Abstract
Spinal muscular atrophy (SMA) is an autosomal recessive genetic disorder resulting in degeneration of α-motor neurons of the anterior horn and proximal muscle weakness. It is the leading cause of genetic mortality in children younger than 2 years. It affects ∼1 in 11,000 live births. In 95% of cases, SMA is caused by homozygous deletion of the SMN1 gene. In addition, all patients possess at least one copy of an almost identical gene called SMN2. A single point mutation in exon 7 of the SMN2 gene results in the production of low levels of full-length survival of motor neuron (SMN) protein at amounts insufficient to compensate for the loss of the SMN1 gene. Although no drug treatments are available for SMA, a number of drug discovery and development programs are ongoing, with several currently in clinical trials. This review describes the assays used to identify candidate drugs for SMA that modulate SMN2 gene expression by various means. Specifically, it discusses the use of high-throughput screening to identify candidate molecules from primary screens, as well as the technical aspects of a number of widely used secondary assays to assess SMN messenger ribonucleic acid (mRNA) and protein expression, localization, and function. Finally, it describes the process of iterative drug optimization utilized during preclinical SMA drug development to identify clinical candidates for testing in human clinical trials.
Introduction to Spinal Muscular Atrophy
Disease Pathophysiology
Spinal muscular atrophy (SMA) is a genetic condition with autosomal recessive inheritance that presents with proximal muscle weakness, caused by the dysfunction and loss of α-motor neurons of the anterior horn.1 The pan-ethnic disease incidence is ∼1 in 11,000 live births.2,3 In its most severe form, SMA is the leading cause of infant genetic death. However, the clinical presentation of the disease is quite variable. SMA patients are typically classified into four subgroups based on the age of onset and highest achieved motor milestones.4–6 SMA type I (Werdnig–Hoffman disease) is the most common form of the disease, with an incidence of about 60% of newly diagnosed patients. It is characterized by the appearance of disease symptoms before 6 months of age, with these patients never gaining the ability to sit. Infants with type I SMA characteristically die before the age of 2 years if not assisted with respiratory and nutritional support. SMA type II manifests between 6 and 18 months of age, with patients achieving the ability to sit but not walk. The incidence is about 30% of newly diagnosed patients. SMA type III (Kugelberg–Welander disease) patients first display symptoms in childhood. These patients achieve the ability to walk independently and typically have normal life expectancies. SMA type IV has the lowest incidence and is characterized by adult-onset of symptoms.
Genetics of SMA
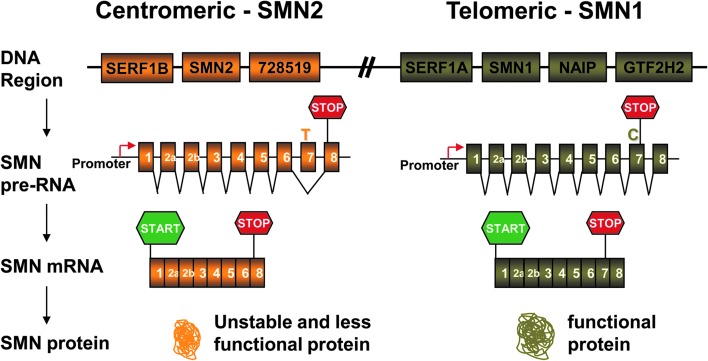
SMA is caused by low levels of survival of motor neuron (SMN) protein, resulting from mutation of the survival of motor neuron 1 (SMN1) gene.7 In fact, 95% of SMA patients have homozygous deletions of the SMN1 gene.7,8 Moreover, all patients possess at least one copy of a nearly identical gene called SMN2. The SMN2 gene predominately produces a messenger ribonucleic acid (mRNA) that is alternatively spliced with skipping of exon 7, due to a single point mutation within the exon.9,10 This single nucleotide change prevents the binding of the SR protein and splicing activator ASF/SF2, in addition to creating an inhibitory binding element for proteins such as hnRNPA1 and Sam68 that regulate SMN2 pre-mRNA splicing patterns.11–16 The resulting SMN transcript lacking exon 7 (called SMNΔ7) produces a truncated protein, which is unstable and cannot functionally compensate for the loss of the SMN1 gene (Fig. 1).17–19 Nevertheless, a small amount of the full-length mRNA and functional SMN protein are still produced by the SMN2 gene, and the observed clinical spectrum of disease severity is known to correlate with the SMN2 copy number.20,21 In fact, several nonsymptomatic adults with homozygous SMN1 mutations and four or five copies of the SMN2 gene have been identified.22,23 Therefore, enhancing the expression from the SMN2 gene has become an obvious therapeutic strategy for SMA.
Fig. 1.
Splicing of SMN1 and SMN2. The genomic regions of the SMN1 and SMN2 genes are drawn, as shown at www.ncbi.nlm.nih.gov/gene/. The major difference between the two SMN gene copies is the C (SMN1) or T (SMN2) nucleotide change at position 6 in exon 7 of the two genes. This single-nucleotide change prevents the binding of the SR protein and splicing activator ASF/SF2, in addition to creating an inhibitory binding element for proteins such as hnRNPA1 and Sam68. Because of this, SMN2 primarily produces messenger ribonucleic acid (mRNA) that excludes exon 7 and results in truncated and unstable SMN protein. However, SMN1 mostly produces mRNA that includes exon 7 and results in stable full-length SMN protein. Adapted with permission from the Families of SMA publication “The Genetics of Spinal Muscular Atrophy.” ASF/SF2, alternative splicing factor 1/pre-mRNA-splicing factor 2; SMN, survival of motor neuron.
Studies in SMA mouse models have also indicated that increased SMN2 copy number correlates with milder disease course. Mice have a single Smn gene.24 Homozygous loss of Smn results in preimplantation death of the embryo.25 This can be rescued by expressing two copies of a transgene containing the human SMN2 locus. These rescued transgenic mice display severe symptoms.26,27 The expression of eight copies of SMN2 fully rescues the animals.27 Disease severity can also be modified by transgenic expression of mutated versions of the SMN gene. For instance, two copies of SMN2 and an intronless SMN allele lacking exon 7 (Smn−/−; SMN2/SMN2; SMNΔ7/SMNΔ7) result in a mouse strain commonly referred to as SMNΔ7.28 SMNΔ7 mice display symptoms days after birth with a median survival time of about 14 days.28 It has been utilized as the primary mouse model for testing SMA drug candidates.29–37 Several other models, including those with milder phenotypes, are also currently in use for drug testing.38–42
SMN Protein Function
SMN is a ubiquitously expressed 38 kDa protein found in both the nucleus and the cytoplasm. It has a well-documented role in small nuclear ribonucleic particle (snRNP) assembly.43,44 As extensively reviewed, during snRNP formation, SMN functions as part of a protein complex containing Gemin proteins 2–8 and Unrip in all cell types and tissues. This protein complex promotes the assembly of Sm proteins with U small nuclear RNAs (snRNAs) into snRNPs, which function in the process of pre-mRNA splicing.45,46 Several studies have shown that snRNP assembly is reduced in tissues with lowered SMN levels.47,48 Moreover, the expression of SMN transgenes possessing missense mutations that restore snRNP assembly also prolong survival in mice with severe SMA.49,50
The mechanisms underlying selective vulnerability of motor neurons in SMA remain debated. Theories invoke either impaired splicing mechanisms resulting from defective snRNP assembly or specific novel functions for SMN protein in motor neurons.51 Currently, studies differ on how widespread mis-splicing is in SMA tissues.52,53 One theory proposes that only a limited number of genes are affected until the end stages of the disease. In addition, some findings have suggested that SMN deficiency results in a selective reduction of the U11 and U12 snRNPs of the minor spliceosome.54,55 The minor spliceosome is involved in processing the pre-mRNA of a small number of genes, some of which are known to be involved in motor neuronal function.56,57 The mis-splicing of these genes due to reduced levels of minor snRNPs may lead to motor neuron vulnerability in SMA. Recently, several genes, both with and without minor introns, have been shown to be specifically mis-spliced in SMN-deficient tissues, including Chondrolectin, Stasimon, and Neurexin2a.53,58,59 In addition, several researchers are using laser capture microdissection of motor neurons from SMA mice followed by next generation RNA sequencing analysis to search for additional gene expression changes. In Drosophila models of SMA, this technique did not identify significant alterations in genes processed by the minor spliceosome.60 In a recent study using mouse models of SMA, RNA sequencing results showed aberrant mRNA splicing in 348 genes in motor neurons at pre- and early symptomatic states of the disease.61 There was no evidence that the minor splicing pathway was selectively perturbed at early stages of SMA. Importantly, only about 3% of genes expressed in motor neurons were affected, indicating that SMN deficiency did not cause widespread transcriptome changes at early disease stages. Gene expression misregulation included complete skipping of the Z exons of Agrin (a gene critical for neuromuscular junction [NMJ] maintenance), upregulation of synapse pruning–promoting complement factor C1q, and downregulation of Etv1/ER81 (a transcription factor required for establishing sensory-motor circuitry). The importance of mis-spliced genes identified from SMN-deficient tissues to SMA pathology should be thoughtfully investigated in established mouse models of SMA before they can be considered as viable drug targets or as downstream biomarkers of disease state.
In contrast to the mis-splicing hypothesis, several lines of evidence also support a neuronal-specific function of SMN. For instance, SMN protein has been detected in granules moving along motor axons62 and shown to colocalize with actin mRNA and other mRNAs within axons.63–65 In neurons, SMN has also been shown to interact with multiple mRNA-binding proteins, such as hnRNP R,66 KSRP,67 HuD,68–70 FMRP,71 and IMP1,72 and proteins known to interact with mRNA-binding proteins, such as COPI.73,74 Therefore, SMN could potentially regulate the assembly of mRNA-binding proteins and mRNAs into ribonucleoprotein particles in motor axons and nerve terminals, affecting local mRNA transport, processing, and translation. However, the definitive function of SMN protein in motor neurons remains to be determined.
Therapeutic Approaches
The goal of many SMA drug discovery programs has been to identify small molecules or antisense oligonucleotides (ASOs) that increase the level of SMN protein produced from the SMN2 gene, which is present in all patients. Multiple mechanisms have been targeted to drive higher expression of the full-length SMN protein from the SMN2 gene and have been reviewed extensively elsewhere.75–80 Briefly, these include increasing exon 7 inclusion in the SMN2 mRNA, increasing transcription from the SMN2 promoter, and stabilizing SMN protein. A number of high-throughput drug screens looking for small molecule or ASO modulators of SMN expression or function have been completed.33,81–93 Several of the efforts by industry groups remain unpublished to date. Other non-SMN2 modulating approaches have been or are currently being assessed for SMA, including stem cell therapy for neurotrophic support of remaining motor neurons,94–96 neuroprotection,97 muscle-enhancing molecules,98–104 and most notably, gene transfer therapy to replace the missing SMN1 gene, which dramatically increases the survival in severe mouse models of SMA.105–108
The identification of additional molecular drug targets for SMA beyond SMN2 is being investigated. The existence of other genetic modifiers for SMA is well accepted. Examples of siblings with identical genotypes but with different phenotypes strongly suggest that non-SMN2 gene modifiers for SMA exist. Several candidate genes and pathways have been suggested.109 For example, Plastin 3, an actin bundling protein,110 is reported to be a protective modifier in human SMA.111 The expression of Plastin 3 has been shown to be protective in zebrafish models of SMA112 and to improve NMJ function but not survival in SMA mice.113 In addition, modulation of pathways regulating actin dynamics, such as RhoA/ROCK, have been shown to provide survival benefit in mouse models of SMA38,114 and could potentially represent possible disease modifiers. Genetic modifiers of the phenotypes arising from SMN deficiency in Drosophila and Caenorhabditis elegans have also been identified.115,116 The identification of genetic modifiers and downstream targets of SMN, once validated and shown to improve phenotypes in established mouse models of SMA, will uncover a wide array of potential new drug targets for SMA to be directly assayed and screened. To date, however, most cell-based assays to identify novel SMA drugs have been designed to assess SMN2 expression. Therefore, this review focuses on approaches that have been utilized to identify modulators of SMN2 production.
SMA Drug Pipeline
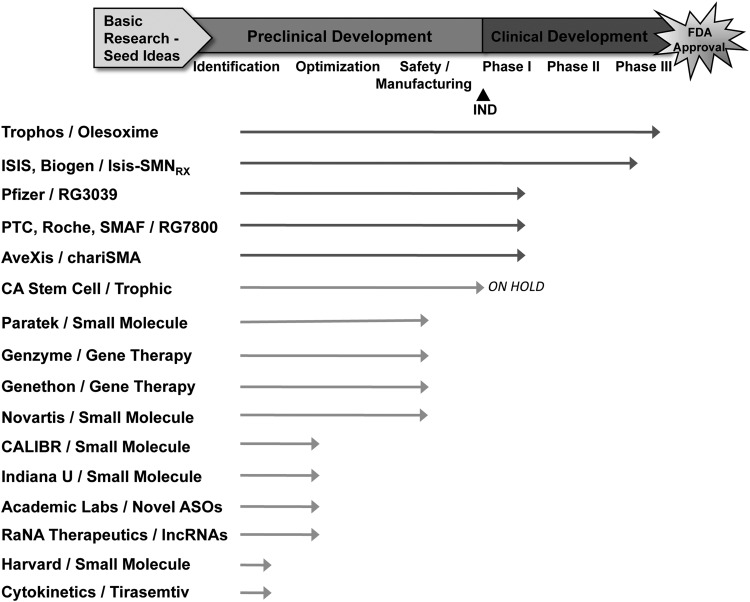
Currently, there are 16 known SMA therapeutic programs in various stages of preclinical and clinical development, a large increase from just a decade ago (Fig. 2). The specifics of each of these programs have been reviewed extensively.75,77,79,80,117 Briefly, there are five novel SMA drug candidates actively being testing in clinical trials for SMA, including (i) olesoxime (Trophos), a neuroprotectant compound that has just completed testing in a pivotal trial from Trophos,97 (ii) RG3039 (Pfizer), a small molecule that has recently completed Phase Ia and Phase Ib safety trials,32,37,39,118 (iii) ISIS-SMNRx (Isis Pharmaceuticals and Biogen Idec), an ASO that is currently being tested in Phase III trials in SMA patients,35,40,83,119,120 (iv) RG7800 (a joint research program between Roche, PTC Therapeutics, and the SMA Foundation), an oral small molecule that corrects the splicing of SMN2 that entered clinical development in early 2014, and (v) chariSMA (AveXis and Nationwide Children's Hospital in Columbus, OH), an intravenously delivered AAV9/SMN1 gene transfer therapy, that is being tested in a Phase I safety in nine infants with type I SMA.105–108 Several more programs are actively working toward Investigational New Drug (IND) applications to the FDA to begin Phase I trials over the next year. These programs illustrate the diversity of approaches being pursued for SMA, including (i) small molecules, (ii) neuroprotectants, (iii) ASOs, (iv) stem cell therapies, (v) gene transfer therapies, and (vi) regulators of muscle function. Today, there is a high level of industry interest in SMA drug development, with about a dozen known companies actively investing in novel SMA drug development and research, including Trophos, Isis Pharmaceuticals, Biogen Idec, Pfizer, PTC Therapeutics, F. Hoffmann-La Roche, AveXis, California Stem Cell Incorporated, Paratek Pharmaceuticals, Genzyme Corporation, Novartis, RaNA Therapeutics, and Cytokinetics (Fig. 2).
Fig. 2.
SMA drug pipeline. The status of known candidate therapies being assessed for the treatment of SMA. Status of the compounds is estimated. The academic laboratories working on novel ASOs include those of Dr. Ravindra Singh at Iowa State University, Dr. Christian Lorson at University of Missouri, and Dr. Arthur Burghes at Ohio State University. lncRNAs, long noncoding RNAs; IND, Investigational New Drug; ASO, antisense oligonucleotide; SMAF, Spinal Muscular Atrophy Foundation; U, University; SMA, spinal muscular atrophy.
Drug discovery programs involving cross-disciplinary partnerships are essential for effective drug development in orphan diseases such as SMA, where it has been traditionally difficult to attract the interest of large pharmaceutical companies. SMA, in particular, has benefited from collaborative industry, advocacy, and government partnerships in both basic research and drug development, and the advances that have occurred in SMA drug development over the past decade directly reflect this collaborative activity. In fact, such collaborations have provided funding for many of the drug development programs, as well as helped build a collection of drug discovery assets and tools.
Assays for SMA Drug Discovery
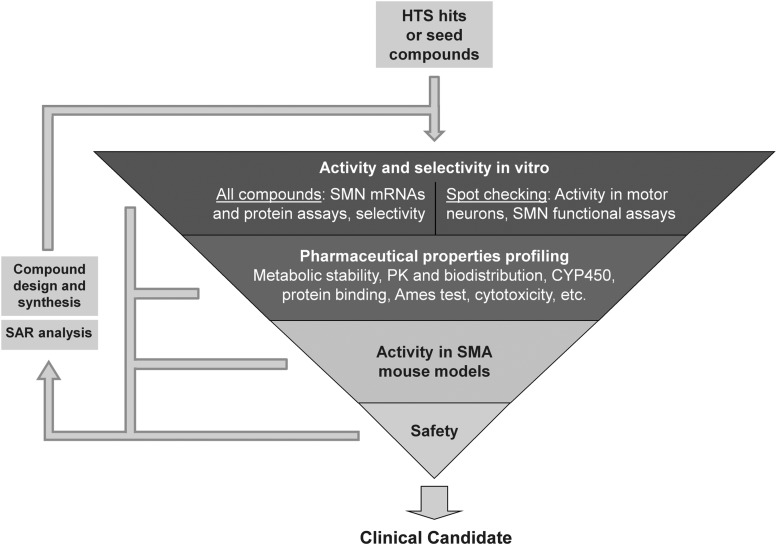
This article describes the preclinical drug discovery assets developed by the SMA research community. Because most cell-based therapeutic assays to date have been designed to evaluate SMN2 gene expression, our review focuses on the approaches that have been utilized to identify modulators of SMN2. Methods have been developed to assess SMN mRNA expression, SMN splicing ratios, SMN protein levels, SMN protein function, and survival, behavioral, and anatomical benefits in animal models of SMA. Moreover, a typical set of secondary screening assays has also been adopted to prioritize hit compounds arising from primary drug screens, as well as to enable hit to lead and lead optimization activities. The methods for performing each of these assays are described here, and the pros and cons of current technology utilized in SMA drug discovery are also discussed.
SMN Transcript Assays
SMN gene transcript analysis, together with SMN protein quantification, is commonly considered the first-level approach in SMA therapeutic development and translational research since increasing the production of the full-length SMN2 mRNA and SMN protein from the SMN2 gene in patients addresses the molecular deficit underlying the disease. Extensive studies of SMN transcript abundance have been performed and can be assigned to two categories: (i) those evaluating possible correlations between SMN2 transcript levels and disease severity and (ii) those assessing the in vitro and in vivo responses to different therapeutic approaches. The studies belonging to the first group have been mainly performed ex vivo, from blood samples of patients and control individuals. The different technical approaches utilized are analyzed in detail throughout this section.121–127 A greater number of studies belong to the second category, as the majority of therapeutic approaches have aimed to increase SMN protein levels by increasing the level of SMN transcripts. An exception to this transcript “rule” is the modification of SMN protein stability,117 where a change in SMN transcript levels is not expected.
When identifying drugs that modulate SMN2 expression, SMN gene reporter assays, which are described in more detail later in this review article, are often utilized as the first tier approach. The results obtained are then generally confirmed by evaluating the effect of a given treatment on the endogenous SMN2 gene expression in disease-relevant cells such as patient-derived cells and/or in murine models of SMA. In this case, transcript analysis is typically paired with SMN protein quantification. While a simple correlation between increased full-length SMN transcript and protein expression is conceptually presumed, ideally the relationship should be independently confirmed. This correlation may not be automatic because the dynamics and the half-life of mRNAs and proteins are not necessarily comparable.17,128
Both SMN1 and SMN2 genes give rise to several mRNA isoforms via alternative splicing. SMN1 mostly produces SMN full-length (SMN-FL) or lacking exon 5 isoforms, whereas SMN2 transcripts may lack exon 7 (SMNΔ7) or both exons 5 and 7.129,130 More recently, an axonal specific isoform (aSMN) has been reported that retains part of intron 3 and is transcribed from the SMN1 gene.131 To our knowledge, most assays utilized in drug development for quantification of SMN transcripts target the more abundant isoforms, SMN-FL and SMNΔ7, due to their proven therapeutic relevance.
Here, we focus on four different technical approaches to quantify mRNA: conventional, relative, absolute, and digital real-time reverse transcription–polymerase chain reaction (RT-PCR). We do not discuss RNA extraction as it is well standardized by different commercially available extraction kits. We do not discuss northern blotting as a tool for mRNA quantification due to its limitations in sensitivity. Finally, we provide some clues on SMN primer design, due to the unusual constitution/splicing of the human SMN genes.
Conventional Semiquantitative RT-PCR
The more classical approach for mRNA quantification is based on endpoint RT-PCR and post-PCR analysis of the amplification products. This approach was most commonly used during the pre-real-time PCR era but still continues to be used today alongside other methodologies.121,132 For the quantification of SMN transcripts, densitometric analysis of PCR products is used, followed by normalization to the levels of a reference gene, often a house keeping gene (such as glyceraldehyde-3-phosphate dehydrogenase and hypoxanthine-guanine-phosphorybosil-transferase). Subsequently, SMN levels in cells treated with test compound are compared with those in control/untreated sample(s). SMN and the reference transcript of choice can be coamplified in a multiplex PCR assay or can be amplified separately, but ideally in the same amplification reaction. SMN-FL and SMNΔ7 isoforms can be coamplified using the same primer pair, and the two resulting amplification products can be discriminated based on their size. This approach has also been used for the evaluation of the ratio between SMN-FL and SMNΔ7 transcripts.121,132,133
Conventional RT-PCR has the advantage of low costs and the use of equipment standard to most molecular biology laboratories. However, its use has been limited by several considerations: (i) it is not readily scalable for high-throughput applications; (ii) it is generally recognized that amplicons when measured together cannot exceed a 10% difference in size to avoid preferential amplification of the smaller product; and (iii) it is influenced by abundance, which can be of particular importance when selecting reference genes. For instance, housekeeping genes are often used as references, and they can be expressed at higher levels compared to SMN and in this case be preferentially amplified.
The use of conventional agarose gel electrophoresis and ethidium bromide staining in some studies could potentially introduce some biases.132 Due to the low sensitivity of this dye (1–5 ng),134 two possible solutions are available to obtain analyzable bands: increasing the number of cycles of PCR amplification and/or increasing the amount of starting materials.130 In both cases, there is a substantial risk that one or more amplicons fall outside the log-linear phase of the amplification curve, and thus, the amount of deoxyribonucleic acid (DNA) obtained is not proportional to that of the starting material. To overcome this issue and to reduce the number of amplification cycles and/or starting amount of complementary DNA (cDNA), different DNA staining tools can be used, such as radioactive isotopes/silver staining coupled with polyacrylamide electrophoresis133 or fluorescent PCR and capillary gel electrophoresis.121 In addition, the analysis of different dilutions of the same cDNA sample may lead to more robust results. Finally, in the case of all semiquantitative approaches, the assumption that reference gene transcript levels do not vary in response to the treatment or are comparable in all samples analyzed can confound the interpretation of results.
Relative Real-Time RT-PCR
Due to the current availability of real-time PCR instruments in most molecular biology laboratories, this approach is most commonly used for transcript quantification. The majority of studies published so far have been performed using this technique. Real-time PCR assays, in general, have several advantages compared with the conventional PCR approach, including sensitivity (down to single molecules) and high reproducibility. The main limitation is the semiquantitative nature of this approach, similar to that of conventional PCR. SMN transcript levels are determined using different techniques, which include the ΔΔCt method135 that uses a housekeeping transcript as an endogenous control and one or more untreated/control samples used as calibrators, and the Liu and Saint methods that simulates PCR reaction efficiency from kinetic curves.136
The two main approaches most commonly used for the visualization of the PCR products are as follows: SYBR® green or similar intercalating dyes that emit fluorescence on excitation when bound to double-strand DNA137 and the TaqMan® probe system, based on the use of unlabeled primers and fluorescent probes. While the two techniques do not differ in terms of sensitivity and efficiency, there are some advantages to each. SYBR green has no additional cost for probe synthesis and can be more easily adapted to different amplicons. The main limitation is that it can be biased by primer dimers and the formation of nonspecific amplification products that cannot be differentiated from the target. Without careful primer design and optimization, this technique may not distinguish SMN1 and SMN2 mRNAs in the same reaction when multiplexed. For both SYBR green and the TaqMan probe system, SMN-FL and SMNΔ7 PCR products should be amplified with caution in the same assay due to the potential formation of hybrid amplicons from the cross-binding of partners. Some of the possible biases of relative real-time PCR are related to the wide variations in SMN gene expression in samples from different control individuals, as well as to differences in the endogenous transcript expression in both patients and controls.126,138 Similarly, a given treatment may alter the reference gene expression, and this could lead to incorrect interpretations of the effect of the treatment on SMN levels.
Absolute Real-Time RT-PCR
In this approach, transcript levels of the target gene are determined by extrapolation from a standard curve, constructed using serial dilutions of an external standard with known quantities of the target amplicon. At least three different kinds of external standards can be used: (i) a control sample (or a pool of different controls), (ii) RNA, or (iii) DNA external standards. For the SMN transcript analysis, the first127 and third126 approaches have been used, whereas to our knowledge, the second has not.
The use of serial dilution of a control sample is more properly indicated for relative standard curves since results are expressed as folds of variation compared to control. In this method, two main approaches are more commonly used for the preparation of serial dilutions to construct standard curves: (i) a single control sample (unaffected, untreated, or mock) and (ii) a mix of equimolar amounts of different control samples. Because the main bias of relative standard curves is caused by variations in SMN levels in the samples used for the construction of the standard curves, the mix of a pool of control samples may be useful to balance possible sample-to-sample differences. Indeed, it has been shown that in control individuals, SMN-FL levels vary widely and do not show the Gaussian distribution.126
The other two approaches exploit techniques that are widely used to determine the plasma load of some viruses, such as HIV or HCV, or for prion genes.139–141 Transcript levels are expressed as number of mRNA molecules per nanogram of total RNA and are determined independently of the use of endogenous controls. Both approaches provide comparable results, with some differences. RNA external standards can serve as good controls for RT-PCR since they undergo reverse transcription together with RNA samples. However, they are less stable compared to DNA standards (especially in the case of the lower concentrations), while not offering consistent advantages in sample quantification.142 Absolute real-time PCR can be biased by different issues. First, the efficiency of amplification of the external standards may vary between preparations and/or can be affected by repeated cycles of freezing and thawing. To circumvent this issue, new preparations of external standards should be compared with the previous, and single-use aliquots should be prepared. Second, PCR efficiency needs to be calculated and evaluated using the various available methodologies. One method is to determine the slope of the standard curves, which ideally should be as close as possible to −3.33, corresponding to a dilution factor of 10-fold. A second technique calculates PCR efficiency without a standard curve using reaction kinetics.136 Large differences in the slope of the standard curve and/or low PCR reaction efficiency often indicates nonoptimized reaction conditions, such as improper preparation of serial dilutions, poorly designed primers, and/or degradation of one or more dilutions. Conversely, the extrapolation of SMN levels from inappropriate standard curves yields inaccurate quantification of mRNA levels and poor reproducibility from one experiment to another. Third, while reference genes can be used as loading controls, it is difficult to assess the quality/quantity of RNA samples with this technique.
In our opinion, certain steps may mitigate (but not remove) these issues. First, RNA extraction should be standardized. In our experience, in-column extraction/purification is preferable to phenolic extraction and should be coupled with DNase I treatment (or other tools for the removal of contaminating genomic DNA). Second, quality and quantity of RNA should be accurately evaluated: the ratios of absorbance at 260/280 nm and at 230/260 nm provide essential information. Spectrometer or fluorescent dye-based techniques evaluation should be coupled with techniques that disclose the presence of contaminating genomic DNA and/or RNA degradation products, such as agarose gel electrophoresis or fluorescent dye-based approaches (using the Agilent Bio-analyzer instrument or similar tools that allow simultaneous quantification of RNA samples). Third, the use of a particular RT-PCR kit should be validated. Indeed, it should not be assumed that different reverse transcriptase or RT-PCR kits will yield comparable results, and thus, the efficiency of different commercially available kits should be initially evaluated.
Digital PCR
Digital PCR is a recent technology that allows the quantification/detection of target nucleic acids by fractioning a single sample into multiple PCR reactions occurring in the same tube. Some of these reactions produce efficient amplification while others do not based on the abundance of the target nucleic acid. The DNA of interest is quantified by comparing the number of effective and noneffective reactions. This approach has the advantage of the ability to quantify samples independently of both endogenous and external controls, and very high sensitivity, down to variations of single molecules of the target, and it has been used successfully to measure DNA copy number and to detect rare mutant alleles in a 100,000-fold excess of wild-type background.143 The main limitations are costs, which are much higher compared to those of traditional real-time PCR. Digital PCR allows for highly sensitive SMN transcript measurements, and its use may ultimately help to illuminate the biological import of single molecule changes in expression levels. This approach has been recently utilized for the quantification of SMN levels in mouse model of SMA, including studies for the quantification of SMN1 and SMN2 mRNA levels36 and of SMN1/2 gene copy number.144
SMN Primer Design
This review does not cover general considerations for PCR primer design since (i) this matter is extensively covered by PCR handbooks, (ii) is generally performed by software tools, and (iii) the SMN cDNA does not have particular concerns (i.e., long stretches of repeated nucleotides, prominent GC-rich domains, and repeated sequences). On the other hand, certain characteristics of the SMN1 and SMN2 genes, as well as the alternative splicing of exon 7, require specific consideration during PCR assay development. There are two areas of key relevance. The first is that SMN1 and SMN2 transcripts can be differentiated only by a single SNP in the coding sequence of exon 7 and one additional SNP in the 3′-UTR. Second, exon 7 is only 54 base pairs long. Thus, both SMN-FL and SMNΔ7 isoforms can be easily coamplified. Being able to distinguish between SMN1 and SMN2 transcripts is relevant in a few circumstances, such as in the case of control samples or in the case of patients bearing point mutations of the SMN1 gene (about 2%–3% of the total since most patients lack both SMN1 alleles). In such cases, the two genes may be differentiated on the basis of the C–T transition in exon 7, either by means of specific oligos (allele-specific oligonucleotide PCR) or by different probes. To our knowledge, the first approach has been followed by Feldkotter et al. for the determination of SMN1/SMN2 gene copy number but may theoretically be applied also for transcript analysis as well.20 In this approach, SMN1 and SMN2 transcripts must be quantified in separate reactions. An alternative approach is to label SMN1 or SMN2 allele-specific probes with different fluorophores (in the context of TaqMan probe-based assays).126 In this case, both transcripts are amplified by the same primer pair. The primary issue with both approaches is the possible cross-hybridization of SMN1 primer or probe with SMN2 transcripts and vice versa.
The SMN-FL and SMNΔ7 transcripts are readily distinguished on the basis of their size difference. Different approaches can be utilized to do this, such as a single primer pair amplifying both isoforms (e.g., the forward primer located in exon 6 and the reverse in exon 8), a commonly used approach in conventional PCR. However, this approach might be biased by the preferential amplification of the SMNΔ7 transcripts (due to both relative abundance and smaller size of this isoform, compared to the full-length form) and consequently lead to an overestimation of this isoform.
Considerations
Independent of which of the above-described techniques is used, SMN transcript analysis is a necessary step in the identification and evaluation of effectiveness of candidate therapies for SMA aimed at modulating SMN levels. It should be coupled with SMN protein analysis, which is described in the following section. The advantages and disadvantages of each approach for SMA transcript analysis are listed in Table 1. SMN transcript analysis also provides information on the molecular mode of action of candidate compounds. The quantification of the two main SMN2 isoforms can help to differentiate the mechanism of therapeutic agents, for instance whether they act at the SMN2 gene promoter level, and/or influence the alternative splicing of exon 7, based on the variation of the total SMN transcript levels and/or the SMN-FL/Δ7 ratio. Some researchers have utilized evaluation of the SMN-FL/Δ7 ratio only, with the tradeoff that putative effects on promoter activation/transcript stabilization cannot be evaluated.132
Table 1.
Comparison of Different Polymerase Chain Reaction Methodologies
| Technique | Suitable for HTS screening | Validated for SMA | Pros | Cons |
|---|---|---|---|---|
| Conventional semiquantitative PCR | No | Yes | Low costs | Low sensitivity |
| Plateau PCR | ||||
| Diversity of amplicon size | ||||
| Relative real-time PCR | Yes | Yes | Less biased by RNA/cDNA quantity/quality | Hampered by variation of expression of endogenous controls |
| Not suitable for comparison between different subjects | ||||
| Absolute real-time PCR | Yes | Yes | Quantification independent of endogenous controls | Biased by RNA and cDNA quality and quantity |
| Allows for comparison between different subjects | Requires external standards | |||
| Digital PCR | Yes | No | Quantification independent of endogenous controls and external standards | Biased by RNA and cDNA quality and quantity |
cDNA, complementary deoxyribonucleic acid; HTS, high-throughput screening; PCR, polymerase chain reaction; RNA, ribonucleic acid; SMA, spinal muscular atrophy.
SMN transcript analysis is not without limitations, particularly when utilized in clinical trials on patient samples. First, it is not feasible to sample neuronal cells and tissues from human patients. Therefore, peripheral blood mononucleated cells (PBMCs) and whole blood are most widely sampled during clinical trials due to their accessibility. In this case, it must be understood that SMN levels are not being assessed in the primary target tissues of the disease, that is, spinal cord and skeletal muscle. Thus, an important preclinical step is to assess drug efficacy in disease-relevant tissues in animal models of SMA to relate SMN levels in the central nervous system (CNS) and muscle to levels in blood, recognizing that drug effects in mice and men may differ for multiple reasons. Second, baseline SMN transcript and protein from patient PBMC samples have not always shown high correlation.122,125 Despite this, the hope is that drugs designed for SMN2 modulation will induce an increase in transcript expression with resultant SMN protein increases, prerequisite for therapeutic effectiveness in SMA. Third, SMN protein and transcript analysis is not indicated for the evaluation of therapeutic compounds that are not intended to modify SMN levels, such as neuroprotecting compounds.
In the context of clinical trials, some technical advantages support the use of SMN transcript analysis as the primary biomarker/surrogate measure for SMA. These include the availability of several stabilization buffers that allow preserving samples from RNA degradation and gene expression variations, as well as the small amount of patient blood necessary for the assay. These technical aspects become very relevant in selecting a biomarker readout in the context of multicenter trials when dealing with severely hypotonic patients and young children, for whom sampling and obtaining adequate amounts of blood can be very challenging. The most appropriate techniques in the context of a clinical trial are absolute real-time RT-PCR or digital RT-PCR. These assays have the advantage that they are not biased by the use of endogenous transcripts, which may be subject to longitudinal variations, in response to treatment itself.
To our knowledge, real-time PCR in general (relative, absolute, or digital) has not yet been described in the literature for primary high-throughput screens on hundreds of thousands of compounds. These tools may prove to be suitable for such applications in the near future because (i) nanogram amounts of RNA are sufficient for transcript analysis, and thus, it is possible to use a limited number of cells or of small amounts of biological samples; (ii) the availability of robotized liquid handling systems found in many laboratories allows for standardization and the manipulation of a high number of samples per unit time; and (iii) the availability of instruments that allow for miniaturization has enabled marked reductions in the reaction volume required for real-time PCR, resulting in a significant decrease in the consumption of reagents.
SMN Protein Quantification
The SMN protein is another key pharmacodynamic measure for drug development programs focused on therapeutic upregulation or replacement of SMN. There are several approaches available for the quantitative analysis of SMN protein in fluids, cells, and tissues, all with distinct uses and limitations (for a general reference on protein quantification technologies such as fluorescence resonance energy transfer [FRET], electrochemiluminescence [ECL], and surface plasmon resonance [SPR], see Inglese et al.).145 Importantly, SMN protein extraction is influenced by its interactome, SMA biology, and reagents used and should be considered in the context of which assays are used for quantification—some of these issues are discussed in this section.
SMN Protein
As previously discussed, SMN protein is ubiquitously expressed across tissues, cells, and cellular compartments (including as punctate accumulations in the nucleus called gems) and exists in a complex that includes multiple SMN molecules and Gemins 2–8.45 There is also a large and growing interactome of additional SMN binding partner proteins, which corroborates the array of functions ascribed to SMN, and also speaks to the effector structure of the protein itself, which recognizes an array of binding motifs.73,146,147 Given these factors, the discussion of SMN protein quantification necessitates consideration of which form of SMN is evaluated, as well as the extraction, denaturation, and stabilization of the protein for each assay platform.
SMN Antibodies, Epitopes, and Extraction
Most SMN protein measurements employ some form of immunoassay. Although several dozens of SMN monoclonal antibodies have been developed, a few selected antibodies are regularly used: 2B1, 8, MANSMA1, H195, and more recently 11708 (Table 2). Most antibodies recognize human and mouse SMN148; however, the 4F11 and 60154 clones are reported to be human specific (ProteinTech, pers. comm.). Notably, SMN is expressed in full-length or truncated forms, by the SMN1 and SMN2 genes, and there is no available antibody against the shorter protein generated by skipping of exon 7 (SMNΔ7). The full-length SMN has also been described as being a substrate for proteolysis by calpain, and depending on epitope and assay, some antibodies capture information on total versus truncated forms of SMN.149,150 While SMN has been described to be post-translationally modified via phosphorylation and ubiquitination, no antibodies for these specific forms have been described.17,151
Table 2.
Selected Antibodies Used for Survival of Motor Neuron Quantification Assays
| Clone | Type | Epitope (human) | Ref. |
|---|---|---|---|
| 2B1 | Mouse IgG1 | aa 14–20 | 155 |
| 8 | Mouse IgG1 | Unknown | 226 |
| 11F3 (MANSMA1) | Mouse IgG1 | aa 42–48 | 153 |
| 62E7 | Mouse IgG1 | Unknown | 43 |
| 11708 | Rabbit polyclonal | aa 197–204 | 154 |
| H195 | Rabbit polyclonal | aa 1–195 | 177 |
| 4F11 | Mouse monoclonal | SMN exon 4 | 148 |
| 60154 | Mouse IgG2a | Unknown | NA |
SMN, survival of motor neuron.
Epitope sequence and epitope accessibility are major considerations in the use and development of quantitative SMN immunoassays. Recently, Lam et al. described the overlap for epitope from the widely used MANSMA1 antibody and the SMN Gemin 2 binding site.152 This also supports the need for extraction and stabilization of SMN proteins before quantification in immunoassays. There is also evidence that some antibodies (e.g., MANSMA1 and 11708) preferentially bind human SMN protein sequences.153,154 Their use in some assays influence the results—especially in assessing SMN protein in transgenic mouse tissues, which may have both human and mouse proteins present.
The extensive network of SMN binding partners as well as oligomerization forms also impacts the selection of new SMN immunogens and use of SMN standards for assay quantification. Generation of new SMN antibodies has generally been achieved through immunization with recombinant SMN, which may have different available epitopes than endogenous SMN.148,153–155 Ideally, assays using these antibodies should test the immunogens as quantification standards—although this is not always possible. In developing new SMN immunoassays, care must be taken to ascertain the relationship between antibody binding to native SMN in cell lysates versus purified or recombinant standards. Although there may be discrepancies between antibody binding of recombinant protein used for standards and endogenous SMN protein, as long as there is a linear relationship between them, a quantitative assay can be developed from those reagents.
Finally, reagents for the lysis of cells and homogenization of tissues for subsequent SMN protein measurements must be adequate to extract and dissociate SMN from its many binding partners. Beyond the multitude of SMN binding partners, SMN forms oligomers of various sizes, which are stabilized by Gemin 2.19,156,157 Although it is unclear whether any antibodies bind preferentially to any oligomers of SMN, or whether stabilized conformations of the protein may be more accessible to epitopes, the use of extraction buffers with higher concentrations of salts has been reported to improve signal in some immunoassays.154 Detergents, such as sodium dodecyl sulfate and Tween-20, perform well in buffers for western blot analysis to make SMN accessible for antibody binding. However, high concentrations may interfere with plate-based assays, such as sandwich enzyme-linked immunosorbent assays (ELISAs).154 Tissue homogenization buffers, such as RIPA, M-PER, T-PER, and ER4, have all been reported to work effectively in western blot, ELISA, FRET, and ECL.
Western Blots
Since the mid-1990s, western blotting has been the most widely used method of SMN protein analysis reported across species, sample type, and laboratories in the SMA literature.27,28,33,133,155,158 Since western blots only require one antibody, they can more reliably give signals of total SMN protein and also provide information on truncated species. Earlier western blotting methods necessitated the use of darkroom and densitometry. However, new devices improving gel to blot transfer and increasingly sophisticated fluorescence and chemiluminescence imaging systems now allow semiquantitative or quantitative determination of SMN levels. While the dynamic range of this assay format has been improved up to 1,000-fold with more sensitive imagers and Z′-factors of >0.5 are achievable (indicating an assay that can exquisitely differentiate between negative and positive values and may be suitable for screening), western blotting is still 4- to 400-fold less sensitive than other comparable quantitative immunoassays for SMN, depending on the manufacturer. Throughput also remains a challenge as western blots are limited to ∼12–16 samples in traditional gel formats or 48–96 samples per microwestern arrays, capillary, or in-cell westerns, whereas other assay types allow for miniaturization to several hundred well formats. In addition, western blots have relatively narrow dynamic signal ranges and are susceptible to positional bias depending on the placement of samples in lanes close to the edge of gels. Although western blots are not optimal for analysis of samples from large screening campaigns, they remain as a workhorse platform for academic research and are useful in the cross-platform validation of new SMN immunoassays and/or spot-check confirmation of drug hits.
Enzyme-Linked Immunosorbent Assay
ELISAs are attractive for use in drug screening because they are simple assays, require minimal instrumentation to perform, can be scaled to 384 and 1,536 well formats, and can yield Z′-factors of >0.80. Multiple research-grade SMN ELISAs, ranging from simple sandwich ELISAs to unique in-cell immunoassays that can serve as models for screening-grade assays, have been published.124,154,159,160 Sensitivity and dynamic ranges are greater than that of western blots: 25–3,200 pg/mL. Many of the SMN ELISAs reported have been developed for measuring the protein in blood cells (typically peripheral blood mononuclear cells and lymphocytes) and can effectively and sensitively quantitate signals in an array of human and mouse cells and tissues.124,159–161 While the goal of creating an ELISA has been achieved for all the reported assays, subsequent studies have not demonstrated a clear relationship between disease severity and SMN protein levels in blood cells.122,125
There are notable limitations to sandwich ELISAs as drug screening assays that relate to the format as well as their reliance on multiple SMN antibodies, and these should be considered when designing any screening ELISAs. Additionally, ELISAs are subject to greater matrix effects than other quantitative protein assay formats and can require substantial validation and optimization. In contrast to western blots, SMN ELISAs reflect total protein without an ability to differentiate between SMN isoforms. Also, ELISAs utilizing antibody pairs with different preferences for human or mouse epitopes (e.g., 11708) can yield results that bias the measurement toward detection of one species over another.41,154 Overall, the careful selection of antibodies and buffer conditions would enable the creation of ELISAs suitable for initial to downstream screens for SMN in cells, depending on the therapeutic concept for a given drug program.
Fluorescence Resonance Energy Transfer
FRET assays are immunoassays in which two fluorescently labeled antibodies produce a signal when they are brought to close proximity by complexing with their antigen. For example, one SMN antibody is conjugated to a donor dye (e.g., europium chelate or cryptate), whereas the other antibody is covalently linked to an acceptor fluorophore. When both donor and acceptor-labeled antibodies bind to SMN, the FRET process is enabled and the excitation of the donor results in acceptor emission, which is proportional to donor–acceptor complex formation or SMN abundance. Another key element of some FRET-based assays is gating to remove noise from short-lived background fluorescence (time-resolved or TR_FRET). A FRET-based assay has all the positive attributes of ELISAs as well as the benefits of matrix effects and being homogenous, with no need for removal of solutions from previous steps. The assay can be particularly useful in reliably measuring changes in cells treated with compounds in a typical drug screening campaign, with the possibility of Z′-factors of >0.8. Although there are no publications currently describing research with SMN FRET-based assays, there is a commercial assay available (Cisbio). However, the same issues concerning species bias of the antibody pair chosen in ELISAs are applicable for FRET-based assays. In addition, the steric requirements of the donor and acceptor-labeled antibodies impose limitations on which SMN antibody pairs can be utilized in this assay format as overlapping epitopes may not be suitable for FRET. While the assay is sensitive, the relatively narrow dynamic range (3–100 pg/mL, as reported by Cisbio) necessitates some validation to ensure the sample is in range at testing dilutions. Using a permissive antibody set, a FRET SMN assay can be a useful primary to tertiary screen for drug-induced SMN changes in cells or in treated tissues from animal studies.
Electrochemiluminescence
ECL assays, such as those provided by Meso Scale Discovery, are a modification of the classical plate-based sandwich immunoassay, with the distinction that their detection signal is based on reading a light signal produced by an electrical current. In this format, the capture antibody immobilized to the surface of an electrode binds the antigen, which in turn binds a detection antibody conjugated to a Ruthenium tag. Upon exposure of the plate to an electric current, the Ruthenium tag undergo a chemical process that emits light. The commercially available PharmOptima SMN ECL assay has a very broad dynamic range and is less subject to matrix effects; sensitivity of the assay can be 3–100,000 pg/mL, exceeding the range for ELISAs (manuscript in preparation). Studies with the assay show that it is capable of measuring SMN in a variety of tissues, including whole blood samples (manuscript in preparation). This 96-well format ECL assay is amenable for use as a secondary or tertiary screen as well as for testing in a range of animal model and human biological samples.
Other SMN Assays
While immunoassays dominate SMA research, other approaches have been reported and have utility in some aspects of assay development or other research. Masson et al. reported an effort to develop an SPR method for quantifying SMN.162 SPR relies on the shifting of a refraction signal when light is beamed across a gold film layer that can be coated, for example, with antibodies that can be exposed to antigen flowing in a solution across the coated surface. Generally SPR has a wide dynamic range and requires only about 15 min per sample to perform. Using SPR to characterize the binding of a MANSMA2 antibody and recombinant SMN, Masson et al. reported a limit of detection of 0.99 ng/mL. However, up to a third of the signal was potentially due to nonspecific interactions. At a minimum SPR, approaches are of immediate usefulness in quantifying the binding properties of SMN reference materials and antibodies.
Mass spectrometry-based techniques have been used for decades in drug discovery to quantitate the levels of small-molecule drugs in samples. More recently, several quantitative mass spectrometry techniques have been developed and used for proteomics.163 Techniques such as isobaric tagging for relative and absolute quantification (iTRAQ) allow a peptide-based approach for evaluating concentrations of proteins in samples and have been tried in SMA research for finding new binding partners to SMN protein and for a proteomics biomarker discovery campaign.164,165 While these techniques can be costly and are generally of low-throughput, they also can provide details on truncation species, oligomerization, and post-translational modifications in the same sample reaction without reliance on expensive antibodies.
Finally, bead-based immunoassays, such as the Luminex® platform, are another possibility for screening SMN assays. The assay format has many of the same sensitivity features as an ELISA with the additional benefits of having reduced false-positive rates and being amenable to extensive multiplexing for reducing sample requirements and long-term cost.166 No bead-based SMN assays have been published. In addition, validation for any multiplexed bead-based assays can be extensive as all antibody pairs must perform well in the presence of several other antibody pairs against other analytes.
Table 3 outlines the details of the assays discussed, as well as their potential for use in drug discovery. Most of the assays profiled in the table have been validated to varying degrees for use in a variety of human primary cells and cell lines, and a multitude of mouse tissues, including the brain, spinal cord, liver, skeletal muscle, heart, pancreas, skin, PBMCs, and in the case of the ECL, whole blood.
Table 3.
Comparison of Quantitative Survival of Motor Neuron Protein Assays
| Approach | Dynamic range | Throughput | Equipment costs | Cost/sample | Usable matrices | Screening use |
|---|---|---|---|---|---|---|
| Western blota (e.g., LI-COR Odyssey) | 1.3–5,000 ng/mL | Low | >$50K | $1–5 | Whole blood, cell lysates, PBMCs, Br, L, M, SC, SK | New assay validation, spot-check hit confirmation |
| ELISA | 25–3,200 pg/mL | Medium–high | <$2K | <$2–15 | Cell lysates, PBMCs, Br, L, M, SC, SK | Secondary to tertiary screen |
| FRET (Cisbio) | 3–100 ng/mL | Medium–high | <$50K | <$2 | Cell lysates, PBMCs, Br, L, M, SC, SK | Primary to tertiary screen |
| ECL (MSD) | 3–10,000 pg/mL | Medium–high | >$100K | $20–30 | Whole blood, cell lysates, PBMCs, Br, L, M, SC, SK | Secondary to tertiary screen |
| SPR | 25–10,000 ng/mL | High | >$100K | <$1 | Cell lysates | Characterizing new antibodies and SMN standards |
| Mass-speca | 10–100,000 ng/mL | Low | >$100K | >$100 | Cell lysates? | Characterization of SMN, biomarker discovery |
| Bead-baseda (Luminex) | 25–50,000 pg/mL | Medium | >$100K | <$1 | Cell lysates, PBMCs, Br, L, M, SC, SK | Secondary to tertiary screen |
The details reported in this table are for the quantitative versions of these assays, for example, LI-COR Odyssey systems.
aIn some cases, the performance criteria displayed relate to the theoretical ranges reported by researchers and vendors rather than from SMN-specific experiments.
ECL, electrochemiluminescence; ELISA, enzyme-linked immunosorbent assay; FRET, fluorescence resonance energy transfer; PBMC, peripheral blood mononucleated cell; SPR, surface plasmon resonance.
Considerations
While the focus thus far has been on the numerous assays available for quantitating SMN protein in screens or other experiments, it is vital to consider the primary cell, cell line, or tissue type that will be evaluated. Overall, there is modest correspondence between SMA type or severity and SMN protein levels.21 The modest correlation is thought to reflect the impact of SMA modifiers and/or the inability to test disease-relevant tissues, such as muscle and spinal cord.109,122,125,133 SMN is expressed ubiquitously, but its levels vary greatly across tissues, with the lowest reported levels being in the muscle, nerve, and spinal cord and with the skin and blood at up to 50-fold higher levels.154 The broad range of SMN protein levels by tissue is further complicated by data on differences in cellular subpopulations such as motor neurons, which have been reported to be inherently deficient in the full-length protein due to inefficient splicing of SMN exon 7.154,167
Another factor that can impact SMN protein levels in screening from cell lines or testing clinical specimens is age. Lines of evidence using quantitative assays suggest that SMN expression across species and tissue type usually decline with age.154,161,168 Whether this decline is potentiated by disease stage and severity in patients remains unclear, without larger quantitative studies.
In summary, factors influencing the measurement of SMN protein in samples and specimens include tissue and cell type, buffer conditions, and age of the specimen or cell line donor should also be weighed when designing any new screening assays. The choice of which assay and sample a drug development program employs will depend greatly on the mechanism of action (e.g., putting a protein assay later in the screening process if screening for a transcript modulator or putting it first if screening for and SMN protein stabilizer). Regardless of approach and scope of program, there are many protein quantification assays available for SMN drug development, and also, many reagents are available for the generation of new assays in other platforms.
Reporter Assays and High-Throughput Screens
Since one of the most common strategies to treat SMA is to exploit the presence of the SMN2 gene, high-throughput screens of various types have been developed to screen for small molecules that increase SMN2 expression. However, changes in the protein expression of endogenous SMN2 genes can sometimes be masked by the presence of even a single copy of the SMN1 gene, which complicates the assessment of SMN2 expression. One method used to overcome this challenge is to use cells that lack SMN1, such as those derived from SMA patients. SMA patient cells that are cultured in vitro can be used to measure changes in the levels of SMN protein and mRNA derived from the endogenous SMN2 gene. These techniques have been used effectively to validate new therapeutic approaches and are discussed in greater detail in other sections of this review but are less suited to high-throughput applications. As discussed in the previous section, there are also technical considerations when measuring SMN expression in primary cells, due to cell-to-cell variability of SMN expression in response to changes in age, cell density, composition of culture medium, passage number, and cell cycle.78,169 These considerations have made it difficult to use primary cell models to develop assays that are amenable to high-throughput screening of hundreds of thousands of compounds, although recent advances are making this more feasible and are discussed in the Motor Neuron Assays and High-Content Screens section.
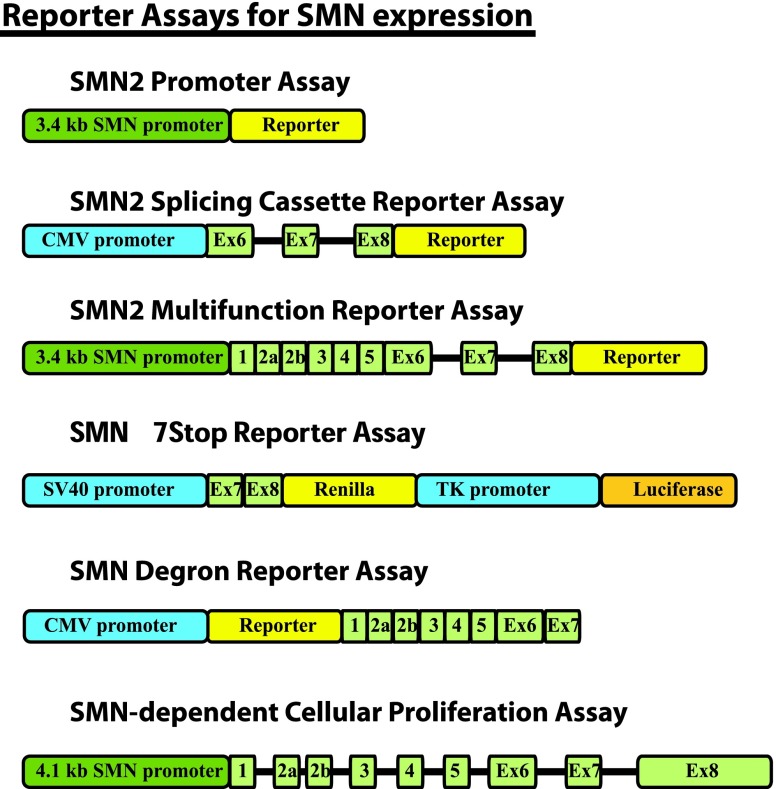
To overcome such difficulties, assays have been developed in immortalized cell lines that use surrogates to predict changes in endogenous SMN2 expression levels. In this section, we discuss the reporter assays that have been developed to identify small molecules that can modify SMN2 expression (Fig. 3 and Table 4).
Fig. 3.
Schematic representation of SMN2 reporter constructs. SMN sequences are colored green. Non-SMN promoter elements are blue. Reporter genes (yellow and orange) vary in different versions of each construct (see Table 4 for details).
Table 4.
Survival of Motor Neuron Reporter Assays
| Assay type | Mechanistic target | Reporter | Host cells | Compounds | Ref. |
|---|---|---|---|---|---|
| Promoter | SMN2 promoter | β-Lactamase | NSC-34 | Diaminoquinazolines | 84 |
| SMN1 promoter | SEAP | 16.4 cells | Taxol | 78 | |
| Splicing | Splicing cassette | Luciferase | C33a | Sodium vanadate | 93 |
| Indoprofen | 92 | ||||
| NSC-34 | Aclarubicin | 169 | |||
| Stability | Read-through | Renilla luciferase | AD293 | G418 | 181 |
| Degron | Luciferase | HEK293 | — | 183 | |
| Multifunction | SMN2 promoter/splicing cassette/stability | Luciferase | HEK293 | 4-arylthiazolyl piperidines | 89 |
| Heterocyclic carboxamides | 33 | ||||
| Dihydroquinolones | 33 | ||||
| Viability | Cellular proliferation | Cell numbers | NIH3T3 | — | 90 |
SEAP, secreted alkaline phosphatase.
SMN2 Promoter-Based Assays
One of the first methods explored to identify compounds that can increase SMN protein levels is based on the regulation of SMN transcription. The promoters for SMN1 and SMN2 are nearly identical and have been shown to have similar levels of expression.170,171 As discussed previously, the SMN2 gene primarily produces a truncated mRNA isoform that lacks exon 7 and just 10%–20% of SMN2 mRNA includes exon 7 and encodes for the stable full-length SMN protein. Even so, enhancing transcription of SMN2 should increase the overall amount of full-length SMN mRNA, without changing the ratio of exon 7 splicing in these transcripts.
The first SMA reporter assay to take advantage of this method used the SMN2 promoter to identify compound that could increase SMN2 transcription.84 In this assay, beta-lactamase (BLA) was used as the surrogate for SMN2 expression. The reporter was cloned downstream of 3.4 kb of the SMN2 promoter. The reporter was transfected into the NSC-34 cell line hybrid of mouse spinal cord cells and a mouse neuroblastoma.172 Treatment with either trichostatin A (TSA) or sodium butyrate, two histone deacetylase inhibitors (HDACi) known to increase SMN expression,30,173 increased BLA expression twofold.
This assay was used to identify the C5-substituted 2,4-diaminoquinazoline series of compounds.84 These compounds increase full-length SMN mRNA, exon 7 inclusion, and SMN protein levels. The quinazoline compounds bind to and inhibit the scavenger decapping enzyme, DcpS174, which is involved in release and recovery of the 7 mG cap following mRNA turnover, and DcpS inhibition could impact mRNA stability, mRNA processing, and protein translation. Newer and more potent analogs of this series were orally bioavailable, had excellent CNS penetration, and promoted improvements to motor function in an SMA mouse.32,118 RG3039, the lead in this series, promotes a significant increase in survival in two mouse models of SMA, increases NMJ maturation, and improves motor function.37,39
A similar assay was developed by the pharmaceutical company, Trophos.78 In this version of the promoter assay, the 3.4 kb SMN1 promoter was used to drive the expression of the secreted alkaline phosphatase (SEAP) reporter, which was transiently transfected into cells (a fusion of mouse neuronal N18TG2 cells and rat primary motor neurons) and levels of SEAP were then assayed from the cell culture medium. This reporter system was validated using the HDACis valproic acid (VPA), sodium butyrate, and TSA and was used to screen a library of 45,000 compounds. Of the ∼100 hit compounds identified, the most potent was taxol, which increases SEAP levels more than 3.5-fold and had a 50% effective concentration (EC50) in the high nanomolar range. The hits were tested in the reporter cells using quantitative RT-PCR but could only reproduce a slight increase in full-length SMN2 mRNA levels in SMA patient fibroblasts and SMN2 transgenic mouse cortical neurons. VPA and sodium butyrate were similarly ineffective in this secondary assay. The authors expressed concerns about specificity and toxicity with these compounds and suggested that the results could vary widely based on choice of cell types for screening and validation assays. None of the compounds identified in this screen was selected for development.
SMN2 Exon 7 Splicing Reporters
Increasing the amount of the full-length SMN protein produced from the SMN2 gene by enhancing the efficiency of exon 7 inclusion is a viable therapeutic approach for SMA. In fact, both the ASO therapeutic ISIS-SMNRx and splicing modifier from the Roche/PTC Therapeutics/SMA Foundation program work in this manner and are currently being tested in clinical trials. Therefore, multiple screens have been developed to identify candidate therapeutics that modulate SMN2 splicing. While the RT-PCR-based approaches discussed earlier have been critical in validating drug candidates, they have not been routinely used to perform primary high-throughput screens on hundreds of thousands of drug compounds, due to time and cost constraints. To address this need, an in vivo reporter system was constructed that simplified identification and quantification of exon 7 inclusion in the context of either the SMN1 or the SMN2 gene.93 Splicing cassettes from either SMN1 or SMN2 were fused to a reporter gene. In this reporter, the endogenous stop codon in exon 7 was disrupted by a single-nucleotide insertion, creating a frame shift in the reporter transcript so that it would be in frame only when exon 7 was included. This entire construct was expressed under the control of the CMV promoter and was tested by transient transfection into C33a cells.93 As expected, the SMN1 constructs expressed higher levels of the reporters and displayed nearly 100% inclusion of exon 7. SMN2 reporter expression was threefold lower than that observed with the SMN1 reporter, had <30% inclusion of exon 7, and was increased threefold with hTra2-β overexpression. C33a cells were stably transfected using the SMN1-luciferase or SMN2-luciferase constructs and were used to screen a small panel of known drugs. The phosphatase inhibitor, sodium vanadate, was shown to selectively increase luciferase fivefold in the SMN2-luciferase cells. These stable cell lines were then used to screen a library of about 47,000 compounds.92 Indoprofen was identified as a hit and shown to increase SMN2-luciferase expression by nearly threefold. In 2005, the internal National Institute of Neurological Disorders and Stroke (NINDS) SMA Project embarked on medicinal chemistry to make more potent and safer analogs of indoprofen, but the project is no longer being actively pursued within NINDS.
A modified version of this reporter was used to confirm that aclarubicin increases exon 7 inclusion in the SMN2 transcript.169 In this format, the SMN1 and SMN2 constructs were fused with BLA and introduced into the NSC-34 cell line. Aclarubicin was able to increase BLA levels by fourfold in the SMN2-BLA reporter cells. The activity of aclarubicin was confirmed with endogenous SMN protein and mRNA in SMA-derived patient fibroblasts. Although aclarubicin was an effective modulator of SMN2 splicing and SMN protein expression, it is a chemotherapeutic drug and expected to be too toxic for the treatment of infants and children.
Assays for SMN2 Protein Stability
Another strategy to increase the amount of functional SMN protein in a cell is to salvage the truncated SMN protein already expressed from the SMN2 gene. The exclusion of exon 7 from the SMN2 transcript results in the loss of the C-terminal 16 amino acids and the addition of four amino acids (EMLA) encoded by exon 8.7 This produces an unstable truncated SMNΔ7 protein. The SMNΔ7 protein has a shorter half-life and impaired ability to oligomerize and bind other proteins.19,175 Expression of the truncated SMNΔ7 protein may retain some functional ability.28,51,176
Further addition of amino acids to the N-terminus of SMNΔ7 appeared to stabilize the protein and corrected its cellular distribution.177 It was hypothesized that compounds that enabled the translational read-through of the stop codon in the SMNΔ7 transcript would stabilize the resulting SMN protein. Anti-terminator compounds have been shown to change the localization, stability, and function of the protein.178–181 The Δ7Stop reporter screen was created to identify novel compounds that promote read-through.181 In this reporter, the stop codon of the SMN transcript was placed upstream of Renilla luciferase. Renilla luciferase would only be expressed if termination was suppressed. Firefly luciferase was included as an internal control for normalization. For validation studies, the Δ7Stop reporter was transfected transiently into the AD293, a derivative of the HEK293 human embryonic kidney cell line that may have some neuronal characteristics.182 The aminoglycosides gentamicin, amikacin, tobramycin, and G418 and the HDACi VPA were tested in this assay. G418 and, to a lesser extent, amikacin were able to induce read-through. G418 increased read-through of the “UAG A” stop codon of the SMN gene by more than fivefold. This activity was also observed in transiently transfected C2C12 mouse muscle cells, Neuro-2a mouse neuroblastoma, and GM09677 SMA patient fibroblasts. Although G418 treatment in SMNΔ7 mice resulted in increases in SMN protein in the kidney, spinal cord, and brain and showed improved gross motor function, it did not elicit an increase in survival.181 The ideal small-molecule therapeutics would display specificity to the molecular target with minimal off-target activity or toxicity. Many antiterminators are known to be toxic, have poor bioavailability, and can affect multiple stop codons, including multiple stop codons within the same transcripts. This assay provides a means to identify new compounds that could safely target the stop codon in SMNΔ7 to stabilize this truncated form of the SMN protein.
An alternative explanation for the lack of stability and diminished function of the truncated SMN2 protein is the formation of a protein degradation signal, or degron, in the SMNΔ7 transcript.183 The juxtaposition of the final 10 amino acids (YG-box) in exon 6 with the EMLA in exon 8 may form a degradation signal for the SMNΔ7 protein.183 To detect changes in the stability of SMN and SMNΔ7, a degron reporter assay was constructed. A reporter gene was fused to the N-terminus of the cDNA encoding for SMN and SMNΔ7 proteins and transfected into HEK293 cells. C-terminal elongation of the SMNΔ7 protein, either through the addition of five amino acids or through the activity of antitermination compounds, increased the half-life of the protein, suggesting that the degron must be exposed at the end of the SMNΔ7 protein to be active. The half-lives of the SMNΔ7 and SMN reporters were increased in the presence of MG132 and lactacystin, but not NH4Cl, 3-methyladenine, or calpeptin. This is consistent with the observation that SMN is degraded by the proteasome.17,184 This assay can be expanded to identify compounds that promote SMN protein stability as a new strategy for treating SMA.
Multifunctional SMN Reporter
The reporters described above are designed to quantify specific steps in SMN2 gene expression separately. The next generation reporter was designed to detect simultaneous changes in SMN2 expression through multiple mechanisms. To achieve this, a new reporter was designed using the corresponding SMN promoter to drive the expression of either an SMN1 or an SMN2 construct that includes the cDNA for exons 1–5 fused in frame to the 6-7-8-luciferase splicing cassette.81 Expression from this reporter results in the production of the full-length SMN protein fused to luciferase. As with the splicing construct described above, luciferase is only in frame and expressed if exon 7 is included. The 3.4 kb promoter will respond to the treatment that modulates transcription. The splicing cassette will respond to the treatment that increases exon 7 inclusion. Inclusion of the entire SMN protein sequence allows for the detection of compounds that stabilize SMN mRNA or protein. This construct was cloned into an EBNA-based episomal expression vector and transfected into HEK293 cells. The selected reporter cell lines were responsive to overexpression of hTra2-β and treatment with a variety of compounds previously reported to increase SMN2 expression.81
These cells have been used to screen over one million compounds at four separate screening centers. The results of two of these screens have been reported, whereas hits from the others are still at the validation and development stages.33,81,89 This reporter assay was used to identify a series of 4-arylthiazolyl piperidines in a screen at the NIH Chemical Genomics Center.89 The lead compounds in this series have a half maximal response or EC50 values in the nanomolar range and increase the SMN protein expression up to twofold in SMA patient fibroblasts. These compounds also have favorable ADME profiles, oral bioavailability, and display CNS penetrance. RT-PCR analysis revealed only a slight increase in total SMN2-luciferase mRNA with no change in exon 7 inclusion, leading the authors to propose that the compounds are acting through a post-translational mechanism.
This assay was also used in a screen at the Laboratory for Drug Discovery in Neurodegeneration at the Harvard NeuroDiscovery Center.81 Two compounds were identified that increase SMN2 expression, the heterocyclic carboxamide LDN-75654 and the dihydroquinolone LDN-76070. These compounds had EC50s in the low micromolar range and increased SMN protein levels in SMA patient fibroblasts. RT-PCR analysis confirmed that LDN-76070 increased transcription of SMN2, but LDN-75654 did not change the amount of either transcription or exon 7 inclusion. Pulse chase studies with LDN-75654 demonstrated a threefold increase in the half-life of SMN protein (B. Burnett, the Uniformed Services University, pers. comm.). Preclinical pharmacokinetic analysis showed that both these compounds had good cell permeability and CNS exposure, but only LDN-76070 had favorable metabolic stability in mouse liver microsomes (K. Hodgetts, Harvard NeuroDiscovery Center, Laboratory for Drug Discovery in Neurodegeneration, pers. comm.). LDN-76070 was active in SMNΔ7 mice, increasing median survival over twofold and increasing SMN protein levels in the brain and spinal cord to 75% of that found in asymptomatic heterozygous littermates.33 Of note, the correlation between overall SMN expression and survival might not always be obvious. Survival in each specific mouse model of SMA is likely not driven just by overall expression levels of SMN, but by expression levels in specific cells types, which could differ from model to model.
The value of this screen is illustrated by the identification of novel SMN2-inducing compounds that function through multiple mechanisms: transcription, translation, and protein stabilization. This value is enhanced by the ability to use this screen to examine the efficacy of multiple compounds simultaneously and in combination. Compounds that increase SMN2 expression through distinct mechanisms could act synergistically to increase the activity with this reporter, whereas compounds that function through similar mechanisms would not. This quality might be used to identify compounds that could be used in combination to improve their therapeutic effects. It could also be used as a preliminary mechanistic screen by combining unknown compounds with compounds that have known molecular targets to determine if they act through similar or independent pathways.
Cellular Viability and Proliferation
The expression of SMN protein is required for cell proliferation.185 The Smn null mutation is embryonic lethal in mice.25 This is presumably due to the loss of the essential function of the SMN complex in snRNP biosynthesis.43,44 Despite this requirement for the SMN protein, decreased expression of the full-length functional SMN has little effect on the cell viability of most cells in the culture. However, inducible Smn knockdown in NIH3T3 mouse fibroblasts by RNAi has been shown to cause growth arrest and induce senescence.58 This cell line was used to develop a novel phenotypic screen for compounds that increase cell proliferation by increasing SMN expression or by inducing a change that compensates for the reduction of functional SMN protein.90
NIH3T3 cells were stably transfected with a cosmid containing the SMN2 gene and two clonal cell lines were selected, one with a high SMN2 copy number and a second expressing a lower copy number. Knockdown of Smn in the low copy number clone resulted in a growth arrest similar to that seen in the parental cell line. This growth arrest was partially corrected in the high copy number SMN2 cell line, confirming a correlation between SMN2 expression and proliferation in these cell lines.
The low copy number cell line was used to develop a high-throughput assay. This assay was scaled for use in 96-well plates, and an automated imaging technique was used to determine the cell number per well. These cells responded to lentiviral overexpression of SMN with nearly a threefold increase in the number of viable cells. Treatment with VPA, a small molecule previously shown to increase SMN expression from the SMN2 gene,186,187 also increased the number of cells 2.5-fold relative to the untreated Smn-depleted cells. The authors proposed that this assay is fit for high-throughput screening, but the results from such efforts have not been reported.
Considerations
Assays have been developed in immortalized cell lines that use surrogates to measure changes in endogenous SMN2 expression levels to identify small molecules that can enhance SMN protein expression (Fig. 3 and Table 4). A number of different approaches have been discussed, including reporter systems to quantify the effects on the SMN2 promoter, to determine the extent of exon 7 inclusion in the SMN2 transcript, and to measure SMN2 mRNA translation and/or protein stability, as well as reporters that determine multiple readouts simultaneously.
The value of any reporter system is dependent on the dynamic range, reproducibility, ability to capture relevant aspects of target biology, and stability of its response to treatment. With the exception of the NIH3T3 proliferation assay, each of the reporter assays described above were designed to be quantified independent of the level of SMN1 expression in the cells. As a result, the assays are not limited to cells that have homozygous inactivation of the SMN1 gene, allowing for the use of immortalized cell lines. These qualities have allowed the SMN2 reporter assays discussed in this review to be suitable for use in high-throughput screening.
Despite the advantages that accompany reporter assays, it is important to remember that these assays measure surrogates for SMN expression, not endogenous SMN expression itself. In the case of all reporter systems, changes in readout can result from reporter-specific artifacts. This should be tested for during secondary screening to confirm the activity of the primary screening hits arising from the use of reporter assays.188,189 Finally, it is critical to remember that while the use of immortalized cell lines may be more suitable to high-throughput screening, these are not the cells that are affected in SMA, which are primarily motor neurons and possibly skeletal muscle. Thus, reporters may not be able to fully recapitulate the proper regulation of SMN expression and function either in central motor neuron synapses or at the NMJ. It is therefore necessary to confirm the activity of molecules identified with these reporters using disease-relevant secondary assays, including those directly measuring SMN expression in appropriate cell types, such as the high-content motor neuron assays described in the following section.
Motor Neuron Assays and High-Content Screens
As described in the previous section, most high-throughput drug screening for SMA has been performed with reporter assays in easily-used immortalized cell lines, which typically are not from disease-relevant tissues. It is generally accepted that a primary manifestation of SMA is motor neuron loss.51 Ideally, both drug discovery and studies of disease mechanism should be performed in primary motor neurons from human patients or animal models, but this is technically challenging. Motor neurons are terminally differentiated cells, and as such, do not proliferate. It is not possible to easily isolate motor neurons from human patients, and isolation of motor neurons from murine or rat embryos results in a low cell yield. Because of these technical challenges, performing a drug discovery screen using motor neurons is extremely difficult. For these reasons, drug screening in SMA has rarely been performed in a disease-relevant cell type, and the lead compounds have not always been tested on human motor neurons before going into the clinic. It is likely that SMN function and stability are regulated uniquely in motor neurons, and therefore, utilizing motor neurons in drug discovery may yield new lead compounds to be tested in SMA patients. In this section, new technical advances will be discussed that now allow for SMA drug screening to be completed directly in motor neurons, either isolated from mice or differentiated from murine or human pluripotent stem cells.
High-content screening, or the measurement of multiple cell-based parameters simultaneously (most commonly by image analysis), goes hand in hand with the development of motor neuron screens for SMA. High-content screening can compensate for small numbers of motor neurons by using image analysis to count motor neurons and to measure parameters associated with motor neuron health, such as neurite length. High-content imaging is also advantageous in SMA-specific screens, as SMN is located within both the cytoplasm and the nucleus. High-content imaging allows for the measurement of SMN in each of these intracellular compartments and the measurement of SMN intensity in gems. Although it is not known whether increasing SMN in the nucleus, cytoplasm, and/or gems is the most therapeutically relevant, understanding how a lead compound changes SMN distribution in the cell is potentially important for understanding its mechanism of action. Therefore, an image-based approach has been undertaken using SMA patient fibroblasts.85 During this screen, Makhortova et al. found that inhibition of GSK-3 with small molecules leads to an increase in SMN protein stability, measured as an increase in SMN in the cytoplasm. Compounds that increased SMN in the nucleus and gems were also identified. Thus, high-content analysis maximizes the data obtained in a screen, which are particularly useful when cell number is limiting, such as in the case of motor neuron screening. By combining high-content screening with motor neuron culture, it is the hope that disease-relevant phenotypic assays can be established and utilized to discover novel therapeutics for SMA.
Screening in Primary Motor Neurons
Despite the challenges of screening in primary motor neurons from rodent models, this is an approach that has led to the identification of a small molecule currently in the clinic for SMA. Bordet et al. screened 40,000 compounds using spinal cord motor neurons purified from embryonic rats.97 Although the throughput of a primary motor neuron screen is limited by cell yield, the authors were able to use high-content imaging to offset this disadvantage. Imaging of calcein dye-stained motor neurons enabled the concurrent measurement of several parameters, including neurite outgrowth, the number of neurite branches, and motor neuron survival. In this case, high-content imaging both increased the data obtained from the primary screen and decreased the cell numbers needed per compound. It also allowed the prioritization of compounds that both kept motor neurons alive and maintained a healthy morphology. This screening approach led to the identification of olesoxime, which increases motor neuron survival in vitro, and extends lifespan in amyotrophic lateral sclerosis (ALS) mice.97,190 Although olesoxime treatment did not meet primary endpoints in a clinical trial for ALS,191 its efficacy in SMA patients is currently being evaluated. The pivotal trial has been completed, and publication of results is pending.
Screening in Murine Embryonic Stem Cell-Derived Motor Neurons
Although primary culture of motor neurons is challenging, advances in stem cell and developmental biology have led to the development of protocols for differentiating motor neurons from murine and human pluripotent stem cells.192–195 Because pluripotent stem cells readily expand in vitro, they can provide a continuous source of motor neuron cultures to be used in drug screening and phenotypic assays. Wichterle et al. first demonstrated that murine embryonic stem (ES) cells can be differentiated into motor neurons using the morphogens sonic hedgehog and retinoic acid (RA), which are known to be involved in motor neuron differentiation during development.192 These motor neurons are electrophysiologically active and can integrate into the chick spinal cord.192,196 The protocol is relatively short and scalable and thus provides an alternative to primary motor neuron culture for drug screening. Additionally, the starting population of ES cells can be derived from genetically modified mice, enabling the use of reporter genes and/or disease models.
This approach has been undertaken by Yang et al. in a murine ES cell-derived motor neuron survival screen.195 ES cells generated from both wild-type and ALS (SODG93A) mice were differentiated into motor neurons. The cells contained an HB9-GFP reporter, which allows for the rapid identification of the motor neurons. Generation of motor neurons from murine ES cells yields cultures that are 20%–30% GFP positive, so using a reporter and high-content imaging allowed the authors to count the number of motor neurons per well after 3 days of trophic factor withdrawal. The authors identified a small molecule, kenpaullone, which dramatically increases the number of motor neurons in vitro. Kenpaullone-treated motor neurons maintained a healthy morphology, have increased synapses compared with untreated motor neurons, and are electrophysiologically active. When compared to olesoxime and dexpramipexole, two compounds that have not been successful in ALS clinical trials, kenpaullone was more effective in promoting human motor neuron survival in the culture. It remains to be seen whether kenpaullone is effective in SMA or ALS animal models and/or patients, but if this proves to be the case, then it would support the use of in vitro motor neuron testing as a prerequisite for clinical trials in motor neuron diseases.
Another advantage of the stem cell-derived motor neuron model is the ability to test hit compounds across multiple disease models. Interestingly, kenpaullone promotes the survival of human pluripotent stem cell-derived motor neurons from wild-type, ALS, and SMA patients.195 Early information that a lead compound is effective in multiple diseases, which can be obtained by this kind of in vitro testing, can inform the preclinical approach and may lead to more rapid development of drugs that are effective in multiple diseases.
Screening in Human Induced Pluripotent Stem-Derived Motor Neurons
Another major technical advance with implications for SMA drug screening is the production of induced pluripotent stem (iPS) cells from human fibroblasts or other cell types isolated from SMA patients.197 iPS cells, like ES cells, are pluripotent and can be differentiated into multiple cell types, including motor neurons.193–195 A combination of dual SMAD inhibition to promote neuralization and RA/sonic hedgehog treatment will, over the course of 3–4 weeks, induce motor neuron differentiation of human iPS cells.194 As with murine ES-derived motor neurons, this is a continuous source of stem cell-derived motor neurons that can be used for drug discovery and phenotypic assays and has enabled the testing of lead compounds in motor neurons derived from patient cells for the first time.
While the protocol for producing motor neurons from human iPS cells is longer and more technically challenging than producing motor neurons from murine cells, it has two major advantages. First, it is most efficient way to study motor neurons derived from human cells and thus should provide new insights about human motor neuron biology. Second, iPS cells can be efficiently produced from relatively accessible patient cells, such as fibroblasts. This allows for the production of iPS cells from SMA patients95,198–202 and also facilitates the building of iPS cell collections that represent a range of patient genotypes and disease severities (L. Rubin, Harvard University, pers. comm.). This makes it possible to test drug candidates on motor neurons representing different patients. It is the hope of the field that in vitro testing of lead compounds across many patient genetic backgrounds will be predictive of efficacy in subsequent clinical trials and will perhaps even provide insight into which patient populations might benefit from a particular therapy.
Importantly, the ability to produce patient-specific neurons from iPS cells makes it feasible to conduct therapeutic screens in motor neurons. This approach has been undertaken in the laboratory of Dr. Lee Rubin at the Harvard University for neurodegenerative diseases, such as ALS, and is currently being explored for SMA. Proof of principle for small-molecule testing on human iPS-derived motor neurons has been established in both SMA and ALS. Egawa et al. differentiated iPS cells from familial ALS patients carrying TDP-43 mutations into motor neurons and found several disease-related phenotypes, such as shorter neurites and aggregated TDP-43.203 The authors tested four small molecules on TDP-43 mutant motor neurons and found that anacardic acid reverses the pathology of the TDP-43 motor neurons. It is easy to imagine how this assay could be used in an unbiased, high-throughput, and high-content screening approach.
In SMA, Garbes et al. investigated whether a link might exist between the response of an initial patient in a clinical trial and the in vitro response of iPS-derived neurons from the same patient.199 In this case, VPA was tested both on patient fibroblasts and on GABAergic neurons differentiated from SMA iPS cells. VPA, which has failed in SMA clinical trials, has been shown to increase SMN2 transcription in a subset of patients. When fibroblasts and iPS-derived neurons were tested from both responders and nonresponders from the clinical trial, there was a 66% concordance between the patients who responded to VPA in the clinic and the ability of VPA to increase SMN in cells derived from that patient. While it remains unclear why cells from some nonresponders did exhibit elevated SMN in vitro, it is possible that the concordance would have been higher if motor neuron cultures were used. The ability to compare clinical trial results with in vitro results, while in its infancy may yield insights into why some clinical trials are more successful than others. The goal is that eventually in vitro testing on iPS-derived motor neurons can predict patient response in a clinical trial.
Currently, in iPS-derived motor neuron, high-content screening approach is being used to identify small molecules that modulate SMN levels in the culture (Rubin, unpublished data) (Fig. 4). iPS cells from a type II SMA patient are differentiated into motor neurons over 28 days, treated with small molecules, and immunostained for SMN and the motor neuron marker Islet1. Quantitative image analysis is used to count the number of Islet1-positive motor neurons per well and to measure the SMN intensity in the nucleus and cytoplasm of motor neurons. In this case, high-content imaging is particularly advantageous since it allows SMN levels to be measured in Islet1-positive motor neurons, which are a subset of the total culture. Furthermore, the approach has led to the identification of compounds that increase SMN in the cytoplasm, nucleus, and throughout the cell. High-content imaging enables the prioritization of compounds that increase both total SMN levels and the number of motor neurons in the culture. It can also be used to prioritize lead compounds based on their ability to preserve a healthy neuronal morphology, thereby allowing for the exclusion of toxic compounds. Thus far, several promising compounds have been identified, and several of these are effective in motor neurons differentiated from multiple patient iPS lines (L. Rubin, Harvard University, pers. comm.).
Fig. 4.
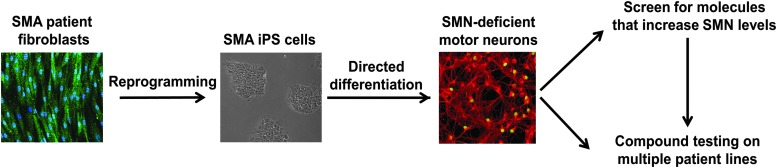
Schematic outline of the use of iPS cells in drug screening for SMA. SMA patient fibroblasts (green, SMN; blue, DAPI) can be reprogrammed to iPS cells (shown in brightfield). These pluripotent cells can be differentiated into motor neurons (red, neuronal marker Tuj1; green, motor neuron marker Islet1). These SMA patient-derived motor neurons can then be used for drug screening as well as for testing therapeutic candidates across multiple patient lines. iPS, induced pluripotent stem.
Considerations
It is widely believed that a primary manifestation of SMA is motor neuron loss.51 Ideally, drug discovery screens would be performed in primary motor neurons from patients or animal models. Traditionally, this has been technically challenging. However, advances in screening technology and in the ability to differentiate motor neurons in vitro has made this more feasible.
To date, motor neuron screens have yielded several novel drug candidates for SMA and other motor neuron diseases, although the field is still in its infancy and the numbers will only increase with time. In particular, the use of human SMA iPS-derived motor neurons in both drug screening and candidate drug testing is particularly promising. It is not, however, without its drawbacks: motor neuron assays are long, expensive, and labor intensive, and differentiation protocols yield cultures with variable quality and number of motor neurons. Fortunately, some of the technical challenges of working with primary or stem cell-derived motor neurons can now be overcome with the use of high-content imaging, and these techniques will be further refined in the years to come. Moreover, the ability to compare clinical trial results with in vitro results could provide important insights into why drugs are successfully in clinical trials or not. New screening technologies and greater ability to generate motor neurons in culture will provide greater insight to SMN biology and to identify therapeutics with the best chance of being efficacious in the treatment of SMA.
Secondary Assays to Assess the Effects of Compounds On SMN Function
Once the activity of a compound has been established in assays quantifying the levels of full-length and SMNΔ7 mRNAs and/or of full-length SMN protein, described in the preceding sections, the next step is the evaluation of the biological activity of the newly generated SMN protein. The main goal of this evaluation is to establish not only that SMN is expressed at a higher level but also that it is biologically active. Several types of assays can be used to assess SMN biological function. A brief overview of each assay is presented below.
snRNP Assembly
Of the several described or proposed functions of SMN protein, its role in snRNP biogenesis is the best characterized.43,44,204 SMN nucleates the assembly of all snRNPs and provides a specificity determinant for the assembly of the Sm protein core on U snRNAs. Formation of a splicing-competent spliceosome is dependent on SMN. When SMN levels drop below a certain threshold snRNP assembly is diminished.47,48,50 Increasing levels of SMN using genetic50 or pharmacologic approaches30,37 results in the restoration of snRNP assembly, with the degree of restoration correlating with the extent of SMN protein rescue. Therefore, quantification of snRNPs abundance provides a functional readout of SMN activity in the cell.
The snRNP assembly assay has been described for both cell extracts,44,47,48 as well as tissue homogenates.30–35,37,54,55,205 In addition, a version of the protocol compatible with high-throughput screening has been developed.206 The method includes the following main steps: (i) preparation of a cell extract or tissue homogenate and incubation with a synthetic radiolabeled U snRNA (typically at 10,000 cpm or between 100 pM and 1 nM RNA in a 20 μL reaction) in the presence of a test compound, (ii) immunoprecipitation against a protein component of the snRNP (typically using the anti-Sm antibody Y12), (iii) denaturation to release the RNA, and (iv) gel electrophoretic separation followed by autoradiography. The assay has been miniaturized down to 384-well microtiter plates to increase throughput. In this assay, a synthetic tagged RNA is captured by a tag–antitag interaction (e.g., biotin–streptavidin pair), and the amount of assembled snRNP is quantified using an anti-Sm antibody bound to a secondary antibody conjugated to a signal-generating moiety, such as horseradish peroxidase.206 Challenges in measuring snRNP assembly in a robust and reproducible manner include the multistep preparation of whole cell or nuclear extract, the temperature sensitivity of the extracts during storage, and variability in snRNP assembly capacity between different preparations of the extract.
Gems Count Assay
Once functional snRNPs have been assembled, they are imported, with SMN still bound, from the cytoplasm into the nucleus first appearing in Cajal bodies. Here, SMN is thought to disengage from the snRNPs and localize in adjacent separate structures termed gems (Gemini of coiled bodies), thus completing the snRNP maturation pathway.155,207,208 The formation of gems is nearly completely abolished in type I SMA fibroblasts in which the level of SMN protein is ∼30% of that in unaffected SMA carrier cells.153 Therefore, the number of gems may serve as a metric of the amount of SMN in the cell and perhaps also of SMN's functional state. The assay typically involves staining compound-treated cells with an SMN-specific antibody, co-staining the nucleus with a DNA dye, such as Hoechst 33258, followed by immunofluorescence microscopy.155,209 Results are expressed as the number of gems per 1 nucleus or the number of gems per 100 nuclei. Several drug discovery programs have incorporated the gems count assay in cultured cells or mouse tissues.39,84,180,210 The main challenge in the gems count assay is the appearance of SMN-immunoreactive gem-like granules in the nucleus of cells elicited as a result of stress,211 which can be induced by compound treatment (N. Naryshkin, unpublished results). Another possible confounding factor with the assay is the masking of weakly immunoreactive gems in the presence of high levels of SMN protein (S. Paushkin, SMA Foundation, pers. comm.). In most cases, an increase in the number of gems correlates with improved SMN function, but to be prudent, this assumption should also be confirmed with assays directly showing an increase in SMN protein function and/or SMN protein levels.
In Vitro Splicing
For expression of modulators that influence the alternative splicing of SMN2 exon 7, an in vitro splicing assay can be used to demonstrate the direct effect of a test compound on the generation of full-length and SMNΔ7 mRNAs. Typically, a radiolabeled pre-mRNA is prepared using in vitro runoff transcription, incubated with a nuclear extract from HeLa or another cell line of choice in the presence of the compound, and then the spliced RNA products are resolved and visualized by denaturing gel electrophoresis followed by autoradiography.12,212–214 This methodology has been applied to the evaluation of ASO-based and small molecule SMN2 expression modulators.12,82,83 Several challenges exist for this in vitro splicing assay, including the need to prepare a whole-cell or nuclear extract, the temperature sensitivity of the extracts during storage, and the possibility that not all mechanisms controlling the splicing of SMN2 exon 7 would be fully operational in the extract, outside the intact cell.
Cell Viability Assays
Another type of assay for evaluating SMN function is the cell viability assay, described in the Reporter Assays and High-Throughput Screens section. This assay relies on the use of mouse fibroblasts that carry the human SMN2 gene in which the mouse Smn gene is under the control of an inducible shRNA.58 Upon the knockdown of mouse Smn, these cells become proliferation impaired. Compounds that increase the expression of the full-length protein arising from the SMN2 gene rescue cell replication, an outcome that can be easily quantified.
Considerations
A number of different assays exist to measure the biological function of SMN protein. By their nature, functional assays provide information linking SMN gene expression to a biochemical process, providing a functional readout for the SMN activity. Assays such as snRNP assembly, gems count, and cell proliferation are best used in conjunction with the methods to quantify SMN mRNAs and protein directly. SMN functional assays provide an orthogonal validation of compound activity, helping to demonstrate not only that the level of SMN expression has been increased but also that the SMN protein produced is biologically active. For earlier stage compounds undergoing lead optimization, the ability to assess the biological activity of nascent SMN is critical. For more advanced drug programs assessing molecules that have already been optimized for both activity and biodistribution, the use of in vitro functional assays may be of lower importance. At later stages in the drug optimization process, the focus often shifts to testing compounds directly in animal models of SMA for survival and pharmacokinetics–pharmacodynamics (PK-PD) correlations. Such considerations play an important role in designing an integrated screening cascade used in drug optimization, as described in the next section.
Screening Cascade for the Identification of Optimized SMN2 Modulators
The goal of any preclinical drug program is the identification of a clinical candidate compound, which drug regulatory agencies would consider safe and efficacious for human clinical trials. For modulators of SMN2 gene expression, a clinical candidate needs to (i) demonstrate sufficient activity and selectivity against the target of SMN2 gene expression, (ii) show increased SMN protein levels, (iii) demonstrate an improvement in SMN-dependent biological processes, (iv) have appropriate pharmaceutical properties (e.g., blood–brain barrier penetration, metabolic stability, pharmacokinetics, and biodistribution), (v) alleviate the SMA phenotype in animal models, and (vi) be free of specific safety liabilities (e.g., neurotoxicity, cardiotoxicity, and carcinogenicity) with an overall acceptable general safety and toxicity profile.
At the start of a drug discovery program, a drug product profile is generated with a comprehensive set of desired properties and parameters that guide all drug development activities. Some of the descriptors can be calculated using chemoinformatic approaches,215–217 but the majority of the properties have to be experimentally measured in an array of biological profiling assays. The activities involved in characterizing compounds are organized into a screening cascade, which is then used to test and filter up to several thousand unique compounds to arrive ultimately at a clinical candidate. A generalized screening cascade for an SMN2 expression modulator program is shown in Figure 5.
Fig. 5.
A screening cascade for the identification of SMN2 expression modulators. Testing is organized in a sequential manner, with high-throughput in vitro activity and selectivity assays residing in the top tier, followed by profiling for drug-like properties and penetration into CNS and other target tissues in the second tier, determination of activity in SMA mouse models in the third tier, and assessment of in vivo safety in the fourth tier of testing. Predefined metrics need to be satisfied for progression between individual tiers. Data are collected at every tier and analyzed to build SAR models, as indicated by the information flow arrows on the left. New structural analogs with improved properties are generated via medicinal chemistry approaches and enter the screening funnel from the top. When a compound satisfying most of the product profile criteria is identified, the selected clinical candidate can progress into human clinical trials. CNS, central nervous system; SAR, structure–activity relationship.
With SMN2 modulators, cell-free and cell-based assays are utilized in the top tier of the cascade to enable a quick characterization of several hundred to several thousand compounds generated during structure–activity relationship (SAR) studies. Here, the emphasis is on the improvement of compound potency, biological activity, and selectivity. Depending on the targeted mechanism of SMN2 gene expression, the assay(s) of choice should provide a direct readout of that mechanism. For instance, for compounds that modulate alternative splicing of SMN2 and shift the balance of the splicing reaction toward the production of the SMN-FL mRNA, an RT-qPCR assay to determine the levels of SMN-FL and SMNΔ7 mRNAs would need to be performed (described in the SMN Transcript Assays section), as well as a protein assay (described in the SMN Protein Quantification section) and selectivity assays.
All newly made compounds would be tested for activity and selectivity in these assays, with just small sets of compounds also being tested in motor neuron assays and SMN functional assays. These latter assays are more laborious but generate richer information, providing a higher degree of confidence in biological activity and thus are utilized in spot-checking compounds of particular interest.
As more active and selective molecules are identified, they progress further down the screening cascade into pharmaceutical profiling assays (e.g., blood–brain barrier penetration, metabolic stability, PK, and biodistribution).218 Also at this stage, standard in vitro safety risk assessment assays are initiated, including cytotoxicity assays, Ames test for mutagenicity, hERG inhibition, and a receptor binding panel for off-target effects. The data-driven generation of structural analogs continues until advanced lead candidates with the desired balance of all of these properties are identified. At this stage, compounds are often tested in mouse models of SMA for efficacy.
Mouse models of SMA have been used extensively in SMA drug discovery research to assess the efficacy of small molecules. Due to the dedicated efforts of several groups, there is a diversity of available mouse models to test drug candidates (see Table 5 for details). These mouse models of SMA cover a wide range of pathological phenotypes from very severe mice that survive only a few days after birth,27 to those that exhibit overt progressive motor circuit and neuromuscular pathology,28,219,220 to intermediate SMA models,42,221,222 and to very mildly affected animals.26,41,49 A variety of compounds with different modes of action have been tested in mouse models of SMA, and the protocols for both SMN mRNA and protein expression as well as phenotypic readouts (e.g., survival, motor circuit structure and function, muscle mass, strength, and peripheral tissue necrosis) are well documented.30,37,39,82,106,119,205,223
Table 5.
Mouse Models of SMA Utilized in Candidate Drug Assessment
| Model | Genotype and JAXa Catalog No. | Phenotype | Endpoints | Advantages | Disadvantages | Refs. |
|---|---|---|---|---|---|---|
| Δ7 | Tg(SMN2)89Ahmb Smn1tm1Msd Tg(SMN2*delta7)4299Ahmb/J | Severe phenotype with median survival time ∼14 days. Mice die by postnatal day ∼21. |
SMN2 FL mRNA SMN protein Survival Muscle atrophy Motor behavior Central circuits and peripheral innervation Electrophysiology Tissue and organ pathology |
Carries human SMN2 allowing for facile quantification of the FL mRNA Useful to assess a variety of SMA endpoints, for example, electrophysiology Extensive literature on all therapeutic modalities |
Severe phenotype requiring early pharmacological intervention Presence of high level of Δ7 mRNA from the cDNA gene complicates quantification of Δ7 mRNA from SMN2 |
28,220 |
| 005025 | ||||||
| 006964 | ||||||
| 007952 | ||||||
| C/C-allele | Smn1tm5(Smn1/SMN2)Mrph/J | Mild phenotype with most pronounced phenotype of peripheral necrosis of ears, eyelids, and tail and paw edema. | Both SMN2 FL and Δ7 mRNAs using primers specific to SMN2 SMN protein Prevention of peripheral tissue necrosis Potential for select electrophysiology endpoints, for example, EIM Feasibility of additional functional endpoints is being studied |
All ages are accessible for evaluation Ease of oral dosing postweaning Both FL and Δ7 mRNA can be quantified Some electrophysiology endpoints may be feasible, for example, EIM |
Very mild phenotype, not allowing for full phenotypic assessment Paw edema/necrosis of ears, eyelids, and tail confounds treatment effects Pre-mRNA splicing from the hybrid Smn1–SMN2 gene differs from human SMN (skewed to Δ7 mRNA at 99% of product) |
41 Seward Rutkove of Beth Israel Deaconess Medical Center, pers. comm. N. Naryshkin and A. Dakka of PTC Therapeutics, unpublished data (on hybrid pre-mRNA splicing) |
| 008604 | ||||||
| 008714 | ||||||
| 008384 | ||||||
| Burgheron | Smn1tm5(Smn1/SMN2)Mrph/Tg(SMN2)89Ahmb Smn1tm1Msd | Intermediate phenotype with median survival time ∼50 days. Peripheral necrosis of ears, eye lids, and tail. |
SMN2 both FL and Δ7 mRNAs using primers specific to SMN2 SMN protein Survival Electrophysiology Tissue and organ pathology Peripheral tissue necrosis Motor behavior endpoints may be feasible |
Both FL and Δ7 SMN2 mRNAs can be quantified Useful to study intermediate SMA response to potential therapies |
Relatively complex genotype Pre-mRNA splicing from the hybrid Smn1–SMN2 gene differs from human SMN (skewed to Δ7 mRNA at 99% of product) Recent model, needs more characterization of survival times and phenotype |
Cathleen Lutz of JAXa, pers. comm.; Reference Guide to Mouse Models of Spinal Muscular Atrophyb |
| 2B/− | Smn1tm(Smn1-2B)/− | Intermediate phenotype with median survival time ∼30 days. | Mutant murine Smn1 splicing—both FL and Δ7 mRNAs Smn protein Survival Muscle atrophy Motor behavior Peripheral innervation Electrophysiology Tissue and organ pathology |
A model of intermediate SMA Useful to test SMN expression modifiers with mechanism of action common to human and murine Smn gene expression Useful to assess a variety of SMA endpoints, for example, electrophysiology Large published reference base |
Does not carry the human SMN2 gene and may not be suitable for testing of drugs working via SMN2-specific nucleotide sequences | 38,39 |
| Taiwanese | Smn1tm1Hung Tg(SMN2)2Hung/J | Varies from severe to very mild. Mice of all three types of SMA can be obtained. | Endpoints depend on which subpopulation of mice is studied Both SMN2 FL and Δ7 mRNAs SMN protein Survival Motor behavior Tissue and organ pathology Peripheral tissue necrosis |
Carries only full SMN2 gene Both FL and Δ7 mRNA can be quantified Severe, intermediate, and mild mice can all be born in the same litter |
Requires multistep breeding to isolate a single desired phenotype Severe, intermediate, and mild mice can be born from the same litter, complicating interpretation of treatment effects |
26,227 |
| 005058 | ||||||
| SMNRT | Tg(SMN2)89Ahmb Smn1tm1Msd Tg(SMN2*delta7-RT) | Intermediate phenotype with median survival time ∼20 days. |
SMN2 FL mRNA Survival Motor behavior endpoints may be feasible |
Carries human SMN2 allowing for facile quantification of the FL mRNA Useful to study intermediate SMA response to potential therapies |
Presence of Δ7 mRNA from the cDNA complicates quantification of Δ7 mRNA from SMN2 SMN protein measurement is complicated by stabilized SMNRT protein Recent model, needs more characterization of survival times and phenotype |
42 |
aJAX, The Jackson Laboratory (www.jax.org).
bLink to the reference guide can be found at https://forms.jax.org/121
CMAP, compound muscle action potential; EIM, electrical impedance myography; mRNA, messenger RNA; MUNE, motor number unit estimation.
Advanced lead compounds that have demonstrated sufficient activity, selectivity, and safety in vitro and in vivo move forward into formal good laboratory practice safety studies required by drug regulatory agencies for an IND application submission in the United States or a Clinical Trial Authorization (CTA) in Europe to begin human clinical trials. An in-depth evaluation of compound safety and toxicokinetics for an IND application typically requires testing in two different species, one of which is required to be a nonrodent. The theory and practice of preclinical drug safety assessment are well documented.224,225 Compounds with acceptable safety profiles are selected as clinical candidates and advanced to testing in human clinical trials.
Considerations
The SMA screening cascade integrates all relevant assays for potency, activity, selectivity, and pharmaceutical properties with the goal of identifying a clinical candidate via the iterative lead optimization process, which involves traditional medicinal chemistry, computational modeling, and empirical testing.218 Lead optimization is pursued until a suitable molecule with the best possible balance of characteristics and closest to that described in the original product target profile is identified.
The cascade described here is aimed to present a generalized case that would be applicable to most drug discovery and preclinical development programs with the goal of identifying modulators of SMN2 gene expression. Since the specific aims of a particular program may differ, the actual assays that populate the cascade for that program and the weight given to each assay in prioritizing compounds should be tailored to the goals of the specific program.
Conclusions
The discovery of the SMN2 gene and its ability to modulate SMN protein expression has led to an obvious therapeutic target for SMA, spurring a robust interest in the development of SMA therapies. Fifteen years ago, there were no ongoing drug discovery and development programs for SMA. Today, there are more than a dozen drug discovery projects being advanced for the treatment of SMA, with many representing strong collaborations between academia, government, industry, and nonprofit/advocacy groups. The drug discovery assets described in this review have been generated through the collective efforts of these stakeholders. These assets represent a collection of cellular assays, biochemical assays, and animal models that will facilitate further discoveries and innovation in SMA drug development.
Abbreviations Used
- AAV9
adeno-associated virus serotype 9
- ALS
amyotrophic lateral sclerosis
- ASF/SF2
alternative splicing factor1/pre-mRNA-splicing factor 2
- ASO
antisense oligonucleotide
- BLA
beta-lactamase
- cDNA
complementary DNA
- CMV
cytomegalovirus
- CNS
central nervous system
- cpm
counts per minute
- CTA
Clinical Trial Authorization
- DNA
deoxyribonucleic acid
- ECL
electrochemiluminescence
- ELISA
enzyme-linked immunosorbent assay
- EMLA
glutamic acid, methionine, leucine, alanine
- ES
embryonic stem
- FDA
Food and Drug Administration
- FRET
fluorescence resonance energy transfer
- HDACi
histone deacetylase inhibitors
- hERG
human Ether-à-go-go related gene
- HTS
high-throughput screening
- IND
Investigational New Drug
- iPS
induced pluripotent stem
- iTRAQ
isobaric tagging for relative and absolute quantification
- lncRNAs
long noncoding RNAs
- mRNA
messenger RNA
- NINDS
National Institute of Neurological Disorders and Stroke
- NMJ
neuromuscular junction
- PBMC
peripheral blood mononucleated cell
- PCR
polymerase chain reaction
- PK-PD
pharmacokinetics–pharmacodynamics
- RA
retinoic acid
- RNA
ribonucleic acid
- RNAi
RNA interference
- RT-PCR
reverse transcription–polymerase chain reaction
- RT-qPCR
reverse transcription–quantitative polymerase chain reaction
- SAR
structure–activity relationship
- SEAP
secreted alkaline phosphatase
- shRNA
small hairpin RNA
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleic particle
- SMA
spinal muscular atrophy
- SMN
survival of motor neuron
- SMN-FL
SMN full-length
- SMNΔ7
SMN delta exon 7
- SPR
surface plasmon resonance
- TR
time-resolved
- TSA
trichostatin A
- VPA
valproic acid
Acknowledgments
We are grateful to Shannon Taylor, Douglas E. Decker, and Roger A. Poorman of PharmOptima for assistance in providing data regarding the ECL SMN assay and for their review of the section on SMN protein quantification; to Amal Dakka, Joseph Colacino, and Paul Martin of PTC Therapeutics; to Claudia Mitchell of the LCMD2i Fund; and to Sergey Paushkin of the SMA Foundation for discussion and guidance on the article. We thank Jesse L. Mull for use of the patient fibroblast image in Figure 4. Families of SMA expresses our sincere gratitude to the SMA families, our regional chapters, and our supporters for their dedication in finding a treatment for SMA.
Disclosure Statement
Jonathan Cherry is an employee of Pfizer and is the lead biologist for the RG3039 program. Jill Jarecki is an employee of Families of SMA and has no financial interest in any SMA drug program or assay. Families of SMA has financial interests in SMA drug programs and assays. Dione T. Kobayashi is a consultant to for-profit and nonprofits in the CNS and rare disease space, holds options for Annexon (a company engaged in developing therapeutics for CNS disorders), and was engaged in the development of a commercial SMN ELISA and SMA biomarker panel; she has no financial interests in any SMA assays. Nikolai Naryshkin is an employee of PTC Therapeutics, Inc., which has a collaboration in the SMA area with the SMA Foundation and F. Hoffmann-La Roche, Inc. Research, and writing of this article was performed as part of Nikolai Naryshkin's duties as an employee of PTC Therapeutics, Inc. As an employee of PTC Therapeutics, Inc., Nikolai Naryshkin holds stock and options in PTC Therapeutics, Inc.
References
- 1.Crawford TO: From enigmatic to problematic: the new molecular genetics of childhood spinal muscular atrophy. Neurology 1996;46:335–340 [DOI] [PubMed] [Google Scholar]
- 2.McAndrew PE, Parsons DW, Simard LR, et al. : Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet 1997;60:1411–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugarman EA, Nagan N, Zhu H, et al. : Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens. Eur J Hum Genet 2012;20:27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearn J: Autosomal dominant spinal muscular atrophy: a clinical and genetic study. J Neurol Sci 1978;38:263–275 [DOI] [PubMed] [Google Scholar]
- 5.Wang CH, Finkel RS, Bertini ES, et al. : Consensus statement for standard of care in spinal muscular atrophy. J Child Neurol 2007;22:1027–1049 [DOI] [PubMed] [Google Scholar]
- 6.Zerres K, Rudnik-Schoneborn S: Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol 1995;52:518–523 [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre S, Burglen L, Reboullet S, et al. : Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995;80:155–165 [DOI] [PubMed] [Google Scholar]
- 8.Wirth B: An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat 2000;15:228–237 [DOI] [PubMed] [Google Scholar]
- 9.Lorson CL, Hahnen E, Androphy EJ, Wirth B: A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A 1999;96:6307–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monani UR, Lorson CL, Parsons DW, et al. : A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet 1999;8:1177–1183 [DOI] [PubMed] [Google Scholar]
- 11.Cartegni L, Krainer AR: Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 2002;30:377–384 [DOI] [PubMed] [Google Scholar]
- 12.Cartegni L, Krainer AR: Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol 2003;10:120–125 [DOI] [PubMed] [Google Scholar]
- 13.Kashima T, Manley JL: A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 2003;34:460–463 [DOI] [PubMed] [Google Scholar]
- 14.Kashima T, Rao N, David CJ, Manley JL: hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet 2007;16:3149–3159 [DOI] [PubMed] [Google Scholar]
- 15.Kashima T, Rao N, Manley JL: An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci U S A 2007;104:3426–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedrotti S, Bielli P, Paronetto MP, et al. : The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J 2010;29:1235–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH: Regulation of SMN protein stability. Mol Cell Biol 2009;29:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorson CL, Androphy EJ: An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet 2000;9:259–265 [DOI] [PubMed] [Google Scholar]
- 19.Lorson CL, Strasswimmer J, Yao JM, et al. : SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet 1998;19:63–66 [DOI] [PubMed] [Google Scholar]
- 20.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B: Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 2002;70:358–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefebvre S, Burlet P, Liu Q, et al. : Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 1997;16:265–269 [DOI] [PubMed] [Google Scholar]
- 22.Zheleznyakova GY, Kiselev AV, Vakharlovsky VG, et al. : Genetic and expression studies of SMN2 gene in Russian patients with spinal muscular atrophy type II and III. BMC Med Genet 2011;12:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prior TW, Swoboda KJ, Scott HD, Hejmanowski AQ: Homozygous SMN1 deletions in unaffected family members and modification of the phenotype by SMN2. Am J Med Genet A 2004;130A:307–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiDonato CJ, Chen X-N, Noya D, Korenberg JR, Nadeau JH, Simard LR: Cloning, characterization, and copy number of the murine survival motor neuron gene: homolog of the spinal muscular atrophy-determining gene. Genome Res 1997;7:339–352 [DOI] [PubMed] [Google Scholar]
- 25.Schrank B, Gotz R, Gunnersen JM, et al. : Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A 1997;94:9920–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh-Li HM, Chang JG, Jong YJ, et al. : A mouse model for spinal muscular atrophy. Nat Genet 2000;24:66–70 [DOI] [PubMed] [Google Scholar]
- 27.Monani UR, Sendtner M, Coovert DD, et al. : The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(-/-) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet 2000;9:333–339 [DOI] [PubMed] [Google Scholar]
- 28.Le TT, Pham LT, Butchbach ME, et al. : SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 2005;14:845–857 [DOI] [PubMed] [Google Scholar]
- 29.Butchbach ME, Edwards JD, Burghes AH: Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol Dis 2007;27:207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avila AM, Burnett BG, Taye AA, et al. : Trichostatin A increases SMN expression and survival in a mouse model of spinal muscular atrophy. J Clin Invest 2007;117:659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butchbach ME, Edwards JD, Schussler KR, Burghes AH: A novel method for oral delivery of drug compounds to the neonatal SMNDelta7 mouse model of spinal muscular atrophy. J Neurosci Methods 2007;161:285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butchbach ME, Singh J, Thorsteinsdottir M, et al. : Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum Mol Genet 2010;19:454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherry JJ, Osman EY, Evans MC, et al. : Enhancement of SMN protein levels in a mouse model of spinal muscular atrophy using novel drug-like compounds. EMBO Mol Med 2013;5:1103–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Khodor BF, Edgar N, Chen A, et al. : Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp Neurol 2008;212:29–43 [DOI] [PubMed] [Google Scholar]
- 35.Passini MA, Bu J, Richards AM, et al. : Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med 2011;3:72ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porensky PN, Mitrpant C, McGovern VL, et al. : A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum Mol Genet 2012;21:1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Meerbeke JP, Gibbs RM, Plasterer HL, et al. : The DcpS inhibitor RG3039 improves motor function in SMA mice. Hum Mol Genet 2013;22:4074–4083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowerman M, Beauvais A, Anderson CL, Kothary R: Rho-kinase inactivation prolongs survival of an intermediate SMA mouse model. Hum Mol Genet 2010;19:1468–1478 [DOI] [PubMed] [Google Scholar]
- 39.Gogliotti RG, Cardona H, Singh J, et al. : The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum Mol Genet 2013;22:4084–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hua Y, Sahashi K, Hung G, et al. : Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 2010;24:1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborne M, Gomez D, Feng Z, et al. : Characterization of behavioral and neuromuscular junction phenotypes in a novel allelic series of SMA mouse models. Hum Mol Genet 2012;21:4431–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cobb MS, Rose FF, Rindt H, et al. : Development and characterization of an SMN2-based intermediate mouse model of spinal muscular atrophy. Hum Mol Genet 2013;22:1843–1855 [DOI] [PubMed] [Google Scholar]
- 43.Liu Q, Fischer U, Wang F, Dreyfuss G: The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 1997;90:1013–1021 [DOI] [PubMed] [Google Scholar]
- 44.Pellizzoni L, Yong J, Dreyfuss G: Essential role for the SMN complex in the specificity of snRNP assembly. Science 2002;298:1775–1779 [DOI] [PubMed] [Google Scholar]
- 45.Kolb SJ, Battle DJ, Dreyfuss G: Molecular functions of the SMN complex. J Child Neurol 2007;22:990–994 [DOI] [PubMed] [Google Scholar]
- 46.Pellizzoni L: Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep 2007;8:340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabanella F, Carissimi C, Usiello A, Pellizzoni L: The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum Mol Genet 2005;14:3629–3642 [DOI] [PubMed] [Google Scholar]
- 48.Wan L, Battle DJ, Yong J, et al. : The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol 2005;25:5543–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monani UR, Pastore MT, Gavrilina TO, et al. : A transgene carrying an A2G missense mutation in the SMN gene modulates phenotypic severity in mice with severe (type I) spinal muscular atrophy. J Cell Biol 2003;160:41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Workman E, Saieva L, Carrel TL, et al. : A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum Mol Genet 2009;18:2215–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burghes AH, Beattie CE: Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 2009;10:597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z, Lotti F, Dittmar K, et al. : SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008;133:585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumer D, Lee S, Nicholson G, et al. : Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 2009;5:e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonne R: Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet 2011;20:641–648 [DOI] [PubMed] [Google Scholar]
- 55.Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L: Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2007;2:e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Q, Krainer AR: Splicing of a divergent subclass of AT-AC introns requires the major spliceosomal snRNAs. RNA 1997;3:586–601 [PMC free article] [PubMed] [Google Scholar]
- 57.Wu Q, Krainer AR: AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol Cell Biol 1999;19:3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lotti F, Imlach WL, Saieva L, et al. : An SMN-dependent U12 splicing event essential for motor circuit function. Cell 2012;151:440–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.See K, Yadav P, Giegerich M, et al. : SMN deficiency alters Nrxn2 expression and splicing in zebrafish and mouse models of spinal muscular atrophy. Hum Mol Genet 2014;23:1754–1770 [DOI] [PubMed] [Google Scholar]
- 60.Garcia EL, Lu Z, Meers MP, Praveen K, Matera AG: Developmental arrest of Drosophila survival motor neuron (Smn) mutants accounts for differences in expression of minor intron-containing genes. RNA 2013;19:1510–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, Pinto AM, Wan L, et al. : Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci U S A 2013;110:19348–19353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ: Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci 2003;23:6627–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fallini C, Bassell GJ, Rossoll W: Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res 2012;1462:81–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rossoll W, Jablonka S, Andreassi C, et al. : Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol 2003;163:801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piazzon N, Schlotter F, Lefebvre S, et al. : Implication of the SMN complex in the biogenesis and steady state level of the signal recognition particle. Nucleic Acids Res 2013;41:1255–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, Sendtner M: Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet 2002;11:93–105 [DOI] [PubMed] [Google Scholar]
- 67.Tadesse H, Deschenes-Furry J, Boisvenue S, Cote J: KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum Mol Genet 2008;17:506–524 [DOI] [PubMed] [Google Scholar]
- 68.Akten B, Kye MJ, Hao le T, et al. : Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci U S A 2011;108:10337–10342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fallini C, Zhang H, Su Y, et al. : The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci 2011;31:3914–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hubers L, Valderrama-Carvajal H, Laframboise J, Timbers J, Sanchez G, Cote J: HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum Mol Genet 2011;20:553–579 [DOI] [PubMed] [Google Scholar]
- 71.Piazzon N, Rage F, Schlotter F, Moine H, Branlant C, Massenet S: In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J Biol Chem 2008;283:5598–5610 [DOI] [PubMed] [Google Scholar]
- 72.Fallini C, Rouanet JP, Donlin-Asp PG, et al. : Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev Neurobiol 2014;74:319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Custer SK, Todd AG, Singh NN, Androphy EJ: Dilysine motifs in exon 2b of SMN protein mediate binding to the COPI vesicle protein alpha-COP and neurite outgrowth in a cell culture model of spinal muscular atrophy. Hum Mol Genet 2013;22:4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peter CJ, Evans M, Thayanithy V, et al. : The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum Mol Genet 2011;20:1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arnold WD, Burghes AH: Spinal muscular atrophy: development and implementation of potential treatments. Ann Neurol 2013;74:348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cherry JJ, Androphy EJ: Therapeutic strategies for the treatment of spinal muscular atrophy. Future Med Chem 2012;4:1733–1750 [DOI] [PubMed] [Google Scholar]
- 77.Lorson MA, Lorson CL: SMN-inducing compounds for the treatment of spinal muscular atrophy. Future Med Chem 2012;4:2067–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pruss RM, Giraudon-Paoli M, Morozova S, Berna P, Abitbol JL, Bordet T: Drug discovery and development for spinal muscular atrophy: lessons from screening approaches and future challenges for clinical development. Future Med Chem 2010;2:1429–1440 [DOI] [PubMed] [Google Scholar]
- 79.Van Meerbeke JP, Sumner CJ: Progress and promise: the current status of spinal muscular atrophy therapeutics. Discov Med 2011;12:291–305 [PubMed] [Google Scholar]
- 80.Lewelt A, Newcomb T, Swoboda K: New therapeutic approaches to spinal muscular atrophy. Curr Neurol Neurosci Rep 2012;12:42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cherry JJ, Evans MC, Ni J, Cuny GD, Glicksman MA, Androphy EJ: Identification of novel compounds that increase SMN protein levels using an improved SMN2 reporter cell assay. J Biomol Screen 2012;17:481–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hastings ML, Berniac J, Liu YH, et al. : Tetracyclines that promote SMN2 exon 7 splicing as therapeutics for spinal muscular atrophy. Sci Transl Med 2009;1:5ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR: Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 2007;5:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jarecki J, Chen X, Bernardino A, et al. : Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet 2005;14:2003–2018 [DOI] [PubMed] [Google Scholar]
- 85.Makhortova NR, Hayhurst M, Cerqueira A, et al. : A screen for regulators of survival of motor neuron protein levels. Nat Chem Biol 2011;7:544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Titus S, Marugan J, Southall N, et al. : High throughput screening for SMA. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD, 2010 [Google Scholar]
- 87.Wang J, Dreyfuss G: A cell system with targeted disruption of the SMN gene: functional conservation of the SMN protein and dependence of Gemin2 on SMN. J Biol Chem 2001;276:9599–9605 [DOI] [PubMed] [Google Scholar]
- 88.Xiao J, Marugan JJ, Zheng W, et al. : Discovery SA R and biological evaluation of aryl-thiazol-piperidines as SMN modulators. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD, 2010 [Google Scholar]
- 89.Xiao J, Marugan JJ, Zheng W, et al. : Discovery, synthesis, and biological evaluation of novel SMN protein modulators. J Med Chem 2011;54:6215–6233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li DK, Tisdale S, Espinoza-Derout J, Saieva L, Lotti F, Pellizzoni L: A cell system for phenotypic screening of modifiers of SMN2 gene expression and function. PLoS One 2013;8:e71965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Letso RR, Bauer AJ, Lunn MR, Yang WS, Stockwell BR: Small molecule screen reveals regulation of survival motor neuron protein abundance by Ras proteins. ACS Chem Biol 2013;8:914–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lunn MR, Root DE, Martino AM, et al. : Indoprofen upregulates the survival motor neuron protein through a cyclooxygenase-independent mechanism. Chem Biol 2004;11:1489–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang ML, Lorson CL, Androphy EJ, Zhou J: An in vivo reporter system for measuring increased inclusion of exon 7 in SMN2 mRNA: potential therapy of SMA. Gene Ther 2001;8:1532–1538 [DOI] [PubMed] [Google Scholar]
- 94.Corti S, Nizzardo M, Nardini M, et al. : Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy. J Clin Invest 2008;118:3316–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corti S, Nizzardo M, Simone C, et al. : Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med 2012;4:165ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wyatt TJ, Keirstead HS: Stem cell-derived neurotrophic support for the neuromuscular junction in spinal muscular atrophy. Expert Opin Biol Ther 2010;10:1587–1594 [DOI] [PubMed] [Google Scholar]
- 97.Bordet T, Buisson B, Michaud M, et al. : Identification and characterization of cholest-4-en-3-one, oxime (TRO19622), a novel drug candidate for amyotrophic lateral sclerosis. J Pharmacol Exp Ther 2007;322:709–720 [DOI] [PubMed] [Google Scholar]
- 98.Russell AJ, Hartman JJ, Hinken AC, et al. : Activation of fast skeletal muscle troponin as a potential therapeutic approach for treating neuromuscular diseases. Nat Med 2012;18:452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bosch-Marce M, Wee CD, Martinez TL, et al. : Increased IGF-1 in muscle modulates the phenotype of severe SMA mice. Hum Mol Genet 2011;20:1844–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Murdocca M, Malgieri A, Luchetti A, et al. : IPLEX administration improves motor neuron survival and ameliorates motor functions in a severe mouse model of spinal muscular atrophy. Mol Med 2012;18:1076–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rindt H, Buckley DM, Vale SM, et al. : Transgenic inactivation of murine myostatin does not decrease the severity of disease in a model of spinal muscular atrophy. Neuromuscul Disord 2012;22:277–285 [DOI] [PubMed] [Google Scholar]
- 102.Tsai LK, Chen YC, Cheng WC, et al. : IGF-1 delivery to CNS attenuates motor neuron cell death but does not improve motor function in type III SMA mice. Neurobiol Dis 2012;45:272–279 [DOI] [PubMed] [Google Scholar]
- 103.Rose FF, Jr., Mattis VB, Rindt H, Lorson CL: Delivery of recombinant follistatin lessens disease severity in a mouse model of spinal muscular atrophy. Hum Mol Genet 2009;18:997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sumner CJ, Wee CD, Warsing LC, et al. : Inhibition of myostatin does not ameliorate disease features of severe spinal muscular atrophy mice. Hum Mol Genet 2009;18:3145–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dominguez E, Marais T, Chatauret N, et al. : Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum Mol Genet 2011;20:681–693 [DOI] [PubMed] [Google Scholar]
- 106.Foust KD, Wang X, McGovern VL, et al. : Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol 2010;28:271–274 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 107.Passini MA, Bu J, Roskelley EM, et al. : CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest 2010;120:1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Valori CF, Ning K, Wyles M, et al. : Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci Transl Med 2010;2:35ra42. [DOI] [PubMed] [Google Scholar]
- 109.Wirth B, Garbes L, Riessland M: How genetic modifiers influence the phenotype of spinal muscular atrophy and suggest future therapeutic approaches. Curr Opin Genet Dev 2013;23:330–338 [DOI] [PubMed] [Google Scholar]
- 110.Shinomiya H: Plastin family of actin-bundling proteins: its functions in leukocytes, neurons, intestines, and cancer. Int J Cell Biol 2012;2012:213492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oprea GE, Krober S, McWhorter ML, et al. : Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 2008;320:524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hao le T, Wolman M, Granato M, Beattie CE: Survival motor neuron affects plastin 3 protein levels leading to motor defects. J Neurosci 2012;32:5074–5084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ackermann B, Krober S, Torres-Benito L, et al. : Plastin 3 ameliorates spinal muscular atrophy via delayed axon pruning and improves neuromuscular junction functionality. Hum Mol Genet 2013;22:1328–1347 [DOI] [PubMed] [Google Scholar]
- 114.Bowerman M, Shafey D, Kothary R: Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J Mol Neurosci 2007;32:120–131 [DOI] [PubMed] [Google Scholar]
- 115.Sen A, Dimlich DN, Guruharsha KG, et al. : Genetic circuitry of survival motor neuron, the gene underlying spinal muscular atrophy. Proc Natl Acad Sci U S A 2013;110:E2371–E2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dimitriadi M, Sleigh JN, Walker A, et al. : Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet 2010;6:e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pruss RM: Developments in the discovery of drugs for spinal muscular atrophy: successful beginnings and future prospects. Expert Opin Drug Discov 2011;6:827–837 [DOI] [PubMed] [Google Scholar]
- 118.Thurmond J, Butchbach ME, Palomo M, et al. : Synthesis and biological evaluation of novel 2,4-diaminoquinazoline derivatives as SMN2 promoter activators for the potential treatment of spinal muscular atrophy. J Med Chem 2008;51:449–469 [DOI] [PubMed] [Google Scholar]
- 119.Hua Y, Sahashi K, Rigo F, et al. : Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 2011;478:123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rigo F, Hua Y, Krainer AR, Bennett CF: Antisense-based therapy for the treatment of spinal muscular atrophy. J Cell Biol 2012;199:21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vezain M, Saugier-Veber P, Melki J, et al. : A sensitive assay for measuring SMN mRNA levels in peripheral blood and in muscle samples of patients affected with spinal muscular atrophy. Eur J Hum Genet 2007;15:1054–1062 [DOI] [PubMed] [Google Scholar]
- 122.Crawford TO, Paushkin SV, Kobayashi DT, et al. : Evaluation of SMN protein, transcript, and copy number in the biomarkers for spinal muscular atrophy (BforSMA) clinical study. PLoS One 2012;7:e33572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Simard LR, Belanger MC, Morissette S, Wride M, Prior TW, Swoboda KJ: Preclinical validation of a multiplex real-time assay to quantify SMN mRNA in patients with SMA. Neurology 2007;68:451–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sumner CJ, Kolb SJ, Harmison GG, et al. : SMN mRNA and protein levels in peripheral blood: biomarkers for SMA clinical trials. Neurology 2006;66:1067–1073 [DOI] [PubMed] [Google Scholar]
- 125.Tiziano FD, Lomastro R, Di Pietro L, et al. : Clinical and molecular cross-sectional study of a cohort of adult type III spinal muscular atrophy patients: clues from a biomarker study. Eur J Hum Genet 2013;21:630–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tiziano FD, Lomastro R, Pinto AM, et al. : Salbutamol increases survival motor neuron (SMN) transcript levels in leucocytes of spinal muscular atrophy (SMA) patients: relevance for clinical trial design. J Med Genet 2010;47:856–858 [DOI] [PubMed] [Google Scholar]
- 127.Brichta L, Holker I, Haug K, Klockgether T, Wirth B: In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann Neurol 2006;59:970–975 [DOI] [PubMed] [Google Scholar]
- 128.Heier CR, Gogliotti RG, DiDonato CJ: SMN transcript stability: could modulation of messenger RNA degradation provide a novel therapy for spinal muscular atrophy? J Child Neurol 2007;22:1013–1018 [DOI] [PubMed] [Google Scholar]
- 129.Gennarelli M, Lucarelli M, Capon F, et al. : Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem Biophys Res Commun 1995;213:342–348 [DOI] [PubMed] [Google Scholar]
- 130.Singh NN, Seo J, Rahn SJ, Singh RN: A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS One 2012;7:e49595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Setola V, Terao M, Locatelli D, Bassanini S, Garattini E, Battaglia G: Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc Natl Acad Sci U S A 2007;104:1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH: Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann Neurol 2005;58:194–202 [DOI] [PubMed] [Google Scholar]
- 133.Patrizi AL, Tiziano F, Zappata S, Donati MA, Neri G, Brahe C: SMN protein analysis in fibroblast, amniocyte and CVS cultures from spinal muscular atrophy patients and its relevance for diagnosis. Eur J Hum Genet 1999;7:301–309 [DOI] [PubMed] [Google Scholar]
- 134.Bonasera V, Alberti S, Sacchetti A: Protocol for high-sensitivity/long linear-range spectrofluorimetric DNA quantification using ethidium bromide. Biotechniques 2007;43:173–174, 176 [DOI] [PubMed] [Google Scholar]
- 135.Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408 [DOI] [PubMed] [Google Scholar]
- 136.Liu W, Saint DA: A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Anal Biochem 2002;302:52–59 [DOI] [PubMed] [Google Scholar]
- 137.Morrison TB, Weis JJ, Wittwer CT: Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 1998;24:954–958, 960, 962 [PubMed] [Google Scholar]
- 138.Tricarico C, Pinzani P, Bianchi S, et al. : Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem 2002;309:293–300 [DOI] [PubMed] [Google Scholar]
- 139.Gueudin M, Simon F: Plasma RNA viral load in HIV-1 group O infection by real-time PCR. Methods Mol Biol 2005;304:221–228 [DOI] [PubMed] [Google Scholar]
- 140.Castelain S, Descamps V, Thibault V, et al. : TaqMan amplification system with an internal positive control for HCV RNA quantitation. J Clin Virol 2004;31:227–234 [DOI] [PubMed] [Google Scholar]
- 141.Tichopad A, Pfaffl MW, Didier A: Tissue-specific expression pattern of bovine prion gene: quantification using real-time RT-PCR. Mol Cell Probes 2003;17:5–10 [DOI] [PubMed] [Google Scholar]
- 142.Bustin SA: Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 2000;25:169–193 [DOI] [PubMed] [Google Scholar]
- 143.Hindson BJ, Ness KD, Masquelier DA, et al. : High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 2011;83:8604–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhong Q, Bhattacharya S, Kotsopoulos S, et al. : Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab Chip 2011;11:2167–2174 [DOI] [PubMed] [Google Scholar]
- 145.Inglese J, Johnson RL, Simeonov A, et al. : High-throughput screening assays for the identification of chemical probes. Nat Chem Biol 2007;3:466–479 [DOI] [PubMed] [Google Scholar]
- 146.Sprangers R, Groves MR, Sinning I, Sattler M: High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. J Mol Biol 2003;327:507–520 [DOI] [PubMed] [Google Scholar]
- 147.Sarachan KL, Valentine KG, Gupta K, et al. : Solution structure of the core SMN-Gemin2 complex. Biochem J 2012;445:361–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mattis VB, Butchbach ME, Lorson CL: Detection of human survival motor neuron (SMN) protein in mice containing the SMN2 transgene: applicability to preclinical therapy development for spinal muscular atrophy. J Neurosci Methods 2008;175:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fuentes JL, Strayer MS, Matera AG: Molecular determinants of survival motor neuron (SMN) protein cleavage by the calcium-activated protease, calpain. PLoS One 2010;5:e15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Walker MP, Rajendra TK, Saieva L, Fuentes JL, Pellizzoni L, Matera AG: SMN complex localizes to the sarcomeric Z-disc and is a proteolytic target of calpain. Hum Mol Genet 2008;17:3399–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.La Bella V, Kallenbach S, Pettmann B: Post-translational modifications in the survival motor neuron protein. Biochem Biophys Res Commun 2004;324:288–293 [DOI] [PubMed] [Google Scholar]
- 152.Lam le T, Fuller HR, Morris GE: The gemin2-binding site on SMN protein: accessibility to antibody. Biochem Biophys Res Commun 2013;438:624–627 [DOI] [PubMed] [Google Scholar]
- 153.Young PJ, Le TT, thi Man N, Burghes AH, Morris GE: The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp Cell Res 2000;256:365–374 [DOI] [PubMed] [Google Scholar]
- 154.Kobayashi DT, Olson RJ, Sly L, et al. : Utility of survival motor neuron ELISA for spinal muscular atrophy clinical and preclinical analyses. PLoS One 2011;6:e24269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu Q, Dreyfuss G: A novel nuclear structure containing the survival of motor neurons protein. EMBO J 1996;15:3555–3565 [PMC free article] [PubMed] [Google Scholar]
- 156.Martin R, Gupta K, Ninan NS, Perry K, Van Duyne GD: The survival motor neuron protein forms soluble glycine zipper oligomers. Structure 2012;20:1929–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ogawa C, Usui K, Aoki M, et al. : Gemin2 plays an important role in stabilizing the survival of motor neuron complex. J Biol Chem 2007;282:11122–11134 [DOI] [PubMed] [Google Scholar]
- 158.Mitrpant C, Porensky P, Zhou H, et al. : Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PLoS One 2013;8:e62114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nguyen thi M, Humphrey E, Lam LT, et al. : A two-site ELISA can quantify upregulation of SMN protein by drugs for spinal muscular atrophy. Neurology 2008;71:1757–1763 [DOI] [PubMed] [Google Scholar]
- 160.Piepers S, Cobben JM, Sodaar P, et al. : Quantification of SMN protein in leucocytes from spinal muscular atrophy patients: effects of treatment with valproic acid. J Neurol Neurosurg Psychiatry 2011;82:850–852 [DOI] [PubMed] [Google Scholar]
- 161.Kobayashi DT, Decker D, Zaworski P, et al. : Evaluation of peripheral blood mononuclear cell processing and analysis for Survival Motor Neuron protein. PLoS One 2012;7:e50763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Masson JF, Barnhart M, Battaglia TM, et al. : Monitoring of recombinant survival motor neuron protein using fiber-optic surface plasmon resonance. Analyst 2004;129:855–859 [DOI] [PubMed] [Google Scholar]
- 163.Elliott MH, Smith DS, Parker CE, Borchers C: Current trends in quantitative proteomics. J Mass Spectrom 2009;44:1637–1660 [DOI] [PubMed] [Google Scholar]
- 164.Finkel RS, Crawford TO, Swoboda KJ, et al. : Candidate proteins, metabolites and transcripts in the biomarkers for spinal muscular atrophy (BforSMA) clinical study. PLoS One 2012;7:e35462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fuller HR, Man NT, Lam le T, et al. : The SMN interactome includes Myb-binding protein 1a. J Proteome Res 2010;9:556–563 [DOI] [PubMed] [Google Scholar]
- 166.Baker HN, Murphy R, Lopez E, Garcia C: Conversion of a capture ELISA to a Luminex xMAP Assay using a multiplex antibody screening method. J Vis Exp 2012;65:e4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ruggiu M, McGovern VL, Lotti F, et al. : A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol Cell Biol 2012;32:126–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Le TT, McGovern VL, Alwine IE, et al. : Temporal requirement for high SMN expression in SMA mice. Hum Mol Genet 2011;20:3578–3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Andreassi C, Jarecki J, Zhou J, et al. : Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 2001;10:2841–2849 [DOI] [PubMed] [Google Scholar]
- 170.Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J: The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet 1999;64:1365–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Monani UR, McPherson JD, Burghes AH: Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT). Biochim Biophys Acta 1999;1445:330–336 [DOI] [PubMed] [Google Scholar]
- 172.Cashman NR, Durham HD, Blusztajn JK, et al. : Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn 1992;194:209–221 [DOI] [PubMed] [Google Scholar]
- 173.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H: Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci U S A 2001;98:9808–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Singh J, Salcius M, Liu SW, et al. : DcpS as a therapeutic target for spinal muscular atrophy. ACS Chem Biol 2008;3:711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pellizzoni L, Charroux B, Dreyfuss G: SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A 1999;96:11167–11172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Frugier T, Tiziano FD, Cifuentes-Diaz C, et al. : Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet 2000;9:849–858 [DOI] [PubMed] [Google Scholar]
- 177.Hua Y, Zhou J: Modulation of SMN nuclear foci and cytoplasmic localization by its C-terminus. Cell Mol Life Sci 2004;61:2658–2663 [DOI] [PubMed] [Google Scholar]
- 178.Carrel TL, McWhorter ML, Workman E, et al. : Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J Neurosci 2006;26:11014–11022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wolstencroft EC, Mattis V, Bajer AA, Young PJ, Lorson CL: A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum Mol Genet 2005;14:1199–1210 [DOI] [PubMed] [Google Scholar]
- 180.Mattis VB, Bowerman M, Kothary R, Lorson CL: A SMNDelta7 read-through product confers functionality to the SMNDelta7 protein. Neurosci Lett 2008;442:54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Heier CR, DiDonato CJ: Translational readthrough by the aminoglycoside geneticin (G418) modulates SMN stability in vitro and improves motor function in SMA mice in vivo. Hum Mol Genet 2009;18:1310–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shaw G, Morse S, Ararat M, Graham FL: Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J 2002;16:869–871 [DOI] [PubMed] [Google Scholar]
- 183.Cho S, Dreyfuss G: A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev 2010;24:438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chang HC, Hung WC, Chuang YJ, Jong YJ: Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem Int 2004;45:1107–1112 [DOI] [PubMed] [Google Scholar]
- 185.Grice SJ, Liu JL: Survival motor neuron protein regulates stem cell division, proliferation, and differentiation in Drosophila. PLoS Genet 2011;7:e1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Brichta L, Hofmann Y, Hahnen E, et al. : Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet 2003;12:2481–2489 [DOI] [PubMed] [Google Scholar]
- 187.Sumner CJ, Huynh TN, Markowitz JA, et al. : Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann Neurol 2003;54:647–654 [DOI] [PubMed] [Google Scholar]
- 188.Cheng KC, Inglese J: A coincidence reporter-gene system for high-throughput screening. Nat Methods 2012;9:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thorne N, Shen M, Lea WA, et al. : Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem Biol 2012;19:1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sunyach C, Michaud M, Arnoux T, et al. : Olesoxime delays muscle denervation, astrogliosis, microglial activation and motoneuron death in an ALS mouse model. Neuropharmacology 2012;62:2346–2352 [DOI] [PubMed] [Google Scholar]
- 191.Lenglet T, Lacomblez L, Abitbol JL, et al. : A phase II-III trial of olesoxime in subjects with amyotrophic lateral sclerosis. Eur J Neurol 2014;21:529–536 [DOI] [PubMed] [Google Scholar]
- 192.Wichterle H, Lieberam I, Porter JA, Jessell TM: Directed differentiation of embryonic stem cells into motor neurons. Cell 2002;110:385–397 [DOI] [PubMed] [Google Scholar]
- 193.Boulting GL, Kiskinis E, Croft GF, et al. : A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol 2011;29:279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L: Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 2009;27:275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yang YM, Gupta SK, Kim KJ, et al. : A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell 2013;12:713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Miles GB, Yohn DC, Wichterle H, Jessell TM, Rafuse VF, Brownstone RM: Functional properties of motoneurons derived from mouse embryonic stem cells. J Neurosci 2004;24:7848–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Takahashi K, Tanabe K, Ohnuki M, et al. : Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–872 [DOI] [PubMed] [Google Scholar]
- 198.Ebert AD, Yu J, Rose FF Jr., et al. : Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 2009;457:277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Garbes L, Heesen L, Holker I, et al. : VPA response in SMA is suppressed by the fatty acid translocase CD36. Hum Mol Genet 2013;22:398–407 [DOI] [PubMed] [Google Scholar]
- 200.McGivern JV, Patitucci TN, Nord JA, Barabas ME, Stucky CL, Ebert AD: Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia 2013;61:1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sareen D, Ebert AD, Heins BM, McGivern JV, Ornelas L, Svendsen CN: Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS One 2012;7:e39113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Chang T, Zheng W, Tsark W, et al. : Brief report: phenotypic rescue of induced pluripotent stem cell-derived motoneurons of a spinal muscular atrophy patient. Stem Cells 2011;29:2090–2093 [DOI] [PubMed] [Google Scholar]
- 203.Egawa N, Kitaoka S, Tsukita K, et al. : Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med 2012;4:145ra104. [DOI] [PubMed] [Google Scholar]
- 204.Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G: The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell 2006;23:273–279 [DOI] [PubMed] [Google Scholar]
- 205.Porensky PN, Burghes AH: Antisense oligonucleotides for the treatment of spinal muscular atrophy. Hum Gene Ther 2013;24:489–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Wan L, Ottinger E, Cho S, Dreyfuss G: Inactivation of the SMN complex by oxidative stress. Mol Cell 2008;31:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Morris GE: The Cajal body. Biochim Biophys Acta 2008;1783:2108–2115 [DOI] [PubMed] [Google Scholar]
- 208.Sleeman JE, Lamond AI: Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol 1999;9:1065–1074 [DOI] [PubMed] [Google Scholar]
- 209.Coovert DD, Le TT, McAndrew PE, et al. : The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 1997;6:1205–1214 [DOI] [PubMed] [Google Scholar]
- 210.Mattis VB, Rai R, Wang J, Chang CW, Coady T, Lorson CL: Novel aminoglycosides increase SMN levels in spinal muscular atrophy fibroblasts. Hum Genet 2006;120:589–601 [DOI] [PubMed] [Google Scholar]
- 211.Hua Y, Zhou J: Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett 2004;572:69–74 [DOI] [PubMed] [Google Scholar]
- 212.Kataoka N, Dreyfuss G: Preparation of efficient splicing extracts from whole cells, nuclei, and cytoplasmic fractions. Methods Mol Biol 2008;488:357–365 [DOI] [PubMed] [Google Scholar]
- 213.Mayeda A, Krainer AR: Mammalian in vitro splicing assays. Methods Mol Biol 1999;118:315–321 [DOI] [PubMed] [Google Scholar]
- 214.Kataoka N, Dreyfuss G: A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J Biol Chem 2004;279:7009–7013 [DOI] [PubMed] [Google Scholar]
- 215.Moroy G, Martiny VY, Vayer P, Villoutreix BO, Miteva MA: Toward in silico structure-based ADMET prediction in drug discovery. Drug Discov Today 2012;17:44–55 [DOI] [PubMed] [Google Scholar]
- 216.Summerfield SG, Dong KC: In vitro, in vivo and in silico models of drug distribution into the brain. J Pharmacokinet Pharmacodyn 2013;40:301–314 [DOI] [PubMed] [Google Scholar]
- 217.Langowski J.1, Long A: Computational methods for the prediction of ADME and Toxicity. Adv Drug Deliv Rev 2002;54:253–254 [Google Scholar]
- 218.Di L, Kerns EH: Profiling drug-like properties in discovery research. Curr Opin Chem Biol 2003;7:402–408 [DOI] [PubMed] [Google Scholar]
- 219.Ling KK, Gibbs RM, Feng Z, Ko CP: Severe neuromuscular denervation of clinically relevant muscles in a mouse model of spinal muscular atrophy. Hum Mol Genet 2012;21:185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ling KK, Lin MY, Zingg B, Feng Z, Ko CP: Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One 2010;5:e15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bowerman M, Murray LM, Beauvais A, Pinheiro B, Kothary R: A critical smn threshold in mice dictates onset of an intermediate spinal muscular atrophy phenotype associated with a distinct neuromuscular junction pathology. Neuromuscul Disord 2012;22:263–276 [DOI] [PubMed] [Google Scholar]
- 222.Hammond SM, Gogliotti RG, Rao V, Beauvais A, Kothary R, DiDonato CJ: Mouse survival motor neuron alleles that mimic SMN2 splicing and are inducible rescue embryonic lethality early in development but not late. PLoS One 2010;5:e15887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mentis GZ, Blivis D, Liu W, et al. : Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron 2011;69:453–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bass A, Kinter L, Williams P: Origins, practices and future of safety pharmacology. J Pharmacol Toxicol Methods 2004;49:145–151 [DOI] [PubMed] [Google Scholar]
- 225.Valentin JP, Hammond T: Safety and secondary pharmacology: successes, threats, challenges and opportunities. J Pharmacol Toxicol Methods 2008;58:77–87 [DOI] [PubMed] [Google Scholar]
- 226.Wang IF, Reddy NM, Shen CK: Higher order arrangement of the eukaryotic nuclear bodies. Proc Natl Acad Sci U S A 2002;99:13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Gogliotti RG, Hammond SM, Lutz C, Didonato CJ: Molecular and phenotypic reassessment of an infrequently used mouse model for spinal muscular atrophy. Biochem Biophys Res Commun 2010;391:517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]