Abstract
Significance: Hypoxia is a hallmark of the tumor microenvironment and represents a major source of failure in cancer therapy. Recent Advances: Recent work has generated extensive evidence that microRNAs (miRNAs) are significant components of the adaptive response to low oxygen in tumors. Induction of specific miRNAs, collectively termed hypoxamiRs, has become an accepted feature of the hypoxic response in normal and transformed cells. Critical Issues: Overexpression of miR-210, the prototypical hypoxamiR, is detected in most solid tumors, and it has been linked to adverse prognosis in many tumor types. Several miR-210 target genes, including iron-sulfur (Fe-S) cluster scaffold protein (ISCU) and glycerol-3-phosphate dehydrogenase 1-like (GPD1L), have been correlated with prognosis in an inverse fashion to miR-210, suggesting that their down- regulation by miR-210 occurs in vivo and contributes to tumor growth. Additional miRNAs are modulated by decreased oxygen tension in a more tissue-specific fashion, adding another level of complexity over the classic hypoxia-regulated gene network. Future Directions: From a biological standpoint, hypoxamiRs are emerging modifiers of cancer cell response to the adaptive challenges of the microenvironment. From a clinical perspective, assessing the status of these miRNAs may contribute to a detailed understanding of hypoxia-induced mechanisms of resistance and/or to the fine-tuning of future hypoxia-modifying therapies. Antioxid. Redox Signal. 21, 1220–1238.
Introduction
Tumor microenvironment and hypoxia: implications for therapy
Tissue hypoxia arising from a rapidly growing tumor mass with inadequate/dysfunctional blood supply is a feature of virtually all solid cancers (22, 147). The adaptive response to low oxygen encompasses complex biochemical and cellular processes, such as energy metabolism, cell survival and proliferation, angiogenesis, adhesion, motility, and resistance to oxidative stress (140). It is currently widely accepted that hypoxia represents an independent adverse prognostic factor in many tumor types and contributes to the ultimate failure of most anticancer therapies (12, 132). Therefore, a complete understanding of cellular adaptation to oxygen deprivation is key for developing more efficient therapeutic strategies (175).
Cells respond to hypoxia, in part, via a transcriptional program that is orchestrated by an oxygen-monitoring machinery, centered on the hypoxia-inducible factors (HIFs) (168–170). When oxygen tension falls below a critical threshold, HIF-prolyl hydroxylase activity is inhibited, leading to HIFα stabilization and heterodimerization with the β subunit, followed by transcriptional activation of hypoxia-inducible genes (148). Among the few hundred HIF targets identified to date, many are mechanistically involved in cancer formation and progression. Multiple studies have also identified elevated levels of HIF-1α and HIF-2α or both in primary tumors and their metastases (154, 193). In addition to lack of oxygen, HIF up-regulation can be the result of oncogenic pathway activation, loss of tumor suppressor genes such as Von Hippel-Lindau (VHL) (106), or increased abundance in reactive oxygen species (ROS) (26), all of which are constant features of tumor biology. Elevation of HIF-1 and HIF-2 is associated with increased tumor growth in the majority of human tumors analyzed to date, including breast, head and neck squamous cell carcinoma, and ovarian cancers (97, 143).
From a clinical perspective, the importance of hypoxia signaling in tumor progression and prognosis has spurred multidisciplinary efforts to more effectively identify highly hypoxic tumors, as well as to identify patients who are most likely to benefit from hypoxia-modifying therapy. Such methods, including radiological and nuclear medicine markers, immunohistochemistry for HIF-1 or its targets, represent undeniable progress toward this end; however, they are also recognized as having significant limitations (6, 18, 166).
During the past few years, the “classic” protein coding hypoxia-regulated genes have been joined by specific microRNAs (miRNAs), thus adding a new layer of regulation in an already complex response (83). These miRNAs, collectively termed hypoxamiRs, and their role in cancer as biomarkers and potentially biological players in their own right, will be the subject of this perspective.
miRNAs: regulation and roles in cancer
miRNAs are short single-stranded oligoribonucleotides (∼22 nucleotides in length) that regulate gene expression by inhibiting mRNA translation or by triggering cleavage of the target mRNA (162). miRNAs are recognized as important regulators in physiological and pathological settings, including tumorigenesis (17). Genes encoding miRNAs are initially transcribed as longer primary transcripts (pri-miRNAs) (108), which are processed by the nuclear RNase III Drosha, leading to hairpin-shaped pre-miRNAs. Pre-miRNAs are subsequently exported to the cytoplasm and cleaved by the Dicer RNase III into a short miRNA duplex. One strand of this duplex is degraded, while the other is retained as mature miRNA and incorporated into the RNA-induced silencing complex (RISC) in complex with proteins from the Argonaute (AGO) family (144). The mature miRNA guides the RISC to recognize mRNAs based on sequence complementarity, in particular between the “seed region” and the 3′-untranslated regions (3′UTRs) of the target, which generally leads to translation inhibition and/or mRNA degradation (41, 42). Due to the relative shortness of the seed region, the 3′ UTR of a given mRNA may contain multiple miRNA recognition sequences. Conversely, any given miRNA can, at least theoretically, regulate a large number of mRNAs, often hundreds, thus posing significant challenges for the efforts to identify biologically relevant targets.
Deregulated miRNA expression has been demonstrated in virtually all neoplasms. Interestingly, different cancer types tend to exhibit specific miRNA signatures (20, 120), including cancers of colon (125), breast (82), brain (33), liver (127), and lung (181). While the mechanisms behind the specific shifts of profiles in tumors are still being dissected, recent data on miRNA responses to microenvironment stresses and oncogenic alterations have provided critical clues.
During the past 6 years, multiple reports have demonstrated that miRNAs respond to low oxygen challenge and contribute to the regulation of specific genes under hypoxia (21, 47, 54, 56, 77, 102). In the next section, we discuss the current knowledge about the involvement of miRNAs in the hypoxic response in tumors, and debate on potential opportunities for cancer diagnosis, prognosis, and treatment.
Hypoxia-regulated miRNAs: a new paradigm of cellular response to the tumor microenvironment
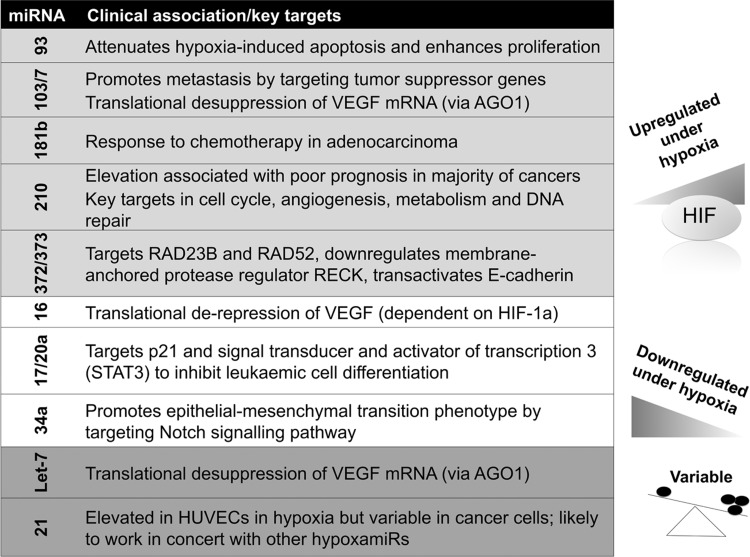
Taking advantage of novel miRNA profiling techniques, over the past 6 years, groups from diverse fields searched for hypoxia-regulated miRNAs in a variety of cellular contexts. A rather large set of miRNAs, including miR-210, 21, 23, 24, 26, 103/107, and 373, was found to be induced under hypoxic conditions (Fig. 1) (21, 27, 36, 47, 77, 102, 141). Although the miRNAs described earlier were reported in at least two publications, there have been significant differences between the lists reported. Moreover, a large number of miRNAs were found to be down-regulated in hypoxia (101). The complexity was further highlighted by “deep-sequencing” of endothelial cells exposed to hypoxia (164). More than 400 annotated microRNA/microRNA* species were identified with a broad abundance range.
FIG. 1.
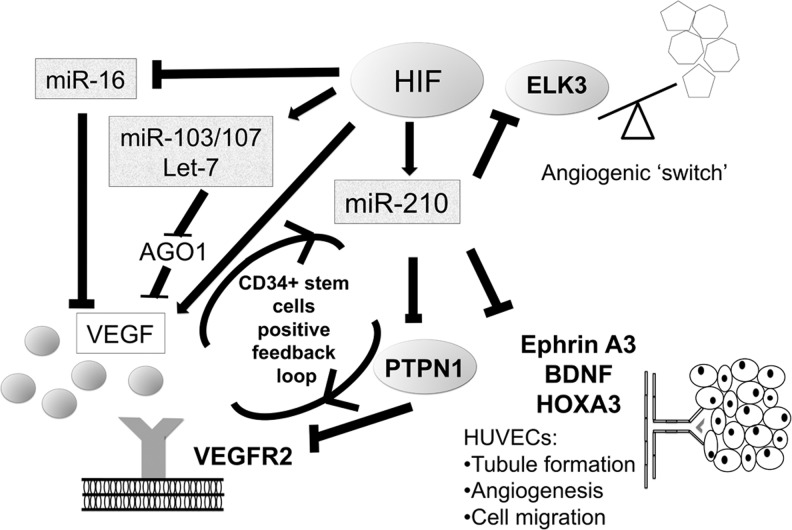
Key or experimentally validated miRNAs up- and down-regulated in hypoxia. A large number of hypoxamiRs have been described (>400 to date in various experimental set-ups)—this figure summarizes those discussed in the text. AGO1, argonaute 1; miRNA, microRNA; VEGF, vascular endothelial growth factor; HUVEC, human umbilical vein endothelial cell.
Apart from miR-210, which will be discussed in detail later in this review, there is very little overlap between hypoxia-induced miRNA profile between different cell lines and experimental set-ups, perhaps due to protocol and intrinsic cell line differences (69, 76). With the caveat of differences in the technologies employed by the different groups, the variability of hypoxamiR responses suggests a tissue-specific component of miRNA regulation under low oxygen. In turn, this may contribute to variations in the magnitude of gene expression changes in hypoxia, as well as to differences in cellular viability under these conditions.
Interestingly, while miR-21 is abundantly expressed in response to hypoxia in normal cells such as human umbilical vein endothelial cells (HUVECs), and it is frequently up-regulated in human tumors (82, 121, 156, 159, 165), its status as a hypoxamiR has been debated. Induction of miR-21 under hypoxia has been variably reported in normal and cancer cells (102, 142). When induced, most likely in an HIF-independent fashion, miR-21 should act as a pro-survival gene in the cancer microenvironment, especially in combination with other miRNAs. Polytarchou et al. found that only cancer cells expressing the protein kinase Akt2 had miR-21 induction during hypoxia. This was dependent on the binding of NF-κB, cAMP responsive element-binding protein, and CBP/p300 to the miR-21 promoter, in addition to the regional acetylation of histone H3K9, all of which were under the control of Akt2, and led to hypoxia resistance (134).
The miR-181 family, miR-372/373 and miR-93 have been reported in several studies (36, 100, 178), and they have intriguing clinical implications. For example, miR-181b was identified in a microarray analysis of hypoxia-regulated miRNAs in retinoblastoma cells, and administration of an miR-181b inhibitor suppressed proliferation (178). miR-181b is strongly associated with response to the 5-fluorouracil-based antimetabolite S-1 in colon cancer (79) and gemcitabine in pancreatic ductal adenocarcinoma (19). The induction of miR-373 by hypoxia has been reported in HeLa, MCF-7, and squamous cell carcinoma cell lines (36, 72). Similar to miR-210, the increase in miR-373 levels by hypoxia appears to be HIF-1α-dependent. Forced expression of miR-373 leads to a reduction in the RAD23B nucleotide excision repair protein, as well as in RAD52 (36); therefore, it may synergize with miR-210 in generating DNA damage and genetic instability in the tumor microenvironment.
Hypoxia and the HIF-signaling pathway play an important role in the regulation and sustenance of cancer stem cells and the epithelial-mesenchymal transition (EMT) phenotype. Under hypoxic conditions, the tumor microenvironment generates and sustains major EMT-triggering pathways, such as transforming growth factor-β and Notch signaling pathways (87), and hypoxamiRs are likely to be involved. Mechanistically, this pathway(s) appears to be even more complex than the pathways involving hypoxia-inducible hypoxamiRs, as both positively and negatively regulated miRs have been assigned to it. For example, hypoxia-induced down-regulation of miR-34a has been shown to promote EMT by targeting the Notch signaling pathway in tubular epithelial cells (44).
Conversely, miR-373 has also been found to transactivate E-cadherin gene expression through pairing with complementary promoter sequences (133), although the net effect of this regulatory mechanism on epithelial differentiation specifically under low oxygen remains unclear.
Recent “deep-sequencing” data have confirmed that miR-103/107 are hypoxamiRs (105) which are strongly induced in vascular endothelial cells. These hypoxamiRs are induced by HIF-1α and target AGO1, which anchors the miRNA-induced silencing complex. Interestingly, hypoxamiR targeting of AGO1 resulted in the translational desuppression of vascular endothelial growth factor (VEGF) mRNA and increased angiogenesis (29). Finally, hypoxia-induced miR-103/107 targets tumor suppressors such as death-associated protein kinase and Kruppel-like factor 4 to promote metastasis of colorectal cancer (27). This may be a contributing arm for the well-recognized effect of hypoxia in promoting invasion and metastasis.
Similar to miR-34a, many other miRNA are down-regulated in hypoxia, releasing suppressed genes that may be critical for adaptation to low oxygen. For example, miR-16, a prototypical tumor suppressor miRNA in leukemia and lymphoma, is down-regulated by HIF-1α, and it contributes to overexpression of VEGF in anaplastic lymphoma kinase-positive anaplastic large-cell lymphomas (37). These findings are summarized in Figure 1. Several groups have found that hypoxia and/or HIF-1α down-regulates miR-17/20a and the miR-17-92 cluster; however, the role of c-myc in this process has been directly contradicted by two studies (71, 180), and the functional significance has been complicated by a study showing that miR-20a is up-regulated by HIF-1α (115). It is likely that a balance of miRNAs in vivo is critical for an overall response, which may be cell-type dependent.
While the majority of work to date has focused on HIF as a transcriptional regulator of hypoxamiRs, recent evidence also implicates other transcription factors. For example, the NFκB subunit p50 was shown to contribute to miR-210 up-regulation in hypoxic trophoblasts, suggesting a role for other factors under oxidative and other stresses (190).
Interaction between HIF and miRNA in tumors: feedback loops with potential implications
HIF-1α is itself regulated by multiple miRNAs, creating complex positive and negative feedback loops. Those described so far include the miR-17-92 cluster mentioned earlier (153) and miR-20b, which is induced under hypoxia, implying negative feedback loops to fine-tune the hypoxia response (23, 110); additional players in such feedback include miR-138 (185); and miR-519c (24), but to date, they remain restricted to one publication.
As discussed earlier, Puissegur et al. as well as Kelly et al. showed that high miR-210 levels participate in HIF-1α stabilization during hypoxia (91, 136), which propagates a feed-forward loop of HIF amplification, and may account for a significant proportion of the total HIF protein expressed at any given point in hypoxia. In contrast to this amplification loop, miR-155 induction contributes to an isoform-specific negative-feedback loop for the resolution of HIF-1α activity in cells exposed to prolonged hypoxia (14). MiR-107 induction, in contrast to miR-210, seems to repress the expression of HIF-1α (179); while miR-145 inhibits HIF-2α (188), both with the effect of decreasing tumor growth and angiogenesis.
Other regulatory mechanisms have been described that act on HIF regulators, rather than HIF itself. For example, FIH, the asparagyl hydroxylase inhibitor of HIF activity, is a target of miR-31. Overexpression of this latter miR leads to normoxic stabilization of HIF, for example, in squamous cell carcinoma (121). A novel mechanism of regulation of HIF has been described by Ghosh et al., who found miR-424 induced in hypoxic endothelial cells targeted cullin 2, a scaffolding protein that is critical to the assembly of the ubiquitin ligase system, thereby stabilizing HIF isoforms (55). Given the increasing number of HIF regulators being reported in the literature, it is highly anticipated that the number of miRNAs which can affect the HIF pathway, directly or indirectly, increases significantly.
miR-210: a mirror of HIF in vitro and in tumors
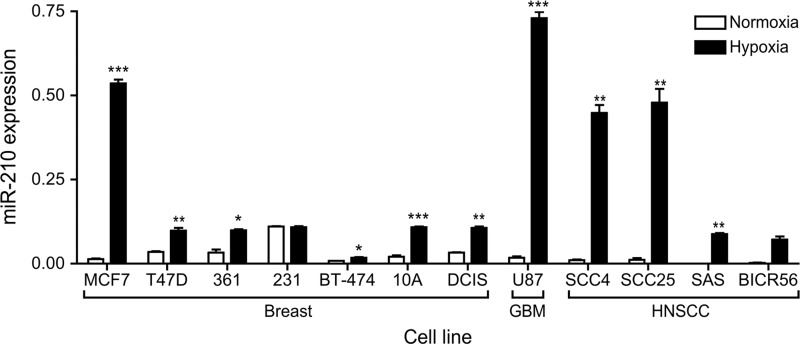
Among the hypoxiamiRs, miR-210 stands out as the only miRNA that all the studies to date agree on (78), being induced in a wide range of cell types in reponse to hypoxia (see Fig. 2 for a range of cancer cells in vitro). Indeed, its induction has been reported in all cells studied to date, except the PEO1 ovarian cancer cell line (and in our hands, this too had induction, albeit at a very low level). This is also drastically different from the case of classic protein-coding genes in which a plethora of mRNAs with diverse functions are induced by hypoxia, with a relatively good overlap between different cell types (39). While miR-210 seems to be a rather HIF-1-specific target (21, 36, 77, 92), HIF-2-dependent regulation of miR-210 has also been reported (191). As is the case for the classic genes, HIF-1 directly binds to a hypoxia-responsive element (HRE) on the proximal miR-210 promoter (77). When the miR-210 core promoter is compared across species, this HRE site is highly conserved, indicating the importance of hypoxia/HIF in regulating miR-210 expression during evolution (34).
FIG. 2.
Comparison of miR-210 levels in normoxia and induction in hypoxia across a range of cancer cell lines. 231—MDA-MB-231; 361—MDA-MB-361; 10A—MCF-10A; DCIS—MCF-10A-DCIS. ***<0.001; **<0.01; *<0.05; paired two tailed t-test. miR-210 levels measured by real-time-polymerase chain reaction after 24 h at 1% oxygen or paired normoxic control, three biological replicates. All cell lines maintained under standard culture conditions. DCIS, MCF-10A-DCIS.
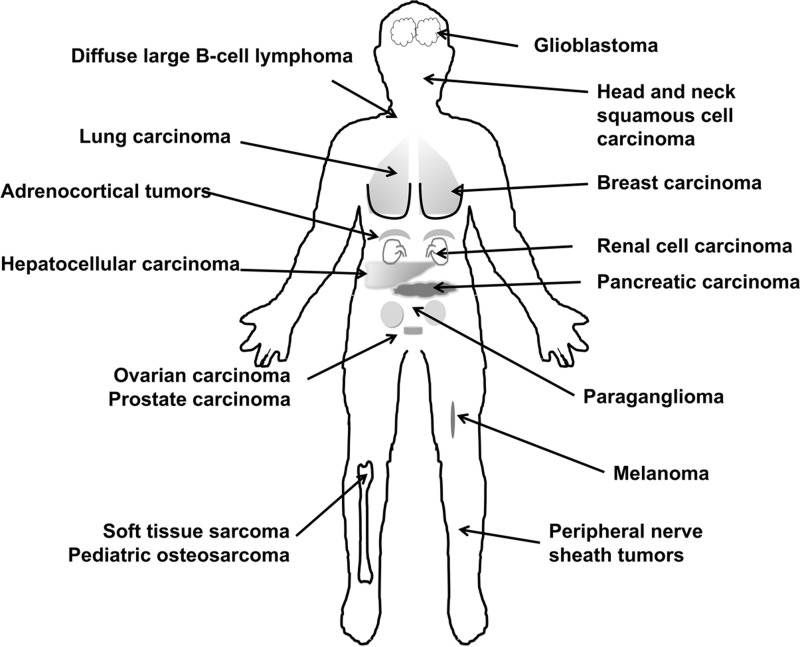
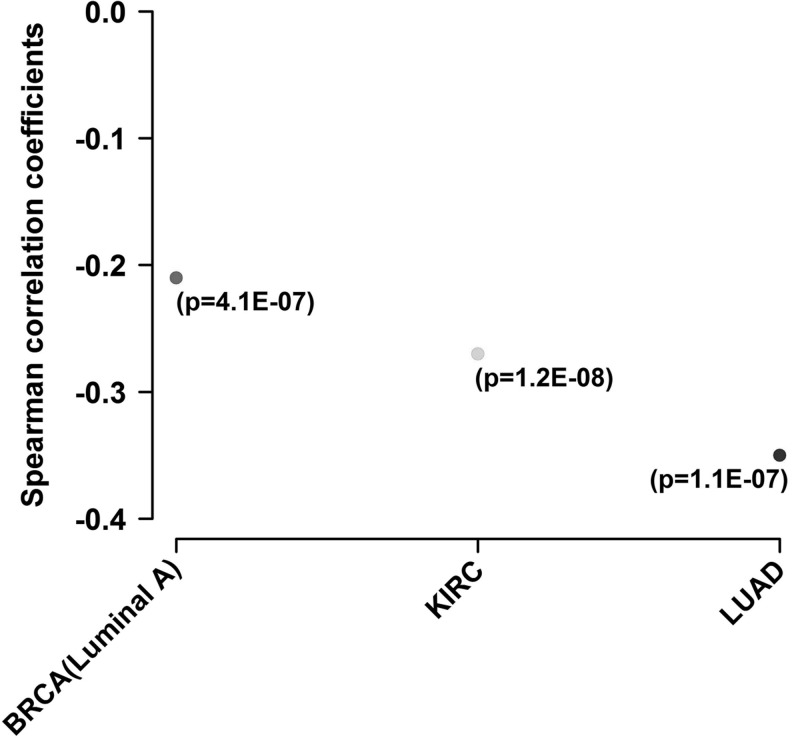
In cancer, in vivo, miR-210 level is correlated with a gene expression signature of hypoxia, termed hypoxia metagene (54, 77). Based on these data, miR-210 expression appears to be an accurate readout of HIF activity in vivo (40, 47, 77), thus opening the door for the use of miRNAs as markers of hypoxia in tumors (Fig. 3 summarizes cancers in which miR-210 is associated with clinicopathological variables; Table 1 includes details and references).
FIG. 3.
Summary of tumor types with which aberrant expression of miR-210 has been described. References and details are given in Table 1.
Table 1.
Association Between miR-210 and Clinicopathological Features in Human Cancer
| Cancer | Role | References |
|---|---|---|
| Adrenocortical tumours | Higher in adrenocortical carcinoma than benign adenomas | Tombol et al. (156a) |
| Breast | High level—poor prognosis in early breast cancer | Camps et al. (21) |
| Foekens et al. (51) | ||
| High level—poor prognosis in lymph-node negative: estrogen receptor (ER)+ve, ER−ve and triple negative subtypes | Rothe et al. (139), Hong et al. (74) | |
| Diffuse large B-cell lymphoma (DLBCL) | Elevated in serum of patients with DLBCL compared with normal | Lawrie et al. (107) |
| Glioblastoma | Elevation associated with poor prognosis | Qiu et al. (136a) |
| Head and neck | High level—poor prognosis | Gee et al. (54) |
| Hepatocellular carcinoma | Upregulated in hepatocellular carcinoma | Ying et al. (186a) |
| Lung | Elevated in primary small cell and adenocarcinoma compared with normal | Cho et al. (32a), Miko et al. (125a), Xing et al. (177a) |
| Detection in sputum differentiated normal from patients with lung squamous cell carcinoma | ||
| Melanoma | Elevated in tumor compared with melanocytic naevi but not associated with prognosis | Satzger et al. (141a) |
| Ovarian | Deleted in many epithelial ovarian cancers | Giannakakis et al. (56), Vaksman et al. (160a) |
| Elevated in effusions compared with primary tumor | ||
| Paraganglioma | Identification of HIF-1a/miR-210 axis independent of SDHD mutation—subgroup of head and neck paragangliomas | Merlo et al. (123a) |
| Pancreatic | Elevated—poor prognosis | Greither et al. (62), Wang et al. (171), Ho et al. (73) |
| Circulating miR-210 elevated in serum and plasma | ||
| Pediatric osteosarcoma | Elevatation associated with poor prognosis | Cai et al. (19) |
| Peripheral nerve sheath tumors | Elevated in malignant tumors compared with benign tumors (neurofibromas) | Presneau et al. (134b) |
| Prostate | Overexpressed in prostate cancer | Porkka et al. (134a) |
| Renal | Classified tumor subtypes | Fridman et al. (51a), Juan et al. (88), McCormick et al. (123) |
| Elevated in clear cell compared with normal | ||
| Associated with improved prognosis | ||
| Soft tissue sarcoma | Expression associated with poor survival and age of tumor onset | Greither et al. (63) |
ER, estrogen receptor; HIF, hypoxia-inducible factor; SDHD, succinate dehydrogenase complex subunit D.
miR-210 targets: a growing and diverse list
Identification of targets is an essential step toward a mechanistic understanding of hypoxamiRs in cancer. Most approaches include computational predictions using one or more of the online programs, including miRanda (8), TargetScan (113), and Pictar (98). These searches identify complementarity between 3′ UTR sequences of annotated coding genes and the “seed region” sequence of the miRNA (4). In general, the lists of candidates generated by individual programs exhibit a rather limited overlap, and none stands out as a perfectly accurate predictor of real targets. Interestingly, two widely employed algorithms, PicTar and TargetScan, predict relatively few targets for human miR-210, and, conversely, most of the experimentally validated targets are not predicted by any of these programs with a high score. It is also becoming increasingly apparent that “seed” binding is not always sufficient, as other features of the surrounding sequences can affect binding efficacy (112). To further complicate matters, in the case of miR-210, there is recent experimental evidence for a “seedless” target (49), which the prediction programs do not routinely address. Therefore, the addition of an experimental component to the bioinformatic search is crucial for the screening strategy.
In the case of hypoxamiR's targets, the approaches have been based on miRNA manipulation using mimics and antagomirs, in both hypoxia and normoxia, followed by expression profiling and comparisons with the results of computational predictions. This approach is well suited to identify targets that are regulated at the level of mRNA abundance (65, 114). However, since miRNAs frequently regulate the targets primarily by inhibiting protein translation (145), mRNA profiling will certainly miss many authentic miRNA targets. To address this limitation, AGO protein immunoprecipitation methods have been developed that capture the mRNAs recruited to the RISC complex which is enriched for a specific miRNA. Pulldown is followed by microarray or RNA-Seq, leading to targets that are regulated by both translational blockade and message degradation (90). Several groups have successfully pursued this approach in cells overexpressing miR-210 as a part of more integrative strategies in order to identify targets (48, 77). Notably, there were very few targets in common between these two studies, leading to the hypothesis that miR-210 regulates different genes in various cell types.
These results are not necessarily surprising, as comprehensive proteomic studies indicated that miRNAs act as rheostats by performing fine-scale adjustments to the output of hundreds of proteins (3, 145). Thus, only minor changes in protein and/or mRNA levels are expected for most real miRNA targets (e.g., 1.2–1.5-fold). Since microarray or RNA-Seq analyses carry a certain level of noise, these approaches are expected to miss a significant percentage of real targets. On the bright side, as the accuracy of “next-generation” RNA sequencing increases (and the cost decreases), an (almost) complete identification of physiologically relevant hypoxamiR targets becomes a highly realistic goal. Next, we discuss a selection of miR-210 targets that have been independently validated by multiple groups and debate their relevance for cancer biology.
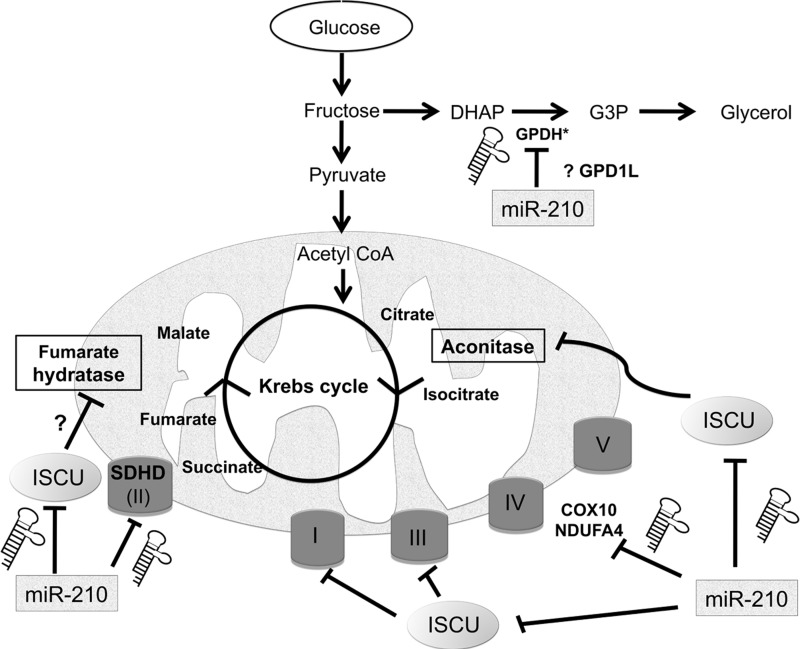
Mitochondrial Metabolism in Cancer: A “Favorite” Target of miR-210?
Under normoxic conditions, mitochondria represent the “cellular energy factories” by generating the majority of ATP through the oxidative phosphorylation pathway using oxygen as a final electron acceptor. When oxygen supply is limited, cells switch to glycolysis for ATP production (the Pasteur effect). HIF-1 plays a critical role in this switch, by up-regulating the expression of most glycolytic enzymes and actively down-regulating mitochondrial respiration and biogenesis (38, 187). Results from several groups have demonstrated that hypoxic induction of miR-210 significantly contributes to this metabolic shift by down-regulating the activity of the mitochondrial electron transport chain (ETC) (Fig. 4). A well-dissected branch of this mechanism is based on the targeting by miR-210 of the iron-sulfur (Fe-S) cluster scaffold protein, ISCU (25, 30, 48, 50). This has been shown in a variety of cell types, including colon cancer, breast cancer, and human pulmonary arterial endothelial cells. ISCU is at the center of an ancient machinery that catalyzes the assembly of iron-sulfur clusters which are critical for the function of aconitase (member of the tricarboxylic acid cycle), and of mitochondrial ETC complexes I, II, and III (157). The relationship between miR-210 and ISCU is robust enough to be detected in cancer samples, a rather rare case for miRNA-mRNA pairs. Indeed, ISCU levels are inversely correlated with miR-210 in multiple tumor data sets (50, 123). Moreover, high ISCU level is generally associated with good prognosis in multiple tumor types, which is the opposite of miR-210, further underlining the biological relevance of this miRNA-target pathway.
FIG. 4.
A model of how miR-210 may coordinate down-regulation of key targets in mitochondrial complexes, leading to a glycolytic phenotype. COX10, cytochrome c oxidase assembly protein; DHAP, dihydroxyacetone phosphate; G3P, glycerol 3-phosphate; GPDH, glycerol-3-phosphate dehydrogenase, GPD1L, glycerol-3-phosphate dehydrogenase-like 1; SDHD, succinate dehydrogenase complex subunit D; ISCU, iron-sulfur cluster scaffold homolog; NDUFA4, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4; miR-210—hsa-miR-210, Roman numerals refer to mitochondrial electron transport chain complexes. Several steps in Krebs cycle have been left off out for clarity. *Protein encoded by GPD1L contains a GPDH (NAD+) motif and shares 72% sequence identity with GPD1.
In addition to ISCU, which affects mitochondrial function indirectly, several integral components of the mitochondrial ETC have been found to be miR-210 targets: NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (NDUFA4) and succinate dehydrogenase complex, subunit D (SDHD), in lung cancer cells, (136), and cytochrome c oxidase assembly homolog 10 (COX10) in colon cancer cells (30). Interestingly, SDHD, a subunit of Complex II, is a well-documented tumor suppressor gene (5, 60), suggesting a pro-tumorigenic effect of miR-210.
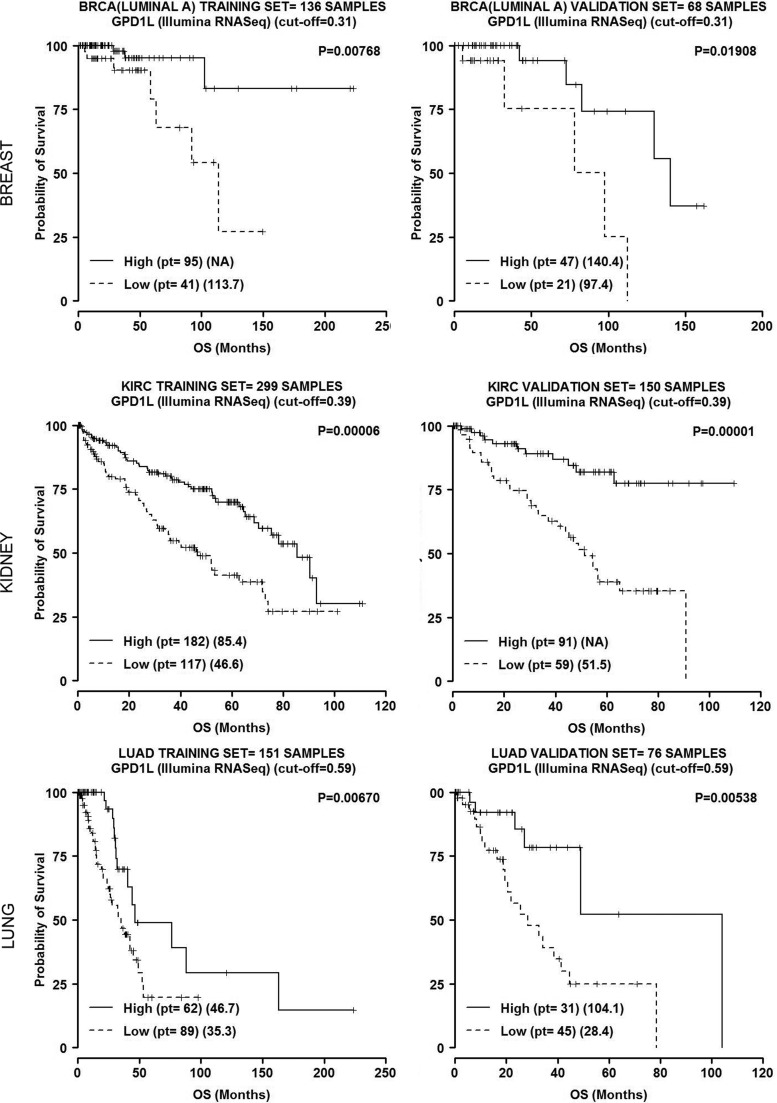
An intriguing metabolic target experimentally confirmed is glycerol-3-phosphate dehydrogenase 1-like (GPD1L) (48). GPD1L is highly homologous to glycerol-3-phosphate dehydrogenases that transfer electrons from the cytoplasmic NADH to the mitochondrial ETC (16), and it may be itself a key regulator of NAD+/NADH ratio (116). Work by Kelly et al. has proposed a feedback mechanism in HEK293 and HeLa cells based on GPD1L repression by miR-210, which inactivates HIF prolyl hydroxylase activity, leading to stabilization of HIF (91). In this model, indirectly supported by data from Puissegur et al. (136), miR-210 is both downstream and upstream of HIF-1. A preliminary analysis of GPD1L expression in vivo shows that miR-210 is inversely correlated with GPD1L (Fig. 5). Lower levels of expression are an adverse prognostic factor in breast, clear cell renal carcinoma (CCRC), and lung adenocarcinoma (Fig. 6). This result remains to be validated at the protein level but suggests that miR-210 regulation of GPD1L is important in vivo. If true, this result stands in contrast to a recent study showing that miR-210 was inversely correlated with ISCU, but was a favorable prognostic marker in CCRC (123). This would need to be further investigated, particularly the impact of VHL mutation on miR-210 regulation. Further details on the methods used here are available in Supplementary Data (Supplementary Data are available online at www.liebertpub.com/ars).
FIG. 5.
Spearman's rank-order correlation test shows a negative correlation between miR-210 and GPD1L in breast, CCRC, and lung adenocarcinoma (data from The Cancer Genome Atlas). Data publicly available from the Cancer Genome Atlas Project (TCGA; http://tcga-data.nci.nih.gov/) were analyzed for mRNA and miRNA expression and correlation coefficient (between miR-210 and GPD1L) was calculated. BRCA, breast cancer (luminal A subtype); CCRC, clear cell renal carcinoma; KIRC, kidney cancer; LUAD, lung adenocarcinoma. Further details in Supplementary Data.
FIG. 6.
GPD1L expression in breast, CCRC, and lung adenocarcinoma (data from The Cancer Genome Atlas). Data publicly available from the Cancer Genome Atlas Project (TCGA; http://tcga-data.nci.nih.gov/) were analyzed for mRNA, miRNA expression. Log-rank test was employed to determine the relationship between expression and overall survival and the Kaplan–Meyer method was used to generate survival curves. The entire population in training/validation cohorts was randomly split (2/3, 1/3) and for each miRNA, we checked for a relation with the survival as follows. In both cohorts, patients were divided into percentiles according to miRNA expression. Using the training set, we considered any cut-off between 25th and 75th to significantly split the samples into two groups and checked for statistical significance in the validation set. We then chose the cut-off value to optimally split the samples in both cohorts. Further details in Supplementary Data.
Since HIF is also induced in tumors as a result of overactive oncogenic signaling even in the absence of hypoxia (94, 194), it is tempting to speculate that elevated miR-210 may also contribute to the Warburg effect in these tumors. This may be achieved by contributing to HIF-1α stabilization to promote aerobic glycolysis, as well as by down-regulating mitochondrial metabolism. Co-ordinated down-regulation of multiple mitochondrial genes has been noted in hypoxic tumors (103), and miR-210 is frequently elevated in highly glycolytic tumors, including glioblastoma (122), and pancreatic cancer. The relative contribution of miR-210 to the Pasteur and/or Warburg effects remains to be elucidated.
Generation of mitochondrial ROS is a well-recognized consequence of electron leakage during electron transport (128). Increased ROS production has been reported in hypoxia, potentially as a result of ETC dysfunction (66). The effect of miR-210 on ROS levels has been the subject of recent debate. Several groups have reported that miR-210 increases oxidative stress, in part, by ISCU suppression (25, 50). In addition, there appears to be a positive feed-forward loop between ROS generation and miR-210, mediated though pathways such as NF-κB (95). Conversely, other groups have reported a protective role of miR-210 against ROS production in a non-cancer context, particularly in human pulmonary artery endothelial cells, raising several possibilities that include cell-specific effects, level of experimental manipulation, and depth and duration of hypoxia.
miR-210: Regulator of Tumor Angiogenesis?
Angiogenesis is a complex multistep process that usually occurs during embryonic development and rarely in the adult under normal conditions (146). Cancer growth is highly dependent on neo-vessel formation to establish nutrient and oxygen supplies for cell viability and proliferation, and tumor hypoxia is a well-recognized trigger of this process (117).
Multiple miRNAs are known or suspected to be involved in the various steps of the angiogenic response, as either positive or negative regulators (172, 177). miR-210 expression was found to correlate closely with VEGF expression, hypoxia, and angiogenesis in breast cancer patients (51). Functionally, transduction of miR-210 in HUVECs using miRNA mimics stimulates the formation of capillary-like structures, as well as VEGF-induced cell migration (47, 119), while inhibition had the opposite effect. Ephrin-A3 (EFNA3) (47, 48, 62, 77) was identified as a candidate mediator for these effects and validated as a target in HUVECs (47). However, regulation of EFNA3 appears to be more complex, as its transcription is usually induced by hypoxia (47), suggesting that a model based on simple miR-210-mediated repression under low oxygen does not hold true. As of early 2013, there is limited information about the roles of EFNA3 in cancer or tumor angiogenesis; therefore, it is premature to state whether or not an axis hypoxia-miR-210-EFNA3 plays a key role in these processes.
More information potentially relevant for cancer angiogenesis was generated in cardiovascular biology models. The tyrosine phosphatase Ptp1b was identified as miR-210 target in a mouse model of myocardial infarction (75). Its human homolog PTP1B is documented to negatively regulate activation of the VEGF receptor VEGFR2, as well as to stabilize cell–cell adhesions through reducing tyrosine phosphorylation of vascular endothelial cadherin (131). Thus, by limiting the expression of EFNA3 and PTP1B, both negative regulators of angiogenesis, miR-210 could positively regulate angiogenesis in hypoxic regions of tumors. Interestingly, a recent study also suggests a positive feedback between VEGF, the best-documented and pharmacologically relevant angiogenic factor, and miR-210 (1). If confirmed in cancer-relevant experimental systems, the studies described earlier may have implications for increasing the efficacy of anti-VEGF therapy, for example by adding miR-210 inhibitors. The major caveat regarding a link between miR-210 and tumor angiogenesis is that a direct correlation between miR-210 expression and tumor angiogenesis (for example, by quantification of microvessel density in miR-210-defective or overexpressing tumors) is still missing. Some of these emerging connections are outlined in Figure 7.
FIG. 7.
A model of ways in which miR-210 and other miRNAs regulated by HIF may regulate angiogenesis. BDNF, brain-derived neurotrophic factor; ELK3, ETS-domain protein (SRF accessory protein 2); HIF, hypoxia inducible factor; HOXA3, homeobox A3; PTPN1 (PTP1B), protein tyrosine phosphatase, non-receptor type 1; VEGF, vascular endothelial growth factor; VEGFR2, vascular endothelial growth factor receptor 2.
miR-210 and the Response to DNA Damage
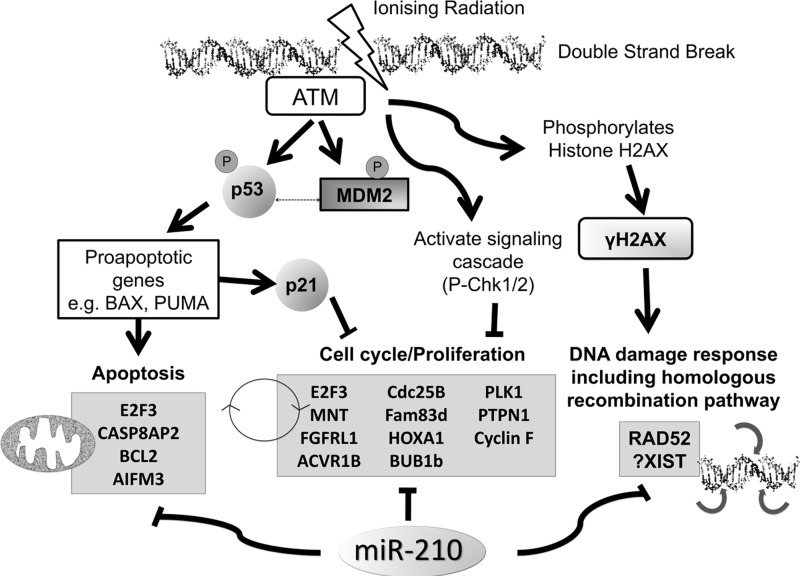
Genome integrity is challenged by diverse stresses, including mutagens, ROS, ultraviolet light, and chemo- or radio-therapeutic agents. Cellular responses to DNA damage involve a complex network of processes that detect and repair genomic lesions. miRNAs have been demonstrated to participate in these processes (104, 151, 167). Zhang et al. provided direct evidence that more than 20% of examined miRNAs are significantly induced on DNA damage (189). While not standing out as robustly induced by irradiation, at least based on the available data, miR-210, nevertheless, appears to have an impact on this complex process, as it down-regulates RAD52 in breast cancer cells (36, 48), a key component in the homologous recombination-mediated repair of double-strand breaks (7, 150). In addition to suppression of RAD51 in an HIF-dependent fashion (10) in hypoxic regions of tumors, suppression of RAD52 by miR-210 may provide an additional mechanism to help explain compromised homologous recombination repair in hypoxic cells (9). Consistent with this hypothesis, overexpression of miR-210 leads to double-strand DNA breaks in cultured diploid fibroblasts (46), although a possible involvement of RAD52 in this system was not investigated. Conversely, high levels of miR-210 may also be involved in lung cancer cells' radioresistance (64), suggesting that induction of this hypoxamiR both triggers error accumulation in the cancer cell genome and diminishes the effectiveness of radiotherapy. Obviously, the current knowledge is far from sufficient to allow us to speculate the relative contribution of its targets to such effects. Targets with possible relevance on this subject keep emerging. For example, a very recent study reported an intriguing role of a novel type of miR-210 target, which may be involved in response to radiotherapy. The first noncoding RNA candidate target of miR-210, X-inactive specific transcript (XIST) is a potent suppressor of hematological cancer in mice, as its inactivation leads to up-regulation of multiple oncogenes and down-regulation of tumor suppressors (186). This suggests that up-regulation of miR-210 may lead to widespread gene expression changes and even genomic instability, in ways not previously considered, and may be especially relevant in response to radiotherapy (see Fig. 8 for an overview of miR-210 relevance to genomic instability and response to radiotherapy).
FIG. 8.
A model of ways in which miR-210 may affect the response to radiotherapy, based on currently known target genes. ACVR1B, activin receptor 1B; AIFM3, apoptosis-inducing factor, mitochondrion-associated, 3; ATM, Ataxia telangiectasia mutated; BCL2, B-cell lymphoma 2; BHB1b, mitotic checkpoint serine/threonine-protein kinase BUB1 beta; CASP8AP2, caspase-8-associated protein 2; Cdc25B, cell division cycle 25 homolog B; E2F3, E2F transcription factor 3; Fam83d, family with sequence similarity 83, member D; FGFRL1, fibroblast growth factor receptor-like 1; H2AX, histone H2AX; HOXA1, homeobox A1; MDM2, mouse double minute 2 homolog; MNT, MAX-binding protein; PLK1, polo-like kinase 1; PTPN1, tyrosine-protein phosphatase non-receptor type 1.
miR-210 and Apoptosis: Boosting Cancer Cell Viability in Hypoxia?
Tumor microenvironmental stresses, including hypoxia and nutrient deprivation, are well-known triggers of cell death, both apoptosis and necrosis. Evasion from death responses is critical for tumor progression and a well-recognized hallmark of cancer (68). Thus, it is only fitting that miR-210 has been investigated in the context of survival responses, and, in particular, as a possible anti-apoptotic component of the hypoxic response. Generally, the available evidence suggests a predominantly anti-death role of miR-210 in a variety of cell types, with overexpression protecting cells from apoptosis, (75, 92, 102, 129, 176) and down-regulation of miR-210 during hypoxia promoting apoptosis (31, 47, 61, 102, 118, 182). These experiments have been performed in a variety of cells, including non cancerous (such as bone marrow-derived mesenchymal stem cells) and cancerous (such as breast and liver cancer). While significant gaps remain in our understanding of this process, multiple targets have been identified to help explain this effect: PTP1B (75), caspase-8-associated protein-2 (CASP8AP2) (92), and apoptosis-inducing factor, mitochondrion-associated, 3 (AIFM3) (182). However, the major caveat is that with the exception of CASP8AP2 (93), none of the other genes has been confirmed independently. In a recent report, while AIFM3 was found to be regulated by miR-210, its overexpression did not overcome the cytoprotective effects of the miRNA, suggesting that repression of other targets may be necessary (129). Moreover, despite the predominant evidence for an anti-apoptotic role, recent data suggested that miR-210 may also exhibit a pro-apoptotic function, at least in neuroblastoma cells, by targeting the anti-apoptotic gene B-cell lymphoma 2 (BCL2) (32).
In summary, while information supporting an miR-210-mediated blockade of apoptosis in hypoxia is accumulating for various cell types, it is still premature to state that this hypoxamiR represents a major protector against hypoxia-induced cell death in the tumor microenvironment.
miR-210: A Pleiotropic Regulator of the Cell Cycle
A common feature of extended exposure to hypoxia is down-regulation of various cell cycle genes, including cyclins and other positive regulators of cell cycle transition (67). However, this statement needs to be taken with particular caution, as especially in mild hypoxic conditions (5% or 10% oxygen), many cell types tend to proliferate better (99). Over the past few years, down-regulation of cell cycle by overexpressed miR-210 has been one of the more consistent themes about the biological effects of this hypoxamiR. Some of the cell cycle targets of miR-210 are also significant players in cancer biology.
One of the better-characterized cell cycle targets of miR-210 is E2F3, a promoter of G1/S transition (61, 109, 111). E2F3 was first reported and validated as an miR-210 target in ovarian cancer (56); however, the context reported by the authors was unique in the fledgling miR-210 field. Thus, the authors reported genomic loss of miR210 in ovarian cancer, and a resulting de-repression of cell cycle under hypoxic condition as a result of this event. However, this has not been found in other cell lines such as SKOV3 and A2780, which retain miR-210 expression and inducibility in hypoxia. Subsequent studies confirmed E2F3 as an miR-210 target (11, 48, 130), but the relative contribution of E2F3 to tumor cell cycle responses within the hypoxic microenvironment in tumors in vivo remains largely unknown. In addition to E2F3, fibroblast growth factor receptor-like 1 (FGFRL1) was also identified as an miR-210 target involved in cell cycle control in human esophageal cancer and derived cell lines (160), which was consistent with the earlier observation that FGFRL1 is robustly repressed by miR-210 (77). De-repression of FGFRL1 after miR-210 blockade accelerates cell cycle progression, while overexpression of miR-210 leads to cell cycle arrest in G1/G0 and G2/M phases (160). Again, as stated earlier with regard to E2F3, the relative contribution, if any, of this target for the viability of cancer cells in the hypoxic niches remains elusive. The effects of miR-210 on the cells cycle may, in fact, be significantly broader, to include a group of mitosis-related genes, such as Plk1, Cdc25B, Cyclin F, Bub1B, and Fam83D (70). Whether all these represent direct targets or more indirect responders downstream of the genes discussed earlier remains unclear.
Under some circumstances, miR-210 may promote cell cycle progression, for example, by down-regulating MAX-binding protein (MNT) (191), a member of the MYC/MAX/MAD network with a basic-Helix-Loop-Helix-zipper domain, and a well-characterized antagonist of c-MYC (80, 124). Since HIF-1 regulates cell proliferation under hypoxia, in part, by interacting with c-MYC (59), miR-210 may fine-tune cycle progression in hypoxic regions by fine-tuning the balance between major pathways that are involved in the response to the microenvironment. The net outcome (i.e., increased proliferation or cell cycle repression) may vary depending on the cell context, severity of hypoxia, and associated stresses.
miR-210: candidate cancer biomarker
A wealth of studies has firmly established that miRNAs are frequently dysregulated in human cancers (163), and that expression signatures can classify cancer subtypes (53). When used to classify poorly differentiated tumors, miRNA expression profiling outperformed mRNA expression profiling (120), pointing toward considerable biomarker potential. Expression of miR-210 has been consistently associated with poor clinical outcome in several solid tumor types (Table 1), including soft-tissue sarcoma, breast, head and neck, and pancreatic tumors (21, 51, 54, 62, 63, 74, 139).
A rather different scenario with regard to the impact of miR-210 expression seems to unfold in CCRC. This is the prototypical malignancy that is associated with a genetically up-regulated HIF pathway due to common inactivation of the VHL tumor suppressor (13, 135). Mutation and loss of heterozygosity of the VHL gene have been found in 57% and 98% of sporadic RCC cases, respectively (58). The VHL gene product functions as the adaptor subunit of the E3 ubiquitin ligase that targets hydroxylated HIFα for ubiquitination and degradation by the 26S proteasome (84, 85). Therefore, it is not surprising that miR-210 is particularly overexpressed in CCRC (88, 137, 174). However, contrary to the other tumors studied to date, expression of miR-210 in CCRC correlates with a favorable prognosis (123). This finding is certainly surprising but perhaps reflects the special biology of CCRC, given the role of VHL mutation or inactivation. The authors [McCormick et al. (123)] speculated that the association of miR-210 with good clinicopathological factors may be due to a shift to HIF-2 predominance or a further loss of cell differentiation coupled with ongoing mutations, but further work will be required to investigate.
Since hypoxic tumors display innate resistance to radiation and chemotherapy, a multitude of approaches have been tested to increase the treatment efficacy, with rather limited success. First, radiotherapy is often fractionated to take advantage of reoxygenation of surviving cells. Radiotherapy has also been combined with a hyperoxic gas and a vasoactive agent (86). Cytotoxins that specifically target hypoxic cells such as tirapazamine had shown promise in phase II trials (138). However, a phase III trial failed to show a benefit, perhaps because patients were not stratified on hypoxia status before treatment (138). Furthermore, effective biomarkers have yet to be identified that can reliably identify this subset of resistant tumors, enabling targeted treatment for patients at high risk, or “de-escalation” of therapy for patients at lower risk, sparing toxic side-effects on the quality of life.
The overall disappointing translational results using hypoxia-modifying therapies to date highlight the critical importance of novel validated markers of hypoxia for clinical practice, and of an improved understanding of the biology behind hypoxia-induced treatment resistance. Expression of miR-210 (in combination or not with the other hypoxamiRs discussed next) could help select patients at a high clinical risk, as it has been linked to aggressiveness in breast cancer, and, in particular, in the therapeutically challenging “triple negative” (estrogen receptor−ve/progesterone receptor−ve/human epidermal growth factor receptor 2−ve) subgroup. In a study of Japanese patients with triple negative breast cancer, patients whose breast cancers showed low miR-210 expression experienced significantly better disease-free and overall survival than those with high miR-210 expression (158). The impact of miR-210 on the efficacy of various types of therapy may, of course, vary. In particular, radiotherapy could benefit from an in-depth knowledge of miR-210 status. Thus, miR-210 overexpression has been shown to increase radioresistance in human lung cancer cell lines (64), while its down-regulation enhances radiosensitivity in hypoxic human hepatoma cells in vitro (182), and, most recently, in vivo (184). These findings have not yet been translated in humans in vivo but intriguingly, in a series of head and neck patients treated with post-operative radiotherapy, the expression of miR-210 was highly prognostic, suggesting that miR-210 may be a marker of radiotherapy resistance (54). However, whether miR-210 only serves as an “beacon” of tumor hypoxia, or actively promotes a more aggressive phenotype remains unclear (78).
There is an early indication that expression of multiple hypoxamiRs may be a viable alternative as a clinical biomarker of hypoxia as gene expression profiles. For example, a signature of hypoxia-related miRNAs derived by direct data mining of breast cancer data was shown to be a significant independent prognostic factor in breast cancer in multivariate analysis correcting for clinicopathological variables (15). This remains to be prospectively validated. The status of miR-210 expression in tumors may also help drive the choice of targeted therapy. Preliminary in vitro evidence was provided by Chen et al. (30), who reported that overexpressing miR-210 rendered cells significantly more susceptible to killing by 3-bromo-pyruvate, an inhibitor of the glycolytic pathway. Molecules of this class, such as 2-deoxyglucose or dichloroacetate, have been considered promising therapeutic agents; however, they are yet to fulfill their promise in clinical settings. Therefore, miR-210 may help identify subsets of patients who can benefit from such agents in the future.
There seems to be a general agreement that miRNAs are exceptionally stable and can be readily detected in the systematic circulation and other body fluids of healthy subjects and patients with malignant diseases (28, 57, 107, 126, 155, 173). It has been suggested that the high stability of miRNAs may be partially attributed to the exosomal miRNA packaging (161). Pilot studies assessing the use of circulating miRNAs as cancer biomarkers have attracted broad interest in the field and to date, at least 79 miRNAs have been reported as plasma or serum biomarker candidates for solid and hematologic tumors (2). miR-210 has been reported to be increased in the serum from patients with diffuse large B-cell lymphoma (107), CCRC (192), and pancreatic cancer (73, 171). Interestingly, hypoxia has been demonstrated as promoting the release of exosomes from cultured breast cancer cells (96); therefore, one can speculate that the elevated levels of circulating miR-210 may directly reflect the hypoxic state of tumor cells.
Circulating miR-210 levels have also been correlated with sensitivity to trastuzumab (a human epidermal growth factor receptor [EGFR] 2 monoclonal antibody), tumor presence, and lymph node metastases in breast cancer patients (89). This provides proof of concept that plasma miR-210 may also be used to monitor the response to anticancer therapies (35).
miR-210: a viable cancer therapeutic target?
The major and still unsolved dilemma regarding oncogenic miRs is whether or not they can be efficiently targeted in tumors. This question can certainly be formulated for hypoxamiRs, and miR-210 in particular. Recent development of anti-miRNA agents such as locked nucleic acids or peptide nucleic acids represents significant steps for therapeutic targeting of miRNAs in vivo (45, 52, 81, 105, 152). It is conceivable that inactivation of miRNAs involved in hypoxic adaptation, in combination with other anticancer agents, may be a viable strategy to target a tumor compartment that poses significant therapeutic challenges.
Hypoxia and miRNA machinery
Initial work suggested that hypoxia had a minimal effect on miRNA processing machinery (43), but more recent evidence suggests that hypoxia plays a regulatory role. Several genes such as Exportin 5 and AGO2 had an association with hypoxia in mRNA arrays (54). Hypoxia potentiates miRNA-mediated gene silencing through post-translational modification of AGO2 (176). Very recently, EGFR was shown to suppress the maturation of specific tumour-suppressor-like miRNAs in response to hypoxic stress through phosphorylation of AGO2. The association between EGFR and AGO2 was enhanced by hypoxia, leading to a reduction in the binding of Dicer to AGO2 and the inhibition of miRNA processing from precursor miRNAs to mature miRNAs (149).
Concluding Remarks and Cautionary Note
As a concluding remark of the plethora of studies which have investigated miRNAs in hypoxic cancer cells, one can state without any reservation that one miRNA deserves the name hypoxamiR: miR-210. However, putting the “equivalent” sign between hypoxamiRs and miR-210 would likely mean missing out on miRNA responders that in a given tumor type may be even more important than miR-210. On the other hand, taking into account the complexity of HIF regulation, the existence of HIF-independent responses to hypoxia, as well as the identification of hypoxia-downregulated miRs, one runs the risk of expanding the hypoxamiR family up to a point where the notion becomes “over-diluted.” Many more debates in the field are likely to follow in our effort to define some boundaries for a rather fluid notion. Overall, the top hypoxamiRs discussed earlier are quite possibly modifiers of tumor response to the microenvironmental challenges. Some, in particular miR-210, when measured in tumors and in circulating blood, already show promise as biomarkers of treatment response. By understanding, and potentially modifying, the cellular adaptation to oxygen deprivation, hypoxamiRs offer the possibility of both measuring hypoxia and developing more efficient therapeutic strategies to overcome hypoxia-induced mechanisms of resistance.
Supplementary Material
Abbreviations Used
- 10A
MCF-10A
- 231
MDA-MB-231
- 3′ UTR
3′-untranslated region
- 361
MDA-MB-361
- ACVR1B
activin receptor 1B
- AGO
argonaute
- AIFM3
apoptosis-inducing factor, mitochondrion-associated, 3
- ATM
ataxia telangiectasia mutated
- BCL2
B-cell lymphoma 2
- BDNF
brain derived neurotrophic factor
- BHB1b
mitotic checkpoint serine/threonine-protein kinase BUB1 beta
- BRCA
breast cancer (luminal A subtype); breast invasive carcinoma
- CASP8AP2
caspase-8-associated protein-2
- CCRC
clear cell renal carcinoma
- Cdc25B
cell division cycle 25 homolog B
- COX10
cytochrome c oxidase assembly homolog 10
- DCIS
MCF-10A-DCIS
- DHAP
dihydroxyacetone phosphate
- EFNA3
ephrin-A3
- EGFR
epidermal growth factor receptor
- ELK3
ETS-domain protein (SRF accessory protein 2)
- EMT
epithelial-mesenchymal transition
- ER
estrogen receptor
- ETC
electron transport chain
- Fam83d
family with sequence similarity 83, member D
- FGFRL1
fibroblast growth factor receptor-like 1
- G3P
glycerol 3-phosphate
- GPD1L
glycerol-3-phosphate dehydrogenase 1-like
- GPDH
glycerol-3-phosphate dehydrogenase
- H2AX
histone H2AX
- HER2
human epidermal growth factor receptor 2
- HIF
hypoxia-inducible factor
- HOXA1
homeobox A1
- HOXA3
homeobox A3
- HRE
hypoxia responsive element
- HUVEC
human umbilical vein endothelial cell
- ISCU
iron-sulfur (Fe-S) cluster scaffold protein
- KIRC
kidney cancer
- LUAD
lung adenocarcinoma
- MDM2
mouse double minute 2 homolog
- miR-210
hsa-miR-210
- miRNA
microRNA
- MNT
MAX binding protein
- NDUFA4
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4
- PLK1
polo-like kinase 1
- PR
progesterone receptor
- PTPN1 (PTP1B)
protein tyrosine phosphatase, non-receptor type 1
- PTPN1
tyrosine-protein phosphatase non-receptor type 1
- RISC
RNA-induced silencing complex
- ROS
reactive oxygen species
- SDHD
succinate dehydrogenase complex, subunit D
- VEGF
vascular endothelial growth factor
- VEGFR2
vascular endothelial growth factor receptor 2
- VHL
Von Hippel-Lindau
- XIST
X-inactive specific transcript
Acknowledgments
H.E.G. received support from the Department of Radiation Oncology, Sydney Cancer Center, Royal Prince Alfred Hospital, Sydney. M.I. received support from the American Cancer Society, the NIH/NCI (1R01CA155332-01), and the American Cancer Society (RSG 09-244-01-TBG).
References
- 1.Alaiti MA, Ishikawa M, Masuda H, Simon DI, Jain MK, Asahara T, and Costa MA. Up-regulation of miR-210 by vascular endothelial growth factor in ex vivo expanded CD34+ cells enhances cell-mediated angiogenesis. J Cell Mol Med 16: 2413–2421, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, and Musolino C. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (Review). Int J Oncol 41: 1897–1912, 2012 [DOI] [PubMed] [Google Scholar]
- 3.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, and Bartel DP. The impact of microRNAs on protein output. Nature 455: 64–71, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, Bosch A, Mey Avd, Taschner PEM, Rubinstein WS, Myers EN, Richard CW, Cornelisse CJ, Devilee P, and Devlin B. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287: 848–851, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Beasley NJ, Leek R, Alam M, Turley H, Cox GJ, Gatter K, Millard P, Fuggle S, and Harris AL. Hypoxia-inducible factors HIF-1alpha and HIF-2alpha in head and neck cancer: relationship to tumor biology and treatment outcome in surgically resected patients. Cancer Res 62: 2493–2497, 2002 [PubMed] [Google Scholar]
- 7.Benson FE, Baumann P, and West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature 391: 401–404, 1998 [DOI] [PubMed] [Google Scholar]
- 8.Betel D, Wilson M, Gabow A, Marks DS, and Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–D153, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bindra R, Crosby M, and Glazer P. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev 26: 249–260, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, and Glazer PM. Alterations in DNA repair gene expression under hypoxia: elucidating the mechanisms of hypoxia-induced genetic instability. Ann N Y Acad Sci 1059: 184–195, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Biswas S, Roy S, Banerjee J, Hussain S-RA, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, and Sen CK. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci U S A 107: 6976–6981, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown NS. and Bicknell R. Hypoxia and oxidative stress in breast cancer. Oxidative stress: its effects on the growth, metastatic potential and response to therapy of breast cancer. Breast Cancer Res 3: 323–327, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brugarolas J. Renal-cell carcinoma—molecular pathways and therapies. N Engl J Med 356: 185–187, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, and Taylor CT. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol 31: 4087–4096, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, and Ragoussis J. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res 71: 5635–5645, 2011 [DOI] [PubMed] [Google Scholar]
- 16.Bunoust O, Devin A, Avéret N, Camougrand N, and Rigoulet M. Competition of electrons to enter the respiratory chain: a new regulatory mechanism of oxidative metabolism in Saccharomyces cerevisiae. J Biol Chem 280: 3407–3413, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Bushati N. and Cohen SM. microRNA functions. Annu Rev Cell Dev Biol 23: 175–205, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Busk M, Horsman MR, Jakobsen S, Bussink J, van der Kogel A, and Overgaard J. Cellular uptake of PET tracers of glucose metabolism and hypoxia and their linkage. Eur J Nucl Med Mol Imaging 35: 2294–2303, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Cai H, Lin L, Tang M, Wang Z. Prognostic evaluation of microRNA-210 expression in pediatric osteosarcoma. Med Oncol 30: 499, 2013 [DOI] [PubMed] [Google Scholar]
- 20.Calin GA. and Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, and Ragoussis J. hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14: 1340–1348, 2008 [DOI] [PubMed] [Google Scholar]
- 22.Cardenas-Navia LI, Mace D, Richardson RA, Wilson DF, Shan S, and Dewhirst MW. The pervasive presence of fluctuating oxygenation in tumors. Cancer Res 68: 5812–5819, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Cascio S, D'Andrea A, Ferla R, Surmacz E, Gulotta E, Amodeo V, Bazan V, Gebbia N, and Russo A. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol 224: 242–249, 2010 [DOI] [PubMed] [Google Scholar]
- 24.Cha ST, Chen PS, Johansson G, Chu CY, Wang MY, Jeng YM, Yu SL, Chen JS, Chang KJ, Jee SH, Tan CT, Lin MT, and Kuo ML. MicroRNA-519c suppresses hypoxia-inducible factor-1alpha expression and tumor angiogenesis. Cancer Res 70: 2675–2685, 2010 [DOI] [PubMed] [Google Scholar]
- 25.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, and Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10: 273–284, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, and Schumacker PT. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem 275: 25130–25138, 2000 [DOI] [PubMed] [Google Scholar]
- 27.Chen H-Y, Lin Y-M, Chung H-C, Lang Y-D, Lin C-J, Huang J, Wang W-C, Lin F-M, Chen Z, Huang H-D, Shyy JY-J, Liang J-T, and Chen R-H. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res 72: 3631–3641, 2012 [DOI] [PubMed] [Google Scholar]
- 28.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, and Zhang C-Y. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006, 2008 [DOI] [PubMed] [Google Scholar]
- 29.Chen Z, Lai TC, Jan YH, Lin FM, Wang WC, Xiao H, Wang YT, Sun W, Cui X, Li YS, Fang T, Zhao H, Padmanabhan C, Sun R, Wang DL, Jin H, Chau GY, Huang HD, Hsiao M, and Shyy JY. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. J Clin Invest 123: 1057–1067, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Li Y, Zhang H, Huang P, and Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 29: 4362–4368, 2010 [DOI] [PubMed] [Google Scholar]
- 31.Cheng AM, Byrom MW, Shelton J, and Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 33: 1290–1297, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chio C-C, Lin J-W, Cheng H-A, Chiu W-T, Wang Y-H, Wang J-J, Hsing C-H, and Chen R-M. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch Toxicol [Epub ahead of print]; DOI: 10.1007/s00204-012-0965-5, 2012 [DOI] [PubMed] [Google Scholar]
- 32a.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer 45: 2197–206, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, and Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334: 1351–1358, 2005 [DOI] [PubMed] [Google Scholar]
- 34.Cicchillitti L, Di Stefano V, Isaia E, Crimaldi L, Fasanaro P, Ambrosino V, Antonini A, Capogrossi MC, Gaetano C, Piaggio G, and Martelli F. Hypoxia-inducible factor 1-a induces miR-210 in normoxic differentiating myoblasts. J Biol Chem 287: 44761–44771, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, and Calin GA. MicroRNAs in body fluids—the mix of hormones and biomarkers. Nat Rev Clin Oncol 8: 467–477, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crosby ME, Kulshreshtha R, Ivan M, and Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 69: 1221–1229, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dejean E, Renalier MH, Foisseau M, Agirre X, Joseph N, de Paiva GR, Al Saati T, Soulier J, Desjobert C, Lamant L, Prosper F, Felsher DW, Cavaille J, Prats H, Delsol G, Giuriato S, and Meggetto F. Hypoxia-microRNA-16 downregulation induces VEGF expression in anaplastic lymphoma kinase (ALK)-positive anaplastic large-cell lymphomas. Leukemia 25: 1882–1890, 2011 [DOI] [PubMed] [Google Scholar]
- 38.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer 8: 705–713, 2008 [DOI] [PubMed] [Google Scholar]
- 39.Denko NC, Fontana LA, Hudson KM, Sutphin PD, Raychaudhuri S, Altman R, and Giaccia AJ. Investigating hypoxic tumor physiology through gene expression patterns. Oncogene 22: 5907–5914, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Devlin C, Greco S, Martelli F, and Ivan M. miR-210: more than a silent player in hypoxia. IUBMB Life 63: 94–100, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Djuranovic S, Nahvi A, and Green R. A parsimonious model for gene regulation by miRNAs. Science 331: 550–553, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Djuranovic S, Nahvi A, and Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donker R, Mouillet J, Nelson D, and Sadovsky Y. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum Reprod 13: 273–279, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, Wang H, Huang C, and Sun S. Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the Notch signaling pathway in tubular epithelial cells. PLoS One 7: e30771, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fabbri E, Brognara E, Borgatti M, Lampronti I, Finotti A, Bianchi N, Sforza S, Tedeschi T, Manicardi A, Marchelli R, Corradini R, and Gambari R. miRNA therapeutics: delivery and biological activity of peptide nucleic acids targeting miRNAs. Epigenomics 3: 733–745, 2011 [DOI] [PubMed] [Google Scholar]
- 46.Faraonio R, Salerno P, Passaro F, Sedia C, Iaccio A, Bellelli R, Nappi TC, Comegna M, Romano S, Salvatore G, Santoro M, and Cimino F. A set of miRNAs participates in the cellular senescence program in human diploid fibroblasts. Cell Death Differ 19: 713–721, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, and Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand ephrin-A3. J Biol Chem 283: 15878–15883, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M, Capogrossi MC, and Martelli F. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem 284: 35134–35143, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fasanaro P, Romani S, Voellenkle C, Maimone B, Capogrossi MC, and Martelli F. ROD1 is a seedless target gene of hypoxia-induced miR-210. PLoS One 7: e44651, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, Vojnovic B, Pires das Neves R, Glazer P, Iborra F, Ivan M, Ragoussis J, and Harris AL. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One 5: e10345, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AWM, Klijn JGM, Wiemer EAC, and Martens JWM. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A 105: 13021–13026, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51a.Fridman E, Dotan Z, Barshack I, David MB, Dov A, Tabak S, Zion O, Benjamin S, Benjamin H, Kuker H, Avivi C, Rosenblatt K, Polak-Charcon S, Ramon J, Rosenfeld N, Spector Y. Accurate molecular classification of renal tumors using microRNA expression. J Mol Diagn 12: 687–96, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gambari R, Fabbri E, Borgatti M, Lampronti I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R, and Corradini R. Targeting microRNAs involved in human diseases: a novel approach for modification of gene expression and drug development. Biochem Pharmac 82: 1416–1429, 2011 [DOI] [PubMed] [Google Scholar]
- 53.Garzon R, Calin GA, and Croce CM. MicroRNAs in cancer. Annu Rev Med 60: 167–179, 2009 [DOI] [PubMed] [Google Scholar]
- 54.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, Homer J, Corbridge R, Cox G, West CML, Ragoussis J, and Harris AL. hsa-miR-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer 116: 2148–2158, 2010 [DOI] [PubMed] [Google Scholar]
- 55.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, and Ramakrishnan S. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest 120: 4141–4154, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O'Brien-Jenkins A, Katsaros D, Weber B, Simon C, Coukos G, and Zhang L. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther 7: 255–264, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, and Chajut A. Serum microRNAs are promising novel biomarkers. PLoS One 3: e3148, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 7: 85–90, 1994 [DOI] [PubMed] [Google Scholar]
- 59.Gordan JD, Thompson CB, and Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell 12: 108–113, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottlieb E. and Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer 5: 857–866, 2005 [DOI] [PubMed] [Google Scholar]
- 61.Gou D, Ramchandran R, Peng X, Yao L, Kang K, Sarkar J, Wang Z, Zhou G, and Raj JU. miR-210 has an antiapoptotic effect in pulmonary artery smooth muscle cells during hypoxia. Am J Physiol Lung Cell Mol Physiol 303: L682–L691, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greither T, Grochola LF, Udelnow A, Lautenschläger C, Würl P, and Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer 126: 73–80, 2010 [DOI] [PubMed] [Google Scholar]
- 63.Greither T, Würl P, Grochola L, Bond G, Bache M, Kappler M, Lautenschläger C, Holzhausen HJ, Wach S, Eckert AW, and Taubert H. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int J Cancer 130: 1230–1235, 2012 [DOI] [PubMed] [Google Scholar]
- 64.Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noel A, Pouyssegur J, Barbry P, Mazure NM, and Mari B. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis 4: e544, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo H, Ingolia NT, Weissman JS, and Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guzy RD. and Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol 91: 807–819, 2006 [DOI] [PubMed] [Google Scholar]
- 67.Hammer S, To KK, Yoo YG, Koshiji M, and Huang LE. Hypoxic suppression of the cell cycle gene CDC25A in tumor cells. Cell Cycle 6: 1919–1926, 2007 [DOI] [PubMed] [Google Scholar]
- 68.Hanahan D. and Weinberg Robert A. Hallmarks of cancer: the next generation. Cell 144: 646–674, 2011 [DOI] [PubMed] [Google Scholar]
- 69.Hazarika S, Farber CR, Dokun AO, Pitsillides AN, Wang T, Lye RJ, and Annex BH. MicroRNA-93 controls perfusion recovery following hind-limb ischemia by modulating expression of multiple genes in the cell cycle pathway. Circulation 127: 1818–1828, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He J, Wu J, Xu N, Xie W, Li M, Li J, Jiang Y, Yang BB, and Zhang Y. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res 41: 498–508, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He M, Wang QY, Yin QQ, Tang J, Lu Y, Zhou CX, Duan CW, Hong DL, Tanaka T, Chen GQ, and Zhao Q. HIF-1alpha downregulates miR-17/20a directly targeting p21 and STAT3: a role in myeloid leukemic cell differentiation. Cell Death Differ 20: 408–418, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hebert C, Norris K, Scheper MA, Nikitakis N, and Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer 6: 5, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le Q-T, and Koong AC. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol 3: 109–113, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hong L, Yang J, Han Y, Lu Q, Cao J, and Syed L. High expression of miR-210 predicts poor survival in patients with breast cancer: a meta-analysis. Gene 507: 135–138, 2012 [DOI] [PubMed] [Google Scholar]
- 75.Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, Robbins RC, and Wu JC. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation 122: S124–S131, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, and Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1: e116, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le Q-T, and Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 35: 856–867, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang X, Le Q-T, and Giaccia AJ. MiR-210- micromanager of the hypoxia pathway. Trends Mol Med 16: 230–237, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hummel R, Hussey DJ, and Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumour types. Eur J Cancer 46: 298–311, 2010 [DOI] [PubMed] [Google Scholar]
- 80.Hurlin PJ, Queva C, and Eisenman RN. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev 11: 44–58, 1997 [DOI] [PubMed] [Google Scholar]
- 81.Iorio MV. and Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4: 143–159, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, and Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070, 2005 [DOI] [PubMed] [Google Scholar]
- 83.Ivan M, Harris AL, Martelli F, and Kulshreshtha R. Hypoxia response and microRNAs: no longer two separate worlds. J Cell Mol Med 12: 1426–1431, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, and Kaelin WG., Jr.HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468, 2001 [DOI] [PubMed] [Google Scholar]
- 85.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, and Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292: 468–472, 2001 [DOI] [PubMed] [Google Scholar]
- 86.Janssens GO, Rademakers SE, Terhaard CH, Doornaert PA, Bijl HP, van den Ende P, Chin A, Marres HA, de Bree R, van der Kogel AJ, Hoogsteen IJ, Bussink J, Span PN, and Kaanders JH. Accelerated radiotherapy with carbogen and nicotinamide for laryngeal cancer: results of a phase III randomized trial. J Clin Oncol 30: 1777–1783, 2012 [DOI] [PubMed] [Google Scholar]
- 87.Jiang J, Tang YL, and Liang XH. EMT: a new vision of hypoxia promoting cancer progression. Cancer Biol Ther 11: 714–723, 2011 [DOI] [PubMed] [Google Scholar]
- 88.Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, Ganesan S, Bhanot G, and Liou LS. Identification of a microRNA panel for clear-cell kidney cancer. Urology 75: 835–841, 2010 [DOI] [PubMed] [Google Scholar]
- 89.Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC, Jr., Park ST, Pusztai L, and Calin GA. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 118: 2603–2614, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, and Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci U S A 104: 19291–19296, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kelly TJ, Souza AL, Clish CB, and Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol 31: 2696–2706, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim HW, Haider HK, Jiang S, and Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem 284: 33161–33168, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim HW, Mallick F, Durrani S, Ashraf M, Jiang S, and Haider KH. Concomitant activation of miR-107/PDCD10 and hypoxamir-210/Casp8ap2 and their role in cytoprotection during ischemic preconditioning of stem cells. Antioxid Redox Signal 17: 1053–1065, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim J-W. and Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res 66: 8927–8930, 2006 [DOI] [PubMed] [Google Scholar]
- 95.Kim JH, Park SG, Song SY, Kim JK, and Sung JH. Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. Cell Death Dis 4: e588, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.King HW, Michael MZ, and Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12: 421, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Koukourakis MI, Giatromanolaki A, Sivridis E, Simopoulos C, Turley H, Talks K, Gatter KC, and Harris AL. Hypoxia-inducible factor (HIF1A and HIF2A), angiogenesis, and chemoradiotherapy outcome of squamous cell head-and-neck cancer. Int J Radiat Oncol Biol Phys 53: 1192–1202, 2002 [DOI] [PubMed] [Google Scholar]
- 98.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, and Rajewsky N. Combinatorial microRNA target predictions. Nat Genet 37: 495–500, 2005 [DOI] [PubMed] [Google Scholar]
- 99.Krick S, Hanze J, Eul B, Savai R, Seay U, Grimminger F, Lohmeyer J, Klepetko W, Seeger W, and Rose F. Hypoxia-driven proliferation of human pulmonary artery fibroblasts: cross-talk between HIF-1alpha and an autocrine angiotensin system. FASEB J 19: 857–859, 2005 [DOI] [PubMed] [Google Scholar]
- 100.Kulshreshtha R, Davuluri RV, Calin GA, and Ivan M. A microRNA component of the hypoxic response. Cell Death Differ 15: 667–671, 2008 [DOI] [PubMed] [Google Scholar]
- 101.Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, and Ivan M. Regulation of microRNA expression: the hypoxic component. Cell Cycle 6: 1426–1431, 2007 [PubMed] [Google Scholar]
- 102.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu C-G, Croce CM, Negrini M, Calin GA, and Ivan M. A microRNA signature of hypoxia. Mol Cell Biol 27: 1859–1867, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar K, Wigfield S, Gee HE, Devlin CM, Singleton D, Li JL, Buffa F, Huffman M, Sinn AL, Silver J, Turley H, Leek R, Harris AL, and Ivan M. Dichloroacetate reverses the hypoxic adaptation to bevacizumab and enhances its antitumor effects in mouse xenografts. J Mol Med (Berl) 91: 749–758, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Landau DA. and Slack FJ. MicroRNAs in mutagenesis, genomic instability, and DNA repair. Semin Oncol 38: 743–751, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, and Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327: 198–201, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260: 1317–1320, 1993 [DOI] [PubMed] [Google Scholar]
- 107.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CSR, and Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 141: 672–675, 2008 [DOI] [PubMed] [Google Scholar]
- 108.Lee Y, Jeon K, Lee J-T, Kim S, and Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, and Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol 13: 7813–7825, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lei Z, Li B, Yang Z, Fang H, Zhang GM, Feng ZH, and Huang B. Regulation of HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the alteration of oxygen concentration. PLoS One 4: e7629, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, and Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev 12: 2120–2130, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lewis BP, Burge CB, and Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20, 2005 [DOI] [PubMed] [Google Scholar]
- 113.Lewis BP, Shih Ih, Jones-Rhoades MW, Bartel DP, and Burge CB. Prediction of mammalian microRNA targets. Cell 115: 787–798, 2003 [DOI] [PubMed] [Google Scholar]
- 114.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, and Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773, 2005 [DOI] [PubMed] [Google Scholar]
- 115.Lin SC, Wang CC, Wu MH, Yang SH, Li YH, and Tsai SJ. Hypoxia-induced microRNA-20a expression increases ERK phosphorylation and angiogenic gene expression in endometriotic stromal cells. J Clin Endocrinol Metab 97: E1515–E1523, 2012 [DOI] [PubMed] [Google Scholar]
- 116.Liu M, Liu H, and Dudley SC., Jr.Reactive oxygen species originating from mitochondria regulate the cardiac sodium channel. Circ Res 107: 967–974, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, Cox SR, Morita T, and Kourembanas S. Hypoxia regulates vascular endothelial growth factor gene expression in endothelial cells: identification of a 5′ enhancer. Circ Res 77: 638–643, 1995 [DOI] [PubMed] [Google Scholar]
- 118.Liu Y, Han Y, Zhang H, Nie L, Jiang Z, Fa P, Gui Y, and Cai Z. Synthetic miRNA-mowers targeting miR-183-96-182 cluster or miR-210 inhibit growth and migration and induce apoptosis in bladder cancer cells. PLoS One 7: e52280, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lou YL, Guo F, Liu F, Gao FL, Zhang PQ, Niu X, Guo SC, Yin JH, Wang Y, and Deng ZF. miR-210 activates notch signaling pathway in angiogenesis induced by cerebral ischemia. Mol Cell Biochem 370: 45–51, 2012 [DOI] [PubMed] [Google Scholar]
- 120.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, and Golub TR. MicroRNA expression profiles classify human cancers. Nature 435: 834–838, 2005 [DOI] [PubMed] [Google Scholar]
- 121.Lui W, Pourmand N, Patterson B, and Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 67: 6031–6043, 2007 [DOI] [PubMed] [Google Scholar]
- 122.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stühler K, Meyer HE, and Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 20: 539–550, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McCormick RI, Blick C, Ragoussis J, Schoedel J, Mole DR, Young AC, Selby PJ, Banks RE, and Harris AL. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br J Cancer 108: 1133–1142, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123a.Merlo A, de Quiros SB, Secades P, Zambrano I, Balbin M, Astudillo A, Scola B, Aristegui M, Suarez C, Chiara MD. Identification of a signaling axis HIF-1alpha/microRNA-210/ISCU independent of SDH mutation that defines a subgroup of head and neck paragangliomas. J Clin Endocrinol Metab 97: E2194–200, 2012 [DOI] [PubMed] [Google Scholar]
- 124.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Nigro CL, Messali S, Zollo M, Ledbetter DH, Brent R, Ballabio A, and Carrozzo R. Rox, a novel bHLHZip protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J 16: 2892–2906, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, and James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 1: 882–891, 2003 [PubMed] [Google Scholar]
- 125a.Miko E, Czimmerer Z, Csanky E, Boros G, Buslig J, Dezso B, Scholtz B. Differentially expressed microRNAs in small cell lung cancer. Exp Lung Res 35: 646–64, 2009 [DOI] [PubMed] [Google Scholar]
- 126.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, and Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105: 10513–10518, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, and Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 25: 2537–2545, 2006 [DOI] [PubMed] [Google Scholar]
- 128.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 417: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mutharasan RK, Nagpal V, Ichikawa Y, and Ardehali H. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am J Physiol Heart Circ Physiol 301: H1519–H1530, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M, and Moriyama M. Overexpression of miR-210, a downstream target of HIF1α, causes centrosome amplification in renal carcinoma cells. J Pathol 224: 280–288, 2011 [DOI] [PubMed] [Google Scholar]
- 131.Nakamura Y, Patrushev N, Inomata H, Mehta D, Urao N, Kim HW, Razvi M, Kini V, Mahadev K, Goldstein BJ, McKinney R, Fukai T, and Ushio-Fukai M. Role of protein tyrosine phosphatase 1B in vascular endothelial growth factor signaling and cell–cell adhesions in endothelial cells. Circ Res 102: 1182–1191, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nordsmark M, Bentzen SM, Rudat V, Brizel D, Lartigau E, Stadler P, Becker A, Adam M, Molls M, Dunst J, Terris DJ, and Overgaard J. Prognostic value of tumor oxygenation in 397 head and neck tumors after primary radiation therapy. An international multi-center study. Radiother Oncol 77: 18–24, 2005 [DOI] [PubMed] [Google Scholar]
- 133.Place RF, Li LC, Pookot D, Noonan EJ, and Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A 105: 1608–1613, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Polytarchou C, Iliopoulos D, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, and Tsichlis PN. Akt2 regulates all Akt isoforms and promotes resistance to hypoxia through induction of miR-21 upon oxygen deprivation. Cancer Res 71: 4720–4731, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134a.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res 67: 6130–5, 2007 [DOI] [PubMed] [Google Scholar]
- 134b.Presneau N, Eskandarpour M, Shemais T, Henderson S, Halai D, Tirabosco R, Flanagan AM. MicroRNA profiling of peripheral nerve sheath tumours identifies miR-29c as a tumour suppressor gene involved in tumour progression. Br J Cancer 108: 964–72, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Presti JC, Jr., Rao PH, Chen Q, Reuter VE, Li FP, Fair WR, and Jhanwar SC. Histopathological, cytogenetic, and molecular characterization of renal cortical tumors. Cancer Res 51: 1544–1552, 1991 [PubMed] [Google Scholar]
- 136.Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, Fourre S, Magnone V, Ricci JE, Pouyssegur J, Gounon P, Hofman P, Barbry P, and Mari B. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ 18: 465–478, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136a.Qiu S, Lin S, Hu D, Feng Y, Tan Y, Peng Y. Interactions of miR-323/miR-326/miR-329 and miR-130a/miR-155/miR-210 as prognostic indicators for clinical outcome of glioblastoma patients. J Transl Med 11: 10, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Redova M, Poprach A, Besse A, Iliev R, Nekvindova J, Lakomy R, Radova L, Svoboda M, Dolezel J, Vyzula R, and Slaby O. MiR-210 expression in tumor tissue and in vitro effects of its silencing in renal cell carcinoma. Tumour Biol 34: 481–491, 2013 [DOI] [PubMed] [Google Scholar]
- 138.Rischin D, Peters LJ, O'Sullivan B, Giralt J, Fisher R, Yuen K, Trotti A, Bernier J, Bourhis J, Ringash J, Henke M, and Kenny L. Tirapazamine, cisplatin, and radiation versus cisplatin and radiation for advanced squamous cell carcinoma of the head and neck (TROG 02.02, HeadSTART): a phase III trial of the Trans-Tasman Radiation Oncology Group. J Clin Oncol 28: 2989–2995, 2010 [DOI] [PubMed] [Google Scholar]
- 139.Rothe F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi NM, Majjaj S, Badran B, Fayyad-Kazan H, Desmedt C, Harris AL, Piccart M, and Sotiriou C. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One 6: e20980, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ruan K, Song G, and Ouyang G. Role of hypoxia in the hallmarks of human cancer. J Cell Biochem 107: 1053–1062, 2009 [DOI] [PubMed] [Google Scholar]
- 141.Sarkar J, Gou D, Turaka P, Viktorova E, Ramchandran R, and Raj JU. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am J Physiol Lung Cell Mol Physiol 299: L861–L871, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141a.Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer 126: 2553–62, 2010 [DOI] [PubMed] [Google Scholar]
- 142.Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, and Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of Fas ligand. J Biol Chem 285: 20281–20290, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schindl M, Schoppmann SF, Samonigg H, Hausmaninger H, Kwasny W, Gnant M, Jakesz R, Kubista E, Birner P, and Oberhuber G. Overexpression of hypoxia-inducible factor 1alpha is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin Cancer Res 8: 1831–1837, 2002 [PubMed] [Google Scholar]
- 144.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, and Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208, 2003 [DOI] [PubMed] [Google Scholar]
- 145.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, and Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63, 2008 [DOI] [PubMed] [Google Scholar]
- 146.Semenza GL. Angiogenesis ischemic and neoplastic disorders. Annu Rev Med 54: 17–28, 2003 [DOI] [PubMed] [Google Scholar]
- 147.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 29: 625–634, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell 148: 399–408, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, Hsu JL, Wu Y, Lam YC, James BP, Liu X, Liu CG, Patel DJ, and Hung MC. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497: 383–387, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shinohara A. and Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391: 404–407, 1998 [DOI] [PubMed] [Google Scholar]
- 151.Simone NL, Soule BP, Ly D, Saleh AD, Savage JE, Degraff W, Cook J, Harris CC, Gius D, and Mitchell JB. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS One 4: e6377, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, and Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol 18: 89–102, 2008 [DOI] [PubMed] [Google Scholar]
- 153.Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, and Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res 68: 5540–5545, 2008 [DOI] [PubMed] [Google Scholar]
- 154.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, and Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol 157: 411–421, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Taylor DD. and Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110: 13–21, 2008 [DOI] [PubMed] [Google Scholar]
- 156.Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, Livolsi VA, and Baloch ZW. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol 18: 163–173, 2007 [DOI] [PubMed] [Google Scholar]
- 156a.Tombol Z, Szabo PM, Molnar V, Wiener Z, Tolgyesi G, Horanyi J, Riesz P, Reismann P, Patocs A, Liko I, Gaillard RC, Falus A, Racz K, Igaz P. Integrative molecular bioinformatics study of human adrenocortical tumors: microRNA, tissue-specific target prediction, and pathway analysis. Endocr Relat Cancer 16: 895–906, 2009 [DOI] [PubMed] [Google Scholar]
- 157.Tong WH. and Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab 3: 199–210, 2006 [DOI] [PubMed] [Google Scholar]
- 158.Toyama T, Kondo N, Endo Y, Sugiura H, Yoshimoto N, Iwasa M, Takahashi S, Fujii Y, and Yamashita H. High expression of microRNA-210 is an independent factor indicating a poor prognosis in Japanese triple-negative breast cancer patients. Jpn J Clin Oncol 42: 256–263, 2012 [DOI] [PubMed] [Google Scholar]
- 159.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O'Brien C, and Rose B. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun 358: 12–17, 2007 [DOI] [PubMed] [Google Scholar]
- 160.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, and Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1). J Biol Chem 286: 420–428, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160a.Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. J Cell Mol Med 15: 1593–602, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, and Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9: 654–659, 2007 [DOI] [PubMed] [Google Scholar]
- 162.Valencia-Sanchez MA, Liu J, Hannon GJ, and Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524, 2006 [DOI] [PubMed] [Google Scholar]
- 163.Ventura A. and Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell 136: 586–591, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Voellenkle C, Rooij J, Guffanti A, Brini E, Fasanaro P, Isaia E, Croft L, David M, Capogrossi MC, Moles A, Felsani A, and Martelli F. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA 18: 472–484, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Volinia S, Calin G, Liu C, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt R, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris C, and Croce C. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103: 2257–2261, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vordermark D, Katzer A, Baier K, Kraft P, and Flentje M. Cell type-specific association of hypoxia-inducible factor-1 alpha (HIF-1 alpha) protein accumulation and radiobiologic tumor hypoxia. Int J Radiat Oncol Biol Phys 58: 1242–1250, 2004 [DOI] [PubMed] [Google Scholar]
- 167.Wan G, Mathur R, Hu X, Zhang X, and Lu X. miRNA response to DNA damage. Trends Biochem Sci 36: 478–484, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wang GL, Jiang BH, Rue EA, and Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 92: 5510–5514, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang GL. and Semenza GL. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A 90: 4304–4308, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang GL. and Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem 270: 1230–1237, 1995 [DOI] [PubMed] [Google Scholar]
- 171.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, and Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res 2: 807–813, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang S. and Olson EN. AngiomiRs—key regulators of angiogenesis. Curr Opin Genet Dev 19: 205–211, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Weber JA, Baxter DH, Zhang S, Huang DY, How Huang K, Jen Lee M, Galas DJ, and Wang K. The microRNA spectrum in 12 body fluids. Clin Chem 56: 1733–1741, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.White NM, Bao TT, Grigull J, Youssef YM, Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R, Pace KT, Bjarnason GA, and Yousef GM. miRNA profiling for clear cell renal cell carcinoma: biomarker discovery and identification of potential controls and consequences of miRNA dysregulation. J Urol 186: 1077–1083, 2011 [DOI] [PubMed] [Google Scholar]
- 175.Wilson WR. and Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer 11: 393–410, 2011 [DOI] [PubMed] [Google Scholar]
- 176.Wu C, So J, Davis-Dusenbery BN, Qi HH, Bloch DB, Shi Y, Lagna G, and Hata A. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol 31: 4760–4774, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wu F, Yang Z, and Li G. Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys Res Commun 386: 549–553, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177a.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol 23: 1157–64, 2010 [DOI] [PubMed] [Google Scholar]
- 178.Xu X, Jia R, Zhou Y, Song X, Wang J, Qian G, Ge S, and Fan X. Microarray-based analysis: identification of hypoxia-regulated microRNAs in retinoblastoma cells. Int J Oncol 38: 1385–1393, 2011 [DOI] [PubMed] [Google Scholar]
- 179.Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, Huso D, and Lowenstein CJ. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A 107: 6334–6339, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, and Sun SH. Repression of the miR-17–92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 28: 2719–2732, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, and Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9: 189–198, 2006 [DOI] [PubMed] [Google Scholar]
- 182.Yang W, Sun T, Cao J, Liu F, Tian Y, and Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res 318: 944–954, 2012 [DOI] [PubMed] [Google Scholar]
- 183.This reference has been deleted.
- 184.Yang W, Wei J, Sun T, and Liu F. Effects of knockdown of miR-210 in combination with ionizing radiation on human hepatoma xenograft in nude mice. Radiat Oncol 8: 102, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yeh YM, Chuang CM, Chao KC, and Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int J Cancer 133: 867–878, 2013 [DOI] [PubMed] [Google Scholar]
- 186.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, and Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152: 727–742, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186a.Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, Yao J, Ding J, Bao M, Ge C, Yao M, Li J, He X. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology 54: 2064–75, 2011 [DOI] [PubMed] [Google Scholar]
- 187.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, and Semenza GL. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11: 407–420, 2007 [DOI] [PubMed] [Google Scholar]
- 188.Zhang H, Pu J, Qi T, Qi M, Yang C, Li S, Huang K, Zheng L, and Tong Q. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 alpha. Oncogene [Epub ahead of print]; DOI: 10.1038/onc.2012.574, 2012 [DOI] [PubMed] [Google Scholar]
- 189.Zhang X, Wan G, Berger FG, He X, and Lu X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell 41: 371–383, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhang Y, Fei M, Xue G, Zhou Q, Jia Y, Li L, Xin H, and Sun S. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: new insights into molecular mechanisms for the disease. J Cell Mol Med 16: 249–259, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, Burchard J, Dai X, Chang AN, Diaz RL, Marszalek JR, Bartz SR, Carleton M, Cleary MA, Linsley PS, and Grandori C. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 8: 2756–2768, 2009 [DOI] [PubMed] [Google Scholar]
- 192.Zhao A, Li G, Peoc'h M, Genin C, and Gigante M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol 94: 115–120, 2013 [DOI] [PubMed] [Google Scholar]
- 193.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, and Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res 59: 5830–5835, 1999 [PubMed] [Google Scholar]
- 194.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, Gottschalk AR, Ryan HE, Johnson RS, Jefferson AB, Stokoe D, and Giaccia AJ. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev 14: 391–396, 2000 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.