Abstract
This paper reports the 1H, 13C and 15N NMR experimental study of five benzimidazoles in solution and in the solid state (13C and 15N CPMAS NMR) as well as the theoretically calculated (GIAO/DFT) chemical shifts. We have assigned unambiguously the "tautomeric positions" (C3a/C7a, C4/C7 and C5/C6) of NH-benzimidazoles that, in some solvents and in the solid state, appear different (blocked tautomerism). In the case of 1H-benzimidazole itself we have measured the prototropic rate in HMPA-d18.
Keywords: CPMAS, DNMR, GIAO, proton transfer, tautomerism
Introduction
Of almost any class of heterocycles it can be said that they have relevant biological and medicinal chemistry properties, because, for instance, over 80% of top small molecule drugs by US retail sales in 2010 contain at least one heterocyclic fragment in their structures [1]. Benzimidazoles besides being the skeleton of many relevant drugs (fungicides, anthelmintics, antiulcerative, antiviral,…) [2–3] are also part of some natural products (the most prominent benzimidazole compound in nature is N-ribosyl-5,6-dimethylbenzimidazole, which serves as an axial ligand for cobalt in vitamin B12) and have interesting ferroelectric properties [4]. Particularly relevant for the present work is their proton conducting abilities, based of the 1,3-N–H···N hydrogen bonds, not only in benzimidazole polymers but in molecular compounds [5–6].
Degenerated tautomerism (autotrope) [7–8] simultaneously simplifies and complicates the NMR spectra of molecules in solution to the point that the assignment of some signals that become magnetically equivalent (isochronous) [9–10] by fast proton exchange has been much neglected. With the advent of solid-state NMR spectroscopy and the suppression of prototropic tautomerism, the assignment problem arises anew. In recent years, the use of very pure NMR solvents, particularly DMSO-d6, and highfield instruments has lead to obtain solution spectra where the prototropy has been considerably slowed down.
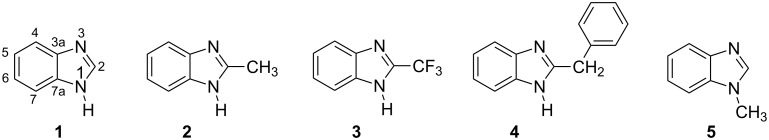
We present in this paper a study of four N-unsubstituted 1H-benzimidazoles (Figure 1) including 2-methyl-1H-benzimidazole (2) that shows in the solid state ferroelectric switching in two dimensions due to its pseudo-tetragonal crystal symmetry [4], and 2-benzyl-1H-benzimidazole (4) a vessel-dilating and spasm-reducing agent known as dibazol or bendazole [3], where the tautomerism has been blocked resulting in the concomitant problem of assignment of some signals. We have selected 1-methyl-1H-benzimidazole (5) as the simplest benzimidazole without tautomerism.
Figure 1.
The five studied compounds.
The use of theoretically calculated chemical shifts has been decisive to solve this problem. We have used from 2001 [at the 6-31+G(d) level] [11] and then, from 2007, at the 6-311++G(d,p) level [12], a statistical approach that consists in comparing GIAO calculated absolute shieldings (σ, ppm) with experimental chemical shifts (δ) determined either in solution or in the solid state:
δ1H = 31.0 – 0.97 σ1H [13]
δ13C = 175.7 – 0.963 σ13C [12]
δ15N = –154.0 – 0.874 σ15N [12]
These equations give excellent results except for atoms (generally, carbon) linked to halogen atoms (I > Br > Cl >> F) [11,14] where relativistic corrections are necessary [15].
Results and Discussion
The experimental data are all original although averaged values of all nuclei have been reported for the three NH-benzimidazoles 1, 2 and 4 [16–21]; besides 13C and 15N NMR chemical shifts of 5 have been published [22–23].
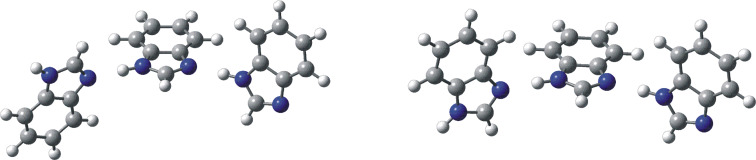
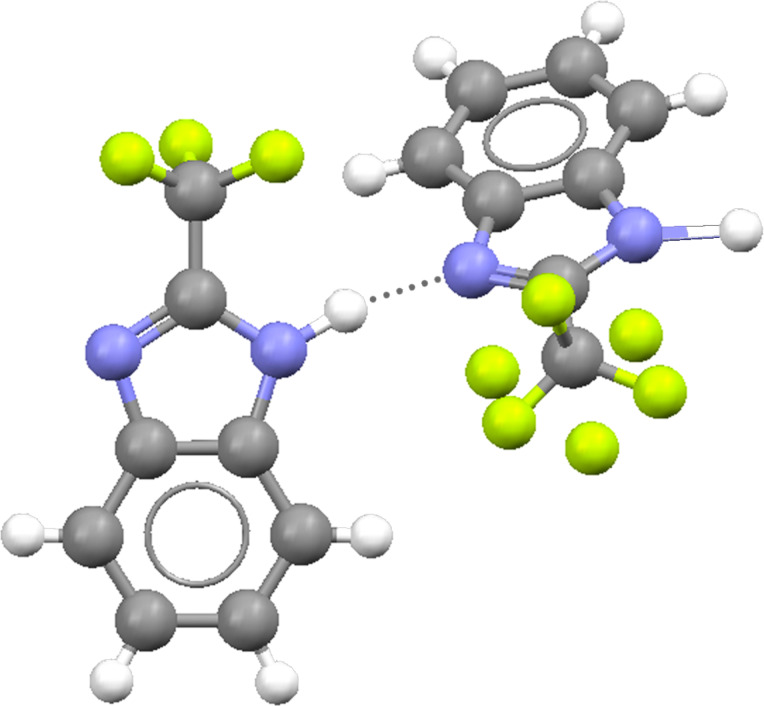
Three kinds of calculations have been done: isolated molecules (gas phase), continuous model solvated molecules in DMSO, and hydrogen-bonded trimers for 1, to simulate the crystal [24]. The central benzimidazole is N–H···N hydrogen bonded to two other benzimidazoles like in the crystal chain (catemer). Two trimers, A and B, that differ in the conformation of the first benzimidazole were calculated for this compound, but the differences in energy are very small, less than 0.1 kJ·mol−1 (Figure 2).
Figure 2.
Trimers A (left) and B (right) of 1.
First, we will discuss the six carbon atoms of the benzene ring of benzimidazole (3a, 4, 5, 6, 7, 7a) and then the remaining atoms (N1, C2, N3 and those of the substituent). In NH-benzimidazoles, when prototropic tautomerism occurs (Figure 3), the signals of the benzimidazole carbons in groups of two coalesce in an average signal (the same happens with the N atoms but the chemical shifts are so different that there are no problems of assignment). Actually, this was a very common occurrence, but in the series of compounds of Table 1, only for benzimidazole itself and for 3, average signals were observed in DMSO-d6, a solvent known to slow down the prototropic exchanges. For these two compounds, we recorded the spectra in HMPA-d18, which is better for this purpose [25] observing the signals of compounds where the prototropic exchange is blocked.
Figure 3.
The tautomerism of 1H-benzimidazole.
Table 1.
Calculated and experimental 1H and 13C chemical shifts of the benzene part.
| Compound |
1H calcd gas |
1H calcd DMSO |
1H exp. HMPA-d18 273 K |
13C calcd gas |
13C calcd DMSO |
13C exp. HMPA-d18 273 K |
13C exp. CPMAS |
| 1 | |||||||
| 4 | 7.81 | 7.79 | 7.54 | 121.67 | 119.58 | 118.7 | 119.4 |
| 5 | 7.20 | 7.34 | 7.11 | 121.68 | 121.79 | 120.1 | 123.0 |
| 6 | 7.24 | 7.41 | 7.11 | 122.88 | 123.04 | 121.2 | 123.8 |
| 7 | 7.32 | 7.67 | 7.42 | 108.23 | 110.99 | 110.9 | 112.9 |
| 3a | – | – | – | 144.67 | 144.22 | 143.4 | 143.1 |
| 7a | – | – | – | 132.70 | 133.54 | 133.7 | 136.4 |
| 1 tri A | |||||||
| 4 | 7.89 | 7.93 | 7.54 | 119.85 | 118.28 | 118.7 | 119.4 |
| 5 | 7.26 | 7.39 | 7.11 | 122.10 | 121.91 | 120.1 | 123.0 |
| 6 | 7.28 | 7.39 | 7.11 | 122.93 | 122.73 | 121.2 | 123.8 |
| 7 | 7.49 | 7.77 | 7.42 | 110.00 | 111.88 | 110.9 | 112.9 |
| 3a | – | – | – | 143.85 | 143.62 | 143.4 | 143.1 |
| 7a | – | – | – | 134.13 | 134.76 | 133.7 | 136.4 |
| 1 tri B | |||||||
| 4 | 7.91 | 7.90 | 7.54 | 119.99 | 118.42 | 118.7 | 119.4 |
| 5 | 7.28 | 7.38 | 7.11 | 122.02 | 121.87 | 120.1 | 123.0 |
| 6 | 7.26 | 7.39 | 7.11 | 122.72 | 122.68 | 121.2 | 123.8 |
| 7 | 7.48 | 7.75 | 7.42 | 110.18 | 112.00 | 110.9 | 112.9 |
| 3a | – | – | – | 143.84 | 143.35 | 143.4 | 143.1 |
| 7a | – | – | – | 134.04 | 134.62 | 133.7 | 136.4 |
| Compound |
1H calcd gas average |
1H calcd DMSO average |
1H exp. DMSO-d6 average |
13C calcd gas average |
13C calcd DMSO average |
13C exp. DMSO-d6 average |
13C exp. CPMAS |
| 1 | |||||||
| 4/7 | 7.695 | 7.825 | 7.58 | 115.08 | 115.21 | 115.3 | |
| 5/6 | 7.27 | 7.385 | 7.17 | 122.37 | 122.28 | 121.7 | |
| 3a/7a | – | 138.94 | 138.98 | 138.4 | |||
| NH | 12.44 | ||||||
| 2 | |||||||
| 4 | 7.70 | 7.68 | 7.47 | 120.78 | 118.54 | 117.8 | 117.4 |
| 5 | 7.11 | 7.23 | 7.11 | 121.22 | 121.09 | 120.7 | 121.7 |
| 6 | 7.06 | 7.21 | 7.10 | 121.91 | 121.86 | 121.2 | 121.7 |
| 7 | 7.21 | 7.54 | 7.40 | 107.79 | 110.32 | 110.5 | 111.6 |
| 3a | – | – | – | 145.08 | 144.60 | 143.5 | 142.9 |
| 7a | – | – | – | 135.53 | 136.25 | 134.3 | 134.7 |
| NH | 12.14 | ||||||
| 3 |
1H calcd gas |
1H calcd DMSO |
1H exp. HMPA-d18 273 K |
13C calcd gas |
13C calcd DMSO |
13C exp. HMPA-d18 273 K |
13C exp. CPMAS |
| 4 | 7.87 | 7.88 | 7.72 | 122.65 | 120.89 | 120.8 | 114.7 118.0 |
| 5 | 7.43 | 7.59 | 7.37 | 124.08 | 124.84 | 123.2 | 120.3 122.1 |
| 6 | 7.37 | 7.57 | 7.42 | 125.89 | 126.68 | 125.1 | 128.1 130.7 |
| 7 | 7.31 | 7.67 | 7.58 | 109.07 | 112.00 | 112.9 | 108.2 109.3 |
| 3a | – | – | – | 144.54 | 143.87 | 142.9 | |
| 7a | – | – | – | 132.27 | 133.24 | 135.1 | |
| NH | 15.05 | ||||||
| 4 |
1H calcd gas |
1H calcd DMSO |
1H exp. DMSO-d6 |
13C calcd gas |
13C calcd DMSO |
13C exp. DMSO-d6 |
13C exp. CPMAS |
| 4 | 7.74 | 7.70 | 7.52 | 120.46 | 118.51 | 118.3 | 120.2 |
| 5 | 7.27 | 7.38 | 7.10 | 121.91 | 122.22 | 120.9 | 123.0 |
| 6 | 7.12 | 7.28 | 7.11 | 122.64 | 122.79 | 121.6 | 124.8 |
| 7 | 6.95 | 7.32 | 7.40 | 107.24 | 109.79 | 110.9 | 113.5 |
| 3a | – | – | – | 144.14 | 143.43 | 143.4 | 143.0 |
| 7a | – | – | – | 134.27 | 135.43 | 134.3 | 135.2 |
| NH | 12.28 | ||||||
| 5 |
1H calcd gas |
1H calcd DMSO |
1H exp. DMSO-d6 |
13C calcd gas |
13C calcd DMSO |
13C exp. DMSO-d6 |
13C exp. CPMAS |
| 4 | 7.80 | 7.77 | 7.64a | 121.81 | 119.62 | 119.2 | 119.1 |
| 5 | 7.23 | 7.36 | 7.19a | 121.55 | 121.73 | 121.3 | 121.2 |
| 6 | 7.20 | 7.36 | 7.25a | 122.32 | 122.53 | 122.1 | 123.2 |
| 7 | 7.24 | 7.59 | 7.54a | 106.50 | 109.36 | 110.1 | 110.7 |
| 3a | – | – | – | 146.49 | 145.60 | 143.3 | 143.4 |
| 7a | – | – | – | 135.58 | 136.21 | 134.5 | 134.7 |
a 3J45 = 7.89, 4J46 = 1.26, 5J47 = 0.78, 3J56 = 7.14, 4J57 = 1.30, 3J67 = 7.99 Hz.
The assignment of the signals of Table 1, Table 2 and Table 3 was straightforward following these steps: i) A NOESY experiment identifies H7 (7.54 ppm) of 5 by its proximity to the N-methyl group; ii) the analysis of the ABCD system of the protons H4, H5, H6 and H7, identifies the remaining protons; iii) a series of 2D experiments assign the CH carbons (HMQC) as well as the quaternary carbons C3a and C7a (HMBC).
Table 2.
Calculated and experimental 13C chemical shifts of the imidazole and substituent parts.
| Compound |
1H calcd gas |
1H calcd DMSO |
1H exp. HMPA-d18 |
13C calcd gas |
13C calcd DMSO |
13C exp. HMPA-d18 |
13C exp. CPMAS |
| 1 | |||||||
| 2 | 7.64 | 7.95 | 8.21 | 136.78 | 140.77 | 141.6 | 143.1 |
| 1 tri A | |||||||
| 2 | 8.01 | 8.32 | 8.21 | 139.05 | 141.86 | 141.6 | 143.1 |
| 1 tri B | |||||||
| 2 | 7.95 | 8.24 | 8.21 | 138.24 | 141.16 | 141.6 | 143.1 |
| Compound |
1H calcd gas |
1H calcd DMSO |
1H exp. DMSO-d6 |
13C calcd gas |
13C calcd DMSO |
13C exp. DMSO-d6 |
13C exp. CPMAS |
| 1 | |||||||
| 2 | 8.20 | 141.9 | |||||
| 2 | |||||||
| 2 | – | – | – | 146.54 | 151.70 | 151.2 | 153.6 |
| Me | 2.41 | 2.50 | 2.47 | 14.37 | 14.42 | 14.6 | 12.6 |
|
13C exp HMPA-d18 |
|||||||
| 3a,b | |||||||
| 2 | – | – | – | 138.96 | 139.54 | 140.8 2JCF = 39.1 |
135.2 137.8 |
| CF3 | – | – | – | 123.53 | 123.96 | 120.0 1JCF = 270.1 |
116.0 |
| 4 | |||||||
| 2 | – | – | – | 151.48 | 155.02 | 153.5 | 157.1 |
| CH2 | 4.18 | 4.22 | 35.0 | 38.73 | 38.42 | 35.0 | 36.0 |
| Cipso | – | – | – | 139.73 | 139.83 | 137.7 | 136.2 |
| Cortho | 7.30 | 7.54 | 7.30 | 129.32 | 129.28 | 128.5 | ~127 |
| Cmeta | 7.38 | 7.49 | 7.33 | 128.76 | 129.44 | 128.8 | ~127 |
| Cpara | 7.34 | 7.38 | 7.22 | 126.76 | 127.15 | 126.5 | 124.3 |
| 5 | |||||||
| 2 | 7.41 | 7.74 | 8.16c | 140.83 | 144.14 | 144.5 | 144.6 |
| NMe | 3.62 | 3.80 | 3.82c | 29.78 | 30.69 | 30.6 | 29.7 |
aIn HMPA-d18 273 K; b 19F calc.: −63.35 (gas), −63.84 (DMSO); 19F exp.: −62.8 (HMPA-d18); −61.2 (CPMAS). c 4JHMe = 0.42 Hz.
Table 3.
Calculated and experimental 15N chemical shifts of the imidazole part.
| Compound | 15N calcd gas | 15N calcd DMSO | 15N exp. HMPA-d18 | 15N exp. CPMAS |
| 1 | ||||
| N1 | −247.63 | −240.09 | −228.4 | −221.8 |
| N3 | −125.53 | −140.75 | −134.0 | −143.9 |
| 1 tri A | ||||
| N1 | −227.89 | −222.08 | −228.4 | −221.8 |
| N3 | −142.23 | −149.24 | −134.0 | −143.9 |
| 1 tri B | ||||
| N1 | −227.80 | −221.92 | −228.4 | −221.8 |
| N3 | −142.21 | −149.18 | −134.0 | −143.9 |
| Compound |
15N calcd gas average |
15N calcd DMSO average |
15N exp. DMSO-d6 average |
15N exp. CPMAS |
| 1 | ||||
| N1/N3 | −186.58 | −190.42 | a | |
| 2 | ||||
| N1 | −246.87 | −240.39 | −230.3 | −219.5, −224.2 |
| N3 | −127.73 | −142.15 | −137.9 | −146.9 |
| 15N exp HMPA-d18 | ||||
| 3b | ||||
| N1 | −247.40 | −241.51 | −230.4 | −225.9 |
| N3 | −120.36 | −132.07 | a | −142.8 |
| 4 | ||||
| N1 | −245.40 | −241.11 | −230.8 | −221.8 |
| N3 | −130.39 | −141.51 | −136.6 | −147.2 |
| 5 | ||||
| N1 | −241.25 | −232.11 | −235.6 | −233.4 |
| N3 | −128.38 | −143.76 | −136.4 | −135.4 |
aNot observed; bIn HMPA-d18 273 K.
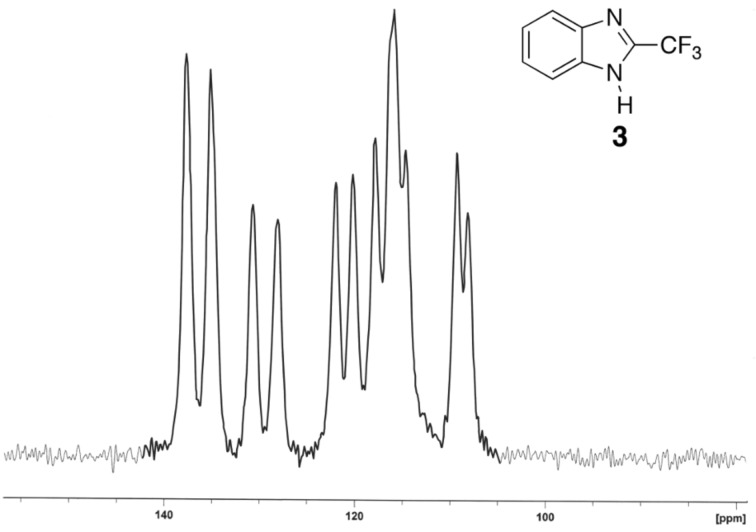
In the case of 3 all signals are considerably split in the solid state (Figure 4).
Figure 4.
The 13C CPMAS NMR spectrum of 3.
The X-ray structure of compound 3 is known and reported in the Cambridge Structural Database (refcode: ZAQRIU01) [4,26]. There are two independent molecules and in one of them, the CF3 is disordered (Figure 5). The most probable explanation of the splittings of Figure 4 is the existence of two independent molecules, a fact that is well documented in the literature [27–30].
Figure 5.
The two independent molecules of 3 drawn with the Mercury program [26].
We have collected in Table 4 the different equations obtained from the data of Table 1, Table 2 and Table 3.
Table 4.
Linear regression equations, intercepts in ppm. If not specified, both trimers yield the same results.
| Eq. | No | Intercept | Slope | R2 | Atom | Exp. | Calcd |
| 1 | 26 | (3.2 ± 0.7) | (0.55 ± 0.10) | 0.563 | 1H | solution | gas phase |
| 2 | 26 | (1.2 ± 0.6) | (0.81 ± 0.08) | 0.797 | 1H | solution | DMSO |
| 3 | 59 | −(0.2 ± 1.3) | (1.00 ± 0.01) | 0.993 | 13C | solution | gas phase |
| 4 | 59 | −(0.7 ± 0.7) | (1.000 ± 0.001) | 0.998 | 13C | solution | DMSO |
| 5 | 59 | −(0.7 ± 2.0) | (1.01 ± 0.02) | 0.987 | 13C | CPMAS | gas phase |
| 6 | 59 | −(0.8 ± 1.2) | (1.01 ± 0.01) | 0.993 | 13C | CPMAS | DMSO |
| 7a | 58 | −(0.2 ± 1.3) | (1.00 ± 0.01) | 0.994 | 13C | solution | gas phase |
| 8a | 58 | −(0.7 ± 0.7) | (1.000 ± 0.005) | 0.998 | 13C | solution | DMSO |
| 9a | 53 | −(0.6 ± 1.9) | (1.01 ± 0.02) | 0.989 | 13C | CPMAS | gas phase |
| 10a | 53 | −(1.1 ± 1.3) | (1.01 ± 0.01) | 0.995 | 13C | CPMAS | DMSO |
| 11 | 6 | (6.6 ± 7.2) | (0.95 ± 0.06) | 0.986 | 13C | CPMAS | monomerb |
| 12 | 6 | (5.2 ± 3.3) | (0.96 ± 0.03) | 0.997 | 13C | CPMAS | trimer Ab |
| 13 | 6 | (3.7 ± 3.4) | (0.98 ± 0.03) | 0.997 | 13C | CPMAS | trimer Bb |
| 14 | 14 | −(22.3 ± 7.3) | (0.86 ± 0.04) | 0.978 | 15N | solution | gas |
| 15 | 14 | (10.5 ± 9.1) | (1.02 ± 0.05) | 0.976 | 15N | solution | DMSO |
| 16 | 14 | −(51.0 ± 7.1) | (0.72 ± 0.04) | 0.970 | 15N | CPMAS | gas |
| 17 | 14 | −(23.5 ± 7.5) | (0.86 ± 0.04) | 0.976 | 15N | CPMAS | DMSO |
| 18 | 10 | −(1.4 ± 6.6) | (0.96 ± 0.03) | 0.991 | 15N | solution | monomerb |
| 19 | 10 | (6.6 ± 10.4) | (1.00 ± 0.05) | 0.979 | 15N | solution | trimerb |
| 20 | 10 | −(29.8 ± 5.3) | (0.82 ± 0.03) | 0.991 | 15N | CPMAS | monomerb |
| 21 | 10 | −(23.8 ± 7.5) | (0.85 ± 0.04) | 0.984 | 15N | CPMAS | trimerb |
aThe 13C signal of the CF3 group has been removed. bDMSO calculations.
i) The 1H chemical shifts are much more consistent with calculations for DMSO as solvent than with those of isolated molecules (gas phase). For 26 points, the R2 coefficient increases from 0.56 (eq. 1) to 0.80 (eq. 2). The use of the monomer or the central part of the trimer has no influence. The worse point is H7 of 4 that appears at 7.40 ppm and fitted with eq. 2 has a value of 7.14 ppm. The origin of this discrepancy is that the theoretical conformation corresponding to the X-ray structure is not stable and reverts to the minimum one, which has the benzyl group rotated.
ii) The 13C chemical shifts (eqs. 3–13) are very well reproduced by the calculations: high R2 values, small intercepts (in several cases, not significant) and slopes close to 1. Systematically, the worse point was the carbon atom of the CF3 substituent (halogen substituents produce effects that are not well reproduced by our calculations that not include relativistic corrections) [15], removing it does not significantly modify the regression values (compare eqs. 3–6 with eqs. 7–10). CPMAS values agree better with calculations including DMSO solvent effect (compare eqs. 9 and 10) and also better with trimer B (in turn, slightly better than with trimer A, compare intercept and slopes of eqs. 12 and 13) than with the monomer (eq. 11).
iii) Concerning 15N NMR (eqs. 14–21), the R2 values are lower than with 13C NMR. Both for solution and for CPMAS, the gas phase and DMSO calculations are comparable in terms of R2 (eqs. 14–17), however, the values of the slopes (the closer to 1, the better) and intercepts (the closer to 0, the better), clearly favored the DMSO calculations. Surprisingly, the monomer appears preferable to the trimer (eqs. 18,19 and 20,21) which is understandable for the solution but not for the solid state. More complex approaches, such as periodic calculations [31], are necessary.
Influence of the substituent at position 2 on the tautomerization rate
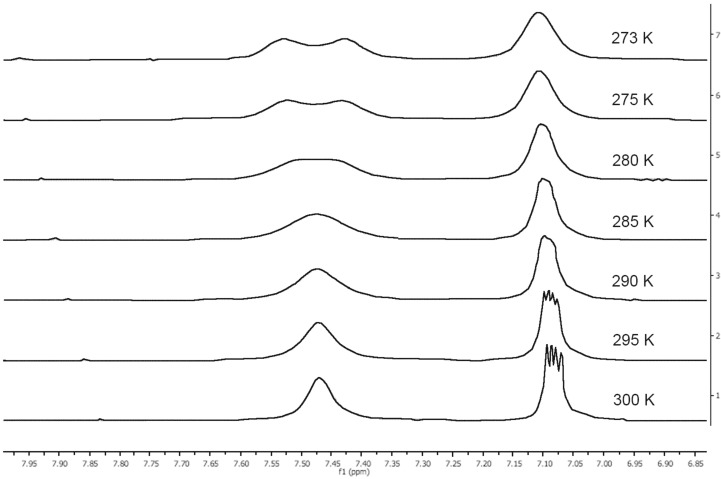
We have observed a different behavior for the four NH-benzimidazoles: 1 and 3 yielded average signals in DMSO-d6 and only in HMPA-d18 the prototropic exchange was slow, on the other hand 2 and 4 behaved as if the tautomerism was blocked in DMSO-d6. In the case of 1 a dynamic NMR (DNMR) study in HMPA-d18 was performed (Figure 6).
Figure 6.
The evolution of the spectrum of 1 with temperature in HMPA-d18.
The relevant data are: TC = 277.5 K and Δν = 0.12 ppm = 48 Hz; from them and using the Eyring equation [ΔG‡TC = 19.12 × TC × (10.32 + log TC/kC)] [32], we obtained: kC = 67.88 s−1 and ΔG‡277.5 = 58.0 kJ·mol−1. This barrier is similar to that of pyrazole in the same solvent (58.6 kJ·mol−1 at 289 K) [33].
The effect of the substituent at position 2 on the rate (roughly, CF3 and H, fast; CH2C6H5 and CH3, slow) is probably the consequence of steric and electronic effects; many years ago, we showed that intramolecular hydrogen bonds also affect the rate [34]. The calculated electrostatic potential minima associated to the lone pair of the N3 follow the tautomerization rate ranking of the molecules (−0.085 au, CF3, −0.103, H, −0.104, CH2C6H5 and −0.105, CH3).
Conclusion
The data reported here for NH-benzimidazoles when the prototropy is blocked should be useful to determine the tautomeric composition when there are substituents at positions 4(7) or 5(6), for instance, in the case of omeprazole, a 5(6)-methoxy-1H-benzimidazole derivative [35–36]. Besides, solid state results as well as GIAO calculations provide new data to characterize this important family of compounds. Finally, by means of DNMR experiments it was possible to determine the barrier to proton transfer of benzimidazole itself in HMPA-d18, thus providing a missing value in heterocyclic tautomerism of azoles and benzazoles [33].
Experimental
Four of the compounds reported in this paper are commercial (Sigma-Aldrich): 1, 2, 3 and 5. We reported the synthesis of the fifth one, 4, in [37].
NMR spectroscopy
Solution NMR spectra were recorded on a Bruker DRX 400 (9.4 Tesla, 400.13 MHz for 1H, 100.62 MHz for 13C and 40.54 MHz for 15N) spectrometer with a 5 mm inverse-detection H-X probe equipped with a z-gradient coil, at 300 K. Chemical shifts (δ) are given from internal solvent, DMSO-d6 2.49 for 1H and 39.5 for 13C. Typical parameters for 1H NMR spectra were spectral width 4800 Hz and pulse width 8.3 μs at an attenuation level of 0 dB. Typical parameters for 13C NMR spectra were spectral width 21 kHz, pulse width 12.5 μs at an attenuation level of −6 dB and relaxation delay 2s, WALTZ-16 was used for broadband proton decoupling; the FIDS were multiplied by an exponential weighting (lb = 1Hz) before Fourier transformation.
Inverse proton detected heteronuclear shift correlation spectra, (1H,13C) gs-HMQC and (1H,13C) gs-HMBC, were acquired and processed using standard Bruker NMR software and in nonphase-sensitive mode. Gradient selection was achieved through a 5% sine truncated shaped pulse gradient of 1 ms.
Selected parameters for (1H,13C) gs-HMQC and (1H,13C) gs-HMBC spectra were spectral width 4800 Hz for 1H and 20.5 kHz for 13C, 1024 × 256 data set, number of scans 2 (gs-HMQC) or 4 (gs-HMBC) and relaxation delay 1s. The FIDs were processed using zero filling in the F1 domain and a sine-bell window function in both dimensions was applied prior to Fourier transformation. In the gs-HMQC experiments, GARP modulation of 13C was used for decoupling.
Selected parameters for (1H,15N) gs-HMQC and (1H,15N) gs-HMBC spectra were spectral width 3500 Hz for 1H and 12.5 kHz for 15N, 1024 × 256 data set, number of scans 4, relaxation delay 1s, 37–60 ms delay for evolution of the 15N,1H long-range coupling. The FIDs were processed using zero filling in the F1 domain and a sine-bell window function in both dimensions was applied prior to Fourier transformation.
For 19F NMR (379.50 MHz) a 5 mm QNP direct-detection probehead equipped with a z-gradient coil, at 300 K was required.
Chemical shifts (δ) are given from internal solvent, DMSO-d6 2.49 and HMPA-d18 2.57 for 1H; 39.5 and 35.8 for 13C, and for 15N and 19F NMR, nitromethane (0.00) and one drop of CFCl3 in CDCl3 (0.00) were used as external references
Typical parameters for 1H NMR were: spectral width 4800 Hz and pulse width 10.25 μs at an attenuation level of −3.0 dB. For 13C NMR: spectral width 21 kHz, pulse width 8.75 μs at an attenuation level of −3 dB and relaxation delay 2 s, WALTZ-16 was used for broadband proton decoupling; the FIDS were multiplied by an exponential weighting (lb = 1Hz) before Fourier transformation. For 19F NMR: spectral width 55 kHz, pulse width 13.57 μs at an attenuation level of −6 dB and relaxation delay 1s.
Variable temperature: A Bruker BVT3000 temperature unit was used to control the temperature of the cooling gas stream and an exchanger to achive low temperatures. To avoid problems at low temperatures caused by air moisture, pure nitrogen was used as bearing, driving and cooling gas. When not stated explicitly, the temperature was 293 K.
We have always used a 1.4 M solution of the compounds either in DMSO-d6 or in HMPA-d18; this corresponds, in the case of 1, to 10 mg in 0.6 mL. To avoid water contamination, a new sealed ampoule was open for each experiment.
Solid state 13C (100.73 MHz) and 15N (40.60 MHz) CPMAS NMR spectra were obtained on a Bruker WB 400 spectrometer at 300 K using a 4 mm DVT probehead. Samples were carefully packed in 4 mm diameter cylindrical zirconia rotors with Kel-F end-caps. Operating conditions involved 2.9 µs 90° 1H pulses and decoupling field strength of 86.2 kHz by TPPM sequence. 13C spectra were originally referenced to a glycine sample and then the chemical shifts were recalculated to the Me4Si [for the carbonyl atom δ (glycine) = 176.1] and 15N spectra to 15NH4Cl and then converted to nitromethane scale using the relationship: δ 15N(nitromethane) = δ1 5N(ammonium chloride) − 338.1.
Typical acquisition parameters for 13C CPMAS were: spectral width, 40 kHz; recycle delay, 5–60 s; acquisition time, 30 ms; contact time, 2–4 ms; and spin rate, 12 kHz. In order to distinguish protonated and non-protonated carbon atoms, the NQS (Non-Quaternary Suppression) experiment by conventional cross-polarization was recorded; before the acquisition the decoupler is switched off for a very short time of 25 μs [38–40]. For 15N CPMAS were: spectral width, 40 kHz; recycle delay, 5–60 s; acquisition time, 35 ms; contact time, 7–9 ms; and spin rate, 6 kHz.
Solid state 19F (376.94 MHz) NMR spectra were obtained on a Bruker WB 400 spectrometer using a MAS DVT BL2.5 X/F/H probe. Samples were carefully packed in 2.5 mm diameter cylindrical zirconia rotors with Kel-F end-caps.
Typical acquisition parameters 19F{1H} MAS were: spectral width, 75 kHz; recycle delay, 10 s; pulse width, 2.5 μs and proton decoupling field strength of 100 kHz by SPINAL-64 sequence; recycle delay, 10 s; acquisition time, 25 ms; 128 scans; and spin rate, 25 kHz. The 19F spectra was referenced to ammonium trifluoroacetate sample and then the chemical shifts were recalculated to the CFCl3 [δ (CF3CO2NH4+) = −72.0].
Computational details
Using the Gaussian 09 facilities [41], GIAO [42–43]/B3LYP [44–46]/6-311++G(d,p) [47–48] calculations were carried out; DMSO effects were calculated using the PCM continuum model [49] also with the Gaussian 09 series of programs.
Supporting Information
Optimized geometry of the systems, and chemical shifts in gas phase and PCM/DMSO environment.
Acknowledgments
This work has been financed by Ministerio de Ciencia e Innovación (CTQ2010-16122) and Ministerio de Economía y Competitividad of Spain (CTQ2012-35513-C02-02) and Comunidad Autónoma de Madrid (Project MADRISOLAR2, ref. S2009/PPQ-1533). One of us (C. I. Nieto) is indebted to UNED for a predoctoral fellowship (FPI “Grupos de Investigación” UNED).
Dedicated to our friend Professor Viktor Milata from the Slovak University of Technology, Bratislava, Slovak Republic.
Contributor Information
Rosa M Claramunt, Email: rclaramunt@ccia.uned.es.
Ibon Alkorta, Email: ibon@iqm.csic.es.
References
- 1.Gomtsyan A. Chem Heterocycl Compd. 2012;48:7–10. doi: 10.1007/s10593-012-0960-z. [DOI] [Google Scholar]
- 2.Velík J, Baliharová V, Fink-Gremmels J, Bull S, Lamka J, Skálová L. Res Vet Sci. 2004;76:95–108. doi: 10.1016/j.rvsc.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Khokra S L, Choudhary D. Asian J Biochem Pharm Res. 2011;1(3):476–486. [Google Scholar]
- 4.Horiuchi S, Kagawa F, Hatahara K, Kobayashi K, Kumai R, Murakami Y, Tokura Y. Nat Commun. 2012;3:No. 1308. doi: 10.1038/ncomms2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pogorzelec-Glaser K, Rachocki A, Ławniczak P, Pietraszko A, Pawlaczyk C, Hilczer B, Pugaczowa-Michalska M. CrystEngComm. 2013;15:1950–1959. doi: 10.1039/c2ce26571k. [DOI] [Google Scholar]
- 6.Rachocki A, Pogorzelec-Glaser K, Ławniczak P, Pugaczowa-Michalska M, Łapinski A, Hilczer B, Matczak M, Pietraszko A. Cryst Growth Des. 2014;14:1211–1220. doi: 10.1021/cg401742b. [DOI] [Google Scholar]
- 7.Elguero J, Katritzky A R, Denisko O V. Adv Heterocycl Chem. 2000;76:1–84. doi: 10.1016/S0065-2725(00)76003-X. [DOI] [Google Scholar]
- 8.Alkorta I, Elguero J. Top Heterocycl Chem. 2008;19:155–202. [Google Scholar]
- 9.Weber U, Thiele H. NMR Spectroscopy: Modern Spectral Analysis, Weinheim: Wiley-VCH; 1998. [Google Scholar]
- 10.Simpson J H. Organic Structure Determination using 2-D NMR Spectroscopy. San Diego: Academic Press; 2012. [Google Scholar]
- 11.Claramunt R M, López C, García M Á, Otero M D, Torres M R, Pinilla E, Alarcón S H, Alkorta I, Elguero J. New J Chem. 2001;25:1061–1068. doi: 10.1039/B103405G. [DOI] [Google Scholar]
- 12.Sanz D, Claramunt R M, Saini A, Kumar V, Aggarwal R, Singh S P, Alkorta I, Elguero J. Magn Reson Chem. 2007;45:513–517. doi: 10.1002/mrc.1992. [DOI] [PubMed] [Google Scholar]
- 13.Silva A M S, Sousa R M S, Jimeno M L, Blanco F, Alkorta I, Elguero J. Magn Reson Chem. 2008;46:859–864. doi: 10.1002/mrc.2272. [DOI] [PubMed] [Google Scholar]
- 14.Seixas R S G R, Silva A M S, Alkorta I, Elguero J. Monatsh Chem. 2011;142:731–742. doi: 10.1007/s00706-011-0473-y. [DOI] [Google Scholar]
- 15.Radula-Janik K, Kupka T, Ejsmont K, Daszkiewicz Z, Sauer S P A. Magn Reson Chem. 2013;51:630–635. doi: 10.1002/mrc.3992. [DOI] [PubMed] [Google Scholar]
- 16.Begtrup M, Elguero J, Faure R, Camps P, Estopá C, Ilavský D, Fruchier A, Marzin C, de Mendoza J. Magn Reson Chem. 1988;26:134–151. doi: 10.1002/mrc.1260260210. [DOI] [Google Scholar]
- 17.Thakuria H, Das G. ARKIVOC. 2008;(xv):321–328. [Google Scholar]
- 18.She J, Jiang Z, Wang Y. Synlett. 2009:2023–2027. doi: 10.1055/s-0029-1217515. [DOI] [Google Scholar]
- 19.Zhang D, Yan L. J Phys Chem B. 2010;114:12234–12241. doi: 10.1021/jp1054606. [DOI] [PubMed] [Google Scholar]
- 20.Alkorta I, Sánchez-Sanz G, Trujillo C, Elguero J, Claramunt R M. ARKIVOC. 2012;(ii):85–106. [Google Scholar]
- 21.Yu B, Zhang H, Zhao Y, Chen S, Xu J, Huang C, Liu Z. Green Chem. 2013;15:95–99. doi: 10.1039/C2GC36517K. [DOI] [Google Scholar]
- 22.Claramunt R M, Sanz D, Boyer G, Catalän J, de Paz J L G, Elguero J. Magn Reson Chem. 1993;31:791–800. doi: 10.1002/mrc.1260310902. [DOI] [Google Scholar]
- 23.Claramunt R M, Sanz D, López C, Jiménez J A, Jimeno M L, Elguero J, Fruchier A. Magn Reson Chem. 1997;35:35–75. doi: 10.1002/(SICI)1097-458X(199701)35:1<35::AID-OMR25>3.0.CO;2-K. [DOI] [Google Scholar]
- 24.Claramunt R M, López C, Sanz D, Alkorta I, Elguero J. Heterocycles. 2001;55:2109–2121. doi: 10.3987/COM-01-9313. [DOI] [Google Scholar]
- 25.Pinto J, Silva V L M, Silva A M S, Claramunt R M, Sanz D, Torralba M C, Torres M R, Reviriego F, Alkorta I, Elguero J. Magn Reson Chem. 2013;51:203–221. doi: 10.1002/mrc.3926. [DOI] [PubMed] [Google Scholar]
- 26. [Jun 3;2014 ];Mercury - Crystal Structure Visualisation, Exploration and Analysis Made Easy. Available from: http://www.ccdc.cam.ac.uk/Solutions/CSDSystem/Pages/Mercury.aspx.
- 27.Foces-Foces C, Trofimenko S, López C, Santa María M D, Claramunt R M, Elguero J. J Mol Struct. 2000;526:59–64. doi: 10.1016/S0022-2860(00)00444-0. [DOI] [Google Scholar]
- 28.Sánchez-Migallón A, de la Hoz A, López C, Claramunt R M, Infantes L, Motherwell S, Shankland K, Nowell H, Alkorta I, Elguero J. Helv Chim Acta. 2003;86:1026–1039. doi: 10.1002/hlca.200390091. [DOI] [Google Scholar]
- 29.Claramunt R M, López C, Lott S, Santa María M D, Alkorta I, Elguero J. Helv Chim Acta. 2005;88:1931–1942. doi: 10.1002/hlca.200590148. [DOI] [Google Scholar]
- 30.Virgili A, Quesada-Moreno M M, Avilés-Moreno J R, López-González J J, García M A, Claramunt R M, Torres M R, Jimeno M L, Reviriego F, Alkorta I, et al. Helv Chim Acta. 2014;97:471–490. doi: 10.1002/hlca.201300395. [DOI] [Google Scholar]
- 31.Webber A L, Emsley L, Claramunt R M, Brown S P. J Phys Chem A. 2010;114:10435–10442. doi: 10.1021/jp104901j. [DOI] [PubMed] [Google Scholar]
- 32.Sandström J. Dynamic NMR Spectroscopy. London: Academic Press; 1982. [Google Scholar]
- 33.Minkin V I, Garnovskii A D, Elguero J, Katritzky A R, Denisko O V. Adv Heterocycl Chem. 2000;76:157–323. doi: 10.1016/S0065-2725(00)76005-3. [DOI] [Google Scholar]
- 34.Elguero J, Llouquet G, Marzin C. Tetrahedron Lett. 1975;16:4085–4086. doi: 10.1016/S0040-4039(00)91243-7. [DOI] [Google Scholar]
- 35.Claramunt R M, López C, Alkorta I, Elguero J, Yang R, Schulman S. Magn Reson Chem. 2004;42:712–714. doi: 10.1002/mrc.1409. [DOI] [PubMed] [Google Scholar]
- 36.Claramunt R M, López C, Elguero J. ARKIVOC. 2006;(v):5–11. [Google Scholar]
- 37.Ribeiro da Silva M A V, Ribeiro da Silva M D M C, Amaral L M P F, Elguero J, Jiménez P, Roux M V, Dávalos J Z, Temprado M, Cabildo P, Claramunt R M, et al. J Chem Thermodyn. 2005;37:1168–1176. doi: 10.1016/j.jct.2005.02.008. [DOI] [Google Scholar]
- 38.Murphy P D. J Magn Reson. 1983;52:343–345. [Google Scholar]
- 39.Murphy P D. J Magn Reson. 1985;62:303–308. [Google Scholar]
- 40.Alemany L B, Grant D M, Alger T D, Pugmire R J. J Am Chem Soc. 1983;105:6697–6704. doi: 10.1021/ja00360a025. [DOI] [Google Scholar]
- 41.Gaussian 09. Wallingford, CT: Gaussian, Inc.; 2009. [Google Scholar]
- 42.London F. J Phys Radium. 1937;8:397–409. doi: 10.1051/jphysrad:01937008010039700. [DOI] [Google Scholar]
- 43.Ditchfield R. Mol Phys. 1974;27:789–807. doi: 10.1080/00268977400100711. [DOI] [Google Scholar]
- 44.Becke A D. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/PhysRevA.38.3098. [DOI] [PubMed] [Google Scholar]
- 45.Becke A D. J Chem Phys. 1993;98:5648–5652. doi: 10.1063/1.464913. [DOI] [Google Scholar]
- 46.Lee C, Yang W, Parr R G. Phys Rev B. 1988;37:785–789. doi: 10.1103/PhysRevB.37.785. [DOI] [PubMed] [Google Scholar]
- 47.Ditchfield R, Hehre W J, Pople J A. J Chem Phys. 1971;54:724–728. doi: 10.1063/1.1674902. [DOI] [Google Scholar]
- 48.Frisch M J, Pople J A, Binkley J S. J Chem Phys. 1984;80:3265–3269. doi: 10.1063/1.447079. [DOI] [Google Scholar]
- 49.Mennucci B, Cammi R, editors. Continuum Solvation Models in Chemical Physics. From Theory to Applications. Chichester: John Wiley & Sons; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Optimized geometry of the systems, and chemical shifts in gas phase and PCM/DMSO environment.