Abstract
Most techniques used to study small molecules, such as pharmaceutical drugs or endogenous metabolites, employ tissue extracts which require the homogenization of the tissue of interest that could potentially cause changes in the metabolic pathways being studied1. Mass spectrometric imaging (MSI) is a powerful analytical tool that can provide spatial information of analytes within intact slices of biological tissue samples1-5. This technique has been used extensively to study various types of compounds including proteins, peptides, lipids, and small molecules such as endogenous metabolites. With matrix-assisted laser desorption/ionization (MALDI)-MSI, spatial distributions of multiple metabolites can be simultaneously detected. Herein, a method developed specifically for conducting untargeted metabolomics MSI experiments on legume roots and root nodules is presented which could reveal insights into the biological processes taking place. The method presented here shows a typical MSI workflow, from sample preparation to image acquisition, and focuses on the matrix application step, demonstrating several matrix application techniques that are useful for detecting small molecules. Once the MS images are generated, the analysis and identification of metabolites of interest is discussed and demonstrated. The standard workflow presented here can be easily modified for different tissue types, molecular species, and instrumentation.
Keywords: Basic Protocol, Issue 85, Mass Spectrometric Imaging, Imaging Mass Spectrometry, MALDI, TOF/TOF, Medicago truncatula, Metabolite, Small Molecule, Sublimation, Automatic Sprayer
Introduction
The growing field of metabolomics has many important biological applications including biomarker discovery, deciphering metabolic pathways in plants and other biological systems, and toxicology profiling4,6-10. A major technical challenge when studying biological systems is to study metabolomic pathways without disrupting them11. MALDI-MSI allows for direct analysis of intact tissues that enables sensitive detection of analytes in single organs12,13 and even single cells14,15.
Sample preparation is a crucial step in producing reproducible and reliable mass spectral images. The quality of the images greatly depends upon factors such as tissue embedding medium, slice thickness, MALDI matrix, and matrix application technique. For imaging applications, ideal section thickness is the width of one cell (8-20 µm depending on the sample type). MALDI requires deposition of an organic, crystalline matrix compound, typically a weak acid, on the sample to assist analyte ablation and ionization.16 Different matrices provide different signal intensities, interfering ions, and ionization efficiencies of different classes of compounds.
The matrix application technique also plays a role in the quality of mass spectral images and different techniques are appropriate for different classes of analytes. Three matrix application methods are presented in this protocol: airbrush, automatic sprayer, and sublimation. Airbrush matrix application has been widely used in MALDI imaging. The advantage of airbrush matrix application is that it is relatively fast and easy. However, the quality of the airbrush matrix application greatly depends on the skill of the user and tends to be less reproducible and cause diffusion of analytes, especially small molecules17. Automatic sprayer systems have similar mechanics to airbrush matrix application, but have been developed to remove the variability seen with manual airbrush application, making the spray more reproducible. This method can sometimes be more time-consuming than traditional airbrush matrix application. Both manual airbrush and automatic sprayer systems are solvent-based matrix application methods. Sublimation is a dry matrix application technique that is becoming more and more popular for mass spectral imaging of metabolites and small molecules because it reduces analyte diffusion; however, it lacks the solvent necessary to extract and observe higher mass compounds18.
Confident identification of metabolites typically requires accurate mass measurements to obtain putative identifications followed by tandem mass (MS/MS) experiments for validation, with MS/MS spectra being compared to standards, literature, or theoretical spectra. In this protocol high resolution (mass resolving power of 60,000 at m/z 400), liquid chromatography (LC)-MS is coupled to MALDI-MSI to obtain both spatial information and confident identifications of endogenous metabolites, using Medicago truncatula roots and root nodules as the biological system. MS/MS experiments can be performed directly on the tissue with MALDI-MSI or on tissue extracts with LC-MS and used for the validation of metabolite identifications.
This protocol provides a simple method to map endogenous metabolites in M. truncatula, which can be adapted and applied to MSI of small molecules in various tissue types and biological systems.
Protocol
1. Instrumentation
MALDI-TOF/TOF MSI. Use a mass spectrometer equipped with a MALDI source for analysis of small molecules (see Table of Materials/Equipment). Perform acquisitions in positive or negative ion mode depending on the analytes of interest. Specify a mass range of interest and collect 500 laser shots/spot at 50 µm intervals in both the x and y dimensions across the surface of the sample to generate ion images. The raster width and number of laser shots can be adjusted to obtain higher spatial resolution and maximum signal intensity respectively. Use DHB matrix peaks or internal standards applied to the slide or directly to the tissue to calibrate the mass spectra.
High Resolution LC-MS. (see Table of Materials/Equipment) Run sample extracts with LC-MS using either reversed phase (RP) LC with a C-18 column, or normal phase (NP) with a HILIC column depending on the analytes of interest. Use mobile phases and gradients as appropriate for the specific sample type. Perform acquisitions in positive or negative ion modes depending on the sample type.
2. Tissue Preparation
Trim the root nodule from the plant, leaving 3-4 mm of root attached to the nodule.
Immediately after dissection, use forceps to place the tissue in a cryostat cup and cover with gelatin (100 mg/ml in deionized water). It is essential for the tissue to be stuck to the bottom of the cup with no air bubbles.
Flash freeze the tissue by placing the cup in a dry ice/ethanol bath until the gelatin hardens and becomes opaque. Store samples at -80 °C until use.
Remove samples from the -80 °C freezer, cut away the plastic cryostat cup and trim away excess gelatin. Mount the embedded tissue to the cryostat chuck with a dime-sized amount of optimal cutting temperature (OCT) media, while not letting the OCT touch the tissue. Place in cryostat box until the OCT solidifies.
Allow the chuck and gelatin to equilibrate in the cryostat box (set to -20 or -25 °C) for approximately 15 min.
Use the cryostat (see Table of Materials/Equipment) to section tissue approximately the thickness of one cell (8-20 µm depending on the tissue type) and thaw mount each slice onto the ITO-coated glass by warming the back (non-ITO-coated side) of an ITO-coated glass slide on the back of your hand. Place the ITO-coated side of the warmed slide near the frozen tissue slice and allow the slice to stick onto the slide. Placing the sections close together on the slide will provide better alignment during MSI.
3. Matrix Application
- Airbrush Application of MALDI Matrix
- All airbrush procedures should be performed in a fume hood.
- Thoroughly clean the airbrush solution container and nozzle (see Table of Materials/Equipment) with methanol and fill the solution container with DHB matrix solution (150 mg/ml in 50% methanol/0.1% TFA v/v).
- Hold the airbrush approximately 35 cm from the sample and apply 10-15 coats of matrix on the surface of the slide with a duration of 10 sec spray and 30 sec drying time in between each coat.
- Thoroughly clean the airbrush again with methanol when finished to avoid clogging from the matrix solution.
- Automatic Sprayer Application of MALDI Matrix
- Follow the start-up instructions provided by the manufacturers of the automatic sprayer system.
- For MSI of metabolites in root nodules using 40 mg/ml (in 50% methanol/0.1% TFA v/v) DHB as the matrix, set the temperature to ~80 °C, velocity to 1,250 mm/min, flow rate to 50 µl/min, and number of passes to 24. For best coverage, it is recommended to rotate the nozzle 90° and/or offset the nozzle 1.5 mm between each pass. Start sprayer method. As a side note, the particular sprayer system used here heats the nozzle for faster evaporation of the solvent. As the solvent evaporates, the concentration of the matrix quickly increases. The matrix applied to the sample with the airbrush and the automatic sprayer have comparable concentrations.
- Follow the shut-down instructions provided by the manufacturers of the automatic sprayer system.
- Sublimation Application of MALDI Matrix
- Weigh out 300 mg DHB into the bottom of the sublimation chamber (see Table of Materials/Equipment).
- Stick the glass slide to the cold finger (top portion of the sublimation chamber) with the tissue sections facing down with double-sided, conductive tape. Cover the entire back of the slide with the double sided tape for even conductivity, producing even matrix deposition.
- Clamp the top and bottom halves of the sublimation chamber together with the C-clamp. Connect the vacuum and add ice and cold water to the top reservoir.
- Place sublimation chamber in a heating mantle that is at room temperature.
- Turn on the vacuum pump. Wait 15 min and turn on the heating mantle. The heating mantle should reach 120 °C over the course of 10 min.
- After 10 min, turn off the heat, close valve to vacuum (so the inside of the chamber remains under vacuum) and turn off the vacuum pump.
- Allow the chamber to come to room temperature, open the valve releasing the vacuum pressure and remove the sample. The size of the sublimation chamber will determine the amount of matrix sublimed to the glass slide. The larger sublimation chambers (the size of a 400 ml beaker) will use approximately 300 mg of DHB while the smaller chambers (the size of a 150 ml beaker) will use approximately 100 mg of DHB and will require cutting the glass slide so that it fits in the chamber.
4. Image Acquisition
Mark a + pattern on each corner of the sample with a WiteOut correction fluid pen to be used as "teach points". Place the glass slide into the MALDI slide adapter plate and take an optical image of the sample using a scanner.
Set up an image acquisition file using the software provided by the instrument company with a raster step size of 50 µm and a laser diameter equal to or smaller than the raster step size. On this particular instrument, the minimum laser setting gives a laser diameter of approximately 10 µm and small laser setting has a 40-50 µm diameter.
Load the optical image into the software and align the plate with the optical image.
Calibrate the instrument before beginning the acquisition using common matrix cluster ions, internal standards, or a calibration mix.
Specify the areas of tissue to be analyzed with MSI, including a spot of pure matrix on the slide to be used as a "blank".
Begin acquisition.
5. Image Generation
Open the imaging file in the commercially available software provided by the vendor and extract the ion images. Other open-source software is available for MSI data processing19.
6. Metabolite Identification
Select a specific m/z of interest from the mass spectrum using the vendor specific software (see Table of Materials/Equipment). An analyte can be distinguished from a matrix ion when an analyte peak is selected from the mass spectrum and ion images are generated specifically localized to the tissue section.
Generate a list of analytes of interest and perform MS/MS experiments. See Table 1 and Table 2 in Representative Results for sample lists of analytes.
Perform targeted LC-MS analysis on a high resolution mass spectrometer (or high resolution MALDI-MS if available) to obtain accurate mass measurements of the analytes of interest and also perform LC-MS/MS of the target analytes to obtain characteristic fragmentation patterns.
Perform database searching to determine putative identifications for the targeted analytes. Examples of metabolite databases include: METLIN, ChemSpider, PubChem, KEGG, and HMDB.
To confirm the putative identifications from accurate mass database searching, match the MS/MS from the targeted analytes to MS/MS spectra of standards, literature, and/or fragmentation prediction software.
Representative Results
An experimental overview of MSI is shown in Figure 1. At the very beginning of the experiment, sample preparation is a critical step. Nodules are trimmed from the plant root and embedded in gelatin. The tissue must be pressed flat against the cryostat cup, with no bubbles, while it is being frozen; this will ensure easier and proper alignment of the tissue while it is being sectioned. When the tissue is being sliced, it is important to cut the tissue at the proper thickness; too thin of sections will tear, ruining the tissue integrity, while too thick of sections will reduce the number of analytes extracted and detected from the tissue. Selection of matrix and application technique will determine the types of analytes detected. Using a combination of matrices could provide complementary results. Three matrix application techniques are presented in this work. The airbrush technique is fast, but typically not suitable for MSI of small molecules because of analyte diffusion. Automatic sprayer systems and sublimation provide smaller matrix crystals, better reproducibility, and less analyte diffusion. Figure 2 shows an optical image of a Medicago truncatula root nodule section. Figure 3 shows an optical image example of the matrix coverage and crystal sizes using airbrush, automatic sprayer, and sublimation respectively.
Conventional matrices, like DHB, produce many ions in the lower mass range (m/z 100-400)20. These matrix ions can interfere with the detection of metabolites in this range. Figure 4 shows MS spectra of just DHB matrix compared to root nodule tissue coated with DHB matrix. DHB matrix peaks are indicated in red and root nodule tissue covered with DHB is shown in blue. Novel matrices such as TiO2 nanoparticles21, 1,5-diaminonapthalene (DAN)22, 2,3,4,5-Tetrakis(3′,4′-dihydroxylphenyl)thiophene (DHPT)23, and 1,8-bis(dimethyl-amino) naphthalene (DMAN)24,25 have been reported that reduce the interference of matrix ions in the low mass range, and also enhance the detection of certain classes of metabolites5. Matrix peaks can be distinguished from real metabolites using the MS images. When a peak is clicked on, the ion image is extracted and displayed overlapping the optical image. Those peaks that generate images with distinct localization to the tissue, and are not present in the matrix only area imaged, are considered metabolites. Figure 5a shows several representative ion images of metabolites found in root nodule tissue, while Figure 5b shows examples of MS images corresponding to matrix related peaks. In Figure 5a the image shows distinct localization to the root nodule tissue and a lack of signal in the matrix only area that was imaged. In Figure 5b the signal shows little localization and is present over the entire tissue; the signal is also seen in the matrix only area that was imaged (square areas in the top right corners). Sample lists of analytes of interest from M. truncatula root nodule tissue is listed in Table 1 (positive mode) and Table 2 (negative mode)4.
The end goal of untargeted metabolomics experiments is to identify the compounds that were detected. When performing MSI on a medium resolution instrument (MRMS), such as a TOF/TOF, it is necessary to obtain accurate mass measurements in a different way. This can be done with a multifaceted MS approach in which MALDI-MSI results are compared to high resolution LC-MS data using tissue extracts. There are many possible tissue extraction protocols to choose from depending on the analytes of interest. High resolution MS (HRMS) can be performed in the positive or negative ionization modes and with normal or reversed phase LC depending upon the analytes of interest. Once an accurate mass is obtained with high resolution LC-MS, the resulting mass can be searched with several databases, listed previously, to obtain a putative identification. Next MS/MS data is collected and the characteristic fragmentation pattern of the analyte of interest can be compared to standards, literature spectra, or theoretical fragmentation patterns. Figure 6 shows an example of one of the metabolites detected with MSI and LC-MS. This metabolite was identified as heme based on the MS/MS spectrum collected with high resolution LC-MS. This MS/MS data was compared to the MS/MS spectra previously published by Shimma and Setou26. The two MS/MS spectra match, therefore the identity of m/z 616.2 was confidently assigned as heme based on the accurate mass database searching and MS/MS data compared to literature MS/MS data.
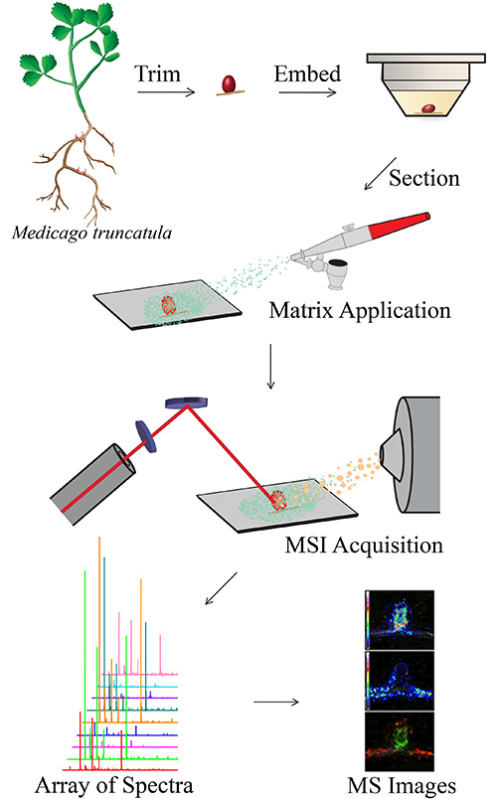 Figure 1. Overview of MSI workflow. Root nodules are trimmed from the plant, embedded in gelatin, sectioned with the cryostat and mounted onto an ITO-coated glass slide. Matrix is applied to the slide using one of three matrix application techniques. MSI acquisition is performed with a MALDI-TOF/TOF mass spectrometer and MS spectra are compiled into images with MSI software.
Figure 1. Overview of MSI workflow. Root nodules are trimmed from the plant, embedded in gelatin, sectioned with the cryostat and mounted onto an ITO-coated glass slide. Matrix is applied to the slide using one of three matrix application techniques. MSI acquisition is performed with a MALDI-TOF/TOF mass spectrometer and MS spectra are compiled into images with MSI software.
 Figure 2. Tissue sample optical image. Representative optical image of a Medicago truncatula root nodule tissue section.
Figure 2. Tissue sample optical image. Representative optical image of a Medicago truncatula root nodule tissue section.
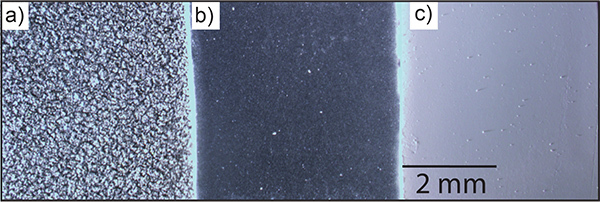 Figure 3. Example matrix depositions. Representative images showing the different consistencies of matrix deposition with a) airbrush, b) automatic sprayer, and c) sublimation techniques. The airbrush application method generates large and small crystals while the automatic sprayer method produces small evenly sized crystals. Sublimation produces one even layer of matrix. Please click here to view a larger version of this figure.
Figure 3. Example matrix depositions. Representative images showing the different consistencies of matrix deposition with a) airbrush, b) automatic sprayer, and c) sublimation techniques. The airbrush application method generates large and small crystals while the automatic sprayer method produces small evenly sized crystals. Sublimation produces one even layer of matrix. Please click here to view a larger version of this figure.
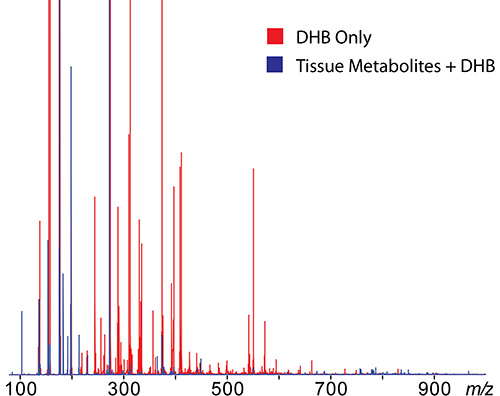 Figure 4. MS spectrum of plain DHB matrix vs. root nodule tissue covered with DHB. DHB matrix peaks are indicated in red and root nodule tissue covered with DHB is shown in blue. Matrix peaks can be distinguished from real metabolites using the MS images. When a peak is clicked on, the ion image is extracted and displayed overlapping the optical image. Those peaks that generate images with distinct localization to the tissue, and are not present in the matrix only area imaged, are considered metabolites. Please click here to view a larger version of this figure.
Figure 4. MS spectrum of plain DHB matrix vs. root nodule tissue covered with DHB. DHB matrix peaks are indicated in red and root nodule tissue covered with DHB is shown in blue. Matrix peaks can be distinguished from real metabolites using the MS images. When a peak is clicked on, the ion image is extracted and displayed overlapping the optical image. Those peaks that generate images with distinct localization to the tissue, and are not present in the matrix only area imaged, are considered metabolites. Please click here to view a larger version of this figure.
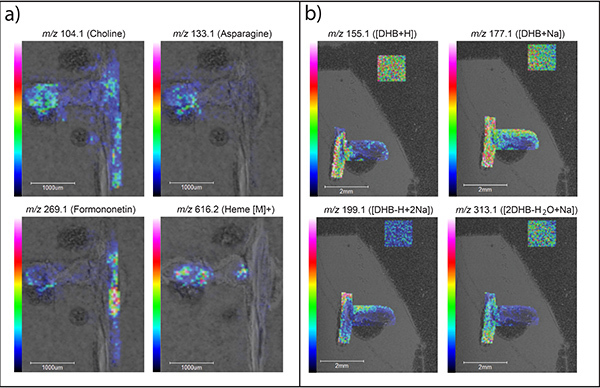 Figure 5. MS images of M. truncatula root nodules.a) Representative ion images of metabolites found in wild-type M. truncatula root nodules. b) Representative ion images of matrix species that would not be considered metabolites. Please click here to view a larger version of this figure.
Figure 5. MS images of M. truncatula root nodules.a) Representative ion images of metabolites found in wild-type M. truncatula root nodules. b) Representative ion images of matrix species that would not be considered metabolites. Please click here to view a larger version of this figure.
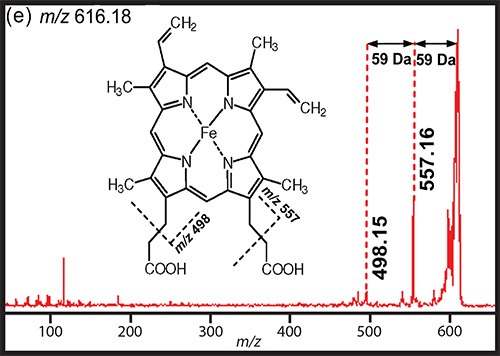 Figure 6. Example MS/MS data for metabolite identification. MS/MS spectrum of m/z 616.2, identified as heme [M+]. The chemical structure is shown and fragmentation structures are assigned. The experimental MS/MS data is compared to the MS/MS spectrum of heme reported in literature by Shimma and Setou26.
Figure 6. Example MS/MS data for metabolite identification. MS/MS spectrum of m/z 616.2, identified as heme [M+]. The chemical structure is shown and fragmentation structures are assigned. The experimental MS/MS data is compared to the MS/MS spectrum of heme reported in literature by Shimma and Setou26.
Table 1. Analytes of interest - positive mode. Sample list of analytes of interest from M. truncatula root nodule tissue in positive ion mode4.
| Name of metabolite | Theoretical [M+H]+ | MRMS Measured [M+H]+ | HRMS Measured [M+H]+ | Δm (mDa) |
| γ-aminobutyric acida | 104.0706 | 104.1 | 104.0706 | 0 |
| cholinea,* | 104.1070 | 104.1 | 104.1071 | 0.01 |
| proline | 116.0706 | 116.07 | 116.0706 | 0 |
| valine | 118.0863 | 118.09 | 118.0865 | 0.02 |
| leucinea | 132.1019 | 132.1 | 132.1022 | 0.03 |
| asparaginea | 133.0608 | 133.06 | 133.0602 | -0.06 |
| adeninea | 136.0618 | 136.07 | NA | NA |
| proling betainea | 144.1019 | 144.1 | 144.1024 | 0.05 |
| glutaminea | 147.0764 | 147.09 | 147.0768 | 0.04 |
| histidinea | 156.0768 | 156.06 | 156.0771 | 0.03 |
| argininea | 175.1190 | 175.13 | 175.1167 | -0.23 |
| sucrose+K | 381.0794 | 381.05 | 381.0791 | -0.03 |
| hemea,* | 616.1768 | 616.15 | NA | NA |
| NADa | 664.1164 | 664.1 | NA | NA |
| formononetina | 269.0808 | 269.08 | 269.0803 | 0.05 |
| chrysoseriol GlcAa | 477.1028 | 477.1 | 477.1008 | 0.2 |
| formononetin MalGlca | 517.1341 | 517.13 | 517.1337 | 0.04 |
| aformosin MalGlca | 547.1446 | 547.16 | 547.1427 | 0.19 |
* [M+] a MS/MS performed for identification.
Table 2. Analytes of interest - negative mode. Sample list of analytes of interest from M. truncatula root nodule tissue in negative ion mode4.
| Name of metabolite | Theoretical [M-H]- | MRMS Measured [M-H]- | HRMS Measured [M-H]- | Δm (mDa) |
| pyruvic acid | 87.0077 | 87 | NA | NA |
| alaninea | 88.0393 | 88.01 | 88.0374 | -0.19 |
| lactic acida | 89.0233 | 89.01 | 89.0296 | 0.63 |
| phosphoric acid | 96.9685 | 96.96 | 96.9625 | -0.6 |
| 2-ketobutyric acid | 101.0233 | 101.02 | 101.0191 | -0.42 |
| γ-aminobutyric acida | 102.055 | 102.04 | 102.0528 | -0.22 |
| serine | 104.0342 | 104.02 | 104.0228 | -1.14 |
| maleic/fumaric acida | 115.0026 | 115.03 | 115 | -0.26 |
| succinic acida | 117.0182 | 117.01 | 117.0125 | -0.57 |
| threonine | 118.0499 | 118.04 | 118.0456 | -0.43 |
| oxalacetic acid | 130.9975 | 131.03 | 130.991 | -0.65 |
| aspartic acida | 132.0291 | 132.03 | 132.0256 | -0.35 |
| malic acida | 133.0132 | 133.02 | 133.0221 | 0.89 |
| salicyclic acid | 137.0233 | 137.02 | 137.0349 | 1.16 |
| α-ketoglutaric acid | 145.0132 | 145.02 | 145.0063 | -0.69 |
| glutamic acida | 146.0448 | 146.01 | 146.0408 | -0.4 |
| pentose | 149.0445 | 149.04 | 149.0414 | -0.31 |
| aconitic acida | 173.0081 | 173.03 | 173.0057 | -0.24 |
| ascorbic acida | 175.0237 | 175.05 | 175.0341 | 1.04 |
| hexose | 179.055 | 179.05 | 179.0484 | -0.66 |
| citric/isocitric acida | 191.0186 | 191.02 | 191.0166 | -0.2 |
| palmitic acid | 255.2319 | 255.22 | 255.2246 | -0.73 |
| hexose-6-phosphatea | 259.0213 | 259.04 | 259.014 | -0.73 |
| stearic acid | 283.2632 | 283.26 | 283.2552 | -0.8 |
| sucrosea | 341.1078 | 341.07 | 341.0972 | -1.06 |
a MS/MS performed for identification.
Discussion
As discussed above, sample preparation is the most critical step in the MSI workflow. Embedding the tissue unevenly will cause sectioning to be difficult or not possible in some cases. The section size and adequate equilibration time are crucial to maintaining the tissue integrity and avoiding folding and tears. Selection of matrix and application technique will play a role in determining the types of analytes to be detected, the spatial resolution, and reproducibility of the results. Using a combination of matrices or application techniques could provide complementary results.
This method was designed specifically for untargeted MSI of endogenous metabolites in M. truncatula root nodule tissue, but can easily be adapted to other tissue types and biological questions. The recommended matrix application methods for small molecule MSI are sublimation and automatic sprayer. Increasing the amount of solvent deposited on the tissue, by adjusting the automatic sprayer method, will increase the extraction of analytes and allow for detection of higher mass compounds should one choose to perform MSI of lipids, peptides, etc. When using other types of tissue, the main adjustment will be the section thickness. Ideally the tissue should be sliced to the thickness of one cell; therefore thicker sections maybe appropriate for plant tissue and thinner sections appropriate for animal tissue. If folding or tearing occurs, typically a longer equilibration time is needed before sectioning or a thicker section size could be necessary. When performing LC-MS to obtain accurate masses, the tissue extraction protocol, mobile phase solvents and gradient, column stationary phase, and MS ionization mode can all be adjusted and optimized for the analytes of interest.
The main advantage of MALDI-MSI is its ability to provide not only mass information, but also spatial information for a given sample without the need for prior knowledge of the target analytes. Other imaging techniques require the use of derivatizations or tags5. As discussed previously, one limitation of this technique is the abundance of interfering matrix ion peaks. Novel matrices have been reported to address this limitation. Alternatively, secondary ion mass spectrometry (SIMS)-MSI is a matrix-free imaging option; however, it has less sensitivity than MALDI-MSI3. Another limitation of this technique is the lower mass resolution of MALDI-TOF/TOF instruments. Because of the low mass resolving power, it is necessary to also perform high resolution MS to obtain the accurate masses for metabolite identification, which means more experiments and more time. This problem could be solved by performing MALDI-MSI on a high resolution instrument platform such as the MALDI-Orbitrap. The final limitation of performing MSI experiments is the lack of software tools available for analysis of MSI data, although some recent advances in MSI software have been made27,28. Typically the MS spectrum for each imaging region must be manually analyzed and ion images extracted by hand. MSI experiments produce an abundance of data and can be incredibly time consuming to analyze. Overall, MALDI-MSI offers unique advantages for obtaining spatial information of many compounds simultaneously within a single experiment that can be extremely useful for the untargeted analysis of small molecules and other compounds with many biological applications.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
The authors would like to acknowledge Dr. Jean-Michel Ané in the Department of Agronomy at UW-Madison for providing Medicago truncatula samples. This work was supported in part by funding from the National Science Foundation (NSF) grant CHE-0957784, the University of Wisconsin Graduate School and the Wisconsin Alumni Research Foundation (WARF) and Romnes Faculty Research Fellowship program (to L.L.). E.G. acknowledges an NSF Graduate Research Fellowship (DGE-1256259).
References
- Seeley EH, Schwamborn K, Caprioli RM. Imaging of Intact Tissue Sections: Moving beyond the Microscope. J. Biol. Chem. 2011;286:25459–25466. doi: 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hove ERA, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. J. Chromatogr. A. 2010;1217:3946–3954. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- Lietz CB, Gemperline E, Li L. Qualitative and quantitative mass spectrometry imaging of drugs and metabolites. Adv. Drug Deliv. Rev. 2013;65:1074–1085. doi: 10.1016/j.addr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, et al. MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula-Sinorhizobium meliloti symbiosis. Plant J. 2013;75:130–145. doi: 10.1111/tpj.12191. [DOI] [PubMed] [Google Scholar]
- Ye H, Gemperline E, Li L. A vision for better health: mass spectrometry imaging for clinical diagnostics. Clin. Chim. Acta. 2013;420:11–22. doi: 10.1016/j.cca.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R. Metabolomics and Its Practical Value in Pharmaceutical Industry. Curr. Drug Metab. 2011;12:345–358. doi: 10.2174/138920011795202947. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, et al. A Novel Serum Metabolomics-Based Diagnostic Approach to Pancreatic Cancer. Cancer Epidem. Biomar. 2013;22:571–579. doi: 10.1158/1055-9965.EPI-12-1033. [DOI] [PubMed] [Google Scholar]
- West PR, Weir AM, Smith AM, Donley ELR, Cezar GG. Predicting human developmental toxicity of pharmaceuticals using human embryonic stem cells and metabolomics. Toxicol. Appl. Pharm. 2010;247:18–27. doi: 10.1016/j.taap.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Spegel P, et al. Time-resolved metabolomics analysis of beta-cells implicates the pentose phosphate pathway in the control of insulin release. Biochem. J. 2013;450:595–605. doi: 10.1042/BJ20121349. [DOI] [PubMed] [Google Scholar]
- Pendyala G, Want EJ, Webb W, Siuzdak G, Fox HS. Biomarkers for neuroAIDS: The widening scope of metabolomics. J. Neuroimmune. Pharm. 2007;2:72–80. doi: 10.1007/s11481-006-9041-3. [DOI] [PubMed] [Google Scholar]
- Prell J, Poole P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006;14:161–168. doi: 10.1016/j.tim.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Kutz KK, Schmidt JJ, Li LJ. In situ tissue analysis of neuropeptides by MALDI FTMS in-cell accumulation. Anal. Chem. 1021;76:5630–5640. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- Stemmler EA, et al. High-mass-resolution direct-tissue MALDI-FTMS reveals broad conservation of three neuropeptides (APSGFLGMRamide, GYRKPPFNGSIFamide and pQDLDHVFLRFamide) across members of seven decapod crustaean infraorders. Peptides. 2007;28:2104–2115. doi: 10.1016/j.peptides.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal. Chem. 1021;78:7267–7272. doi: 10.1021/ac0607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert S, Predel R. Mass spectrometric analysis of single identified neurons of an insect. Biochem. Bioph. Res. Co. 2005;327:640–645. doi: 10.1016/j.bbrc.2004.12.086. [DOI] [PubMed] [Google Scholar]
- Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using. MALDI-TOF MS. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- Baluya DL, Garrett TJ, Yost RA. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal. Chem. 2007;79:6862–6867. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- Hankin JA, Barkley RM, Murphy RC. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 2007;18:1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud G, Garrard KP, Barry JA, Muddiman DC. MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J. Am. Soc. Mass Spectrom. 2013;24:718–721. doi: 10.1007/s13361-013-0607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northen TR, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449 doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Shrivas K, Hayasaka T, Sugiura Y, Setou M. Method for simultaneous imaging of endogenous low molecular weight metabolites in mouse brain using TiO2 nanoparticles in nanoparticle-assisted laser desorption/ionization-imaging mass spectrometry. Anal. Chem. 2011;83:7283–7289. doi: 10.1021/ac201602s. [DOI] [PubMed] [Google Scholar]
- Thomas A, Charbonneau JL, Fournaise E, Chaurand P. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 2012;84:2048–2054. doi: 10.1021/ac2033547. [DOI] [PubMed] [Google Scholar]
- Chen S, et al. 2,3,4,5-Tetrakis(3',4'-dihydroxylphenyl)thiophene: a new matrix for the selective analysis of low molecular weight amines and direct determination of creatinine in urine by MALDI-TOF MS. Anal. Chem. 2012;84:10291–10297. doi: 10.1021/ac3021278. [DOI] [PubMed] [Google Scholar]
- Shroff R, Rulisek L, Doubsky J, Svatos A. Acid-base-driven matrix-assisted mass spectrometry for targeted metabolomics. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10092–10096. doi: 10.1073/pnas.0900914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Svatos A. Proton sponge: a novel and versatile MALDI matrix for the analysis of metabolites using mass spectrometry. Anal. Chem. 2009;81:7954–7959. doi: 10.1021/ac901048z. [DOI] [PubMed] [Google Scholar]
- Shimma S, Setou M. Mass Microscopy to Reveal Distinct Localization of Heme B (m/z 616) in Colon Cancer Liver Metastasis. J. Mass Spectrom. Soc. Jpn. 2007;55:145–148. [Google Scholar]
- Paschke C, et al. Mirion-A Software Package for Automatic Processing of Mass Spectrometric Images. J. Am. Soc. Mass Spectrom. 2013;24:1296–1306. doi: 10.1007/s13361-013-0667-0. [DOI] [PubMed] [Google Scholar]
- Parry RM, et al. omniSpect: an open MATLAB-based tool for visualization and analysis of matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry images. J. Am. Soc. Mass Spectrom. 2013;24:646–649. doi: 10.1007/s13361-012-0572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


