
2xGFP-Nup50 and GFP-Nup98 expression constructs (mouse) were a gift from Jan Ellenberg (EMBL, Heidelberg, Germany) and are described in Rabut et al. (2004b). Truncation mutants of Nup50 were made by PCR amplification of cDNA corresponding to amino acids 1–214 or 214–468 using primers containing restriction sites compatible with ligation into peGFPx2-C1 (derived from Ellenberg plasmids). The Nup50 44KRR46/AAA mutant was made by PCR amplification of the 2xGFP-Nup50 plasmid using Pfu enzyme and primers flanking the mutation site and containing the mutated codons. NLS-GFP is as described in Vargas et al. (2012). Mouse Nup153 was tagged with GFP at its N-terminus by cloning into the pDEST53 vector using the Gateway method. shRNAs against mouse Nup153, Nup98, and Nup50 in the lentiviral pLKO.1 expression vector were obtained from the Sigma-Aldrich MISSION shRNA Library (Sigma-Aldrich, St. Louis, MO). Antibodies were mAb414 (Covance, San Diego, CA), Nup50 (ab151567; Abcam, Cambridge, MA), H3K4me3 (Active Motif), H3K9me3 (Upstate/EMD Millipore, Billerica, MA), Ser-5–phosphorylated RNA Pol II (Covance), and α-tubulin (Sigma-Aldrich). MF-20 antibody to myosin heavy chain was prepared from a hybridoma line (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). A cyclin A antibody was a gift from Tony Hunter (Salk Institute for Biological Studies, La Jolla, CA).
Microscopy
Confocal microscopy and structured illumination microscopy were performed on Zeiss LSM 710 and Elyra microscopes, respectively, using a 63×/1.4 numerical aperture objective. Images were analyzed and prepared for presentation in ImageJ and Photoshop.
FRAP was performed on a Zeiss LSM 710 microscope following conditions described in Mueller et al. (2011). Briefly, cells were grown in eight-well chamber dishes (Ibidi, Martinsried, Germany) and transferred to an environmental chamber at 37°C and 5% CO2 for imaging. For FRAP of NPC pools, a region of interest (ROI) was drawn around a portion of the nuclear periphery and bleached, and 1 frame/s was collected for several minutes. For fast nuclear FRAP, a circular bleach ROI was selected, and imaging was performed in bidirectional scanning mode with 256 × 256 images acquired at ∼0.1-μm pixel size in order to achieve a frame rate of ∼12 frames/s. In both cases, 5–10 prebleach images were acquired and fluorescence values averaged; postbleach intensity values were normalized to the average prebleach intensity value. Each FRAP series was thus expressed as percentage prebleach intensity over time, and FRAP series were averaged across multiple cells for presentation. Data analysis, plotting, and curve fitting were performed in Prism (GraphPad, La Jolla, CA). To determine plateau/mobile fraction and t1/2 values, we fit FRAP series to a single-exponential curve of the form y = y0 + (plateau − y0)(1 − e−kx), where t1/2 = ln(2)/k.
Cell culture, transfections, and RNAi
Low-passage C2C12 myoblasts were obtained from the American Type Culture Collection and grown in DMEM plus 20% fetal bovine serum (FBS) at 5% CO2. C2C12 cells stably expressing a low amount of 2xGFP-Nup50 (Supplemental Figure S1A) were generated by transfection, followed by G418 selection and FACS enrichment of stably expressing cells. U2OS cells stably expressing an amanitin-resistant yellow fluorescent protein–Rpb1 subunit of RNA Pol II were a gift from Robert Singer (Albert Einstein College of Medicine, Bronx, NY) and were grown in DMEM plus 10% FBS supplemented with 25 μg/ml amanitin at 5% CO2. Plasmids were transfected with Lipofectamine 2000 according to the manufacturer's instructions (Life Technologies).
Drugs were administered at the concentrations indicated in Supplemental Table S1 as follows. DRAQ5, actinomycin D, α-amanitin, DRB, triptolide, and Hoechst 33342 were administered for 30 min to 2 h before imaging. Camptothecin, doxorubicin, and cisplatin were administered 2–4 h before imaging. Hydroxyurea and aphidicolin were administered 12 h before imaging. For degradation of RNA Pol II as depicted in Figure 6, 25 μg/ml α-amanitin was administered for 24 h before short-term administration of Act D and imaging.
Stable expression of shRNAs was achieved by lentiviral production in HEK293T cells, followed by infection of C2C12 cells with viral particles in the presence of 6 μg/ml Polybrene. Cells were split into puromycin selection 48 h after infection and maintained in selection thereafter. Knockdown was verified by Western blot and immunofluorescence.
Quiescence induction in C2C12 myoblasts was performed as described in Zhang et al. (2010). Briefly, cells were grown on tissue culture–treated polystyrene dishes in DMEM plus 20% heat-inactivated FBS. When cells reached 40–50% confluency, they were washed extensively with warmed phosphate-buffered saline before switching to DMEM lacking methionine plus 1% FBS for 36–48 h. Quiescent cells were induced to reenter the cell cycle by readdition of DMEM plus 20% FBS.
Differentiation of C2C12 myoblasts into myotubes was achieved by growing C2C12 cells to confluency and transferring them to DMEM plus 2% horse serum for 48–96 h. Cells were then fixed and stained for myosin heavy chain (MHC) to identify differentiated cells.
Supplementary Material
Acknowledgments
We thank the Waitt Advanced Biophotonics Center at the Salk Institute for assistance with superresolution microscopy and members of the Hetzer laboratory for their valuable feedback during preparation of the manuscript. This work was supported by the Glenn Foundation for Medical Research (M.H.), as well as by grants P30CA014195 and R01GM098749 (M.H.) and fellowship F32GM103084 (A.B.) from the National Institutes of Health.
Abbreviations used:
- Act D
actinomycin D
- DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- FLIP
fluorescence lifetime in photobleaching
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- MHC
myosin heavy chain
- NLS
nuclear localization sequence
- NPC
nuclear pore complex
- RNA Pol I/II/III
RNA polymerase I/II/III
- shRNA
short hairpin RNA
- TFIIH
transcription factor II H
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-04-0865) on June 18, 2014.
REFERENCES
- Bensaude O. Inhibiting eukaryotic transcription: which compound to choose? how to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem. 2003;278:20979–20988. doi: 10.1074/jbc.M302061200. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: a hypothesis. Proc Natl Acad Sci USA. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell. 2012;22:1234–1246. doi: 10.1016/j.devcel.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Catez F, Brown DT, Misteli T, Bustin M. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 2002;3:760–766. doi: 10.1093/embo-reports/kvf156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TA, Sterner R, Cozzolino A, Allfrey VG. Reversible and irreversible changes in nucleosome structure along the c-fos and c-myc oncogenes following inhibition of transcription. J Mol Biol. 1990;212:481–493. doi: 10.1016/0022-2836(90)90327-I. [DOI] [PubMed] [Google Scholar]
- Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Cronshaw JM. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in Metazoa. Cell. 2010;141:1030-1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Espinosa JM. Lessons on transcriptional control from the serum response network. Curr Opin Genet Dev. 2011;21:160–166. doi: 10.1016/j.gde.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell. 2003;14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kimura H. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A. Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Macara IG. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell. 2002:110, 349–360. doi: 10.1016/s0092-8674(02)00836-x. [DOI] [PubMed] [Google Scholar]
- Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. The Nup153-Nup50 protein interface and its role in nuclear import. J Biol Chem. 2012;287:38515–38522. doi: 10.1074/jbc.M112.378893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F, Karpova TS, Mazza D, McNally JG. Methods Mol Biol. 833: 2011. Monitoring dynamic binding of chromatin proteins in vivo by fluorescence recovery after photobleaching; pp. 153–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Schéele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res. 2004;292:359–370. doi: 10.1016/j.yexcr.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Pascual-Garcia P, Capelson M. Nuclear pores as versatile platforms for gene regulation. Curr Opin Genet Dev. 2014;25C:110–117. doi: 10.1016/j.gde.2013.12.009. [DOI] [PubMed] [Google Scholar]
- Pumroy RA, Nardozzi JD, Hart DJ, Root MJ, Cingolani G. Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin α5. J Biol Chem. 2012;287:2022–2031. doi: 10.1074/jbc.M111.315838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004a;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Rabut G, Lénárt P, Ellenberg J. Dynamics of nuclear pore complex organization through the cell cycle. Curr Opin Cell Biol. 2004b;16:314–321. doi: 10.1016/j.ceb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Raices M, D'Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- Richard E, Causse S, Spriet C, Fourré N, Trinel D, Darzacq X, Vandenbunder B, Heliot L. Short exposure to the DNA intercalator DRAQ5 dislocates the transcription machinery and induces cell death. Photochem Photobiol. 2011;87:256–261. doi: 10.1111/j.1751-1097.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. Nuclear pores protect genome integrity by assembling a premitotic and mad1-dependent anaphase inhibitor. Cell. 2014;156:1017–1031. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, et al. THO/Sub2p functions to coordinate 3’-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Singer S, et al. Nuclear pore component Nup98 is a potential tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol Cell. 2012;48:799–810. doi: 10.1016/j.molcel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smitherman M, Lee K, Swanger J, Kapur R, Clurman BE. Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol Cell Biol. 2000;20:5631–5642. doi: 10.1128/mcb.20.15.5631-5642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenoien DL, Patel K, Mancini MG, Dutertre M, Smith CL, O'Malley BW, Mancini MA. FRAP reveals that mobility of oestrogen receptor-alpha is ligand- and proteasome-dependent. Nat Cell Biol. 2001;3:15–23. doi: 10.1038/35050515. [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 2009;23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci. 2013;126:2656–2667. doi: 10.1242/jcs.118000. [DOI] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Lapetina DL, Chen W-M, Chiang J-H, Aitchison JD, Wozniak RW. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell. 2013;152:969–983. doi: 10.1016/j.cell.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus. 2012;3:88–100. doi: 10.4161/nucl.18954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CM, Heidenreich O, Nordheim A, Beerman TA. Evaluation of the effectiveness of DNA-binding drugs to inhibit transcription using the c-fos serum response element as a target. Biochemistry. 2000;39:12262–12273. doi: 10.1021/bi001427l. [DOI] [PubMed] [Google Scholar]
- Yu FL. Actinomycin D-binding in vivo: active chromatin preferred. FEBS Lett. 1983;156:83–87. doi: 10.1016/0014-5793(83)80253-1. [DOI] [PubMed] [Google Scholar]
- Zhang K, Sha J, Harter ML. Activation of Cdc6 by MyoD is associated with the expansion of quiescent myogenic satellite cells. J Cell Biol. 2010;188:39–48. doi: 10.1083/jcb.200904144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.