Abstract
Saponins in Platycodi Radix (platycosides) exhibit potent biological activities in mammalian systems, including several beneficial effects such as anti-inflammatory, immunomodulatory and anti-obesity activities. In this study, we developed a new HPLC separation coupled with evaporative light scattering detector (ELSD) for the simultaneous quantitative determination of ten major saponins in Platycodi Radix. Simultaneous separation of these saponins was achieved on a C18 analytical column. The mobile phase consisted of a gradient of aqueous acetonitrile. The method was validated for linearity, precision, accuracy, limit of detection and quantification. Electrospray ionization mass spectrometry (ESI-MS) and liquid chromatography coupled with on-line mass spectrometry (LC-ESI MS/MS) were applied to identify platycosides in the purified fractions and in the crude extract. Under ESI-MS/MS conditions, the fragmentation patterns of [M – H]− ions exclusively show signals corresponding to cleavage of the glycosidic bonds, thus allowing a rapid identification of saponins in the crude extract of Platycodi Radix. The validated HPLC method provides a new basis of overall assessment on quality of Platycodi Radix, and ESI-MS/MS and LC-ESI MS/MS approaches offers analytical tools for a rapid screening of platycosides in the crude extract.
Keywords: Platycodi Radix, Saponin, HPLC-ELSD, Quantification, ESI-MS/MS
1. Introduction
Platycodi Radix, the root of Platycodon grandiflorum A.DC. (Campanulaceae), commonly known as Jiegeng in Chinese, Kilkyong in Korean, and Kikyo in Japanese, has been used as traditional Oriental medicine. Traditionally, Platycodi Radix has been used as an expectorant and to treat bronchitis, tonsillitis, sore throat and other respiratory ailments. Also, Platycodi Radix showed numerous biological activities such as hyperlipidemia, diabetes, inflammatory disease and immunopharmacological reponses [1–9].
Platycosides are the primary constituents of Platycodi Radix, responsible for a diversity of effects including anti-inflammation, anti-allergy, antitumor, augmentation of immune responses, stimulating the apoptosis in skin cells anti-obesity and hyperlipidemia [10–17]. Previously several methods have been reported on the determination of platycosides by high performance liquid chromatography (HPLC) [18,19]. However, they have not shown high sensitivity, complete separation, calibration, and validation. The explanation for this limited number of reports is most likely based on the fact that saponins show very weak absorbance, even in the short wavelength range, which renders sensitive detection by ultraviolet spectroscopy impossible. HPLC-UV analyses of saponins result in a high level of baseline noise, limiting the choice of solvents and the use of mobile-phase modifiers required for improved separation. ELSD is a mass detector that measures the scattered light generated by the non-volatile particles of analytes produced by the nebulization into droplets of the LC effluent. ELSD is a universal, non-specific detector that can provide a stable baseline even with gradient elution [20,21].
In many cases, total triterpenoidal saponins, rather than pure components, are usually treated as the active ingredients in traditional herbal remedies, which may cause trouble in the quality control of natural drugs as well as complicate the search for potential new lead compounds. Recently, much research has focused on the metabolites and pharmacokinetics of saponins due to the biological activities of their metabolites and their absorption profile [22–26]. Traditional analytical protocols for saponins are complicated and time-consuming procedures because of their inherent properties including their high polarity, thermal lability, non-volatility and low content. Therefore, there is an increasing demand for methods that can rapidly identify and characterize known or new structures.
Electrospray ionization (ESI), mass spectrometry has become a powerful tool for structural analysis of saponins in crude extracts of herbal plants due to its high sensitivity, rapid analysis time and low levels of sample consumption [27–30]. It has the potential ability to rapidly determine the bioactive compounds in mixtures and give information on their structures as well as molecular weights. Recent publications on the LC–MS and LC coupled with sequential mass spectrometry (LC–MSn) have been extensively applied on the on-line analysis of saponins [30–41].
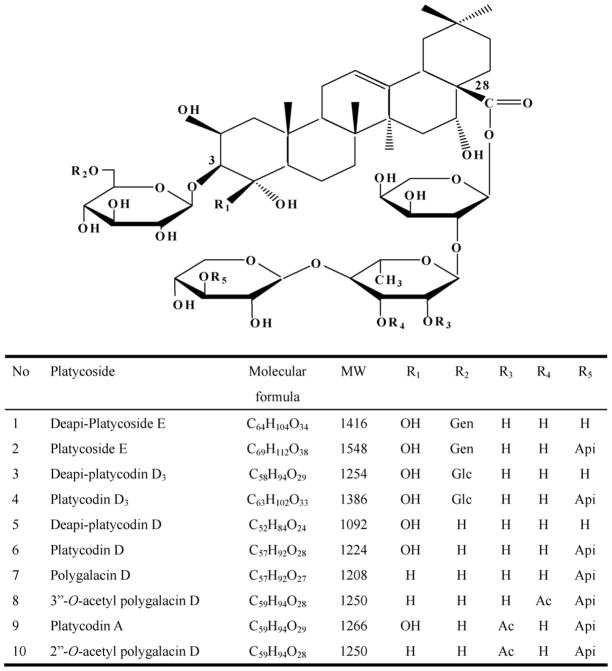
This paper describes, for the first time, a HPLC method coupled with ELSD for quality control allowing the simultaneous determination of ten major triterpenoidal saponins in Platycodi Radix: (1) deapi-platycoside E; (2) platycoside E; (3) deapi-platycodin D3; (4) platycodin D3; (5) deapi-platycodin D; (6) platycodin D; (7) polygalacin D; (8) 3″-O-acetyl polygalacin D; (9) platycodin A; and (10) 2″-O-acetyl polygalacin D (Fig. 1). ESI-MS/MS and LC–MS/MS were employed simultaneously to determine the saponins in the crude extract of Platycodi Radix for rapid structural identification. These studies should provide a method for the quantitative determination and structural characterization of platycosides.
Fig. 1.
Structure of platycosides. Gen: glucose – glucose; Glc: glucose; Api: apiose; Ac: acetyl.
2. Experimental
2.1. Standards and reagents
Ten pure saponins (Fig. 1) were isolated from the aqueous extract from Platycodi Radix as previously described [12]. The structures of the purified saponins were confirmed by comparison of HPLC, FAB-MS and 1H NMR data with references [11,42–45]. The purity of these saponins was determined to be more than 98% by peak areas detected by HPLC with ELSD. Ten saponins were dissolved in 50% CH3CN at 5 mg/ml to prepare their primary stock solutions. These solutions were stored at −20 °C when not in use and they were stable for at least 2 months.
Solid phase extraction columns (Sep-Pak® Vac C18) were obtained from Waters (Milford, MA, USA), and acetonitrile, methanol and water (HPLC-grade) were obtained from Fisher Scientific (Pittsburg, PA, USA). Mobile phases were degassed by sonication for 30 min before use.
2.2. Sample preparation
Approximately 1 g of the finely powdered Platycodi Radix was extracted three times with 10 ml of each solvent system (water, 30% MeOH, 50% MeOH, 70% MeOH and MeOH) by sonication for 10 min, to test the extraction efficiency of these solvent systems. After centrifugation (Union 32R Plus, Hanil, Korea), the combined supernatants were evaporated, and residue was dissolved in 10 ml water. One millilitre of the aqueous sample solution was applied to Sep-Pak® C18 column. The column was eluted with water (3 ml), 30% methanol (5 ml) and 70% methanol (3 ml), sequentially. The 70% methanol elution was filtered through a 0.45 μm syringe filter (Type Millex-HA, Millipore, USA) and 50 μl of the filtrate was injected to the HPLC system. Every sample solution was injected in triplicate, and the contents of the analytes were determined from the corresponding calibration curves.
2.2.1. Analysis of platycosides by HPLC-ELSD
HPLC on platycosides was carried out on a Hitachi L-6200 instrument equipped with a Sedex 75 ELSD and SIL-9A auto injector (Shimadzu, Japan). A Zorbax SB-Aq C18 column (150 mm × 4.6 mm, 5 μm particle size) from Agilent Technologies (Palo Alto, CA, USA) was used for all separations. HPLC conditions were as follows: eluent A, water; eluent B, acetonitrile; gradient, 0–6 min (10–15% B), 6–50 min (15–25% B), 50–60 min (25–47.5% B), and then equilibrated with 10% B for 8 min at a flow of 1 ml/min. ELSD was set to a probe temperature of 70 °C, a gain of 7 and the nebulizer gas nitrogen adjusted to 2.5 bar. Peaks were assigned by comparing their retention times with that of each reference compound eluted in parallel with a series of mobile phases and by spiking sample with reference compounds.
2.2.2. Calibration
The standard solutions containing 2 μg/ml to 400 μg/ml, corresponding to test ranges of ten platycodin saponins, were prepared in triplicate, 50 μl of each sample was injected into the HPLC column, and calibration curves were constructed and their linear ranges determined. Calibration curves were plotted by the peak area versus concentration of each analyte. The linearity was evaluated by linear regression analysis calculated by the least square regression method. Limits of detection (LOD) and quantification (LOQ) under the present chromatographic conditions were determined on the basis of response and slope of each regression equation at a signal-to-noise ratio (S/N) of 3 and 10, respectively.
2.2.3. Validation
The precision of the HPLC method was determined for intra-and inter-day variations. Samples containing approximately 1 g of the pulverized Platycodi Radix were weighed, extracted and analyzed as described in Section 2.2. The intra-day variability was performed in triplicate on the same sample extracted on a single day, while the inter-day precision was carried out in triplicate in another independent sample extracted on three different days. Variations were expressed by the relative standard deviations (RSD).
The recovery test was used to evaluate the accuracy of this method. Accurate amounts of nine saponins were added to 1 g of Platycodi Radix, and then extracted and analyzed as described in Section 2.2. The average recoveries were determined by the formula: recovery (%) = (observed amount − original amount)/spiked amount ×100%, and RSD (%) = (SD/mean) ×100%.
2.2.4. Analysis of pure platycosides by negative ion ESI-MS/MS
Identification of saponins was performed in the negative ion mode using a LCQ DECA XP MS (Thermo Finnigan, San Jose, CA, USA) system equipped with an electrospray source. The source was operated at 4 kV of ion spray voltage, −15 V of capillary voltage, 275 °C of capillary temperature, and under N2 sheath gas set at 35 in arbitrary units. The sample was analyzed by direct injection through a 2 μl direct loop and the elution solvent was a mixture of water and acetonitrile (50:50, v/v). For MS/MS, the [M − H]− of each saponin was selected as a precursor ion and the MS/MS product ions were obtained using the condition of 27% collision energy in the ion trap analyzer.
2.2.5. Analysis of crude extract by LC-ESI MS/MS
The chromatographic separation of a mixture of the pure saponins and the crude extract was performed on a 150 mm ×1 mm i.d., 5 μm Phenomenex Luna-C18 column using a flow rate of 50 μl/min and the gradient composition of the mobile phase was the same described in the Section 2.2.1. The LC system (NANOSPEAC SI-2, Shiseido, Japan) was interfaced to MS described in Section 2.2.4 and a sample of 1 μl was injected into the column using an autosampler. The total ion chromatogram was obtained using a LC-ESI MS/MS at 150–2000 m/z. Using the above conditions, the characteristic MS/MS product ions were obtained at the m/z value of [M − H]− ion for each of 10 saponins sequentially eluting in the chromatograms of crude extracts and pure saponins. Since the crude extracts contain platycosides as well as other unknown compounds, which can interfere with analysis, only the [M − H]− ion of each platycoside was measured using the selective ion monitoring mode.
3. Results and discussion
3.1. Analysis of platycosides by HPLC-ELSD
The important parameters that need to be controlled for the optimization of ELSD response are the flow rate of nebulizer gas (pressure) and drift tube temperature [46]. Under the fixed chromatographic conditions, two parameters was evaluated by the injection of platycoside E, which is the testing saponin for optimizing ELSD conditions, at different detector temperatures from 60 °C to 100 °C and the pressure from 2.0 bar to 3.5 bar. In this study, the drift tube temperature of 70 °C and gas pressure of 2.5 bar were selected for detecting the analytes by comparing peak area values. These optimized parameters allow a complete solvent evaporation and produce negligible baseline noise.
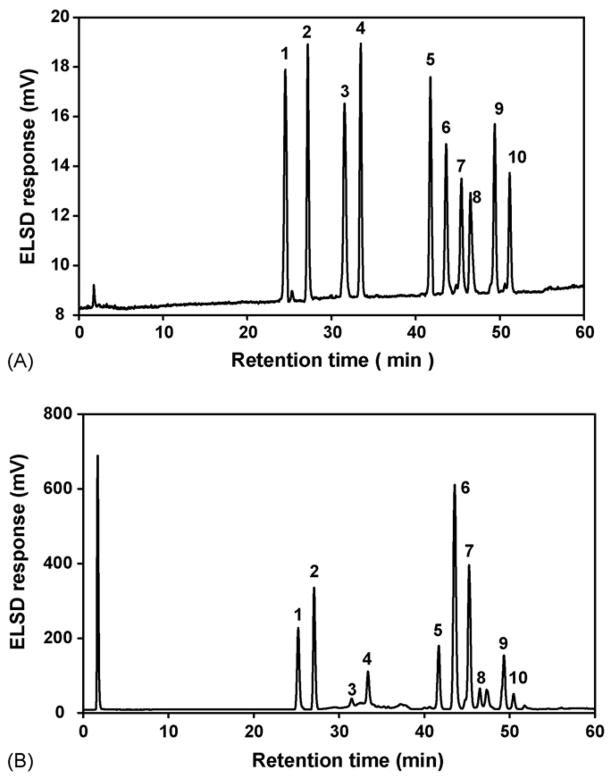
The representative chromatograms for the mixture of ten purified major saponins and a crude saponin fraction (water extract) in Platycodi Radix is shown in Fig. 2A and B. The results indicate that platycosides are clearly separated and their quantitative determination in Platycodi Radix is possible. Table 1 provides equations describing the linear range, correlation coefficient, LOD and LOQ for each compound determined. All calibration curves showed good linearity (R2 > 0.9931) and LODs between 0.12 μg and 0.36 μg, LOQs between 0.28 μg and 0.76 μg were achieved.
Fig. 2.
Representative HPLC chromatograms of mixed standards and extract of Platycodi Radix. Column: Agilent Zorbax SB-Aq C18 column (150 mm ×4.6 mm, 5.0 μm), detector: ELSD, drift tube temperature: 70 °C, nitrogen flow-rate: 2.5 bar. (A) mixed standards; (B) extract of Platycodi Radix. The numbers indicate each platycoside in Fig. 1.
Table 1.
Linearity of calibration curve for ten saponins
| Compounda | Calibration curveb | R2 | Linear range (μg) | LOD (μg) | LOQ (μg) |
|---|---|---|---|---|---|
| 1 | Y = 804.7x −478.1 | 0.9988 | 0.30–20 | 0.16 | 0.34 |
| 2 | Y = 840.7x −448.7 | 0.9998 | 0.28–20 | 0.14 | 0.32 |
| 3 | Y = 828.7x −586.0 | 0.9931 | 0.38–20 | 0.22 | 0.48 |
| 4 | Y = 905.6x −497.4 | 0.9978 | 0.22–18 | 0.12 | 0.28 |
| 5 | Y = 818.8x −499.9 | 0.9999 | 0.28–20 | 0.18 | 0.46 |
| 6 | Y = 743.7x −637.3 | 0.9951 | 0.28–20 | 0.18 | 0.42 |
| 7 | Y = 475.8x −410.4 | 0.9973 | 0.54–20 | 0.36 | 0.76 |
| 8 | Y = 770.5x −721.1 | 0.9977 | 0.42–20 | 0.28 | 0.62 |
| 9 | Y = 945.4x −852.4 | 0.9944 | 0.24–18 | 0.16 | 0.36 |
| 10 | Y = 687.2x −674.3 | 0.9981 | 0.36–20 | 0.28 | 0.74 |
(1) deapi-platycoside E; (2) platycoside E; (3) deapi-platycodin D3; (4) platycodin D3; (5) deapi-platycodin D; (6) platycodin D; (7) polygalacin D; (8) 3″-O-acetyl polygalacin D; (9) platycodin A; and (10) 2″-O-acetyl polygalacin D.
Y: peak area; x: concentration (μg).
The newly developed HPLC-ELSD method was applied to compare the efficiency of solvent system by analyzing ten saponins (Table 2). It was found that there were remarkable differences of the contents of saponins depending on the percentage of methanol used for extraction. The contents of saponins in 5 extracts, except for polygalacin D, 3″-O-acetyl polygalacin D, platycodin A and 2″-O-acetyl polygalacin D, were inversely proportional to the increase of methanol in the solvent system, while the contents of polygalacin D, 3″-O-acetyl polygalacin D, platy-codin A and 2″-O-acetyl polygalacin D remained unchanged. Thus, we decided to extract Platycodi Radix with water in our validation procedure.
Table 2.
Contents of ten saponins in the extract of Platycodi Radix with different solvent systems
| Compounda | Solvent systems
|
||||
|---|---|---|---|---|---|
| Waterb | 30% MeOHb | 50% MeOHb | 70% MeOHb | MeOHb | |
| 1 | 744 ± 13 | 641 ± 8 | 628 ± 11 | 442 ± 4 | 458 ± 8 |
| 2 | 1088 ± 20 | 913 ± 12 | 824 ± 13 | 516 ± 9 | 493 ± 8 |
| 3 | 138 ± 5 | 135 ± 2 | 122 ± 5 | 101 ± 3 | 102 ± 3 |
| 4 | 427 ± 8 | 415 ± 6 | 383 ± 3 | 359 ± 7 | 356 ± 2 |
| 5 | 738 ± 12 | 734 ± 7 | 698 ± 8 | 689 ± 5 | 616 ± 7 |
| 6 | 2431 ± 20 | 2218 ± 8 | 1946 ± 10 | 1924 ± 8 | 1932 ± 11 |
| 7 | 2116 ± 41 | 2182 ± 22 | 2135 ± 12 | 2211 ± 16 | 2194 ± 9 |
| 8 | 282 ± 7 | 280 ± 6 | 276 ± 4 | 293 ± 2 | 296 ± 3 |
| 9 | 590 ± 13 | 573 ± 8 | 577 ± 9 | 592 ± 5 | 605 ± 5 |
| 10 | 294 ± 6 | 296 ± 3 | 302 ± 5 | 302 ± 2 | 302 ± 4 |
(1) deapi-platycoside E; (2) platycoside E; (3) deapi-platycodin D3; (4) platycodin D3; (5) deapi-platycodin D; (6) platycodin D; (7) polygalacin D; (8) 3″-O-acetyl polygalacin D; (9) platycodin A; and (10) 2″-O-acetyl polygalacin D.
Content (μg/g).
As shown in Table 3, this method showed good reproducibility for the quantification of ten saponins in Platycodi Radix with intra- and inter-day variations of less than 4.2% and 3.4%, respectively. As demonstrated in Table 4, the related compounds showed the overall recoveries ranging from 95.9% to 101.1% with RSD ranging from 0.6% to 2.5%. These results demonstrate that this HPLC-ELSD method is precise, accurate and sufficiently sensitive for the quantitative determination of ten major saponins in Platycodi Radix.
Table 3.
Intra- and Inter-day variations of the HPLC method for determination of ten saponins
| Compounda | Intra-day precision
|
Inter-day precision
|
||
|---|---|---|---|---|
| Content (μg/g) | RSD (%) | Content (μg/g) | RSD (%) | |
| 1 | 744 ± 13 | 1.76 | 741 ± 21 | 2.84 |
| 2 | 1088 ± 20 | 1.86 | 1090 ± 24 | 2.22 |
| 3 | 138 ± 5 | 3.41 | 139 ± 3 | 2.44 |
| 4 | 427 ± 8 | 1.94 | 422 ± 11 | 2.70 |
| 5 | 738 ± 12 | 1.61 | 734 ± 13 | 1.78 |
| 6 | 2431 ± 20 | 0.80 | 2419 ± 19 | 0.79 |
| 7 | 2116 ± 41 | 1.94 | 2111 ± 32 | 1.52 |
| 8 | 282 ± 7 | 2.51 | 283 ± 9 | 3.14 |
| 9 | 590 ± 13 | 2.18 | 593 ± 20 | 3.38 |
| 10 | 294 ± 6 | 2.14 | 293 ± 12 | 4.20 |
RSD (%) = (SD/mean) ×100%.
(1) deapi-platycoside E; (2) platycoside E; (3) deapi-platycodin D3; (4) platycodin D3; (5) deapi-platycodin D; (6) platycodin D; (7) polygalacin D; (8) 3″-O-acetyl polygalacin D; (9) platycodin A; and (10) 2″-O-acetyl polygalacin D.
Table 4.
Accuracy of the HPLC method for determination of ten saponins
| Compounda | Original (μg) | Spiked (μg) | Observed (μg) | Recovery (%) | Mean (%) | RSD (%) |
|---|---|---|---|---|---|---|
| 1 | 732 | 700 | 1412 | 97.0 | 99.1 | 2.5 |
| 500 | 1224 | 98.3 | ||||
| 300 | 1038 | 102.0 | ||||
| 2 | 1075 | 1000 | 2081 | 100.6 | 98.7 | 1.9 |
| 700 | 1752 | 96.7 | ||||
| 500 | 1568 | 98.6 | ||||
| 3 | 139 | 100 | 236 | 97.1 | 97.5 | 1.2 |
| 70 | 208 | 98.7 | ||||
| 50 | 187 | 96.4 | ||||
| 4 | 417 | 400 | 819 | 100.6 | 99.5 | 1.4 |
| 300 | 717 | 100.1 | ||||
| 200 | 613 | 97.9 | ||||
| 5 | 736 | 700 | 1451 | 102.1 | 100.2 | 1.6 |
| 500 | 1232 | 99.2 | ||||
| 300 | 1034 | 99.2 | ||||
| 6 | 2425 | 2000 | 4411 | 99.3 | 101.1 | 1.5 |
| 1500 | 3958 | 102.2 | ||||
| 1000 | 3442 | 101.7 | ||||
| 7 | 2099 | 2000 | 4082 | 99.1 | 99.0 | 0.6 |
| 1500 | 3592 | 99.5 | ||||
| 1000 | 3082 | 98.3 | ||||
| 8 | 284 | 300 | 580 | 98.5 | 98.5 | 0.3 |
| 200 | 481 | 98.9 | ||||
| 100 | 380 | 98.3 | ||||
| 9 | 597 | 500 | 1079 | 96.5 | 96.8 | 0.6 |
| 300 | 889 | 97.5 | ||||
| 200 | 790 | 96.4 | ||||
| 10 | 296 | 300 | 582 | 95.3 | 95.9 | 1.2 |
| 200 | 487 | 95.1 | ||||
| 100 | 394 | 97.2 |
Recovery (%) = (Observed amount − original amount)/spiked amount × 100%, RSD (%) = (SD/mean) × 100%.
(1) deapi-platycoside E; (2) platycoside E; (3) deapi-platycodin D3; (4) platycodin D3; (5) deapi-platycodin D; (6) platycodin D; (7) polygalacin D; (8) 3″-O-acetyl polygalacin D; (9) platycodin A; and (10) 2″-O-acetyl polygalacin D.
3.2. Analysis of pure platycodin saponins by ESI-MS and MS/MS
The mass spectra of ten major platycosides were operated in both positive ion and negative ion modes by ESI interface to confirm their molecular weight. Saponins with hydroxyl groups were detected with greater sensitivity in a negative ion mode and showed only peaks corresponding to [M − H]− and [M − H + 2H2O]−. Identification of platycosides was clearer in the negative ion mode than in the positive ion mode (spectra not shown).
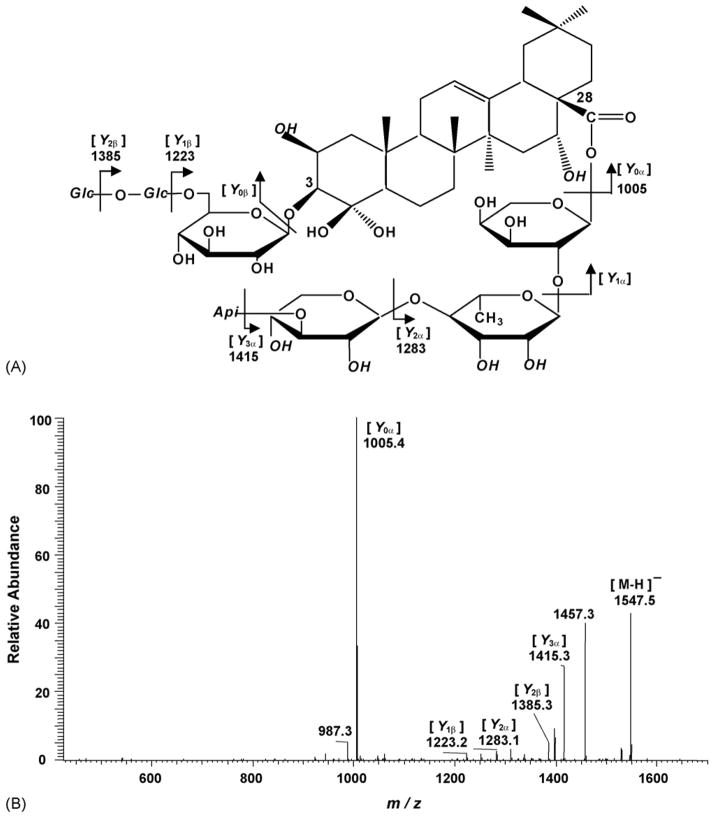
To establish the structure, the deprotonated molecular ion peak of each saponin was selected to obtain the MS/MS spectra using collision energy optimized in the ion trap analyzer. The fragmentation pattern used in this paper is based on that described by Domon and Costello [47], where ions retaining the charge at the reducing terminus are termed Y (glycosidic cleavage). As described in Fig. 3A, the MS/MS data showed product ions corresponding to the sequential loss of sugar from platycoside E. A typical MS/MS spectrum of platycoside E was shown at the Fig. 3B. There are five main product ions from Y cleavage at m/z 1415, 1385, 1283, 1223, and 1005. The MS/MS product ions m/z 1415, 1283 and 1005 represented the loss of apiosyl, xylosyl and rhamnosyl + arabinosyl sugar attached at C-28 of triterpene, respectively. The MS/MS product ions at m/z 1385 and 1223 corresponded to the loss of a hexose and two hexoses located at the β sugar chain termini, respectively. In this ESI-MS/MS spectrum, Y1α and Y0β product ions were not detected under a variety of MS conditions. The above MS/MS data in a negative ion mode confirmed the number and type of sugar unit in oligosaccharide chains, and the type of saponin of platycoside E. Table 5 lists the major Y ions of pure platycosides, obtained by ESI-MS/MS in a negative mode.
Fig. 3.
Fragmentation pattern of the [M − H]− ion of platycoside E (A), and ESI-MS/MS spectrum of the m/z 1547 ion ([M − H]−) of platycoside E (B).
Table 5.
Major Y product ions observed by negative ion ESI-MS/MS analysis of ten saponins (m/z values)
| Compounda | [M − H]− | [Y3α] | [Y2α] | [Y0α] | [Y2β] | [Y1β] |
|---|---|---|---|---|---|---|
| 1 | 1415.4 | 1283.2 | 1005.3 | 1253.2 | 1091.2 | |
| 2 | 1547.2 | 1415.3 | 1283.1 | 1005.3 | 1385.0 | 1223.7 |
| 3 | 1252.8 | 1121.2 | 843.2 | 1091.2 | ||
| 4 | 1385.3 | 1253.2 | 1121.2 | 843.2 | 1223.2 | |
| 5 | 1091.0 | 959.3 | 681.1 | |||
| 6 | 1223.2 | 1091.0 | 959.1 | 681.1 | ||
| 7 | 1207.1 | 1075.0 | 943.0 | 665.1 | ||
| 8 | 1249.0 | 1117.1 | 985.1 | 665.1 | ||
| 9 | 1265.3 | 1133.1 | 1001.1 | 681.2 | ||
| 10 | 1249.0 | 1117.1 | 985.1 | 665.1 |
(1) deapi-platycoside E; (2) platycoside E; (3) deapi-platycodin D3; (4) platycodin D3; (5) deapi-platycodin D; (6) platycodin D; (7) polygalacin D; (8) 3″-O-acetyl polygalacin D; (9) platycodin A; and (10) 2″-O-acetyl polygalacin D.
In deapi-platycoside E, the [M − H]−, Y1β and Y2β ions at m/z 1415, 1253, and 1091 correspond to [M − H]−-132, Y1β-132 and Y2β-132 of platycoside E, but Y0α and Y2α ions are identical. This result indicates that apiose is attached to sugar residue of C-28 of triterpene and that the difference of Y1β and Y2β product ions can distinguish deapiosyl and apiosyl platyodi saponins. The polygalacin D and 2″-O-acetyl polygalacin D are dehydroxylated at R1 position of platycodin D and platycodin A, respectively, yielding ion difference of 16 Da between two groups. For platycodin A, 2″-O-acetyl polygalacin D and 3″-O-acetyl polygalacin D, the mass difference between Y0α and Y2α is 320 Da. But the other saponins show 278 Da as mass difference, which suggests that the acetyl group of platycodin A, 2″-O-acetyl polygalacin D and 3″-O-acetyl polygalacin D is attached to rhamnose or arabinose. As shown in Table 5, 2″-O-acetyl polygalacin D and 3″-O-acetyl polygalacin D are isomers with molecular weights of 1250 Da and produce the same negative ion MS/MS spectra. Using LC-ESI MS/MS analysis, however, two isomeric compounds were successfully separated and identified (see Section 3.3). All the major Y ions resulting from a glycosidic bond cleavage and the expected structural features of platycosides are clearly distinguishable in the resulting spectra.
3.3. Analysis of the crude extract of Platycodi Radix by LC-ESI MS/MS
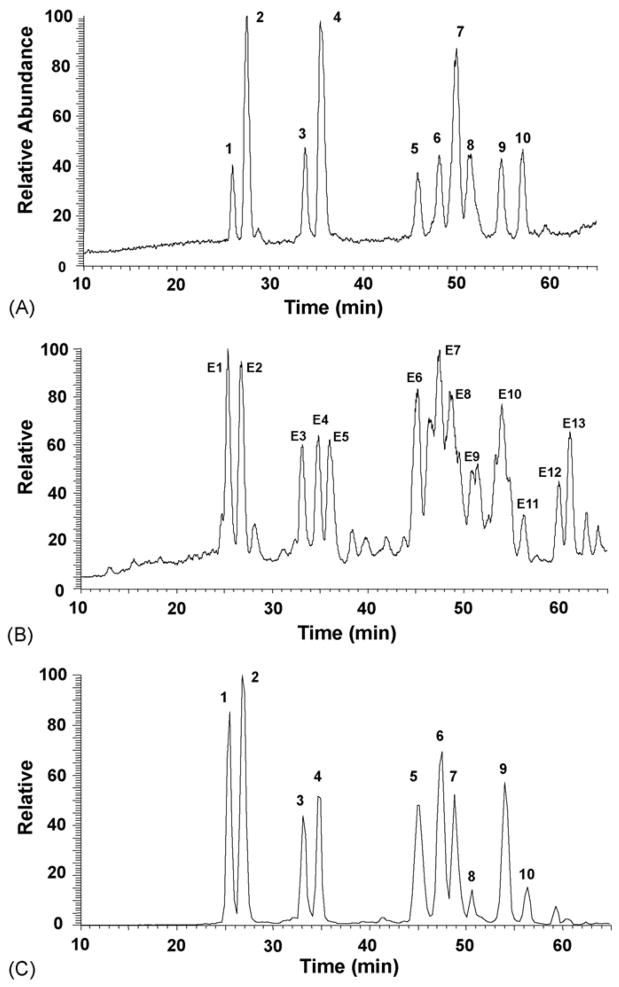
To demonstrate individual platycosides in a complex mixture, the pure platycosides were mixed and analyzed by LC-ESI MS/MS. Fig. 4A shows the negative ion chromatogram of platycosides collected in 60 min. All platycosides were well resolved under the LC conditions used. Platycosides labeled on the reconstructed ion chromatogram were identified on the basis of their retention times, the m/z values of intact molecular ions and the MS/MS product ions and because they present a variety of structural characteristics. Table 6 summarizes the retention times and the set of negative ions used for identification of individual platycosides from the crude extract of Platycodi Radix. To identify major ten saponins of the crude extract from Platycodi Radix, LC-ESI MS/MS analyses were performed and the results are shown in Fig. 4B and C and Table 6. The individual intense peaks labeled in Fig. 4B were characterized by comparing molecular/product ion pairs and retention times of the samples with results obtained for the pure platycosides. Based on the data in Table 5 and Fig. 4A, peaks E1–4 and E6–11 are assigned as deapi-platycoside E (E1), platycoside E (E2), deapi-platycodin D3 (E3), platycodin D3 (E4), deapi-platycodin D (E6), platycodin D (E7), polygalacin D (E8), 3″-O-acetyl polygalacin D (E9), platycodin A (E10), and 2″-O-acetyl polygalacin D (E11).
Fig. 4.
Representative HPLC–MS chromatograms of mixed standards and extract of Platycodi Radix in negative ion ESI mode. (A) mixed standards; (B) extract of Platycodi Radix; (C) selected ion monitoring of extract from Platycodi Radix. Selected ions as intact molecular ions of platycosides (m/z 1415, 1547, 1253, 1385, 1091, 1223, 1207, 1265, 1249) with the retention times of corresponding constituents presented in a chromatogram (C). The numbers indicate each platycoside in Fig. 1.
Table 6.
Retention times and the MS/MS product ions in negative ion mode of LC peaks E1–E13 observed in the crude extract of Platycodi Radix
| Peak | Retention time (min) | Molecular ion (m/z) | Characteristic MS/MS product ions (m/z) |
|---|---|---|---|
| E1 | 25.9 | 1415.4 | 1283.2, 1253.2, 1005.3 |
| E2 | 27.5 | 1547.2 | 1415.3, 1385.0, 1283.2, 1223.7, 1005.3 |
| E3 | 33.7 | 1252.8 | 1121.2, 1091.2, 843.2 |
| E4 | 35.4 | 1385.3 | 1253.2, 1223.2, 1121.2, 843.2 |
| E5 | 37.1 | 1005.5 | 843.3, 681.4 |
| E6 | 45.5 | 1091.0 | 959.3, 681.1 |
| E7 | 47.9 | 1223.2 | 1091.0, 959.1, 681.1 |
| E8 | 49.7 | 1207.1 | 1075.0, 943.0, 665.1 |
| E9 | 51.2 | 1249.0 | 1117.3, 985.1, 665.2 |
| E10 | 53.9 | 1265.3 | 1133.1, 1001.1, 681.2 |
| E11 | 56.3 | 1249.0 | 1117.1, 985.1, 665.1 |
| E12 | 60.4 | 843.3 | 681.3 |
| E13 | 61.6 | 681.3 |
The TIC spectrum from the extract contains some intense peaks, such as E5, E12 and E13, which display different molecular ions and product ions when compared with our purified platycosides. Therefore, we performed LC-ESI MS/MS analysis in both positive ion and negative ion modes on the unidentified peaks from the extract. The [M − H]− ions of E1, E12 and E13 were determined to be m/z 1005, 843 and 681 (Table 6), corresponding to prosapogenins [12], in which the C-28 glycoside was hydrolyzed and the free carboxyl group was exposed. These prosapogenins may be generated during the extraction procedure. In fact, these were present as intense peaks in the TIC spectrum despite of their absence in the HPLC-ELSD spectrum. Fig. 4C shows a chromatogram obtained using selected ion monitoring of [M − H]− for each platycoside. This technique could be used to exclude interference from the extract on the determination of the platycosides and unique peaks were obtained with a good chromatographic resolution and sensitivity. Our HPLC-ESI MS/MS experiments agreed well with our above HPLC-ELSD results.
4. Conclusions
The analytical method described in this paper is the first report of HPLC-ELSD and HPLC-ESI MS/MS method for the quantitative determination and identification of major ten saponins in Platycodi Radix. The method of HPLC-ELSD is accurate and precise and was successfully used to analyze the crude extract of Platycodi Radix. Under ESI-MS/MS conditions, the fragmentation patterns of [M − H]− ions exclusively show signals corresponding to cleavage of the glycosidic bonds, thus allowing a rapid identification of saponins in the crude extract of Platycodi Radix. The results demonstrate that the proposed method could be readily utilized for quality control of Platycodi Radix and an analytical tool for the rapid characterization of platycosides in the crude extract.
Acknowledgments
This work was supported in part by the grants of the Korea Health 21 R&D Project, Ministry of Health & Welfare, Republic of Korea (HMP-01-PJ2-PG6-01NA01-002) and College of Pharmacy, Seoul National University.
References
- 1.Takagi K, Lee EB. Yakugaku Zasshi. 1972;92:951. doi: 10.1248/yakushi1947.92.8_951. [DOI] [PubMed] [Google Scholar]
- 2.Lee EB. Yakugaku Zasshi. 1973;93:1188. doi: 10.1248/yakushi1947.93.9_1188. [DOI] [PubMed] [Google Scholar]
- 3.Kim KS, Ezaki O, Ikemoto S, Itakura H. J Nutr Sci Vitaminol (Tokyo) 1995;41:485. doi: 10.3177/jnsv.41.485. [DOI] [PubMed] [Google Scholar]
- 4.Han LK, Xu BJ, Kimura Y, Zheng Y, Okuda H. J Nutr. 2000;130:2760. doi: 10.1093/jn/130.11.2760. [DOI] [PubMed] [Google Scholar]
- 5.Kim KS, Seo EK, Lee YC, Lee TK, Cho YW, Ezaki O, Kim CH. J Nutr Biochem. 2000;11:420. doi: 10.1016/s0955-2863(00)00098-x. [DOI] [PubMed] [Google Scholar]
- 6.Choi CY, Kim JY, Kim YS, Chung YC, Hahm KS, Jeong HG. Cancer Lett. 2001;166:17. doi: 10.1016/s0304-3835(01)00440-2. [DOI] [PubMed] [Google Scholar]
- 7.Choi CY, Kim JY, Kim YS, Chung YC, Seo JK, Jeong HG. Int Immunopharmacol. 2001;1:1141. doi: 10.1016/s1567-5769(01)00047-9. [DOI] [PubMed] [Google Scholar]
- 8.Lee KJ, Jeong HG. Food Chem Toxicol. 2002;40:517. doi: 10.1016/s0278-6915(01)00104-1. [DOI] [PubMed] [Google Scholar]
- 9.Park DI, Lee JH, Moon SK, Kim CH, Lee YT, Cheong J, Choi BT, Choi YH. Pharmacol Res. 2005;51:437. doi: 10.1016/j.phrs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Kim JY, Kim DH, Kim HG, Song GY, Chung YC, Roh SH, Jeong HG. Toxicol Appl Pharmacol. 2006;210:150. doi: 10.1016/j.taap.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Kim YS, Kim JS, Choi SU, Kim JS, Lee HS, Roh SH, Jeong YC, Kim YK, Ryu SY. Planta Med. 2005;71:566. doi: 10.1055/s-2005-864161. [DOI] [PubMed] [Google Scholar]
- 12.Zhao HL, Sim JS, Shim SH, Ha YW, Kang SS, Kim YS. Int J Obes (Lond) 2005;29:983. doi: 10.1038/sj.ijo.0802948. [DOI] [PubMed] [Google Scholar]
- 13.Ahn KS, Noh EJ, Zhao HL, Jung SH, Kang SS, Kim YS. Life Sci. 2005;76:2315. doi: 10.1016/j.lfs.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Schuller Levis GB, Lee EB, Levis WR, Lee da W, Kim BS, Park SY, Park E. Int Immunopharmacol. 2004;4:1039. doi: 10.1016/j.intimp.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Han LK, Zheng YN, Xu BJ, Okuda H, Kimura Y. J Nutr. 2002;132:2241. doi: 10.1093/jn/132.8.2241. [DOI] [PubMed] [Google Scholar]
- 16.Shin CY, Lee WJ, Lee EB, Choi EY, Ko KH. Planta Med. 2002;68:221. doi: 10.1055/s-2002-23130. [DOI] [PubMed] [Google Scholar]
- 17.Kim YP, Lee EB, Kim SY, Li D, Ban HS, Lim SS, Shin KH, Ohuchi K. Planta Med. 2001;67:362. doi: 10.1055/s-2001-14317. [DOI] [PubMed] [Google Scholar]
- 18.Xu CL, Yang LH, Zheng YN, Liu MX, Xu BJ. J Jilin Agric Univ. 1999;21:35. [Google Scholar]
- 19.Saeki T, Koike K, Nikaido T. Planta Med. 1999;65:428. doi: 10.1055/s-1999-14021. [DOI] [PubMed] [Google Scholar]
- 20.Ganzera M, Bedir E, Khan IA. J Pharm Sci. 2001;90:1752. doi: 10.1002/jps.1124. [DOI] [PubMed] [Google Scholar]
- 21.Fuzzati N, Chromatogr J. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;812:119. doi: 10.1016/j.jchromb.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 22.Wakabayashi C, Murakami K, Hasegawa H, Murata J, Saiki I. Biochem Biophys Res Commun. 1998;246:725. doi: 10.1006/bbrc.1998.8690. [DOI] [PubMed] [Google Scholar]
- 23.Tohda C, Matsumoto N, Zou K, Meselhy MR, Komatsu K. Neuropsychopharmacology. 2004;29:860. doi: 10.1038/sj.npp.1300388. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Reddy MB, Hendrich S, Murphy PA. J Nutr. 2004;134:1867. doi: 10.1093/jn/134.8.1867. [DOI] [PubMed] [Google Scholar]
- 25.Paek IB, Moon Y, Kim J, Ji HY, Kim SA, Sohn DH, Kim JB, Lee HS. Biopharm Drug Dispos. 2006;27:39. doi: 10.1002/bdd.481. [DOI] [PubMed] [Google Scholar]
- 26.Lee HU, Bae EA, Han MJ, Kim DH. Biol Pharm Bull. 2005;28:1992. doi: 10.1248/bpb.28.1992. [DOI] [PubMed] [Google Scholar]
- 27.Cui M, Song F, Zhou Y, Liu Z, Liu S. Rapid Commun Mass Spectrom. 2000;14:1280. doi: 10.1002/1097-0231(20000730)14:14<1280::AID-RCM26>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 28.Li B, Abliz Z, Fu G, Tang M, Yu S. Rapid Commun Mass Spectrom. 2005;19:381. doi: 10.1002/rcm.1803. [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Cui M, Liu Z, Song F, Mo W. J Am Soc Mass Spectrom. 2004;15:133. doi: 10.1016/j.jasms.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Abliz Z, Tang M, Fu G, Yu S. J Chromatogr A. 2006;1101:53. doi: 10.1016/j.chroma.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 31.Berhow MA, Cantrell CL, Duval SM, Dobbins TA, Maynes J, Vaughn SF. Phytochem Anal. 2002;13:343. doi: 10.1002/pca.664. [DOI] [PubMed] [Google Scholar]
- 32.Wolfender JL, Hostettmann K, Abe F, Nagao T, Okabe H, Yamauchi T. J Chromatogr A. 1995;712:155. doi: 10.1016/0021-9673(95)00522-o. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Sakuma T, Asafu-Adjaye E, Shiu GK. Anal Chem. 1999;71:1579. doi: 10.1021/ac980890p. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Gu C, Zhang H, Awang DV, Fitzloff JF, Fong HH, van Breemen RB. Anal Chem. 2000;72:5417. doi: 10.1021/ac000650l. [DOI] [PubMed] [Google Scholar]
- 35.Bao Y, Li C, Shen H, Nan F. Anal Chem. 2004;76:4208. doi: 10.1021/ac0499423. [DOI] [PubMed] [Google Scholar]
- 36.Ji QC, Harkey MR, Henderson GL, Gershwin ME, Stern JS, Hackman RM. Phytochem Anal. 2001;12:320. doi: 10.1002/pca.593. [DOI] [PubMed] [Google Scholar]
- 37.Gu L, Tao G, Gu W, Prior RL. J Agric Food Chem. 2002;50:6951. doi: 10.1021/jf0257300. [DOI] [PubMed] [Google Scholar]
- 38.Dalluge JJ, Eliason E, Frazer S. J Agric Food Chem. 2003;51:3520. doi: 10.1021/jf030036l. [DOI] [PubMed] [Google Scholar]
- 39.Fuzzati N, Gabetta B, Jayakar K, Pace R, Peterlongo F. J Chromatogr A. 1999;854:69. doi: 10.1016/s0021-9673(99)00463-x. [DOI] [PubMed] [Google Scholar]
- 40.Chan TW, But PP, Cheng SW, Kwok IM, Lau FW, Xu HX. Anal Chem. 2000;72:1281. doi: 10.1021/ac990819z. [DOI] [PubMed] [Google Scholar]
- 41.Kite GC, Howes MJ, Simmonds MS. Rapid Commun Mass Spectrom. 2004;18:2859. doi: 10.1002/rcm.1698. [DOI] [PubMed] [Google Scholar]
- 42.Hiroahi I, Kauzuo T, Yohko Y. J Chem Soc Perkin Trans I. 1981:1928. [Google Scholar]
- 43.Hiroahi I, Kauzuo T, Takehiko T, Yohko Y. J Chem Soc Perkin Trans I. 1984:661. [Google Scholar]
- 44.Tada A, Kaneiwa Y, Shoji J, Shibata S. Chem Pharm Bull (Tokyo) 1978;26:668. doi: 10.1248/cpb.23.2965. [DOI] [PubMed] [Google Scholar]
- 45.Tada A, Kaneiwa Y, Shoji J, Shibata S. Chem Pharm Bull (Tokyo) 1975;23:2965. doi: 10.1248/cpb.23.2965. [DOI] [PubMed] [Google Scholar]
- 46.Cardenas S, Gallego M, Valcarcel M. Anal Chim Acta. 1999;402:1. [Google Scholar]
- 47.Domon B, Costello CE. Glycoconjugate J. 1988;5:397. [Google Scholar]