Abstract
The Sleeping Beauty (SB) and piggyBac (PB) DNA transposons represent an emerging new gene delivery technology, potentially suitable for human gene therapy applications. Previous studies pointed to important differences between these transposon systems, depending on the cell types examined and the methodologies applied. However, efficiencies cannot always be compared because of differences in applications. In addition, “overproduction inhibition,” a phenomenon believed to be a characteristic of DNA transposons, can remarkably reduce the overall transgenic rate, emphasizing the importance of transposase dose applied. Therefore, because of lack of comprehensive analysis, researchers are forced to optimize the technology for their own “in-house” platforms. In this study, we investigated the transposition of several SB (SB11, SB32, SB100X) and PB (mPB and hyPB) variants in various cell types at three levels: comparing the excision efficiency of the reaction by real-time PCR, testing the overall transgenic rate by detecting cells with stable integrations, and determining the average copy number when using different transposon systems and conditions. We concluded that high excision activity is not always followed by a higher transgenic rate, as exemplified by the hyperactive transposases, indicating that the excision and the integration steps of transposition are not strongly coupled as previously thought. In general, all levels of transposition show remarkable differences depending on the transposase used and cell lines examined, being the least efficient in human embryonic stem cells (hESCs). In spite of the comparably low activity in those special cell types, the hyperactive SB100X and hyPB systems could be used in hESCs with similar transgenic efficiency and with reasonably low (2–3) transgene copy numbers, indicating their potential applicability for gene therapy purposes in the future.
Introduction
Transposons were first exclusively considered to be genomic parasites, placing a heavy burden on the host genome. Later it was revealed that these mobile elements have played active roles in shaping the genetic materials of organisms (Kazazian, 2004; Feschotte and Pritham, 2007; Goodier and Kazazian, 2008), and additional studies further supported the idea that transposon activity could have a positive impact during evolution even in higher eukaryotic organisms (Singer et al., 2010; Shen et al., 2011). After cloning and isolating active elements from various sources, transposons have been used as molecular tools for genetic experiments, including mutagenesis screens or transgenesis experiments. DNA transposons are especially favorable for such purposes, mainly because most of them are transposed by a nonreplicative “cut and paste” mechanism (Ivics and Izsvák, 2004, 2006). For a long time, however, no DNA transposons were known to be active in vertebrates, which also limited human gene delivery and gene therapy application by these molecular devices. This burden was lifted with the “resurrection” of the artificial transposon Sleeping Beauty (SB), which can actively transpose in all vertebrate genomes tested, including human cells (Ivics et al., 1997). Since then, various other transposons of several species have been shown to be active in vertebrates (Miskey et al., 2003; Ding et al., 2005; Balciunas et al., 2006). In addition, tremendous efforts have been made to develop hyperactive transposase versions (Zayed et al., 2004; Baus et al., 2005; Pledger and Coates, 2005) and with the development of the SB100X system (Mátés et al., 2009), transposon-based gene delivery became a rational alternative to virus-based applications. Indeed, a clinical trial has already been initiated using the SB system (Williams, 2008).
At present, the most efficient and therefore the most widely used transposon-based applications are the SB, piggyBac (PB), and Tol2 systems, the latter two representing naturally active transposons derived from the cabbage looper moth (Trichoplusia ni) (Ding et al., 2005) and the medaka fish (Balciunas et al., 2006) genomes, respectively. All have high transgenic potential and can mobilize large cargos (Rostovskaya et al., 2012; Wang et al., 2014), which is a considerable factor as compared with viral packaging systems. For safety considerations in gene therapy, SB seems to be the most ideal system because of its favorable integration profile: there is no obvious preference at the genomic level, and therefore the delivered transgene is integrated randomly, lowering the risk of undesirable insertional mutagenesis (Galvan et al., 2009; Grabundzija et al., 2010; Izsvák et al., 2010; Meir et al., 2011; Burnight et al., 2012; M.A. Li et al., 2013). On the other hand, the clear advantage of the PB system comes when the traceless removal of the delivered transgene is favorable, as exemplified by removal of the reprogramming cassettes after the generation of induced pluripotent stem cells from somatic cells (Kaji et al., 2009; Woltjen et al., 2009; Yusa et al., 2009). In addition, this genetic manipulation is further enhanced with the development of an integration-deficient PB transposase mutant (X. Li et al., 2013). Nevertheless, there are various other aspects (e.g., transgene copy numbers, applications in difficult-to-transfect cell lines) that require careful adjustments and considerations, especially when developing future gene therapy methodologies, and novel DNA transposons with defined integration profiles may also be considered for such purposes (Kojima and Jurka, 2013).
There are only a few studies addressing systematic comparisons of available transposon-based systems, and they often concentrate on a limited number of cell lines, mainly tumor cell lines, that may not be representative for all cell types or for in vivo applications (Grabundzija et al., 2010; Huang et al., 2010; Doherty et al., 2012; Sharma et al., 2012). Moreover, specific interactions of the transposase proteins with endogenous cell type-specific factors may also influence the overall outcome of transposition, an important aspect that also requires further investigations. In this study, we systematically analyzed SB and PB transposition from several aspects: examining the excision efficiency of the transposase as compared with the transgenic rate and copy numbers, the overall end products of transposition. Moreover, we examined the phenomenon of overproduction inhibition (OI), a transposase dose-dependent inhibition of the reaction, which is believed to be a general characteristic of DNA transposons (Lohe and Hartl, 1996; Hartl et al., 1997; Lampe et al., 1998; Wilson et al., 2005; Claeys Bouuaert et al., 2013). We found that the choice of transgene, as well as the promoter identity, may also have a strong impact on the outcome of the assays. Besides investigating cultured cell lines, we also addressed all the raised issues in a human embryonic stem cell line, which represents a model system for tissue differentiation studies and for regenerative medicine investigations. The important finding of our investigations is that although the SB100X transposon system seemed to be performing better in several respects in cultured cell lines, embryonic stem cells seem to represent an equally less permissive environment for both SB and PB transposons. Our results underlie the importance of careful optimization of transposon-based gene delivery in the targeted cell type, from the choice of the delivered gene expression cassette, through the careful selection of the transposase system, until the thorough setup of the gene delivery protocol. With all these details cautiously addressed, transposon-based gene methodologies clearly represent promising new technologies for future gene therapy applications.
Materials and Methods
Cell lines and maintenance
Human embryonic kidney cells (HEK-293) and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 1% l-glutamine, and 1% penicillin–streptomycin (Life Technologies, Carlsbad, CA). The HUES9 embryonic stem cell line (originally provided by D. Melton, Harvard University, Cambridge, MA) was maintained essentially as described previously, using cells from passage 35 (Apáti et al., 2008).
Plasmid constructs, transfection methods, and transposition assays
Structures of the transposon plasmids used are shown in Fig. 1A. During the comparison studies, the exactly same expression cassette sequences were cloned between the corresponding SB or PB inverted repeat sequences. Transposase proteins were expressed from standard expression plasmids under the control of the cytomegalovirus (CMV) promoter; transposase levels were checked by Western blotting. During the experiments, the following transposase protein variants were used: for the SB system, the SB11, SB32, and SB100X variants (Mátés et al., 2009); for the PB system, the mPB (Cadinanos and Bradley, 2007) and hyPB (Yusa et al., 2011) variants.
FIG. 1.
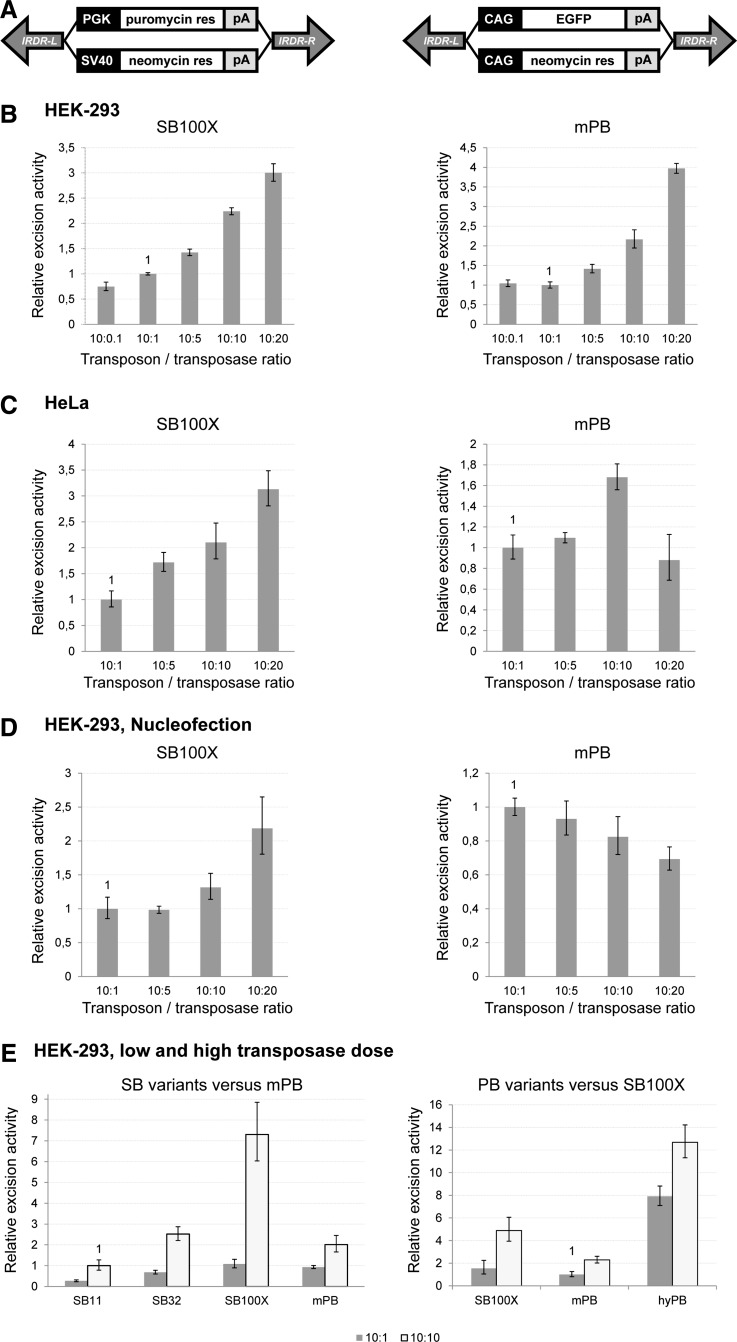
Transposon constructs and their application in studying overproduction inhibition on the level of excision of DNA transposons. (A) Schematic representation of the four DNA transposons used. IRDR-L/-R, inverted repeat-direct repeat sequences of SB and PB transposons (left and right); PGK, phosphoglycerate kinase promoter; SV40, simian virus 40 early promoter; CAG, CMV enhancer–chicken β-actin–rabbit β1-globin promoter; pA, polyadenylation signal. (B–D) Transposase dose dependence of excision efficiency: when using lipid-based transfection in (B) HEK-293 cells or (C) HeLa cells, or (D) when using nucleofection in HEK-293 cells. (E) Effect of the amount of transposase on the excision efficiency in the case of different SB and PB variants in HEK-293 cells.
All transfections were carried out in duplicate. For lipid-based transfections, 2×105 HEK-293 cells or 4×105 HeLa cells were seeded onto 12-well plates, and transfected with FuGENE 6 or FuGENE HD reagent, respectively, according to the manufacturer's instructions (Roche, Basel, Switzerland). The next day, a maximum of 250 ng of transposon plasmid was transfected with various amounts of transposase plasmid, except when the transposon was rate limiting at 25 ng (see Fig. 4C). All transfections were supplemented with 250 ng of pCMV-EGFP expression plasmid to control transfection efficiencies. In every experiment, transfection cocktails were supplemented with pET-41 plasmid if necessary, to ensure the same amount of total DNA in all reactions. At 48 hr posttransfection, cells were harvested and counted; one-tenth of them were analyzed with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) for green fluorescent protein (GFP)-expressing cells to measure transfection efficiency. For antibiotic selection, 1% of the transfected cells were seeded onto cell culture Petri dishes, selected for 2 weeks with either puromycin (1 μg/ml; Sigma-Aldrich, St. Louis, MO) or G418 (600 μg/ml; Sigma-Aldrich), and surviving cells were fixed with methanol and stained with Giemsa (Sigma-Aldrich). Colonies were quantified with a universal hood gel imager model 75S, using Quantity One 4.4.0 software (Bio-Rad, Hercules, CA). Transgenic rates were calculated from the colony numbers, normalizing with the number of seeded cells that was also normalized previously with transfection efficiencies; transgenic rates are therefore defined as a percentage of transfected cells.
FIG. 4.
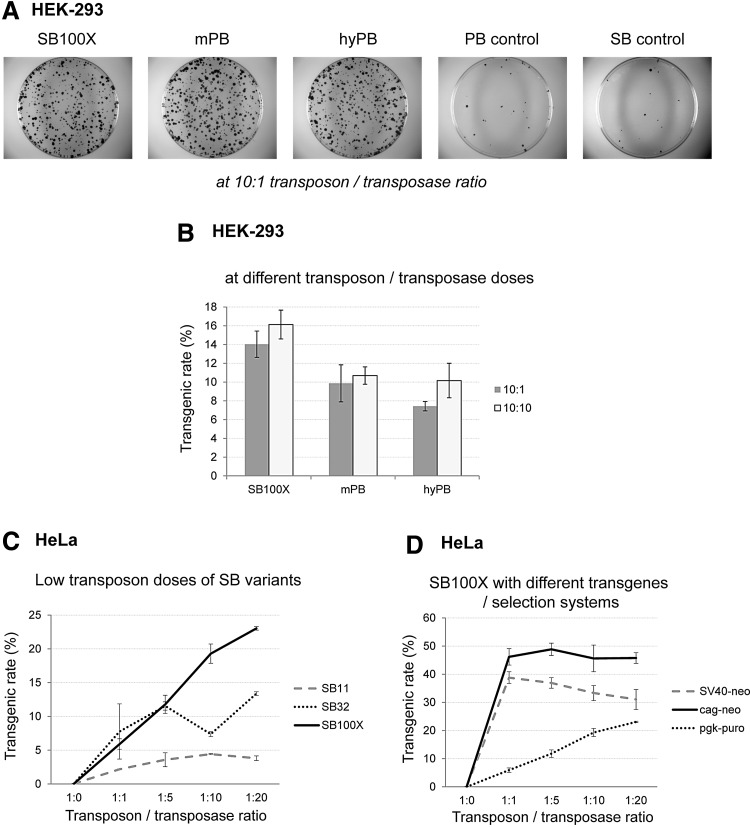
Comparing the transgenic rates of different transposon systems. (A) Representative colony assays and (B) transgenic rates of the PB systems versus SB100X. PB control: transfection carried out with the PB transposon cassette but without the transposase; SB control: transfection carried out with the SB transposon cassette and the mutant SB transposase. (C) Transposase dose dependence of transgenic rates at low transposon dose in HeLa cells. (D) Measured dose dependence of transgenic rates is strongly influenced by the transposon cassette sequences and thereby the selection methodology. Transgenic rates are defined as the transgene-carrying cells, as a percentage of total transfected cells (see also Materials and Methods). Error bars represent SD values.
In nucleofection experiments, 5×105 HEK-293 cells were electroporated, using a Nucleofector kit V (Lonza Group, Basel, Switzerland), according to the manufacturer's instructions. One microgram of transposon and various amounts of transposase, supplemented with 1 μg of transfection control plasmid, were transfected and seeded onto 6-well plates. Electroporations were also filled up to equal the amount of DNA. HEK nucleofections were analyzed 48 hr posttransfection for transfection efficiency by fluorescence-activated cell sorting (FACS) and for transposon excision efficiency by qPCR.
HUES9 cells were grown on feeder cells in 6-well plates until confluency; then ∼2×106 cells (one entire well per nucleofection) were harvested for nucleofection with a human stem cell Nucleofector kit 1 (Lonza Group). The same amounts of DNA were applied as in HEK-293 cells, but because the transposon in these cases contained the CAG-EGFP-expressing cassette, the transfection control plasmid could be omitted. At 48 hr posttransfection, cells were analyzed by FACS to determine transfection efficiency. At two later time points (1 and 2 weeks later), the ratio of GFP-expressing transgenic cells was also determined by FACS. Normalizing this ratio with the transfection efficiency resulted in the “GFP index,” which is proportional to the transgenic rate.
HEK-293 and HeLa transfection efficiencies were in the range of 40–70%, and HUES9 electroporations were within 10–15% efficiencies, but in simultaneous transfection experiments, deviations of transfection efficiencies showed only a small range (3–8%) of alterations.
Quantitative real-time PCR measurements
All quantitative real-time PCR measurements were carried out on a StepOnePlus real-time PCR system platform (Life Technologies); SYBR green and TaqMan reactions were performed according to the standard protocols. Data analysis was done with StepOne software version 2.1 (Life Technologies) and relative quantifications were performed by the ΔΔCt method; error bars represents confidence intervals of 95%. To quantify SB transposase-interacting factors at the mRNA level by SYBR methodology, the following primer pairs were used: for DNAPKcs (DNA-dependent protein kinase, catalytic subunit): sense primer, 5′-TTGAACACCATGTCCCAAGA; antisense primer, 5′-CTGACATTTTTGTCAGCCAATC; for HMGB1 (high-mobility group protein B1): sense primer, 5′-CATTGAGCTCCATAGAGACAGC; antisense primer, 5′-AGGATCTCCTTTGCCCATGT; for Ku70: sense primer, 5′-AGAGTGAAGATGAGTTGACACCTTT; antisense primer, 5′-CCAAGAGATCTCGATCACTGCT; for Ku80: sense primer, 5′-CATCTGATGCTACCAGATTTTGA; antisense primer, 5′-TCCATGCTCACGATTAGTGC; for HSP90 (heat shock protein 90, as endogenous control): sense primer, 5′-TGGATATCCCATTACTCTTTTTGTG; antisense primer, 5′-TTCTTTTTCTTCTTCTTTGTCTTCCT. Methods for determining transposon excision efficiency and copy numbers are described in the following sections in detail.
Quantifying transposon excision efficiency
After processing cells for transposition assays and for measuring transfection efficiency at 48 hr posttransfection, the remaining cells were used for the excision assay. Direct comparisons of various transposases were carried out in simultaneous experiments with similar transfection efficiency values to exclude minor discrepancies due to high transfection efficiency differences. Duplicated transfections were pooled and plasmids were isolated from the cells, using a modified protocol of the Qiagen plasmid miniprep kit, applying 300 μl of 1.2% sodium dodecyl sulfate (SDS) supplemented with 50 μg of proteinase K for the cell lysis step and protein removal from the DNA. Ten nanograms of each isolated plasmid DNA was used in a two-round nested PCR assay in which primers were specific to the plasmid backbone and resulted in a PCR product only from the excised and repaired plasmid copies due to large transposon cassettes. First-round PCR primers were as follows: 5′-GCGAAAGGGGGATGTGCTGCAAGG, 5′-TCTTTCCTGCGTTATCCCCTGATTC. Fifteen cycles were done in a 20-μl reaction volume with 2×PCR master mix (Promega, Madison, WI) and 200 nM primers. The first-round PCR profile was as follows: 94°C for 30 sec, 60°C for 15 sec, 72°C for 1 min. Second-round PCR primers were as follows: 5′-CAGCTGGCACGACAGGTTTCCCG, 5′-CGATTAAGTTGGGTAACGCCAGGG. A five-step fourfold dilution series (1024×) was done from each first-round PCR and 5 μl was introduced to the second-round PCR, which was the real-time PCR step, and it was carried out with 2×Power SYBR green PCR master mix (Life Technologies) using 50 nM concentrations of primers in triplicate 20-μl reactions. Excision PCR was normalized to the PCR from the ampicillin sequence, which was present exclusively in the transposon donor constructs, thus enabling quantification of the excised ratio of transposon plasmids passed into the cells. In this way we could compare excision efficiencies independent of transfection efficiency and cell type. PCR primers for the ampicillin sequence were as follows: 5′-TTTGCTCACCCAGAAACGC, 5′-AGTTGGCCGCAGTGTTATCAC. Ampicillin-specific PCRs were done similarly to the second-round excision PCR from the first-round excision PCR product, using the same serial dilutions. Efficiencies determined for all assays were in the range of 95–105%, so the ΔΔCt method proved to be suitable for quantification.
Determining transposon copy numbers
Average copy numbers of transgenic cell populations were measured from transfected and selected cells. Concerning the HEK-293 cell line, puromycin (1 μg/ml; Sigma-Aldrich) selection was carried out for 3 weeks, and then cells were harvested and duplicate transfections were pooled. Genomic DNA was isolated by the standard phenol–chloroform extraction method. For copy number measurements, the amount of input DNA was 90 ng and the self-designed TaqMan assays were used as previously published (Kolacsek et al., 2011). Briefly, specific assays were applied for the SB inverted repeat-direct repeat sequence-left region (SB-IRDR-L) transposon motif and the GFP-coding sequence; the latter assay was used to determine PB transposon copy numbers. Transposon-specific assays were normalized to RPPH1 endogenous control assay, and genomic DNA of cell clones with one transposon copy were also used as reference samples (Kolacsek et al., 2011). Concerning HUES9 cells, a GFP-expressing transposon cassette was used, and therefore these transfections were sorted at 48 hr for the GFP-expressing cells, and two additional sorts were carried out for the selection of transgenic cells. Average copy numbers of cell populations were measured from these sorted cells approximately 3 weeks after transfection.
Results
Cell type dependence of transposon excision rates: SB100X never exhibits overproduction inhibition at this level
The “cut and paste” mechanism of DNA transposons can be divided into at least two well-defined steps: the excision step from the donor locus and the integration step of the transposon into the host genome. We attempted to characterize these steps separately and to compare the PB and SB variants from these aspects. To avoid variability resulting from different DNA sequences, we cloned exactly the same transgene cassette expressing the puromycin resistance gene between the SB or PB inverted repeat (IR) sequences (Fig. 1A). The same amount of transposon-carrying plasmids was transfected with variable amounts of the given transposase-expressing plasmids into HEK-293 or HeLa cells by lipid-based transfection. Excision efficiencies were measured by real-time PCR-based quantification of the repaired donor plasmids isolated from the cells 48 hr posttransfection. Excision PCR was normalized to the PCR of the ampicillin sequence, which was exclusively present in the transposon donor constructs, thus enabling determination of the ratio of plasmids that underwent transposon excision to the total amount of transposon-carrying plasmids passed into the cell. In this way we can compare excision efficiencies in different cell types. In HEK-293 cells, excision efficiencies increased steadily with the amount of transposase for both the SB100X and mPB systems, whereas in HeLa cells, mPB excision activity showed a decline after a certain amount of transposase, resembling the overproduction inhibition phenomenon (Fig. 1B and C). To further increase the transfection efficiency and thereby the amount of expressed transposase in the cell, we used nucleofection technology, which is known to achieve higher transgene delivery efficiency (Aluigi et al., 2006). Using this method, the mPB transposase indeed showed decreasing excision efficiency even in HEK-293 cells with the increasing amount of transposase, whereas the SB100X excision rate still showed a positive correlation (Fig. 1D). In HEK-293 cells we also examined the OI phenomenon for other, less active SB versions (SB32 and SB11), as well as for a hyperactive PB (hyPB) version in several experiments. None of the transposases showed OI when raising the transposase dose from 10:1 to 10:10, and hyPB showed the highest excision activity, exceeding that of SB100X (Fig. 1E). Similarly, no OI was detected for the SB variants in HeLa cells (data not shown).
In comparing excision efficiencies between the two cell lines at fixed transposon/transposase amounts, the HEK-293 cell line seemed the most permissive among all examined systems (Fig. 2A). It is also notable that the hyperactive transposases highly exceeds all other variants in terms of excision efficiency, whereas from this point of view, mPB showed efficiency comparable to different SB variants in different cell lines: to SB32 in HEK-293 cells and to SB11 in HeLa cells. These correlations suggest that the cell type dependence of mPB might not be similar to that of SB. These results were independent of the transposon sequence because similar excision results were obtained with different transcription units such as CAG or CMV promoter-driven EGFP expression cassettes (data not shown).
FIG. 2.
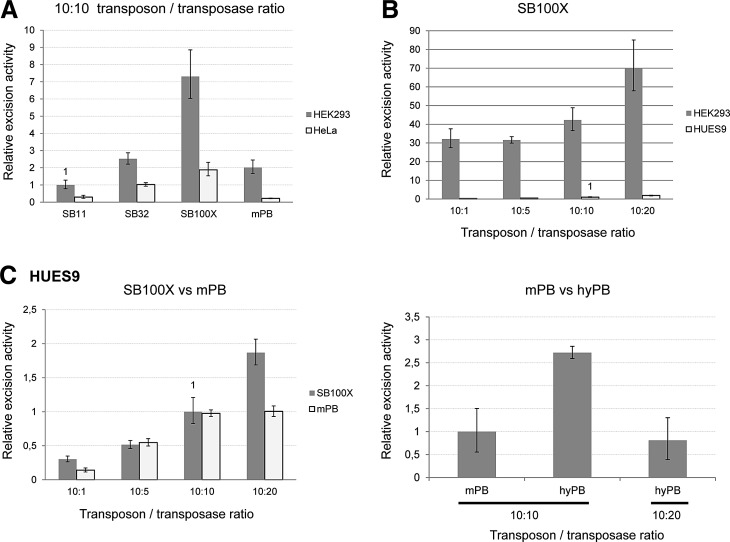
Comparing the excision efficiencies of SB and PB transposon systems in various cell types. (A) Excision efficiencies of various DNA transposons in HEK-293 versus HeLa cells at a fixed transposon-to-transposase ratio. (B) Comparison of SB100X excision efficiency in HEK-293 versus HUES9 cells. (C) Transposase dose dependence and comparison of excision efficiencies of SB100X, mPB, and hyPB in a human embryonic stem cell line, HUES9.
We also examined excision efficiency in the human embryonic stem cell (hESC) line HUES9, another relevant test system for gene therapy applications and tissue differentiation models. Here, increasing the amount of transposase also resulted in a higher excision rate of SB100X, whereas the efficiency of mPB showed more of a saturation-type reaction kinetic profile (Fig. 2C). However, for various transposase concentration points, mPB and SB100X had similar excision efficiencies, although these values were far below those measured in HEK-293 cells (Fig. 2B), or even in HeLa cells. When comparing mPB with hyPB, the hyperactive version had higher excision efficiency but showed OI at a higher transposase dose (Fig. 2C). To summarize this part of the study, the excision step of DNA transposition showed strong cell type dependence, with the hESCs being the least permissive cell type for this event. In addition, the SB100X reaction could not be saturated at this level, whereas under the conditions applied the PB excision rate (both for mPB and hyPB) was more sensitive to the amount of transposase present in the reaction.
Comparing transgenic rates of various transposon systems in various cell types
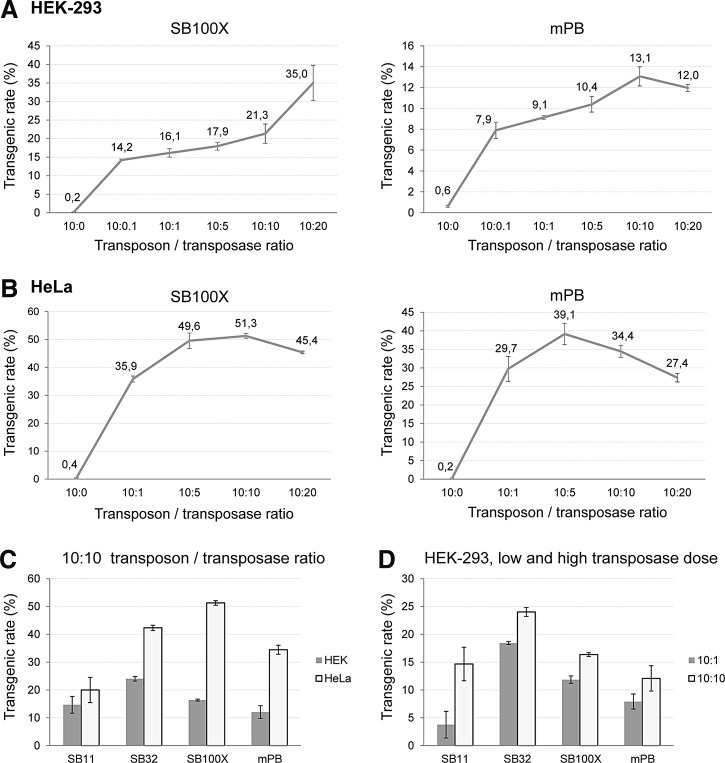
To determine the stable integration of transgenes into the host genomes, we performed colony-forming assays after transfection and puromycin selection of cells. Transgenic rates were determined in the same transfections as the excision assays, and were calculated from the number of detected colonies defined as a percentage of seeded and also transfected cells. The most prominent observation was that high excision efficiencies did not correlate with higher transgenic rates, and overproduction inhibition was also detectable at this level. When examining the HEK-293 cell line, overproduction inhibition could not be detected for either the SB100X transposase or the mPB system (Fig. 3A). In the HeLa cell line, transgenic rates of SB100X showed more of a saturation curve when plotted as a function of increasing transposase concentration (Fig. 3B), in contrast to the strong OI results reported previously (Grabundzija et al., 2010). On the other hand, mPB showed a tendency toward slightly higher OI at higher transposase dose (especially at 10:20; see Fig. 3B), in agreement with the excision assay and also in line with some previous data (Grabundzija et al., 2010) but not with others (Doherty et al., 2012).
FIG. 3.
Transgenic rates of SB and mPB systems and their transposase dose dependence in (A) HEK-293 cells and (B) HeLa cells. (C) Transgenic rates of different transposon systems in the two cell lines at a fixed transposon -to-transposase ratio. (D) Effect of low and high transposase doses on transgenic rates in HEK-293 cells. Transgenic rates are defined as the transgene-carrying cells, as a percentage of total transfected cells (see also Materials and Methods). Error bars represent SD values.
We also compared the two cell types for overall transgenic rate at fixed transposon/transposase amounts (10:10). HeLa cells showed the highest transgenic rates for hyperactive transposases examined, with SB100X having the highest values, and the SB32 variant exceeding the rate of mPB (Fig. 3C). Nevertheless, the HeLa cell line is reported to exhibit overproduction inhibition in terms of transgenic rates for most transposases, although for the SB100X variant our data partially contradict that previously reported, but mPB showed OI at both levels, which was in agreement with previous results (Grabundzija et al., 2010). An interesting feature revealed was that in spite of the high excision efficiencies in HEK-293 cells, transgenic rates for most transposases were found to be significantly lower than in HeLa cells (the only exception was the SB11 variant). Moreover, SB32 seemed to be more efficient in HEK-293 cells than SB100X even at a low transposase dose (Fig. 3D), and a high transposase dose of SB11 resulted in similar transgenic efficiency, like SB100X. In addition, we could not detect inhibition of less active SB variants at the transgenic level (Fig. 3D), and the transgenic rate of SB11 increased to a similar extent as its excision activity when raising the transposase dose (Fig. 1E vs. Fig. 3D). When examining the PB systems, in spite of exhibiting the highest excision activity in HEK-293 cells, the transgenic rate of the hyPB variant was similar to that of mPB even at different transposase doses, and it was lower than that of SB100X (Fig. 4A and B), which is not in agreement with some previous results (Doherty et al., 2012). Taken together, our results pointed out that despite the higher detectable excision rate of DNA transposons, HEK-293 cells seemed to be less permissive for stable genomic integration of transgenes, resulting in lower transgenic rates after transposition.
In previous studies, overproduction inhibition for SB variants was found to be more prominent at low transposon amounts, so we tested these conditions in our assay system with the more susceptible HeLa cell line. However, we could not see a decrease in transgenic rates with increasing amount of transposase and even for less hyperactive variants; rather, a saturation-type curve could be derived when plotting the data (Fig. 4C).
To determine whether the nature of the expression cassette has any effect on the transgenic rate, we tested the neomycin selection system in HeLa cells with different promoters (CAG or SV40) at low transposon dose (Fig. 4D). It turned out that they indeed influenced the kinetics of the transgenic rate, and slight inhibition was observed when exactly the same expression cassette (SV40 promoter-driven neomycin resistance gene) was used as in the previously published study (Grabundzija et al., 2010). These results emphasize the importance of the careful design of the expression cassette (e.g., choice of the promoter) carried by the transposon as it can severely influence the outcome and kinetics of the gene delivery and integration processes.
There are a number of proteins described to interact with the SB transposase, most of which are involved in DNA repair processes or in cell cycle regulation (Zayed et al., 2003; Izsvák et al., 2004; Walisko et al., 2006). As these proteins could differ in their cell type-specific distribution and activity, we measured the mRNA expression levels of four SB-interacting factors (DNAPKcs, HMGB1, Ku70, Ku80) in the HeLa and HEK-293 cell lines. Contrary to our expectations, the expression patterns of all examined factors were strikingly similar in both cell lines (data not shown), so at least their expression level differences cannot be attributed to the sharp differences in excision efficiency and in transgenic rates.
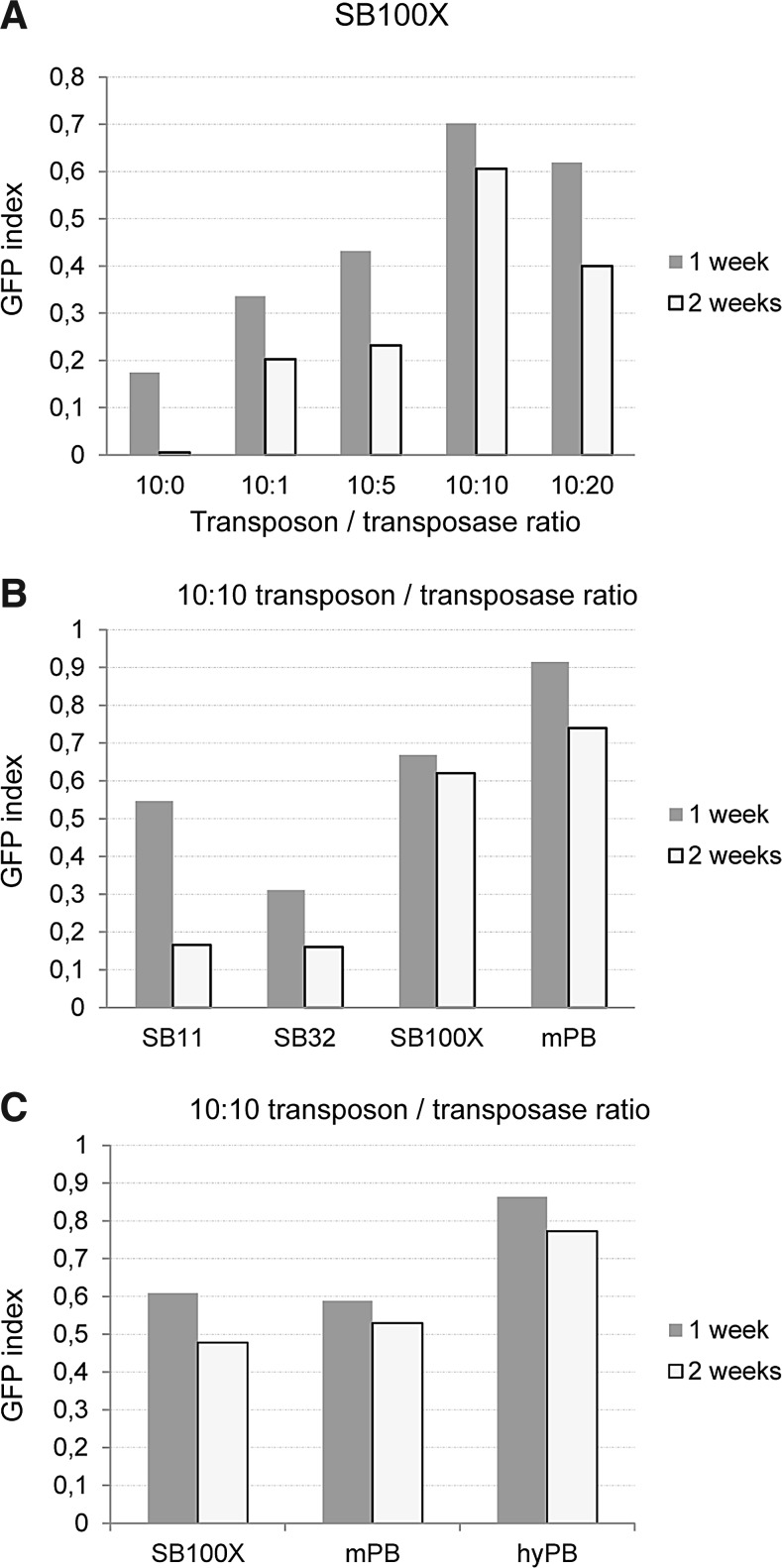
Overproduction inhibition at the level of the transgenic rate was also examined in hESCs. However, puromycin selection cannot be reliably applied to these cells as they express the ABCG2 membrane transporter protein, which can extrude the selecting compound from the cells (Sarkadi et al., 2010). Therefore, for these experiments we used a GFP-expressing transposon cassette. We measured the transfected cell ratio 48 hr posttransfection via FACS, and the ratio of transgene-expressing cells 1 and 2 weeks later. Normalizing the latter with the transfection efficiency resulted in the GFP index, which is proportional to the transgenic rate. SB100X showed a decline in GFP index at a 10:20 transposon/transposase dose in these cells (Fig. 5A), which might be attributable to overproduction inhibition detected only at the level of the transgenic rate. Here again, excision efficiencies cannot reliably predict the overall transposition rate, as the excision rate of SB100X showed a continuous increase in this range of transposase concentration (Fig. 2C). The GFP index of mPB showed more of a saturation-type kinetic curve in hESCs (data not shown), similar to what was measured for its excision efficiency (Fig. 2C). Under conditions in which SB100X and PB showed similar excision efficiency (10:10 ratio) we compared the SB and PB variants for the GFP index (Fig. 5B and C). In these experiments lower activity SB variants were notably less efficient for gene delivery in hESCs compared with SB100X, whereas both mPB and hyPB had higher transgenic rates than SB100X by 2 weeks posttransfection (Fig. 5C), which is in line with some previous data (Yusa et al., 2011). These results indicate that there are certain cell types in which the overall PB transposition rate can outweigh that of the SB100X system, and hESCs seem to be one example of these.
FIG. 5.
GFP indexes: the ratio of transfected cells that retain transgene expression in the HUES9 cell line. (A) Dose dependency of the GFP index when using SB100X, measured at two time points after transfection. (B) GFP indexes at a fixed transposon-to-transposase ratio of different SB variants versus the mPB transposon system. (C) GFP indexes at a fixed transposon-to-transposase ratio of different PB variants versus the SB100X transposon system.
Transgenic copy numbers: SB generally exceeds PB
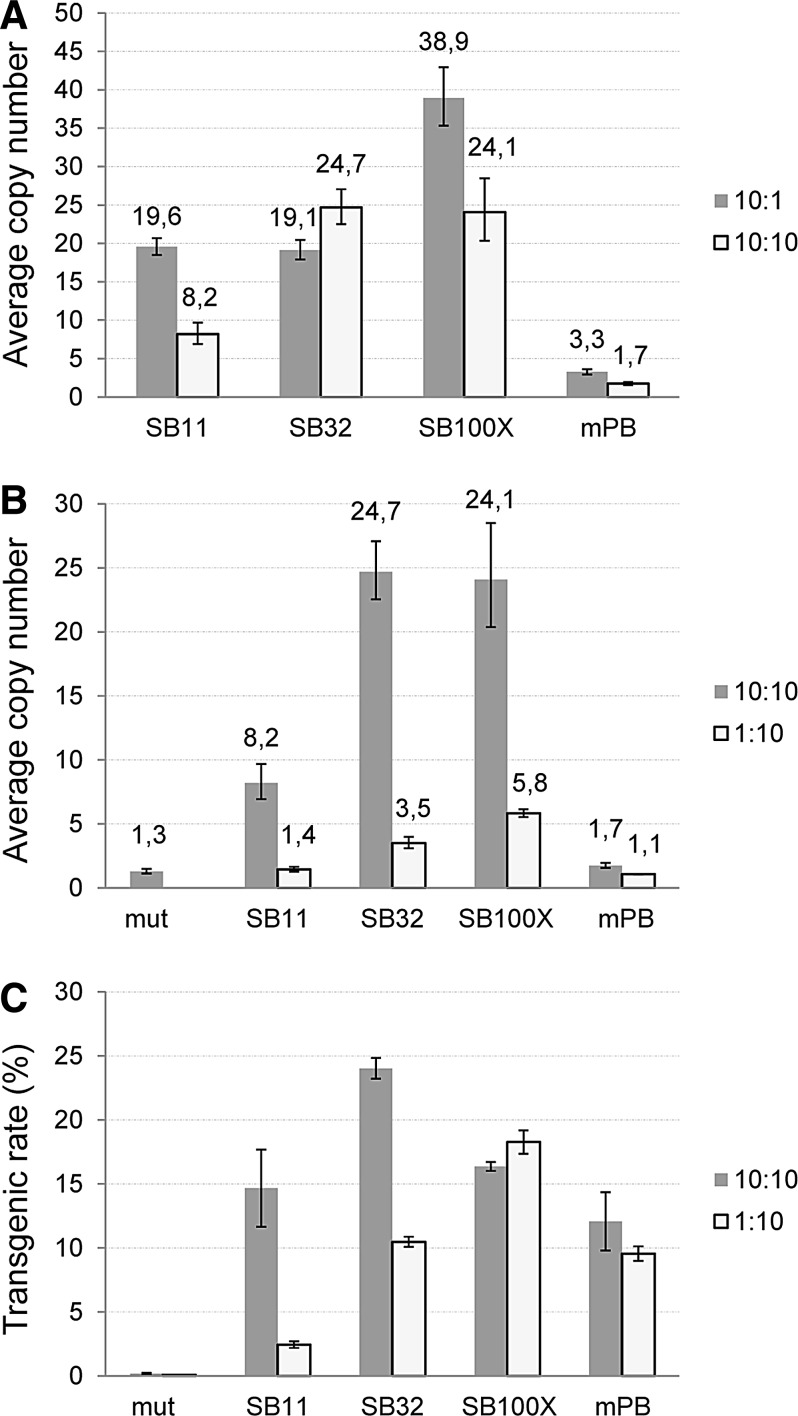
A crucial issue in any gene delivery experiment is to determine the stably integrated transgene copy numbers. In one of the former comparative experiments, in which SB variants and mPB were tested in HEK-293 cells at two different transposase doses (10:1 and 10:10; see Figs. 1E and 3D) we determined the average transposon copy numbers after 3 weeks of selection (Fig. 6A). In general, mPB transposition resulted in the lowest average copy numbers, lower than expected on the basis of excision and the transgenic rate comparisons (Figs. 1E and 3D), a finding that was in agreement with a previous study addressing copy number issues in HeLa cells (Grabundzija et al., 2010). Similar results were obtained with the hyPB version: the copy numbers were comparable to that measured for the mPB version, again showing a clear indication that the high excision activity of a hyperactive transposase may not necessarily indicate higher transgenic rates and/or copy numbers. Using a high amount of transposons with low transposase concentrations (10:1 in Fig. 6A), the SB variants produced high copy numbers (≥10) after 3 weeks of selection, with SB100X resulting in extreme copy numbers of even ≥40 in some of our experiments. In this case, high copy numbers seemed to be associated with the high excision efficiency of SB100X. However, it again does not necessarily mean higher transgenic rates because SB32 could exceed SB100X in this aspect (Fig. 3D).
FIG. 6.
Copy numbers of DNA transposons in HEK-293 cells. (A) Copy numbers resulting from different transposon systems at low versus high transposase dose. Limiting the amount of transposon also influenced (B) copy numbers and (C) transgenic rates when applying different DNA transposon systems. Mut, control transfection/transposition with a catalytically inactive SB transposase.
High transgene copy numbers are clearly unfavorable when considering gene therapy applications because, among other problems, it significantly increases the risk of insertional mutagenesis. It also places a heavy burden on any cellular genome, which is seen in experiments when transgenic cells were kept in culture for 6 weeks and the average copy number dropped significantly. Our experiments with HEK-293 cells indicate that average copy numbers exceeding 20 cannot be stably maintained in long-term culture. The most effective way to avoid such high copy numbers is to substantially lower the amount of transposon in the reaction. This can result in physiologically more acceptable copy numbers of 5–6 when using SB100X, without effectively lowering the transgenic rate (Fig. 6B and C). The same also works for the mPB transposase, lowering its copy number from ∼3 to 1.
Considering hESCs, however, the copy number profiles of DNA transposons were found to be different from the other examined cell types. The mPB system produced the characteristically low transgene copy numbers (∼2–3) that was observed in HEK-293 cells, whereas the SB100X transposase did not result in such extreme copy numbers as in other cell lines; both transposase systems resulted in an average copy number of ∼2 in HUES9 cells. Just like for excision efficiency and transgenic rates, this suggests that PB can work efficiently in hESCs despite the fact that these cell types seem to be less permissive for transgenic manipulations.
Discussion
The rationale behind our study was to carefully examine the two most common and reportedly most efficient vector systems, the Sleeping Beauty and piggyBac transposon vehicles, in normal (HEK-293) and tumorous (HeLa) model cells, as well as in human embryonic stem cells. We also aimed to characterize aspects of the transposition reaction that have not been investigated in detail so far: we compared the excision efficiency of the transposases with their transgenic rates (the overall outcome of transposition), addressed the presence of overproduction inhibition at both levels, and also shed light on the connection between these properties and the average transgene copy numbers resulting from transposon-based gene delivery.
To examine the excision phase of transposition, we studied this reaction by a real-time PCR method using SYBR green technology. As opposed to a previous study in which TaqMan chemistry was used (Jin et al., 2011), the potential occurrence of imprecise excision by the SB transposase (Liu et al., 2004) prompted us to apply a method that was not biased by the sequence variability of the repaired donor locus after excision, so all possible excision events could be confidently detected. Our studies revealed that transposon excision is strongly cell type dependent, indicating that certain cell types represent a more permissive environment for the reaction. In our experiments, the highest excision rates were detected in HEK-293 cells, in which the level could be a magnitude higher than in hESCs. An important finding was that the selection for hyperactive transposases often results in higher excision efficiency as exemplified by the SB100X and hyPB variants. However, overproduction inhibition (Lohe and Hartl, 1996; Hartl et al., 1997) may still affect the excision reaction in a cell type-dependent manner: hyPB was sensitive to the amount of transposase in hESCs, whereas the excision of SB100X could not be saturated under the conditions tested. The high excision efficiency of a hyperactive transposase, on the other hand, is not necessarily accompanied by higher transgenic rates or higher copy numbers: in spite of the highest excision values, the transgenic rates measured in HEK-293 cells were lower than in HeLa cells; moreover, SB32 may achieve higher transgenic rates in certain cell types than the hyperactive SB100X. Cell type-specific differences may be due to different cellular defense mechanisms and/or interacting factors. The higher number of transgenic clones in HeLa cells may be related to its cancerous phenotype and its higher “genomic buffer” capacity due to the higher number of chromosomes. An important conclusion therefore is that testing the excision efficiency of transposases is a good prescreen for activity; however, it may not be representative of the overall transposition activity.
An important result of our study was that in human embryonic stem cells much lower (one magnitude lower) activity of the examined systems was detected, even at the excision level of transposition. On the other hand, at this lower level of activity, there did not seem to be a significant difference between SB100X and the two PB variants, and both mPB and hyPB seemed to outweigh SB100X in transgenic rates. This was also detected on investigating transgene copy numbers: whereas SB100X delivered copy numbers significantly lower as compared with values in other cultured cell lines, use of the PB system resulted in the “expected” number of transgene copies. Nevertheless, we cannot rule out that for the PB system, the more permissive environment in embryonic stem cells is due to specific cell factors; this needs further investigation.
In our experiments, another important result was that the nature of the promoter and the transgene sequence, as well as the nature of the selection method, might influence the phenomenon of overproduction inhibition of the transgenic rate. Changing only the promoter of the neomycin transgene, the transposase dose–response curve showed different, rather saturation-type kinetics in the same cell line. However, when the selection method was changed to puromycin (the resistance gene driven by the housekeeping PGK promoter) the curve changed significantly (Fig. 4D), with lower transgenic rates and showing a constant increase with enzyme activity. These results point to the importance of and need to optimize new transposon constructs, but also indicate that increasing transposition efficiency might be achieved by increasing the transposase concentration when it is not contra-indicated (e.g., minimizing the probability of transposase integration in gene therapy applications).
An important aspect of gene delivery is the transgene copy numbers achieved by a given method. Mutagenesis screens can favor higher copy numbers to maximize the hits, whereas gene therapy applications typically favor low copy numbers to avoid genotoxicity by high gene dose or the unwanted burden of insertional mutagenesis. Our results clearly demonstrate that high excision activity is not necessarily associated with increased copy numbers as hyperactive transposases behave differently: as opposed to hyPB, SB100X can achieve high copy numbers. It can be concluded that excision of DNA transposons might not always be followed by integration, which is more pronounced in the case of the hyPB version. It is possible that for the PB system, the excision and integration phases of transposition are kinetically more distinct than for other DNA transposons. This could explain why the PB transposase could be used to remove the previously integrated transgenes (Kaji et al., 2009; Woltjen et al., 2009; Yusa et al., 2009) and why an integration mutant PB with merely excision activity could be generated (X. Li et al., 2013). For the SB100X variant, the key factor to manipulate copy numbers is to change the amount of the transposon, rather than the transposase (Fig. 6A and B). It is worth noting that the transgenic rate will reflect the transposase ab ovo activity only if the transposon amount is rate limiting (Figs. 3D and 6C), and this is exactly the case with gene delivery into difficult-to-transfect cells; that is why hyperactive transposase versions are substantially more efficient in these applications. The sole disadvantage of low average copy number is that it carries the possibility that transgenic rates may decrease even further due to OI because inhibition will raise the number of cells that will not receive even a single copy of transgene. In the case of higher average copy number, inhibition will result only in a decrease in average copy number but not in transgenic cell number. An alternative solution to the problem could be achieved by a method published by Cai and colleagues (2014): they demonstrated that the hyPB transposase embedded in a lentiviral Gag polypeptide could be used efficiently to generate single-copy clones while still keeping a high average transgenic rate.
In this study, we demonstrated strong cell type differences between SB and PB DNA transposons both at the excision level and in transgenic rate of transposition. We could provide additional data that the overproduction inhibition is not such a widespread phenomenon as previously anticipated and that careful experimental design could maximize the efficiency of gene delivery. An important finding of our study is that hyperactive DNA transposase versions seem to be selected for high excision efficiency, which is not necessarily coupled with either higher transgenic rate or increased transgene copy number. Nevertheless, the hyperactive transposases are definitely the method of choice both for insertional mutagenesis and for general gene delivery purposes. Our results also indicate that in terms of efficiency, the PB system seemed to outweigh SB100X in human embryonic stem cells, although its reported nonrandom integration profile (Wilson et al., 2007; Wang et al., 2008; Galvan et al., 2009; Liang et al., 2009) should be considered when such experiments are designed for gene therapy purposes.
Acknowledgments
Research in our laboratory was supported by grants from TransRat (KMR_12-1-2012-0112) and OTKA (NK83533).
Author Disclosure Statement
No competing financial interests exist.
References
- Aluigi M., Fogli M., Curti A., et al. (2006). Nucleofection is an efficient nonviral transfection technique for human bone marrow-derived mesenchymal stem cells. Stem Cells 24, 454–461 [DOI] [PubMed] [Google Scholar]
- Apáti A., Orbán T.I., Varga N., et al. (2008). High level functional expression of the ABCG2 multidrug transporter in undifferentiated human embryonic stem cells. Biochim. Biophys. Acta 1778, 2700–2709 [DOI] [PubMed] [Google Scholar]
- Balciunas D., Wangensteen K.J., Wilber A., et al. (2006). Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baus J., Liu L., Heggestad A.D., et al. (2005). Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol. Ther. 12, 1148–1156 [DOI] [PubMed] [Google Scholar]
- Burnight E.R., Staber J.M., Korsakov P., et al. (2012). A hyperactive transposase promotes persistent gene transfer of a piggyBac DNA transposon. Mol. Ther. Nucleic Acids 1, e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadinanos J., and Bradley A. (2007). Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 35, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Bak R.O., Krogh L.B., et al. (2014). DNA transposition by protein transduction of the piggyBac transposase from lentiviral Gag precursors. Nucleic Acids Res. 42, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys Bouuaert C., Lipkow K., Andrews S.S., et al. (2013). The autoregulation of a eukaryotic DNA transposon. Elife 2, e00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Wu X., Li G., et al. (2005). Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122, 473–483 [DOI] [PubMed] [Google Scholar]
- Doherty J.E., Huye L.E., Yusa K., et al. (2012). Hyperactive piggyBac gene transfer in human cells and in vivo. Hum. Gene Ther. 23, 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., and Pritham E.J. (2007). DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan D.L., Nakazawa Y., Kaja A., et al. (2009). Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J. Immunother. 32, 837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J.L., and Kazazian H.H., Jr (2008). Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell 135, 23–35 [DOI] [PubMed] [Google Scholar]
- Grabundzija I., Irgang M., Mátés L., et al. (2010). Comparative analysis of transposable element vector systems in human cells. Mol. Ther. 18, 1200–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D.L., Lozovskaya E.R., Nurminsky D.I., et al. (1997). What restricts the activity of mariner-like transposable elements. Trends Genet. 13, 197–201 [DOI] [PubMed] [Google Scholar]
- Huang X., Guo H., Tammana S., et al. (2010). Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol. Ther. 18, 1803–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z., and Izsvák Z. (2004). Transposable elements for transgenesis and insertional mutagenesis in vertebrates: A contemporary review of experimental strategies. Methods Mol. Biol. 260, 255–276 [DOI] [PubMed] [Google Scholar]
- Ivics Z., and Izsvák Z. (2006). Transposons for gene therapy! Curr. Gene Ther. 6, 593–607 [DOI] [PubMed] [Google Scholar]
- Ivics Z., Hackett P.B., Plasterk R.H., et al. (1997). Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91, 501–510 [DOI] [PubMed] [Google Scholar]
- Izsvák Z., Stuwe E.E., Fiedler D., et al. (2004). Healing the wounds inflicted by Sleeping Beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13, 279–290 [DOI] [PubMed] [Google Scholar]
- Izsvák Z., Hackett P.B., Cooper L.J., et al. (2010). Translating Sleeping Beauty transposition into cellular therapies: Victories and challenges. Bioessays 32, 756–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Maiti S., Huls H., et al. (2011). The hyperactive Sleeping Beauty transposase SB100X improves the genetic modification of T cells to express a chimeric antigen receptor. Gene Ther. 18, 849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K., Norrby K., Paca A., et al. (2009). Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 458, 771–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H.H., Jr (2004). Mobile elements: Drivers of genome evolution. Science 303, 1626–1632 [DOI] [PubMed] [Google Scholar]
- Kojima K.K., and Jurka J. (2013). A superfamily of DNA transposons targeting multicopy small RNA genes. PloS One 8, e68260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolacsek O., Krízsik V., Schamberger A., et al. (2011). Reliable transgene-independent method for determining Sleeping Beauty transposon copy numbers. Mob. DNA 2, 5 [Published erratum appears in Mob. DNA 2013;4:11.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe D.J., Grant T.E., and Robertson H.M. (1998). Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.A., Pettitt S.J., Eckert S., et al. (2013). The piggyBac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration sites. Mol. Cell. Biol. 33, 1317–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Burnight E.R., Cooney A.L., et al. (2013). piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. U.S.A. 110, E2279–E2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q., Kong J., Stalker J., et al. (2009). Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis 47, 404–408 [DOI] [PubMed] [Google Scholar]
- Liu G., Aronovich E.L., Cui Z., et al. (2004). Excision of Sleeping Beauty transposons: Parameters and applications to gene therapy. J. Gene Med. 6, 574–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A.R., and Hartl D.L. (1996). Autoregulation of mariner transposase activity by overproduction and dominant-negative complementation. Mol. Biol. Evol. 13, 549–555 [DOI] [PubMed] [Google Scholar]
- Mátés L., Chuah M.K., Belay E., et al. (2009). Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat. Genet. 41, 753–761 [DOI] [PubMed] [Google Scholar]
- Meir Y.J., Weirauch M.T., Yang H.S., et al. (2011). Genome-wide target profiling of piggyBac and Tol2 in HEK 293: Pros and cons for gene discovery and gene therapy. BMC Biotechnol. 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskey C., Izsvák Z., Plasterk R.H., et al. (2003). The Frog Prince: A reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31, 6873–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pledger D.W., and Coates C.J. (2005). Mutant Mos1 mariner transposons are hyperactive in Aedes aegypti. Insect Biochem. Mol. Biol. 35, 1199–1207 [DOI] [PubMed] [Google Scholar]
- Rostovskaya M., Fu J., Obst M., et al. (2012). Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res. 40, e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkadi B., Orbán T.I., Szakacs G., et al. (2010). Evaluation of ABCG2 expression in human embryonic stem cells: Crossing the same river twice? Stem Cells 28, 174–176 [DOI] [PubMed] [Google Scholar]
- Sharma N., Hollensen A.K., Bak R.O., et al. (2012). The impact of cHS4 insulators on DNA transposon vector mobilization and silencing in retinal pigment epithelium cells. PLoS One 7, e48421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Lin L., Cai J.J., et al. (2011). Widespread establishment and regulatory impact of Alu exons in human genes. Proc. Natl. Acad. Sci. U.S.A. 108, 2837–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T., McConnell M.J., Marchetto M.C., et al. (2010). LINE-1 retrotransposons: Mediators of somatic variation in neuronal genomes? Trends Neurosci. 33, 345–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walisko O., Izsvák Z., Szabó K., et al. (2006). Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc. Natl. Acad. Sci. U.S.A. 103, 4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin C., Lu D., et al. (2008). Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 105, 9290–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J., Devaraj A., et al. (2014). Suicidal autointegration of Sleeping Beauty and piggyBac transposons in eukaryotic cells. PLoS Genet. 10, e1004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D.A. (2008). Sleeping Beauty vector system moves toward human trials in the United States. Mol. Ther. 16, 1515–1516 [DOI] [PubMed] [Google Scholar]
- Wilson M.H., Kaminski J.M., and George A.L., Jr (2005). Functional zinc finger/Sleeping Beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 579, 6205–6209 [DOI] [PubMed] [Google Scholar]
- Wilson M.H., Coates C.J., and George A.L., Jr (2007). PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 15, 139–145 [DOI] [PubMed] [Google Scholar]
- Woltjen K., Michael I.P., Mohseni P., et al. (2009). piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458, 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Rad R., Takeda J., et al. (2009). Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods 6, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Zhou L., Li M.A., et al. (2011). A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U.S.A. 108, 1531–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed H., Izsvák Z., Khare D., et al. (2003). The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31, 2313–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed H., Izsvák Z., Walisko O., et al. (2004). Development of hyperactive Sleeping Beauty transposon vectors by mutational analysis. Mol. Ther. 9, 292–304 [DOI] [PubMed] [Google Scholar]