Abstract
Capsids of papilloma and polyoma viruses (papovavirus family) are composed of 72 pentameric capsomeres arranged on a skewed icosahedral lattice (triangulation number of seven, T = 7). Cottontail rabbit papillo mavirus (CRPV) was reported previously to be a T = 7laevo (left-handed) structure, whereas human wart virus, simian virus 40, and murine polyomavirus were shown to be T = 7dextro (right-handed). The CRPV structure determined by cryoelectron microscopy and image reconstruction was similar to previously determined structures of bovine papillomavirus type 1 (BPV-1) and human papillomavirus type 1 (HPV-1). CRPV capsids were observed in closed (compact) and open (swollen) forms. Both forms have star-shaped capsomeres, as do BPV-1 and HPV-1, but the open CRPV capsids are ~2 nm larger in radius. The lattice hands of all papillomaviruses examined in this study were found to be T = 7dextro. In the region of maximum contact, papillomavirus capsomeres interact in a manner similar to that found in polyomaviruses. Although papilloma and polyoma viruses have differences in capsid size (~60 versus ~50 nm), capsomere morphology (11 to 12 nm star-shaped versus 8 nm barrel-shaped), and intercapsomere interactions (slightly different contacts between capsomeres), papovavirus capsids have a conserved, 72-pentamer, T = 7dextro structure. These features are conserved despite significant differences in amino acid sequences of the major capsid proteins. The conserved features may be a consequence of stable contacts that occur within capsomeres and flexible links that form among capsomeres.
Keywords: cryoelectron microscopy, enantiomer, handedness, Papovaviridae, three-dimensional image reconstruction
Introduction
Papovaviruses infect vertebrate animals including mammals (e.g. cattle, elephants, humans, mice, monkeys, and rabbits), birds (e.g. chaffinches, chickens, and parrots), and reptiles (e.g. turtles) (Howley, 1990; Murphy et al., 1995; Olson, 1987; Shah, 1990; Stoll et al., 1993). This virus family is subdivided into the genus Papillomavirus and the genus Polyomavirus (Murphy et al., 1995). At least 70 strains of human papillomavirus have been identified (Van Ranst et al., 1994), and these have been the subjects of intensive study because they cause benign and cancerous diseases (De Villiers, 1989; Howley, 1990). Most humans are infected with the polyomaviruses JC and BK virus (Eckhart, 1990), but these viruses appear to cause disease only when individuals have impaired immune systems (Shah, 1990).
Papovaviruses are spherical particles with capsids made up of 72 morphological units (capsomeres). All papovaviruses are non-enveloped and multiply in the nucleus of the infected cell. They carry a double-stranded, circular DNA genome that associates with host-encoded histones in the virions. However, papillomaviruses (e.g. bovine, cottontail rabbit, and human papillomaviruses) and polyomaviruses (e.g. murine polyomavirus and simian virus 40) differ in diameter, genome size, protein composition and size, and capsomere morphology and size. Papillomaviruses (~60 nm diam.) have an ~8000 bp genome that encodes two structural proteins: the major capsid protein, L1 (~510 amino acid residues and ~58 kDa), and the minor protein, L2 (~470 a.a. and ~51 kDa). Papillomavirus capsomeres are mushroom-like protrusions, 11 to 12 nm in diameter, with pentameric, “star-shaped” heads (Baker et al., 1991). In contrast, the ~50 nm diameter polyomaviruses have an ~5000 bp genome that codes for three structural proteins: the major capsid protein, VP1 (~370 a.a. and ~41 kDa), and two minor proteins, VP2 (~350 a.a. and ~38 kDa) and VP3 (~230 a.a. and ~26 kDa). Polyomavirus capsomeres are also pentamers but are smaller (8 nm in diam.) and have a barrel-shaped morphology (Rayment et al., 1982; Baker et al., 1988; Liddington et al., 1991; Stehle et al., 1994).
Despite these distinguishing differences, electron microscopy and X-ray crystallography studies have shown that all papovavirus capsids exhibit conserved features (Table 1). The 72 capsomeres are pentamers of the major capsid protein and are arranged on a T = 7 icosahedral lattice (Figure 1). Twelve pentavalent pentamers (each surrounded by five other capsomeres) are centered on the icosahedral 5-fold axes and 60 hexavalent pentamers (each surrounded by six other capsomeres) are centered on vertices that lie between the 5-fold axes.
Table 1.
Literature references to conserved features in papovavirus capsids
| Virus | T = 7 icosahedron, 72 capsomeres | Pentameric capsomeres | T = 7l | T = 7d |
|---|---|---|---|---|
| Polyomaviruses | ||||
| Murine polyomavirus | a,f,j,l,o,s,u | c,f,l,s,t,u | j,u | |
| SV40 | b,d,r | d,r | b,r | |
| K virus | o | |||
| Papillomaviruses | ||||
| BPV-1 | e | e | v | |
| CRPV | k,q,v | v | k,q | v |
| HPV-1 | e,m | e,m | v | |
| Human wart virus | g,h,i,o,p,q,w | n | p,q,w |
Human wart virus refers to human papillomavirus of unspecified type.
a, Adolph et al. (1979); b, Anderer et al. (1967); c, Baker et al. (1983); d, Baker et al. (1988, 1989); e, Baker et al. (1991); f, Belnap et al. (1993), Baker (1992); g, Caspar (1966); h, Crowther & Amos (1972); i, Crowther et al. (1970a); j, Finch (1974); k, Finch & Klug (1965); l, Griffith et al. (1992); m, Hagensee et al. (1994); n, Kiselev & Klug (1969); o, Klug (1965); p, Klug & Finch (1965); q, Klug & Finch (1968); r, Liddington et al. (1991); s, Rayment et al. (1982); t, Salunke et al. (1986); u, Stehle et al. (1994); v, this study; w, Yabe et al. (1979).
Figure 1.
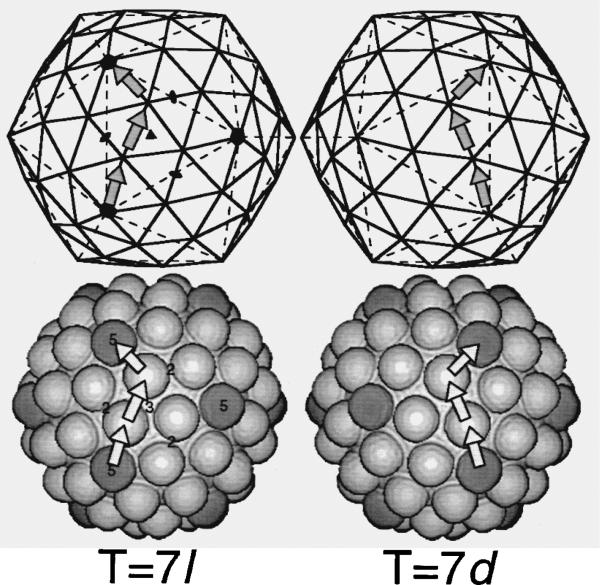
Icosahedra with a triangulation number of seven (T = 7) are left (T = 7laevo) or right (T = 7dextro) handed. Here, T = 7l and T = 7d lattices (top row, continuous lines) and computer-generated models (bottom row) are displayed. Vertices in the lattices are replaced by spheres in the models. For the papovaviruses, each sphere represents a pentameric capsomere. Three adjacent, 5-fold-symmetry axes (filled pentagons, top left) define the vertices of one of the 20 triangular faces in the basic, T = 1 icosahedron (broken lines, top row). Both 2-fold and 3-fold symmetry elements (filled ovals and triangle, respectively) are depicted also within this triangular face. (Filled symbols are replaced by numbers in the lower left figure.) A T = 7 lattice has 72 vertices or lattice points that define the corners of 140 smaller triangles. The 12 pentavalent vertices at the 5-fold axes are each surrounded by five hexavalent vertices, giving a total of 60 hexavalent lattice points (three in each face of the T = 1 lattice). Each hexavalent lattice point is surrounded by one pentavalent and five hexavalent points. Pentava-lent spheres in the computer models (bottom row) are shaded dark grey to distinguish them from hexavalent spheres. The absolute hand of a T = 7 lattice is defined by the arrangement of lattice points between neighboring pentavalent points, analogous to the moves a knight makes in the game of chess. Thus, in a T = 7l lattice (top left) or structure (bottom left) the path between adjacent pentavalent lattice points or capsomeres consists of two “steps” forward onto adjacent hexavalent points followed by one step to the left (arrows). Similarly, a T = 7d lattice (top right) or structure (bottom right) exhibits a “two steps forward followed by one to the right” pattern. The lattices and computer models are enantiomorphs (i.e. mirror related). However, if all spheres in each model were replaced by chemically identical, chiral, pentameric capsomeres as are found in all papovaviruses, the resulting T = 7l and T = 7d structures would not be an enantiomorphic pair.
A T = 7 lattice is a geometrical object (Figure 1). It exists as either a left (T = 7laevo) or a right (T = 7dextro) handed enantiomorph. Handedness is defined by the arrangement of lattice points between neighboring 5-fold vertices. In nature, individual protein molecules exist as a single enantiomorph because they are composed of chiral amino acids. Likewise, oligomeric protein assemblies (from simple dimers to complex icosahedra) are also enantiomorphic structures. The packing of morphological units in icosahedral virus capsids is described in terms of T (triangulation) lattices (Caspar & Klug, 1962). Some T lattices, such as T = 7, are skewed and therefore add yet another level of handedness to virus structure. Though there is no theoretical restriction against a left or right-handed form (Caspar & Klug, 1962), all capsids within a particular virus species or strain are assumed to have the same hand. The chirality of amino acids ensures that one hand is preferred because interactions among protein subunits are more energetically favorable. However, because capsid proteins of related viruses can differ, it is not known whether different viruses within a genus or family must have the same lattice hand. Indeed, murine polyomavirus, simian virus 40 (SV40), and human wart virus are T = 7d structures, whereas cottontail rabbit papillomavirus (CRPV), which was discovered more than 60 years ago by Shope & Hurst (1933), is the only papovavirus reported to have a T = 7l capsid (Table 1).
We investigated the structure of CRPV by means of cryoelectron microscopy (cryoEM) and three-dimensional (3D) image reconstruction. These techniques have been extremely useful for studying a wide variety of icosahedral viruses (Baker, 1992), including papovaviruses such as SV40 (Baker et al., 1988, 1989), bovine papillomavirus type 1 (BPV-1; Baker et al., 1991), and human papillomavirus type 1 (HPV-1; Baker et al., 1991; Hagensee et al., 1994). Initially, our goal was to determine the structure of CRPV to understand the basis of its T = 7l symmetry. Detailed comparisons of CRPV, BPV-1, and HPV-1 reconstructions compelled us to examine the hand of the T = 7 lattice for all three structures by means of tilting experiments. The hand of BPV-1 was also examined with metal-shadowing techniques. We found that, like the other papillomaviruses, the CRPV capsid is composed of 72 pentameric, star-shaped capsomeres arranged on a T = 7d icosahedral lattice. In addition, we discovered that papillomavirus capsids exist in open and closed states similar to those observed in cowpea chlorotic mottle virus, a structure with unexpected similarities to the polyomaviruses (Speir et al., 1995).
Results
CryoEM
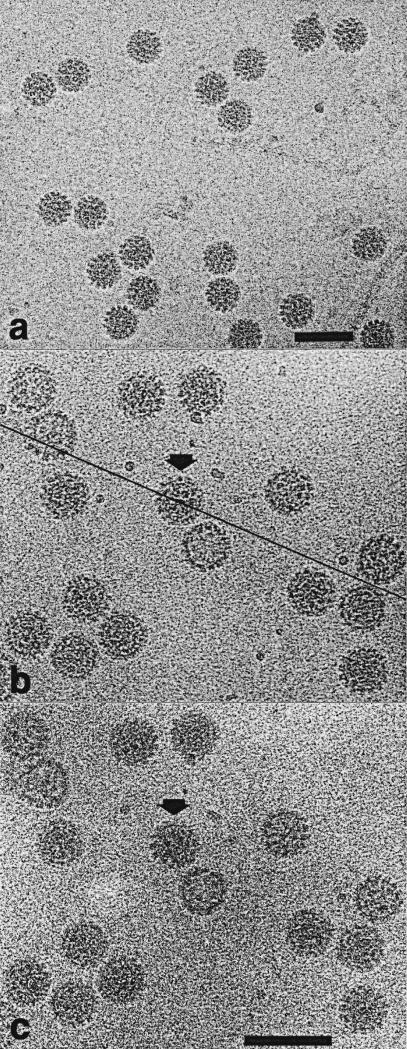
Frozen-hydrated CRPV particles appeared approximately circular in profile, whether the sample was untilted (Figure 2a,b) or tilted (Figure 2c), indicating that the spherical shape of the virions was well preserved. The distinct appearances of different particles showed that the particles were randomly oriented in the vitrified sample. The CRPV images appeared indistinguishable from previously recorded images of two other papillomaviruses, BPV-1 and HPV-1 (Baker et al., 1991). New images of BPV-1, HPV-1 virus-like particle (VLP), and SV40 specimens were recorded for this study (data not shown). These images were nearly identical to those seen previously (Baker et al., 1988, 1991; Hagensee et al., 1994).
Figure 2.
Electron micrographs of vitrified CRPV samples. CRPV particles are distributed in a monodisperse layer indicating that the frozen-hydrated sample is about as thick as the diameter of the particles. Capsomeres appear as “knob-like” structures, which are prominent especially along particle edges. a, CRPV dialyzed against 10 mM Tris-HCl and 1 mM EDTA (pH 8.0). These particle images were used to compute the 3D reconstruction seen in Figure 3. Bar represents 100 nm. b and c, Tilting experiment with CRPV dialyzed against 20 mM Tris-HCl, 1 M NaCl (pH 7.4). View of particles with specimen stage at 0° (b) and –5° (c) tilt. The tilt axis (line in b) is approximately 23° from the horizontal. The particle highlighted with the arrows also appears in Figure 4. Bar represents 100 nm.
Papillomavirus morphology: open and closed forms
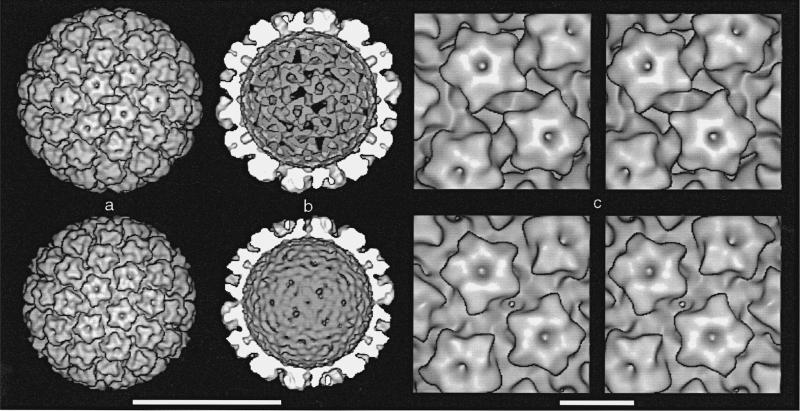
A 3D reconstruction of CRPV at 2.7 nm resolution reveals a capsid structure (63 nm diam.) that has 72 capsomeres arranged with T = 7 icosahedral symmetry (Figure 3a, top). All capsomeres are “mushroom-like” protrusions with pentameric, “star-shaped” heads. Intercapsomere contacts occur within a region of relatively continuous density at the base of each capsomere. This region forms the capsid shell layer, wherein the primary difference between the CRPV and HPV-1 structures occurs.
Figure 3.
Surface-shaded representations of CRPV (top row) and HPV-1 (bottom row) reconstructions viewed along a 2-fold symmetry axis. The CRPV capsid has an “open” form and the HPV-1 has a “closed” form. a, Outside view of virions. b, Inside view of capsids (back half). Chromatin cores were computationally removed from the virion density map (radii < 46 nm for CRPV and < 42 nm for HPV-1). c, Closeup stereo views. In CRPV (top), elongated holes lie on opposite sides of the 2-fold axis. Triangular holes on the 3-fold axes can be seen at the top and bottom center, just behind the protruding point of the capsomere. In HPV-1 (bottom), there is a single, small hole on the 2-fold axis. Bars represent: a,b, 50 nm; c, 10 nm.
The CRPV capsid contains several “holes” that extend through the protein shell and expose the chromatin core to solvent (Figure 3, top). Two elongated holes lie on either side of each 2-fold symmetry axis and smaller, triangular holes are found on each 3-fold axis.
The HPV-1 reconstruction (Figure 3, bottom) was recomputed from the Baker et al. (1991) images. It appears nearly identical to other published HPV-1 structures (Baker et al., 1991; Hagensee et al., 1994). Unlike CRPV, the HPV-1 capsid (59 nm diam.) has no large holes but instead has only small, circular holes on the 2-fold axes. The HPV-1 structure closely resembles BPV-1 (Baker et al., 1991).
Reconstructions from other papillomavirus specimens revealed a “closed” CRPV structure (no large holes) and a possible “open” HPV-1 VLP structure (with large holes) (Table 2). Other HPV-1 VLP specimens revealed closed form structures (Hagensee et al., 1994). Our limited data suggest that the open form occurs when virus is exposed to low ionic strength conditions or when divalent cations are chelated (Table 2). Consistent with our diameter measurements, the maximum radially averaged density within the shell of the open CRPV capsid occurred at a radius ~2 nm higher than that in the closed HPV-1 capsid (data not shown). Thus, the open capsid is ~7% larger in diameter than the closed capsid.
Table 2.
Data collection and processing of papovavirus images
| Hand determination experiments |
||||||||
|---|---|---|---|---|---|---|---|---|
| CRPV | HPV-1 | BPV-1 capsid | BPV-1 | CRPV | CRPV | HPV-1 VLPs | SV40 | |
| Underfocus of objective lens (μm) | 1.0 | 1.0–1.2 | 1.5, 2.0 | 1.5, 2.0 | 1.3–1.4, 1.1 | 1.2–5, 0.8–5 | 4, 4 | 2.0, 1.5 |
| Number of particle images | 76 | 35 | 16 | 18 | 31 | 32 | 14 | 80 |
| Eigenvalues (%) | ||||||||
| >100 | 98.0 | 87.9 | 28.2 | 37.3 | 83.1 | 88.0 | 12.9 | 98.7 |
| 10–100 | 2.0 | 12.1 | 71.7 | 62.5 | 16.8 | 12.0 | 86.6 | 1.3 |
| 1.0–10 | 0.0 | 0.0 | 0.1 | 0.2 | 0.1 | 0.0 | 0.5 | 0.0 |
| <1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Correlation coefficient | 0.47 (0.04) | 0.46 (0.03) | 0.64 (0.04) | 0.60 (0.03) | 0.59 (0.04) | 0.55 (0.06) | 0.67 (0.05) | 0.66 (0.04) |
| Resolution (nm) | 2.7 | 2.5 | 2.9 | 2.9 | 2.8 | 4.4 | 3.6 | 3.5 |
| Origin and orientation refinement method | CCL | PFT | PFT | PFT | CCL | PFT | CCL | PFT |
| Form of capsid | Open | Closed | Closed | Closed | Closed | Open | Open? | — |
| Ionic strength (mM) | 3 | 80 | 170 | 170 | 1000 | 0 | 50 | — |
| EDTA | 1 mM | 0 | 0 | 0 | 0 | 0 | 0 | — |
For the six hand-determination experiments, values of objective lens underfocus for the untilted and tilted specimens are listed on the left and right, respectively Eigenvalue measurements include both real and imaginary components. Correlation coefficients (standard deviations in parentheses) are averages for each set of particle images and were computed over the full particle radius. Resolution of each 3D reconstruction was determined by the point of intersection between the ring correlation data and an expected significance level, as described by Conway et al. (1993) and Saxton & Baumeister (1982). CCL and PFT refer to the cross-correlation/cross-common-lines (Baker et al., 1990; Fuller et al., 1996) and polar-Fourier-transform (Baker & Cheng, 1996) procedures, respectively The first CRPV sample (column one) was purified from virus grown in athymic mice and was dialyzed against 10 mM Tris-HCl, 1 mM EDTA (pH 8.0). Images of BPV-1 empty capsids and full virions (columns three and four) were recorded in the same micrograph. The second CRPV specimen (column five) was purified from virus grown in athymic mice and was dialyzed against 20 mM Tris-HCl, 1 M NaCl (pH 7.4). The last CRPV sample (column six) was purified from rabbit warts and particle images from eight micrographs formed the data set used to compute the 3D reconstruction. Eight micrographs were needed to obtain a satisfactory number of particle images. The maximum resolution of this reconstruction was limited by the lowest-resolution (largest defocus) micrograph. The seven sets of particle images used to compute the seven other reconstructions came from a single micrograph each.
Hands of BPV-1, CRPV, and HPV-1 icosahedral lattices
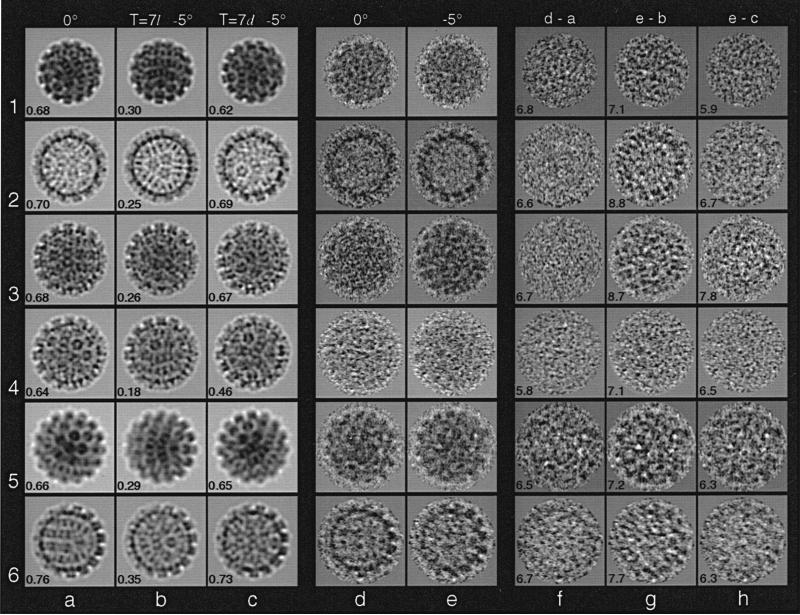
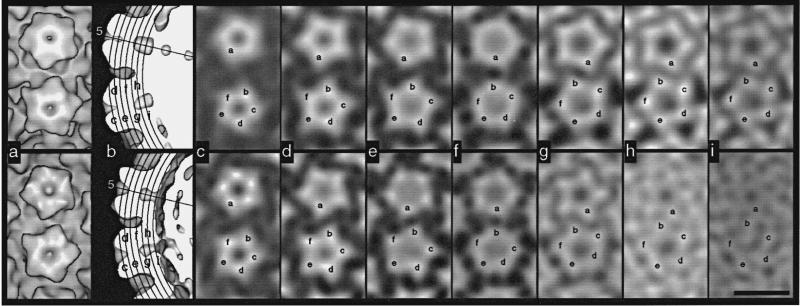
Tilt experiments were used to determine the lattice hands of BPV-1, CRPV, and HPV-1. Enantiomeric reconstructions were rotated to simulate specimen tilt in the microscope and then were projected to simulate the recorded particle images. For the three papillomaviruses we studied, projections of T = 7d reconstructions correlated significantly better with images of tilted specimens than did projections of T = 7l reconstructions (Table 3). Indeed, every particle image correlated better with the T = 7d model (data not shown). Careful visual inspection of some cryoEM data confirmed these comparisons (Figure 4a to e). Difference images, in which projected model images were subtracted from the corresponding particle images, showed the T = 7d model was correct in all cases examined (Figure 4f to h). Difference images obtained with the T = 7l model exhibited higher density variances (Figure 4g) compared to those obtained from the untilted or the T = 7d tilted model (Figure 4f and h).
Table 3.
Hand determinations of BPV-1, CRPV, and HPV-1
| Average correlation coefficients |
||||
|---|---|---|---|---|
| Specimen | Untilted | Tilted versus T = 7l | Tilted versus T = 7d | n |
| BPV-1Capsid | 0.67 (0.05) | 0.33 (0.06) | 0.66 (0.05) | 16 |
| BPV-1 | 0.65 (0.03) | 0.23 (0.04) | 0.64 (0.03) | 18 |
| CRPVClosed form | 0.60 (0.04) | 0.18 (0.05) | 0.47 (0.04) | 30 |
| CRPVOpen form | 0.55 (0.06) | 0.23 (0.09) | 0.49 (0.07) | 32 |
| HPV-1VLPs | 0.68 (0.05) | 0.32 (0.07) | 0.62 (0.06) | 13 |
| SV40 | 0.66 (0.04) | 0.28 (0.07) | 0.54 (0.05) | 80 |
| BackgroundCarbon | 0.00 (0.07) | –0.01 (0.07) | 32 | |
| BackgroundIce | 0.00 (0.06) | 0.00 (0.06) | 80 | |
Images of the tilted specimens were compared to projections from T = 7l or T = 7d structures. Correlation coefficients (1.0 = perfect positive correlation) are the average of n comparisons (standard deviations in parentheses). Comparisons of model data with “background” image data (carbon film and vitreous water) and with images of the untilted specimen serve as control measurements. SV40, known to be T = 7d (Anderer et al., 1967; Liddington et al., 1991), served as a control to establish a consistent set of conventions for the hand determination experiments. Differences between these data and those listed in Table 2 (i.e. n and coefficients for images of untilted particles) arise because only particle images available in both the untilted and tilted views were used for these calculations. Also, the comparisons in this Table were made at the resolution limits of images of the tilted specimen (second micrograph), which usually were lower because of radiation damage than those of the untilted specimen (first micrograph).
Figure 4.
Representative images from the hand determinations of SV40, BPV-1 capsid, BPV-1, CRPV closed form, CRPV open form, and HPV-1 VLPs (rows 1 to 6, respectively). Darkest features represent the highest projected densities. Each panel is 70 nm square. a–c, Projection images of 3D models at orientations corresponding to images of the untilted (d) and tilted (e) particles. Correlation coefficients at the bottom left of each panel quantify the agreement between the model projections (a to c) and particle images (d and e). a, Untilted view and correlation coefficient between a and d. b, Tilted view of T = 7l model and correlation coefficient between b and e. c, Tilted view of T = 7d model and correlation coefficient between c and e. d and e, Electron images of a given virus particle untilted (d) and tilted by –5° (e). The particles in row 4 appear in Figure 2b and c. f to h, Difference images computed by subtracting projection images (a to c) from particle images (d and e), after appropriate scaling (Baker et al., 1990). The average density in each difference map (from the center to the particle edge) is 0.0. Standard deviations are listed at the lower left of each panel. f, Image d – projection a. g, Image e – projection b. h, Image e – projection c.
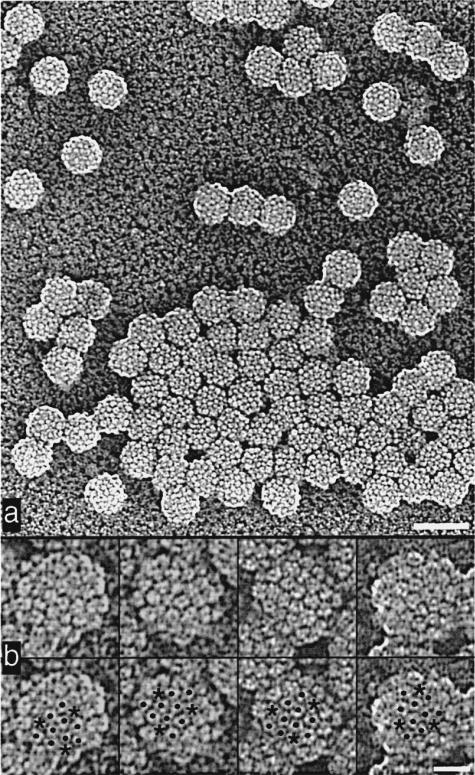
We inspected images of metal-shadowed BPV-1 (Figure 5a) and found 31 particles in which adjacent pentavalent capsomeres and the path between them were clearly resolved. All 31 showed the BPV-1 hand to be T = 7d (Figure 5b).
Figure 5.
Representative images of BPV-1 particles shadowed with Pt-C. Images are shown in reverse contrast. a, Low magnification view. Bar represents 100 nm. b, Unmarked (top) and marked (bottom) views of four shadowed particles. Asterisks identify pentavalent capsomeres and dots indicate hexavalent capsomeres. The path between adjacent capsomeres (see Figure 1) indicates that the surface lattice of BPV-1 has T = 7d symmetry. Bar represents 25 nm.
Capsomere morphology and intercapsomere contacts in papillomaviruses
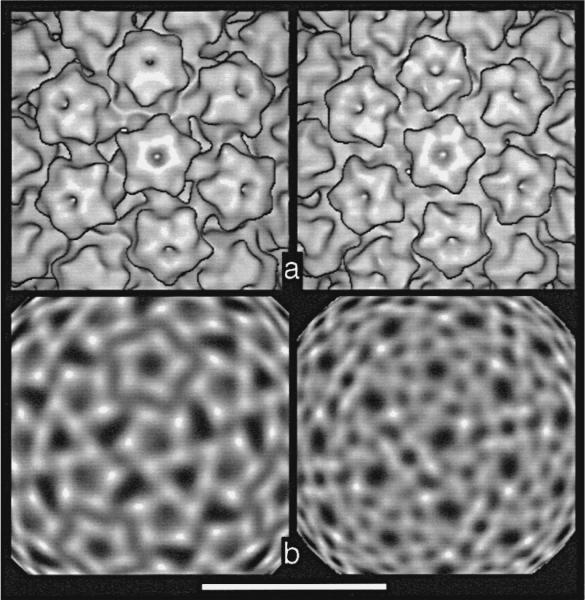
For CRPV and HPV-1 capsids, we examined the morphology of capsomeres and packing of protein subunits by viewing projection maps that only displayed density at specific radii (Figures 6 and 7). Density at the corners of the pentameric capsomeres (Figure 6c to i, symbols a to f) usually corresponded to the maximum density at each radius that may identify individual protein subunits. When viewed at progressively decreasing radii, the high-density features in each capsomere follow a counterclockwise trajectory (Figure 6). Between radii of 30 and 25 nm for CRPV and between radii of 28 and 23 nm for HPV-1, the pentavalent and hexavalent capsomeres skew by 44 to 50° (Figure 6c to h). At the innermost radii of the open CRPV capsid (25 to 24 nm), pentavalent and hexavalent capsomeres skew clockwise by 5 to 8° (Figure 6h and i). A similar clockwise skew was observed in the closed CRPV capsid (24 to 21 nm) but not in HPV-1 (Figure 6h and i) or BPV-1 (Baker et al., 1991).
Figure 6.
Skew in capsomeres of open CRPV (top) and closed HPV-1 (bottom) structures. A pentavalent capsomere lies directly above a hexavalent capsomere in each panel. Bar represents 10 nm. a, Surface-shaded views. b, Surface-shaded, side views of the left half of each structure shown in a. The level of each radial section depicted in c to i is indicated by the labeled arcs. The 5-fold symmetry axis is also identified. The slightly larger radius of the open CRPV form compared with the closed HPV-1 structure is apparent. The reason for the presence (bottom, r ≈ 21 nm) or absence (top, r ≈ 23 nm) of a gap between the capsid shell and the chromatin core is unknown but appears to be unrelated to the open or closed state of the capsid. c to i, Projected density at radii from 30 to 24 nm (CRPV) and 28 to 22 nm (HPV-1) in 1 nm steps, and with contrast reversed relative to that shown in Figure 4. All projections in the CRPV series are at radii 2 nm larger than the corresponding projections of HPV-1. The highest average density occurs at level h. Six strong density features (subunits) are labeled a to f as described by Baker et al. (1991) and are traced through the capsid structure. These six make up an icosahedral asymmetric unit (1/60th of an icosahedron). The path of each subunit becomes somewhat ambiguous at levels where capsomere features blur (e.g. f). Hence, it is impossible at this resolution to follow unambiguously the density unique to a given subunit. However, subtle subunit skewing is observed in capsomeres of a 2.5 nm resolution reconstruction of murine polyomavirus (Belnap, D., Olson, N. & Baker T., unpublished results), and this is confirmed by comparison to the high-resolution structure of VP1 whose elongated mass lies in a nearly radial orientation (Stehle et al., 1994).
Figure 7.
Views of open CRPV (left) and closed HPV-1 (right) structures. The view direction coincides with the axis of a hexavalent capsomere. A pentavalent capsomere appears directly above the central hexavalent capsomere. Bar represents 25 nm. a, Surface-shaded views. b, Density at radii of 25 nm (CRPV) and 23 nm (HPV-1) showing density in center of shell region (Figure 6h) with contrast the same as in Figure 6c to i.
The types of intercapsomere interactions within the shell layer are distinct in the open and closed forms at radii of 25 nm for CRPV and 23 nm for HPV-1 (Figure 7b). The average density is highest in both structures at these radii. Here, the hexavalent capsomeres are about ten nm in diameter, whereas the pentavalent capsomeres of CRPV are slightly larger (~11 nm) and those of HPV-1 are slightly smaller (~9 nm). The subunits are packed tightly in the closed HPV-1 capsid, as Baker et al. (1991) observed. In contrast, the open CRPV capsid exhibits much looser packing, as indicated by the presence of large holes on the 3-fold axes and also near the 2-fold axes.
Papovavirus sequence alignments
We used the program GAP (Genetics Computer Group, Madison, Wisconsin) to compare the L1 and L2 sequences of BPV-1, CRPV, and HPV-1 (Table 4). The L1 sequences are more highly conserved compared to the L2 sequences. Pairwise comparisons showed that CRPV and HPV-1 sequences have the highest identity and similarity. Danos et al. (1984) observed a similar trend in L1 sequences. Comparison of the three L1 sequences to SV40 VP1 and the three L2 sequences to SV40 VP2/VP3 clearly demonstrated far less sequence similarity between the two papovavirus genera (Table 4). The program PILEUP (Genetics Computer Group, Madison, Wisconsin) was used to align L1 sequences of CRPV, deer papillomavirus, European elk papillomavirus, canine oral papillomavirus, multimammate rat papillomavirus, pygmy chimpanzee papillomavirus type 1, 3 types of BPV, and 47 types of HPV (data not shown). All these papillomaviruses showed significant sequence identities and similarities. An analogous alignment of 26 L2 sequences demonstrated less agreement in comparison to the L1 alignment.
Table 4.
Sequence comparisons for the major (L1, VP1) and minor (L2, VP2/VP3) capsid proteins of papovaviruses obtained by use of the program GAP (Genetics Computer Group, Madison, Wisconsin)
| Sequence comparison |
|||
|---|---|---|---|
| HPV-1 L1 | BPV-1 L1 | SV40 VP1 | |
| CRPV L1 | 58 (75) | 48 (67) | 19 (42) |
| HPV-1 L1 | 48 (69) | 16 (37) | |
| BPV-1 L1 | 17 (39) | ||
| L2 | L2 | VP2/VP3 | |
|---|---|---|---|
| CRPV L2 | 42 (61) | 27 (47) | 18 (41) |
| HPV-1 L2 | 30 (52) | 19 (41) | |
| BPV-1 L2 | 18 (41) |
The percentage of identical (left) and similar (in parentheses) residues is given for several L1–L1, L1–VP1, L2–L2, and L2–VP2/VP3 comparisons. Correlation of L1 sequences to random sequences the same length as VP1 gave an identity of 14 to 16% and a similarity of 40 to 42%.
Discussion
Conserved features in papovaviruses
The CRPV capsid exhibits several features in common with all other papovavirus capsids (Table 1). It consists solely of pentameric capsomeres as do the capsids of murine polyomavirus, SV40, BPV-1, and HPV-1 (Table 1, column 2). Seventy-two capsomeres are arranged on a T = 7 icosahedral lattice, as was observed previously in CRPV and also in murine polyomavirus, SV40, K virus, HPV-1, BPV-1, and human wart virus (Table 1, column 1). Our results show that the CRPV capsomeres are arranged on a T = 7dextro skew lattice, just like murine polyomavirus, SV40, BPV-1, HPV-1, and human wart virus (Table 1, column 4). Therefore, the T = 7d, 72-pentamer capsid appears to be conserved in all papovaviruses.
Capsomeres in open and closed CRPV capsids have the same star-shaped morphology. This morphology is conserved among the papillomaviruses (Baker et al., 1991; Hagensee et al., 1994), but is not found in the polyomaviruses.
Open and closed papillomavirus capsids
Papillomavirus capsids exist in open and closed forms. The open capsid has several large holes and the entire capsid appears to have expanded by ~2 nm in radius when compared with the closed capsid. Though the open and closed forms cannot be distinguished directly in electron micrographs, the 3D reconstructions clearly reveal distinct structures. Ionic strength and divalent-cation concentration appear to influence the state of the capsid (Table 2). To our knowledge, open and closed forms of polyomavirus capsids have not been reported.
The reversibility and functional significance of the open and closed states remains unknown. However, structural similarities between papillomaviruses and cowpea chlorotic mottle virus (CCMV), an RNA, T = 3 icosahedral, plant virus, may be relevant. CCMV exists in swollen (open) and unswollen (closed), forms which may occur during distinct stages of viral infection (Bancroft et al., 1967; Speir et al., 1995). At acidic pH or in the presence of divalent metal ions, CCMV is a rigid, compact structure; but at pH 7.0 and with EDTA present, CCMV swells by ~10% in size, large openings appear in the capsid shell, and viral RNA is rendered susceptible to nuclease digestion (Bancroft et al., 1967; Speir et al., 1995). Lowering the pH to 5.0 or raising the Ca2+ or Mg2+ concentration reverses this transition (Speir et al., 1995). The comparison between CCMV and papillomaviruses is even more compelling because the high-resolution structure of the capsid protein of CCMV (Speir et al., 1995) is strikingly similar to the SV40 (Liddington et al., 1991) and murine polyomavirus (Stehle et al., 1994) VP1 structures. Also, CCMV and polyomavirus capsids are composed of capsomeres linked together by extensions of the subunit polypeptide chains (Liddington et al., 1991; Stehle et al., 1994; Speir et al., 1995).
A few studies have shown that papovavirus capsids are stabilized by calcium or high ionic strength. For example, 10 mM EGTA and 3 mM dithiothreitol in 150 mM NaCl at pH 7.5 to 10.5 dissociates murine polyomavirus capsids into capsomeres, though capsids remain stable in 1 M NaCl (Brady et al., 1977). Also, capsomeres self-assemble into capsids in the presence of 1 M NaCl, and subsequent lowering of the salt concentration in the presence of Ca2+ and absence of reducing agent does not affect capsid stability (Salunke et al., 1986). Similar effects have recently been observed for BPV-1 (Paintsil, J., Müller, M., Picken, M., Gissmann, L. & Zhou, J., personal communication). Paintsil et al., found that treatment of purified BPV-1 with 10 mM EGTA, 3 mM dithiothreitol, and 150 mM NaCl at pH 8.0 caused virions to dissociate into capsomeres. Papillomavirus VLPs were found to dissociate into capsomeres when incubated in 2 mM EGTA and 20 mM dithiothreitol (Volpers et al., 1995).
Papillomavirus capsomeres are linked by disulfide bonds (Sapp et al., 1995). Hence, these may stabilize the open structure because reducing agent was not present in the buffers of open capsids (see Materials and Methods).
Hand of CRPV and other papillomaviruses
Papillomavirus capsomeres have a skewed morphology that is apparent when density at different radii are compared (Figure 6; and Baker et al., 1991). When displayed on a T = 7l lattice, capsomeres from our first CRPV reconstruction (2.7 nm resolution) exhibited a skew opposite that of T = 7d BPV-1 and HPV-1 structures (Baker et al., 1991). This skew variation was perplexing given that highly conserved proteins, such as L1 (Table 4), would be expected to adopt similar tertiary structures. However, the openness of the CRPV capsid compared to the closed BPV-1 and HPV-1 capsids did indicate possible differences among papillomavirus structures. A literature search revealed that lattice hands were known for only two papillomaviruses, CRPV and human wart virus (unknown strain), and their hands were opposite (Table 1). The differences in capsomere skew and capsid form that distinguished CRPV from BPV-1 and HPV-1, and the unknown hands of the BPV-1 and HPV-1 lattices, stimulated us to determine the absolute hands of all three viruses. All were found to be T = 7d structures. Handedness remained conserved regardless of whether the capsid was open or closed or whether chromatin was present (i.e. empty capsid or full virion). All 220 particles tested had the same hand. This conserved property is consistent with the notion that all capsids within a particular virus species or strain have the same hand and also that all papovaviruses have the same hand.
Our determination that CRPV is a T = 7d structure contradicted the T = 7l results obtained from negatively stained CRPV samples (Finch & Klug, 1965; Klug & Finch, 1968). Though we have no explanation for this discrepancy (nor do we believe that the distortions and deformations caused by negative stain and drying could invert the lattice hand) other evidence supports the T = 7d answer. L1, the major capsid protein in papillomaviruses, self-assembles into capsids that are indistinguishable from the capsid structure in virions (e.g. see Kirnbauer et al., 1992; Hagensee et al., 1994). Significant sequence identities and similarities among the L1 proteins (Table 4; and Danos et al., 1984; Baker, 1987) suggest that all L1 proteins form similar tertiary and quaternary structures. The similarly skewed morphologies of the papillomavirus capsomeres (Figure 6) lend additional support for a conserved T = 7d structure because conserved morphology correlates with the conserved primary structures of L1 polypeptides. The conserved quaternary (Figure 6; and Baker et al., 1991) and primary (Table 4; and Danos et al., 1984; Baker, 1987) structures strongly suggest that the tertiary structures of all L1 proteins also are highly conserved.
Handedness of the T = 7 lattice is necessarily linked with handedness at all other structural levels, from the l-amino acids that form the polypeptide chain to the quaternary interactions that form the whole structure. As Klug & Finch (1965) discussed, two structures of opposite lattice hand are not enantiomers if they are built from identical, chiral substrates. They explained how, for example, left-handed and right-handed helices can be formed from l-amino acids, but these helices are not enantiomers and each helix has unique interactions between the residues. Enantiomeric helices could only be formed if they were composed of corresponding enantiomeric amino acids. Therefore, if T = 7l and T = 7d structures could be formed from the same or similar proteins, subunit–subunit interactions in the two non-enantiomers must differ, as Finch & Klug (1965) realized. Despite the limited resolution (2.5 to 2.8 nm) of our present or past (Baker et al., 1991) analyses, some differences in interactions were seen in papillomaviruses, most notably between the open and closed forms (Figures 7b and 8b to d). These, however, did not account for any difference in lattice hand because all were shown to be T = 7d.
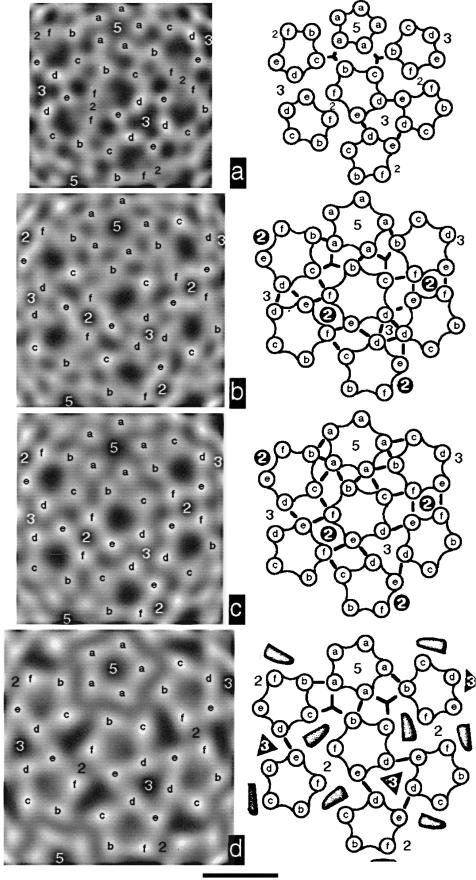
Figure 8.
Intercapsomere interactions in papovavirus capsids viewed as in Figure 7. Capsid density is projected at the radius of maximum average density in the shell (left). Interpretive diagrams of mass arrangements and intercapsomere interactions are shown to the right of each image. Numbers identify icosahedral symmetry axes. Labeling of subunits corresponds to that used in Figure 6. Arcs depict intracapsomere contacts and thick lines represent intercapsomere connections not on 2- or 3-fold axes. Holes seen in the papillomavirus capsid shells are depicted in the diagram as shaded regions. Bar represents 10 nm. a, Murine polyomavirus, r = 20 nm. b, Closed HPV-1, r = 23 nm. Holes in the shell occur on 2-fold symmetry axes and are bounded by e and f subunits. c, Closed BPV-1, r = 23 nm, from Baker et al. (1991). Holes are located in the same positions as in b. d, Open CRPV, r = 25 nm. Holes in the shell are bounded by b, c, d, e and f subunits. They also appear on the 3-fold axes where they are bounded by d and e subunits.
Subunit interactions in papovaviruses
The nature of subunit contacts in papovaviruses has been the subject of several investigations (Rayment et al., 1982; Salunke et al., 1986, 1989; Baker et al., 1988, 1989, 1991; Liddington et al., 1991; Marzec & Day, 1993; Stehle et al., 1994; Tarnai et al., 1995). A complete understanding of both intracapsomere and intercapsomere interactions can only come from high-resolution studies, which, to date, have only been accomplished with SV40 and murine polyomavirus (Liddington et al., 1991; Stehle et al., 1994). However, the complementarity and fidelity between structure determinations by cryoEM and X-ray crystallography (e.g. see Cheng et al., 1994; Speir et al., 1995) lend confidence that general aspects of these interactions can be seen in the cryoEM data (Figure 8) and compared with the higher resolution structures of SV40 and murine polyomavirus.
In the shell, the highest density features within the pentavalent capsomere are similarly oriented in both papilloma and polyoma viruses (Figure 8). The hexavalent capsomeres of the papilloma closed capsids are rotated slightly clockwise relative to polyoma capsomeres, and this difference in orientation necessarily creates a different pattern of intercapsomere contacts (Figure 8a to c). For example, in murine polyomavirus, subunit b participates in a quasi-3-fold interaction with a and c subunits from two other capsomeres (Figure 8a). In papillomaviruses, the b subunit interacts with two a subunits in the pentavalent capsomere (Figure 8b to d). Likewise, the c and f subunits from adjacent capsomeres appear to interact in closed papillomavirus capsids (Figure 8b and c) but are farther apart in murine polyomavirus (Figure 8a) and in open CRPV capsids (Figure 8d). Closed papilloma capsids have a small hole on the 2-fold symmetry axis whereas murine polyomavirus capsids do not. Variations occur in the f–f, e–f, e–d, and d–d interactions (Figure 8a to c). Slight differences are also seen between HPV-1 and BPV-1 closed capsids (e.g. interactions among a, b, and c subunits, Figure 8b,c).
Intercapsomere interactions in the shell of open papillomavirus capsids are like those in murine polyomavirus (Figure 8a and d). The key difference between the papilloma open and closed forms is the position of the c and f subunits. They appear to interact in the closed form but are separated in the open form by a large, elongated hole (Figure 8b to d). Other subunits are in slightly different positions between the two capsid forms, and might be influenced by the locations or interactions of the c and f subunits. Loss of the presumed c–f interaction may allow the hexavalent capsomere to adopt the more counterclockwise orientation resulting in the f–f contacts and the closing of the small hole on the 2-fold symmetry axis (Figures 3c and 8b to d). Also, changes in the c–f interactions could affect e–f contacts, because these appear to be different or absent in the open capsid. Conversely, changes in the e–f interactions could cause the c-f changes. Distances between d subunits increase in the open form leaving large holes on the 3-fold axes. The closed-to-open transition appears to have little if any affect on intercapsomere interactions among the a, b, and c subunits, despite the slight clockwise rotation of the pentavalent capsomere. The capsomere rotations and switching of interactions may be necessary to allow the capsid to expand.
Speir et al. (1995) found that CCMV hexameric capsomeres also adopt a new rotational orientation when the capsid changes from the native to the swollen form, and also that three closely spaced, acidic residues form putative binding sites for divalent metal ions. They deduced that swelling of the capsid and opening of holes likely result from repulsion of these negatively charged amino acids when pH is raised and metal ions are removed. Similar effects may occur in papillomavirus capsids.
Extended carboxy-terminal “arms” interlock capsomeres of murine polyomavirus and SV40 (Stehle et al., 1994; Liddington et al., 1991). Similar intercapsomere links may occur in the shells of papillomaviruses. Yabe et al. (1979) observed “fibrous bridge-like structures” joining capsomeres of human papillomavirus. Such interactions might act to stabilize the open structure and render swelling reversible, as occurs in CCMV (Speir et al., 1995). Intercapsomere connections within the shell, such as c–f and e–f, which change dramatically between the open and closed forms (Figure 8b to d), possibly involve ionic interactions that are dictated by solution conditions. The larger diameter papillomaviruses might require longer linking arms or may have additional links compared to the polyomaviruses. The extra ~140 amino acid residues in the papillomavirus subunits might be responsible for such differences.
Preservation of conserved features in papovaviruses
Papovavirus capsids share many common features even though the major capsid proteins of polyoma (VP1) and papilloma (L1) viruses exhibit only weak sequence similarities (Table 4; and Chen et al., 1982; Danos et al., 1982). The larger size of L1 likely accounts for additional features, such as surface loops that give papillomavirus capsomeres a star shape. L1 subunits also contribute to the larger size of papillomavirus capsomeres (Figure 8) and hence the larger diameter of papillomavirus capsids.
Despite these differences, all papillomavirus and polyomavirus capsids have 72 pentameric capsomeres arranged with T = 7d icosahedral lattice symmetry. Within the shell region, capsomere arrangement and intercapsomere contacts are only slightly different between the two genera (Figure 8). Tarnai et al. (1995) and Marzec & Day (1993) have shown that capsomeres in closed papillomavirus capsids occupy space and minimize electrostatic free energy in the most efficient manner possible for 72 pentamers distributed on a spherical surface. If polyoma and papilloma viruses are evolutionarily unrelated, as Chen et al. (1982) suggest, then perhaps the two genera have evolved convergently and have merely adopted an idealized natural structure. If the Papillomavirus and Polyomavirus genera diverged from a common ancestor, selective pressures must have preserved the pentameric capsomeres and T = 7d lattice despite the differences in intercapsomere interactions, capsid size, capsomere morphology, and amino acid sequence.
Conserved and stable interactions within capsomeres might explain why papovavirus capsomeres remain pentameric while surface loops generate distinct external morphologies. Purified VP1 protein from murine polyomavirus was observed in electron micrographs as either pentamers or assembled capsids under non-denaturing conditions (Salunke et al., 1986). Under certain non-denaturing conditions, polyomavirus and papillomavirus capsids dissociate into intact capsomeres (Brady et al., 1977; Volpers et al., 1995; Paintsil, J., Müller, M., Picken, M., Gissmann, L. & Zhou, J., personal communication). These observations are consistent with the presence of interlocking secondary structure among the VP1 subunits of polyomavirus pentamers (Liddington et al., 1991; Stehle et al., 1994). Perhaps a polyoma-like core exists in papillomavirus L1 subunits that accounts for stable contacts within papillomavirus capsomeres.
Conservation of T = 7d symmetry could be dictated by flexible polypeptide extensions that link capsomeres. Flexible carboxy-terminal ends of SV40 and murine polyomavirus subunits adopt six different conformations to accommodate six unique bonding environments (Liddington et al., 1991; Stehle et al., 1994). Polypeptide extensions, if also present in papillomaviruses, probably adopt conformations different from those occuring in VP1. These extensions would accommodate or account for several features that distinguish polyoma and papilloma viruses, including the star-shaped capsomeres, different intercapsomere connections, larger subunits and capsid, and open and closed forms. However, the flexibility of these polypeptide extensions in papovaviruses might be limited, and, in the presence of stable intracapsomere interactions, a T = 7l structure would not be permitted.
Materials and Methods
Virus purification
Two samples of CRPV were prepared as follows: ear skin fragments (2 mm × 2 mm) from wild cottontail rabbits (courtesy of the Pennsylvania Game Commission) were infected with CRPV (from a viral stock isolated from Kansas cottontail rabbits in 1968) and were implanted subcutaneously in athymic nude mice (Kreider et al., 1979, 1985). After 90 days, the resulting cysts were harvested and the virus-rich keratin cores were removed from the cyst walls. A 15 ml portion of homogenization buffer (1 M NaCl, 0.02 M Tris-HCl, pH 7.4) was added to 5 g of keratin cores. The mixture was homogenized in a Virtis homogenizer at 30,000 rpm and 0°C for five minutes. The slurry was spun for ten minutes at 10,000 rpm in an SA600 rotor. The supernatant was divided into fourths, and each aliquot was layered onto a step gradient consisting of 3 ml CsCl (1.47 g/ml) overlaid with 5 ml CsCl (1.2 g/ml) in SW 41 clear tubes. Tubes were spun for two hours at 25,000 rpm and 20°C. We removed samples from the top of the tube and analyzed them by ELISA using a CRPV monoclonal antibody developed by Christensen & Kreider (1991). Samples with the highest amounts of CRPV were combined, and the total volume was increased to 10 ml with CsCl (1.36 g/ml). The solution was spun in two clear SW 55 tubes for 20 hours at 45,000 rpm and 20°C, resulting in three distinct bands per tube. Isolated bands were analyzed by ELISA. Samples for structural analysis were taken from the low band (1.34 g/ml) which contained full virions. They were dialyzed against 10 mM Tris-HCl and 1 mM EDTA at pH 8.0 or 20 mM Tris-HCl and 1 M NaCl at pH 7.4 (Table 2).
A third sample of CRPV was purified from warts obtained directly from wild cottontail rabbits. Virus was purified in a manner similar to that used previously to extract BPV-1 from cattle warts (Baker et al., 1991).
The BPV-1 sample was the same as the one reported by Baker et al. (1991). SV40 was purified as described (Baker et al., 1988). HPV-1 VLPs were expressed and purified as described by Hagensee et al. (1993) with the addition of a second CsCl gradient purification followed by dialysis against 10 mM potassium phosphate buffer at pH 7.2 (Hagensee, M. & Galloway, D., personal communication).
Electron microscopy
Viral specimens were applied to carbon-coated (perforated or non-perforated), 400-mesh, electron microscope grids and were examined with cryoelectron microscopy techniques (Adrian et al., 1984; Olson & Baker, 1989). Before application to the grid, the SV40 sample was diluted with distilled water, and the HPV-1 VLP specimen was mixed with a sample of bacteriophage ϕX174 in distilled water. The CRPV sample from rabbit warts was washed several times on the grid with distilled water to remove CsCl. The other CRPV specimens and the BPV-1 sample were not diluted or washed. Images were recorded at 80 kV and at a nominal magnification of 36,000× in a Philips EM420 transmission electron microscope (Philips Electronics Instruments, Mahwah, NJ) equipped with a Gatan anticontaminator and a Gatan single-tilt cryotransfer holder (Model 626, Gatan Inc., Warrendale, PA). Each exposure was recorded with an electron dose of 800 to 1500 e–/nm2 at the specimen. Micrographs were recorded 0.8 to 5 μm underfocus (Table 2, Figure 2). For each hand determination experiment, two images of the same field of particles were recorded: the first at 0° tilt and the second at –5° tilt (positive tilt corresponded to a clockwise rotation of the microscope goniometer). The HPV-1 micrograph was that used by Baker et al. (1991).
Metal-shadowed BPV-1 particles were prepared by applying samples of unstained and unfixed BPV-1 to carbon/Formvar-coated, 400-mesh, electron microscope grids. Air-dried specimens were rotary shadowed at a 13° angle with Pt-C in a Balzers BAE 080 vacuum evaporator (Hudson, NH). Micrographs were recorded at 80 kV and at nominal magnifications of 33,000× or 50,000×, in a JEOL 100CX transmission electron microscope (Peabody, MA). The specimen side of the grid faced the electron source.
Three-dimensional image reconstruction
We computed eight different reconstructions (Table 2): one each of SV40, HPV-1, and HPV-1 VLPs; two of BPV-1; and three of CRPV. Six of these structures were used in hand determination experiments. The CRPV and HPV-1 reconstructions at highest resolution (2.7 and 2.5 nm, respectively) were utilized to study fine details of papilloma capsid structure.
Micrographs were digitized at 25 μm intervals (0.69 nm pixels at the specimen) on a rotating-drum microdensito-meter and images were displayed on a raster graphics device. Individual virus images were extracted within a circular mask (Baker et al., 1988) and floated (DeRosier & Moore, 1970). Image intensities were adjusted to remove linear background gradients (Baker et al., 1990; Booy et al., 1991) and to normalize means and variances (Carrascosa & Steven, 1978).
A preliminary 3D reconstruction of CRPV was obtained by means of established cross-correlation (Belnap et al., 1993; Baker et al., 1990) and common-lines procedures (Fuller, 1987; Baker et al., 1988; Fuller et al., 1996). Then, a model-based, polar-Fourier-transform (PFT) method (Cheng et al., 1994; Baker & Cheng, 1996) was used to determine initial orientation (Θ,Φ,Ω) and translation (x,y origin) parameters for all the BPV-1, CRPV, HPV-1, HPV-1 VLP, and SV40 particle images. The models for the PFT procedure were 3D reconstructions previously computed from the same image data or from another micrograph.
We used two iterative methods in refining the origin and orientation parameters (Table 2). Parameters for five data sets were refined by the PFT procedure (Baker & Cheng, 1996). Origins and orientations for the three others were refined by cross-correlation (Baker et al., 1990) and cross-common-lines (Fuller, 1987; Baker et al., 1988; Fuller et al., 1996) routines, respectively. We computed intermediate and final reconstructions by using Fourier Bessel (Crowther, 1971) and 3-fold averaging (Fuller, 1987; Fuller et al., 1996) methods. Data quality and completeness were assessed (Table 2) by cross-common-lines phase residuals (Fuller, 1987; Fuller et al., 1996), correlation coefficients (Dryden et al., 1993), eigenvalue spectra (Crowther et al., 1970a,b; Fuller et al., 1996), and Fourier ring correlation coefficients (Conway et al., 1993; Saxton & Baumeister, 1982).
Densities in the HPV-1 reconstruction were scaled to those in the 2.7 nm resolution, CRPV reconstruction. Surface-shaded renderings were computed at the same density level for both structures (Figures 3, 6 and 7). The contour level chosen was one standard deviation above the average noise level. Densities corresponding to noise, unconnected to the capsid, and occupying less than 0.5% of the reconstruction volume, were erased from the density maps for some views (Conway et al., 1993, 1996).
Hand determinations
We determined the absolute hands of the T = 7 lattice for BPV-1, CRPV, and HPV-1 by means of tilting experiments. Essential aspects of these experiments were adapted from the procedure in which projections from computer-generated models were compared to images of negatively stained viruses (Klug & Finch, 1968). We compared projections of 3D reconstructions with cryoEM images of unstained specimens (Table 3, Figure 4). T = 7l and T = 7d reconstructions were tilted by –5° from each untilted Θ,Φ,Ω orientation to simulate the experiment performed in the microscope. The projected image of each tilted model was correlated against the image of each tilted particle. SV40, whose atomic structure and T = 7d symmetry are known (Anderer et al., 1967; Liddington et al., 1991), served as a standard in establishing a self-consistent set of conventions for defining the tilt direction and angle in the EM420 microscope. As an additional control, each reconstruction was rotated by the untilted Θ,Φ,Ω orientation angles and projected. These projections were correlated against the corresponding images of each untilted particle.
We also determined the hand of BPV-1 from images of metal-shadowed specimens (Figure 5). Because the shadowed surfaces of the particles faced the electron source in the microscope, micrographs were viewed with their emulsion sides up to give a face-on view of the shadowed surfaces.
Sequence analysis
Sequences of papillomavirus and SV40 structural proteins were obtained from the Swiss Protein, GenPeptide, and Protein Identification Resource data banks. We used the programs GAP and PILEUP from the Genetics Computer Group Sequence Analysis Software Package (Version 7.2, release October 1992, Genetics Computer Group, Madison, Wisconsin). GAP was used to align one sequence to another (Table 4). PILEUP was used to align more than two sequences.
Acknowledgements
We thank the following people for providing specimens: C. Olson and L. Cowsert for BPV-1, C. Olson and R. Shope Jr for CRPV, D. Galloway and M. Hagensee for HPV-1 VLP, and M. Bina for SV40. We thank H. Cheng, J. Conway, N. Dilley, T. Dinh-Phung, K. Dryden, E. Eltzroth, W. Grochulski, and B. Trus for computer programs; R. A. Crowther for help in understanding the relationship of handedness to projections; R. Roden, J. Johnson, P. Low, and D. Asai for helpful suggestions; and J. Paintsil and M. Müller for sharing their unpublished data. This work was supported by U.S. Public Health Service grants GM33050 (T.S.B.), CA47622 (J.W.K.), AI1375549-01 (J.C.B.), and AI27713 (Purdue University, AIDS Center Laboratory for Computational Biology); by National Science Foundation grant MCB-941770 (J.C.B.); by a grant from the Lucille P. Markey Charitable Trust for Structural Biology (Purdue University); by a Purdue University, Frederick N. Andrews fellowship (D.M.B.); and by a National Institutes of Health Training Grant (D.M.B.).
Abbreviations used
- 3D
three-dimensional
- a.a.
amino acid
- BPV
bovine papillomavirus
- BPV-1
bovine papillomavirus type 1
- CCL
cross-common-lines
- CCMV
cowpea chlorotic mottle virus
- CRPV
cottontail rabbit papillomavirus
- cryoEM
cryoelectron microscopy
- d
dextro
- diam.
diameter
- EGTA
ethyleneglycol-bis-N,N′-tetraacetic acid
- HPV
human papillomavirus
- HPV-1
human papillomavirus type 1
- l
laevo
- PFT
polar Fourier transform
- SV40
simian virus 40
- T = 7
triangulation number of seven
- VLP
virus-like particle
References
- Adolph KW, Caspar DLD, Hollingshead CJ, Lattman EE, Philips WC, Murakami WT. Polyoma virion and capsid crystal structures. Science. 1979;203:1117–1120. doi: 10.1126/science.218286. [DOI] [PubMed] [Google Scholar]
- Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- Anderer FA, Schlumberger HD, Koch MA, Frank H, Eggers HJ. Structure of simian virus 40: II. symmetry and components of the virus particle. Virology. 1967;32:511–523. doi: 10.1016/0042-6822(67)90303-0. [DOI] [PubMed] [Google Scholar]
- Baker CC. Sequence analysis of papillomavirus genomes. In: Salzman NP, Howley PM, editors. The Papovaviridae, vol. 2, The Papillomaviruses. Plenum Press; New York: 1987. pp. 321–385. [Google Scholar]
- Baker TS. Cryo-electron microscopy and three-dimensional image reconstruction of icosahedral viruses. In: Megías-Megías L, Rodríquez-García MI, Ríos A, Arias JM, editors. Electron Microscopy 92: Proceedings of the 10th European Congress on Electron Microscopy held in Granada, Spain, 7-11 September 1992. Vol. 3. Secretariado de Publicaciones de la Universidad de Granada; Granada: 1992. pp. 275–279. [Google Scholar]
- Baker TS, Cheng RH. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 1996;116:120–130. doi: 10.1006/jsbi.1996.0020. [DOI] [PubMed] [Google Scholar]
- Baker TS, Caspar DLD, Murakami WT. Polyoma virus ‘hexamer’ tubes consist of paired pentamers. Nature. 1983;303:446–448. doi: 10.1038/303446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. Reconstruction of the three-dimensional structure of simian virus 40 and visualization of the chromatin core. Proc. Natl Acad. Sci. USA. 1988;85:422–426. doi: 10.1073/pnas.85.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Drak J, Bina M. The capsid of small papova viruses contains 72 pentameric capsomeres: direct evidence from cryoelectron-microscopy of simian virus 40. Biophys. J. 1989;55:243–253. doi: 10.1016/S0006-3495(89)82799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Newcomb WW, Booy FP, Brown JC, Steven AC. Three-dimensional structures of maturable and abortive capsids of equine herpesvirus 1 from cryoelectron microscopy. J. Virol. 1990;64:563–573. doi: 10.1128/jvi.64.2.563-573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TS, Newcomb WW, Olson NH, Cowsert LM, Olson C, Brown JC. Structures of bovine and human papillomaviruses: analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys. J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft JB, Hills GJ, Markham R. A study of the self-assembly process in a small spherical virus: formation of organized structures from protein subunits in vitro. Virology. 1967;31:354–379. doi: 10.1016/0042-6822(67)90180-8. [DOI] [PubMed] [Google Scholar]
- Belnap DM, Grochulski WD, Olson NH, Baker TS. Use of radial density plots to calibrate image magnification for frozen-hydrated specimens. Ultramicroscopy. 1993;48:347–358. doi: 10.1016/0304-3991(93)90110-j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booy FP, Newcomb WW, Trus BL, Brown JC, Baker TS, Steven AC. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell. 1991;64:1007–1015. doi: 10.1016/0092-8674(91)90324-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JN, Winston VD, Consigli RA. Dissociation of polyoma virus by the chelation of calcium ions found associated with purified virions. J. Virol. 1977;23:717–724. doi: 10.1128/jvi.23.3.717-724.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrascosa JL, Steven AC. A procedure for evaluation of significant structural differences between related arrays of protein molecules. Micron. 1978;9:199–206. [Google Scholar]
- Caspar DLD. An analogue for negative staining. J. Mol. Biol. 1966;15:365–371. doi: 10.1016/s0022-2836(66)80232-2. [DOI] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- Chen EY, Howley PM, Levinson AD, Seeburg PH. The primary structure and genetic organization of the bovine papillomavirus type 1 genome. Nature. 1982;299:529–534. doi: 10.1038/299529a0. [DOI] [PubMed] [Google Scholar]
- Cheng RH, Reddy VS, Olson NH, Fisher AJ, Baker TS, Johnson JE. Functional implications of quasi-equivalence in a T = 3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure. 1994;2:271–282. doi: 10.1016/s0969-2126(00)00029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen ND, Kreider JW. Neutralization of CRPV infectivity by monoclonal antibodies that identify conformational epitopes on intact virions. Virus Res. 1991;21:169–179. doi: 10.1016/0168-1702(91)90031-p. [DOI] [PubMed] [Google Scholar]
- Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J. Struct. Biol. 1993;111:222–233. doi: 10.1006/jsbi.1993.1052. [DOI] [PubMed] [Google Scholar]
- Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. Visualization of three-dimensional density maps reconstructed from cryoelectron micrographs of viral capsids. J. Struct. Biol. 1996;116:200–208. doi: 10.1006/jsbi.1996.0031. [DOI] [PubMed] [Google Scholar]
- Crowther RA. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. Roy. Soc. Lond. B. 1971;261:221–230. doi: 10.1098/rstb.1971.0054. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Amos LA. Three-dimensional image reconstructions of some small spherical viruses. Cold Spring Harbor Symp. Quant. Biol. 1972;36:489–494. doi: 10.1101/sqb.1972.036.01.062. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Amos LA, Finch JT, DeRosier DJ, Klug A. Three dimensional reconstructions of spherical viruses by Fourier synthesis from electron micrographs. Nature. 1970a;226:421–425. doi: 10.1038/226421a0. [DOI] [PubMed] [Google Scholar]
- Crowther RA, DeRosier DJ, Klug A. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc. Roy. Soc. Lond. A. 1970b;317:319–340. [Google Scholar]
- Danos O, Katinka M, Yaniv M. Human papillomavirus 1a complete DNA sequence: a novel type of genome organization among Papovaviridae. EMBO J. 1982;1:231–236. doi: 10.1002/j.1460-2075.1982.tb01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danos O, Giri I, Thierry F, Yaniv M. Papillomavirus genomes: sequences and consequences. J. Inves. Dermatol. 1984;83(1 Supplement):7s–11s. doi: 10.1111/1523-1747.ep12281115. [DOI] [PubMed] [Google Scholar]
- DeRosier DJ, Moore PB. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J. Mol. Biol. 1970;52:355–369. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- De Villiers E. Heterogeneity of the human papillomavirus group. J. Virol. 1989;63:4898–4903. doi: 10.1128/jvi.63.11.4898-4903.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J. Cell Biol. 1993;122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart W. Polyomavirinae and their replication. In: Fields BN, Knipe DM, editors. Virology. 2nd edit. Vol. 2. Raven Press; New York: 1990. pp. 1593–1607. [Google Scholar]
- Finch JT. The surface structure of polyoma virus. J. Gen. Virol. 1974;24:359–364. doi: 10.1099/0022-1317-24-2-359. [DOI] [PubMed] [Google Scholar]
- Finch JT, Klug A. The structure of viruses of the papilloma-polyoma type: III. structure of rabbit papilloma virus. J. Mol. Biol. 1965;13:1–12. doi: 10.1016/s0022-2836(65)80075-4. [DOI] [PubMed] [Google Scholar]
- Fuller SD. The T = 4 envelope of Sindbis virus is organized by interactions with a complementary T = 3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- Fuller SD, Butcher SJ, Cheng RH, Baker TS. Three-dimensional reconstruction of icosahedral particles – the uncommon line. J. Struct. Biol. 1996;116:48–55. doi: 10.1006/jsbi.1996.0009. [DOI] [PubMed] [Google Scholar]
- Griffith JP, Griffith DL, Rayment I, Murakami WT, Caspar DLD. Inside polyomavirus at 25-Å resolution. Nature. 1992;355:652–654. doi: 10.1038/355652a0. [DOI] [PubMed] [Google Scholar]
- Hagensee ME, Yaegashi N, Galloway DA. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J. Virol. 1993;67:315–322. doi: 10.1128/jvi.67.1.315-322.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagensee ME, Olson NH, Baker TS, Galloway DA. Three-dimensional structure of vaccinia virus-produced human papillomavirus type 1 capsids. J. Virol. 1994;68:4503–4505. doi: 10.1128/jvi.68.7.4503-4505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howley PM. Papillomavirinae and their replication. In: Fields BN, Knipe DM, editors. Virology. 2nd edit. Vol. 2. Raven Press; New York: 1990. pp. 1625–1650. [Google Scholar]
- Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl Acad. Sci. USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev NA, Klug A. The structure of viruses of the papilloma-polyoma type: V. tubular variants built of pentamers. J. Mol. Biol. 1969;40:155–171. doi: 10.1016/0022-2836(69)90465-3. [DOI] [PubMed] [Google Scholar]
- Klug A. Structure of viruses of the papillomapolyoma type: II. comments on other work. J. Mol. Biol. 1965;11:424–431. doi: 10.1016/s0022-2836(65)80067-5. [DOI] [PubMed] [Google Scholar]
- Klug A, Finch JT. Structure of viruses of the papilloma-polyoma type: I. human wart virus. J. Mol. Biol. 1965;11:403–423. doi: 10.1016/s0022-2836(65)80066-3. [DOI] [PubMed] [Google Scholar]
- Klug A, Finch JT. Structure of viruses of the papilloma-polyoma type: IV. analysis of tilting experiments in the electron microscope. J. Mol. Biol. 1968;31:1–12. doi: 10.1016/0022-2836(68)90050-8. [DOI] [PubMed] [Google Scholar]
- Kreider JW, Bartlett GL, Sharkey FE. Primary neoplastic transformation in vivo of xenogeneic skin grafts on nude mice. Cancer Res. 1979;39:273–276. [PubMed] [Google Scholar]
- Kreider JW, Howett MK, Wolfe SA, Bartlett GL, Zaino RJ, Sedlacek TV, Mortel R. Morphological transformation in vivo of human uterine cervix with papillomavirus from condylomata acuminata. Nature. 1985;317:639–641. doi: 10.1038/317639a0. [DOI] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. Structure of simian virus 40 at 3.8-Å resolution. Nature. 1991;354:278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Marzec CJ, Day LA. Pattern formation in icosahedral virus capsids: the papova viruses and Nudaurelia capensis [p98] virus. Biophys. J. 1993;65:2559–2577. doi: 10.1016/S0006-3495(93)81313-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD. Virus Taxonomy: Classification and Nomenclature of Viruses: Sixth Report of the International Committee on Taxonomy of Viruses. Springer-Verlag; Vienna: 1995. pp. 136–142. [Google Scholar]
- Olson C. Animal papillomas: historical perspectives. In: Salzman NP, Howley PM, editors. The Papovaviridae. Vol. 2. Plenum; New York: 1987. pp. 39–66. [Google Scholar]
- Olson NH, Baker TS. Magnification calibration and the determination of spherical virus diameters using cryo-microscopy. Ultramicroscopy. 1989;30:281–297. doi: 10.1016/0304-3991(89)90057-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Baker TS, Caspar DLD, Murakami WT. Polyoma virus capsid structure at 22.5Å resolution. Nature. 1982;295:110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salunke DM, Caspar DLD, Garcea RL. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986;46:895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- Salunke DM, Caspar DLD, Garcea RL. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys. J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp M, Volpers C, Müller M, Streeck RE. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J. Gen. Virol. 1995;76:2407–2412. doi: 10.1099/0022-1317-76-9-2407. [DOI] [PubMed] [Google Scholar]
- Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microscopy. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Shah KV. Polyomaviruses. In: Fields BN, Knipe DM, editors. Virology. 2nd edit. Vol. 2. Raven Press; New York: 1990. pp. 1609–1623. [Google Scholar]
- Shope RE, Hurst EW. Infectious papillomatosis of rabbits: with a note on the histopathology. J. Exp. Med. 1933;58:607–624. doi: 10.1084/jem.58.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir JA, Munshi S, Wang G, Baker TS, Johnson JE. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T, Yan Y, Benjamin TL, Harrison SC. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature. 1994;369:160–163. doi: 10.1038/369160a0. [DOI] [PubMed] [Google Scholar]
- Stoll R, Luo D, Kouwenhoven B, Hobom G, Müller H. Molecular and biological characteristics of avian polyomaviruses: isolates from different species of birds indicate that avian polyomaviruses form a distinct subgenus within the polyomavirus genus. J. Gen. Virol. 1993;74:229–237. doi: 10.1099/0022-1317-74-2-229. [DOI] [PubMed] [Google Scholar]
- Tarnai T, Gáspár Z, Szalai L. Pentagon packing models for “all-pentamer” virus structures. Biophys. J. 1995;69:612–618. doi: 10.1016/S0006-3495(95)79938-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ranst MA, Tachezy R, Burk RD. Human papillomavirus nucleotide sequences: what’s in stock? Papillomavirus Rep. 1994;5:65–75. [Google Scholar]
- Volpers C, Unckell F, Schirmacher P, Streeck RE, Sapp M. Binding and internalization of human papillomavirus type 33 virus-like particles by eukaryotic cells. J. Virol. 1995;69:3258–3264. doi: 10.1128/jvi.69.6.3258-3264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe Y, Sadakane H, Isono H. Connection between capsomeres in human papilloma virus. Virology. 1979;96:547–552. doi: 10.1016/0042-6822(79)90110-7. [DOI] [PubMed] [Google Scholar]