Abstract
Mitosis is a cell-cycle stage during which condensed chromosomes migrate to the middle of the cell and segregate into two daughter nuclei before cytokinesis (cell division) with the aid of a dynamic mitotic spindle. The history of mitosis research is quite long, commencing well before the discovery of DNA as the repository of genetic information. However, great and rapid progress has been made since the introduction of recombinant DNA technology and discovery of universal cell-cycle control. A large number of conserved eukaryotic genes required for the progression from early to late mitotic stages have been discovered, confirming that DNA replication and mitosis are the two main events in the cell-division cycle. In this article, a historical overview of mitosis is given, emphasizing the importance of diverse model organisms that have been used to solve fundamental questions about mitosis.
For ∼150 years, mitosis research has involved species with unique cellular and chromosomal characteristics, such as large cells from salamanders, ring-like chromosomes from maize, and cdc mutants in yeast.
Onko Chisin—An attempt to discover new truths by studying the past through scrutiny of the old.
LARGE SALAMANDER CHROMOSOMES ENABLED THE FIRST DESCRIPTION OF MITOSIS
Mitosis means “thread” in Greek. In the 19th century, pioneering researchers who developed light microscopic techniques discovered characteristic thread-like structures in dye-stained cells before cell division. They named this stage “mitosis,” for the appearance of the threads. The threads are now known to be condensed chromosomes, which first become visible with light microscopy during a mitotic stage called prophase. This is followed by prometaphase (later known to be important as this stage is controlled by the spindle assembly checkpoint [SAC]), then metaphase (in which the chromosomes are aligned in the middle of cell), anaphase A (in which identical sister chromatids comprising individual chromosomes separate and move toward opposite poles of the cell), anaphase B (in which the spindle elongates as the chromosomes approach the poles), and telophase (the terminal phase of mitosis during which chromosomes decondense, again becoming invisible with light microscopy, the nuclear membrane reforms, and the spindle disassembles) before cytokinesis (cell division) (see Fig. 1 for terminology related to G1, G2, S, and M phases, and Fig. 2 for a schematic of the progression of mitosis).
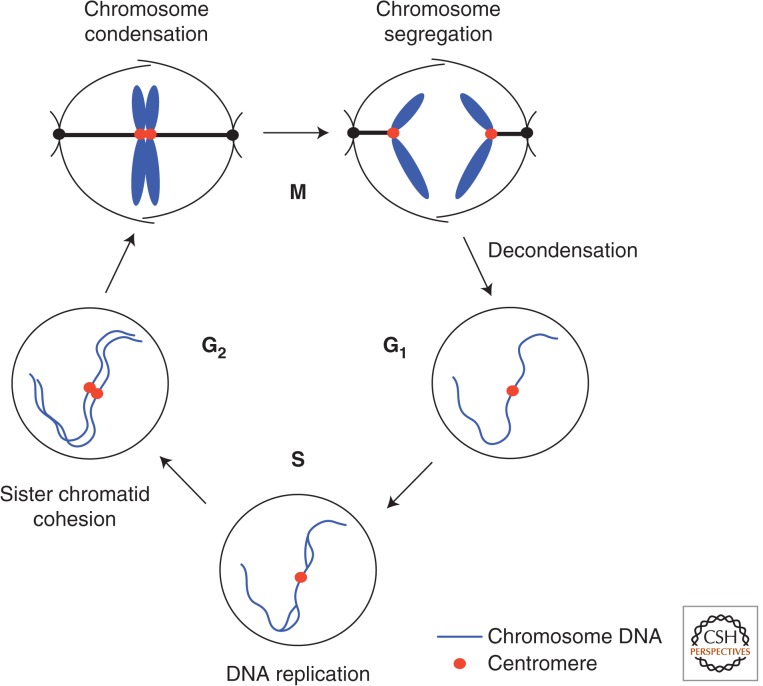
Figure 1.
The cell cycle consists of four phases: G1, S, G2, and M. Mitosis (M phase) is a brief period of the cell-division cycle. Blue denotes chromosomal DNA; red, centromere/kinetochore. S phase, which comprises a period of DNA synthesis, is preceded by a gap (G1 means the first gap) in which there is no DNA synthesis. G1 phase is alternatively called the prereplicative phase. S phase is followed by another gap (G2). Again, there is no synthesis of DNA in G2 unless DNA damage must be repaired by replication. G2 phase eventually enters M phase or mitosis, which completes one cell cycle. Morphological and biochemical events occurring in these phases are described. The cell-cycle concept was developed around 1953. The boundary between G2 and M is somewhat ambiguous because the initiation of prophase is difficult to define in some cells. Although the activation of cyclin-dependent kinase 1 (CDK1) protein kinase and two other protein kinases, polo and aurora B, are generally accepted as biochemical markers for the onset of mitosis, it should be noted that mitosis is a morphological (cell structural) event, and a number of visible cell structural mitotic markers have been proposed, such as nuclear membrane disassembly, chromosome condensation, spindle formation, kinetochore microtubule formation, etc. (see Fig. 2). The end of mitosis is also ambiguous. In telophase, chromosomes are fully segregated, but still condensed. Going into the G1 phase occurs after chromosome decondensation, reformation of the nuclear membrane around daughter nuclear chromatin, then followed by cytokinesis. In many cells, such as Physarum or vertebrate skeletal muscle cells, cytokinesis does not occur, producing multinucleate cells.
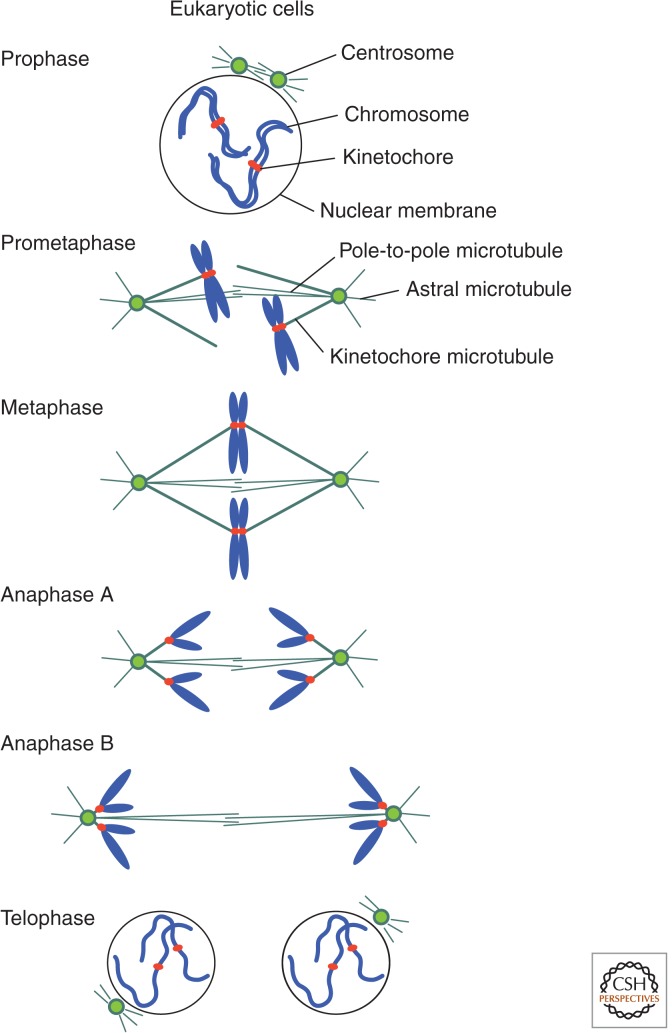
Figure 2.
Higher eukaryotic mitosis. In higher eukaryotic prophase, the nuclear membrane begins to degrade on the onset of chromosome condensation. In fungi, such as yeast, the nuclear membrane remains during mitosis. Centrosomes (called “spindle pole bodies” in yeast) are duplicated and begin to form the mitotic spindle. In prometaphase, the full spindle forms and condensed chromosomes are attached to kinetochore microtubules. In metaphase, chromosomes are aligned at the middle. In anaphase A, sister chromatids are pulled toward opposite poles. In anaphase B, spindle extension occurs. Finally, in telophase, the nuclear membrane reforms.
In comparison with the whole-cell-division cycle, mitosis is a brief period during which condensed chromosomes are accurately segregated into daughter nuclei with the aid of an assemblage of pole-to-pole microtubules called the spindle. In addition, there are short aster microtubules that radiate from the spindle poles toward the cell cortex, and kinetochore microtubules that join the attachment region of chromosomes (called sister kinetochores). This is normally followed by a postmitotic event, cytokinesis, which generates two daughter cells.
The first person to observe mitosis in detail was a German biologist, Walther Flemming (1843–1905), who is the pioneer of mitosis research and also the founder of cytogenetics (see Fig. 3) (Paweletz 2001). Flemming described the behavior of chromosomes during mitosis with amazing accuracy in an 1882 collection entitled, “Cell substance, nucleus and cell division.” For visualization of chromosomes, Flemming used aniline dyes, which bind to chromosomes.
Figure 3.
(A) Walther Flemming (image is part of the public domain and, therefore, is reprinted free from copyright restrictions). (B) Theodor Boveri (image is part of the public domain and, therefore, is reprinted free from copyright restrictions). (C) Barbara McClintock (image courtesy of the Barbara McClintock Papers, American Philosophical Society; copyright holder is unknown).
Chromosome, in Greek, means colored (“chroma”) body (“soma”). A chromosome is an organized structure of DNA, protein, and RNA, which changes its form dramatically during the cell cycle and under different physiological and physicochemical conditions. Significantly, most of Flemming’s work was performed before the rediscovery (1900) of the genetic principles discovered by Gregor Mendel (1822–1884). Flemming had no knowledge of DNA, which was discovered as the “nuclein” substance in 1869 by a Swiss biochemist, Friedrich Miescher, and much later identified as the genetic material by Oswald Avery (Avery et al. 1944).
Flemming used a species of salamander as the source of his material because salamanders have very large chromosomes. The genome (a haploid, or single set of chromosomes) size may be approximately 10 times larger than that of humans, because it contains a large number of repetitious DNAs. In Flemming’s day, large cells containing large chromosomes were an obvious advantage. The large chromosomes of cells such as thin epithelial cells of newt lungs have proven to be extremely useful in making high-resolution movies of mitosis (Rieder and Hard 1990). Detailed behavior of individual chromosomes at the SAC (Li et al. 1993), which regulates prometaphase, was clearly observed by Nomarski-differential interference contrast video microscopy (Rieder and Alexander 1990).
SEA URCHIN EMBRYOS REVEALED RAPID, REPEATED MITOSES
Theodor Boveri (1862–1915), a German biologist, understood the importance of cell and chromosome research and he worked on the early development of sea urchins (Phylum Echinodermata). He discovered fast, repeated mitoses, revealing individual chromosomes and centrosomes at early embryonic stages. Because embryonic sea urchin cells are highly transparent and divide every 20–30 min, they are ideal for observing mitotic events in real time. Boveri’s fundamental contributions to the chromosomal theory of inheritance, mitotic cell cycle, and tumorigenesis are available in English (Baltzer 1964; Boveri 2008).
Baltzer (1964) observed that Boveri’s work established the foundation for the chromosome theory that identified chromosomes as the hereditary material. First, chromosomes are individual bodies in the cell nucleus. Second, different chromosomes carry different hereditary materials. Chromosomes are gene transmitters. Third, there exists a relationship between the location of individual genes on the chromosome and frequencies of crossing over affecting them. In addition, by observing aberrant mitoses, Boveri (2008) proposed that scrambled chromosomes produced by abnormal mitosis might be the cause of carcinogenesis, so that cancer can originate from a single cell that divided abnormally.
MAIZE REVEALED CHROMOSOME PLASTICITY
Barbara McClintock (1902–1992), an American cytogeneticist, made several important discoveries in genetics and chromosome biology, using maize as her experimental organism. She mapped maize genes on chromosomes (McClintock 1929, 1931), discovered ring-shaped chromosomes (McClintock 1932), unstable chromosome ends (McClintock 1941), and transposable genes (McClintock 1950). Although none of these was closely related to mitosis, McClintock’s work made it possible to understand certain behaviors of chromosomes, such as recombination and transposition. For example, if telomeres of two chromosomes were lost during cell division, the resulting daughter cells might produce ring-like chromosomes with two centromeres (special DNA regions in which kinetochores attach to the spindle microtubules during metaphase). McClintock’s studies became very important when one considers the outcomes of unusual chromosome events, such as cases of unequal segregation or transposition. In addition, she rightly pointed out that differentiated cells often arise after asymmetric mitosis. She stated that different gene regulations arise from daughter cells formed by asymmetric cell divisions in which one daughter cell gained or lost something compared with the other (McClintock 1984). Studies of chromosome structure also flourished using plant cells because chromosomes of certain plant cells, such as lily, onion, and Haemanthus, are very large. For example, meiotic behavior of lily kinetochores could be observed in detail with light microscopy (Matsuura 1951). However, chromosome studies at the molecular level using plants are relatively uncommon today.
MITOTIC SPINDLE FOUND IN DIVERSE EUKARYOTES
Polarization microscopy, using birefringence, was invented for observing the mitotic spindle in living cells, as spindle fibers connecting the spindle poles and chromosome are doubly refractive (Inoue and Dan 1951; Inoue 1953). Before observation of the mitotic spindle by polarization microscopy, the existence of the “spindle fibers” was regarded with skepticism. Because microtubules are dynamic ephemeral structures, fibrous mitotic elements were thought, by some, to be a fixation artifact. Shinya Inoue, who used polarization microscopy to observe the mitotic spindle in living cells, resolved the dispute. The spindle was found in dividing cells of various organisms, including grasshoppers, fruit flies, tube worm (Chaetopterus pergamentaceous) oocytes, and pollen mother cells of Lilium longiflorum. Inoue and others also used polarization optics to show the fragile nature of the spindle, which disappears after mitosis and in the presence of microtubule-destroying poisons. Glutaraldehyde, a bifunctional crosslinker, was later found to be a good fixative. Confirmation of the ubiquity of the mitotic spindle among eukaryotes, such as fungi, had to await thin sectioning of cells for electron microscopy (Robinow and Marak 1966).
Mazia and Dan (1952) developed cell-free methods to isolate the mitotic apparatus from dividing sea urchin eggs (Strongylocentrotus franciscanus and Strongylocentrotus purpuratus) in quantity. Their procedures were based on selective solubilization of the cytoplasm surrounding the spindle apparatus. The isolated structures at various stages in mitosis contained chromosomal and nonchromosomal parts and correspond well to the microscopic structures in vivo. The isolated apparatus is birefringent, as seen in polarized microscopy. The entire mitotic apparatus behaved in the test tube as a single physical entity.
It was unclear whether the mitotic spindle and asters were required for cell division. Yukio Hiramoto (1956) bravely removed the spindle apparatus from dividing cells of sea urchin embryos using a micropipette. Cytokinesis (cell division) still occurred when the spindle was removed during anaphase or later, indicating that the spindle apparatus, although essential for chromosomal movement, was not required for cell division per se. In some eggs, initiation of furrowing was even seen despite the removal of both spindle and asters during mitotic metaphase. The position of the cleavage plane was not altered by elimination of the whole spindle apparatus. Hiramoto concluded that furrowing results from an active function of the cell cortex. It is now known that an actomyosin ring formed at the cell equator later in mitosis is essential for cell division (Mabuchi 1973).
CULTURED MAMMALIAN CELLS REVEALED NUCLEOCYTOPLASMIC INTERACTIONS IN MITOSIS
Mammalian cell culture techniques have quite a long history, starting in the 19th century by a German zoologist, Wilhelm Roux. Cells isolated from tissues were grown in primary cultures and then maintained as cell lines or stocked for multiple use. Animal cell culture became a common technique in the 1970s. Rao and Johnson performed key experiments that showed the induction of mitosis by trans-acting (freely diffusing) factors (Johnson and Rao 1970; Rao and Johnson 1970). Using the cell fusion technique, they mixed nuclei at different cell-cycle phases in the same cytoplasm (heterokaryon) and attempted to determine whether these nuclei at different cell-cycle phases could influence one another. Their results were quite striking.
When M-phase cells were mixed with G1, S, or G2 phase cells, premature (inappropriate) mitosis occurred in the interphase nuclei, showing that there were diffusible factors that could promote M phase in the interphase nuclei. When S-phase nuclei were mixed with G1-phase nuclei, G1 nuclei were induced to start S phase, suggesting that S-phase nuclei contain a diffusible factor that induces DNA replication. When S-phase nuclei were mixed with G2-phase nuclei, the G2 nucleus did not reinitiate S phase. G2-phase nuclei were refractory to the diffusible factor from S-phase nuclei. When G1-phase nuclei were mixed with G2-phase nuclei, no S or M phase occurred. These results strongly suggested that diffusible induction factors are produced in the nuclei during S and M phases. The S-phase-promoting factor only works on G1 nuclei. The M-phase-promoting factor works on everything. Rao and Johnson’s experiments using mammalian cells paved the way for understanding mammalian cell-cycle regulations, and later led to the discovery of maturation-promoting factor (MPF) and cyclin-dependent kinase (CDK).
YEAST cdc MUTANTS REVEALED GENETIC CONTROL OF THE CELL CYCLE
For 3000–5000 years, mankind has depended on the budding yeast, Saccharomyces, for making bread and beer. In 1857, Louis Pasteur (1822–1895) discovered the fermentation process using yeast. He showed that alcoholic fermentation was conducted by living yeast cells and not by a chemical catalyst. In the laboratory, yeast has been intensively used by biochemists to study enzymatic roles in various metabolic pathways. Lehland Hartwell, a molecular biologist originally trained in mammalian DNA synthesis, directed his attention to yeast genetics about 1965 and introduced a revolutionarily new approach to cell-cycle research. In his initial yeast genetics paper, he isolated and characterized 400 temperature-sensitive (ts) mutants of Saccharomyces cerevisiae, which were defective in certain aspects of the cell-division cycle (Hartwell et al. 1970, 1971, 1973, 1974, 1991).
These ts mutants were unable to form colonies on rich media at 36°C, but they grew normally, or nearly so, at 23°C. The mutants were tested for loss of viability, change in morphology, cell number increase, and the ability to synthesize protein, ribonucleic acid (RNA), and deoxyribonucleic acid (DNA) after a shift from 23°C (permissive) to 36°C (restrictive temperature). Mutations were found that resulted in a preferential loss of the ability to perform protein synthesis, RNA synthesis, DNA synthesis, cell division, or cell-wall formation. Time-lapse light microscopy was used to detect ts mutants defective in gene functions needed at specific stages of the cell-division cycle.
By characterizing mutants of genes that control different stages of the cell cycle, these ts strains were called cdc (cell-division cycle) mutants. For example, when cells carrying one cdc mutation arrest at a cell-cycle stage (the execution point), most cells end up with a tiny bud that does not develop further. They are arrested at bud emergence. When cells carrying another cdc mutation terminate at mitosis, cells display a large bud and are destined to arrest in mid-nuclear division. Cells carrying another cdc mutation are defective in cell separation. They do not show a definite termination point because other processes of the cell cycle, such as bud initiation and nuclear division, continue, despite the block in cell separation.
After characterization of cdc mutants defective at different cell-cycle stages, particularly at initiation of DNA replication, bud emergence, nuclear division (mitosis), and cell separation (cytokinesis), Hartwell et al. (1974) proposed a model that accounted for the order of cell-cycle events that was deduced from the phenotypes of budding yeast ts mutants. These pioneering genetic studies were performed before the age of DNA cloning and sequencing and recombinant DNA technology. At the time of cdc mutant isolation, there was no concrete hope that genes responsive to mutations and molecular functions of gene products would be elucidated in the near future. However, Hartwell and his colleagues identified CDC28 as the crucial cell-cycle regulator, which later turned out to be the catalytic subunit of CDK1, a fundamental cell-cycle regulator.
The fission yeast, Schizosaccharomyces pombe, is also an excellent model for studying cell-cycle control, mitosis, and genome biology. S. pombe possesses approximately 5000 genes and is believed to have diverged from S. cerevisiae about one billion years ago. Parallel studies are often useful because that which is true in both yeasts often applies to vertebrates. Mitchison and Leupold, respectively, initiated cell physiology and genetics of S. pombe in the 1950s (Mitchison 1957; Leupold 1958). S. pombe vegetative cells are rod-shaped and the organism increases its length by growth. Using this property, Fantes and Nurse (1977) isolated cell-size mutants, later found to be defective in Cdc2, Wee1, and Cdc25, which are important cell-cycle-regulating kinases and a phosphatase, respectively (Nurse 1990). Fission yeast has only three chromosomes, and mitotic chromosome condensation is visualized using a fluorescent probe, DAPI (1,4-diaminidino-2-phenylindole), for staining DNA (Toda et al. 1981). The genome is an attractive system because it contains heterochromatin with histone H3 lysine-9 methylation in centromeres, telomeres, and the mating-type locus and includes numerous noncoding RNAs (Bernard et al. 2001; Volpe et al. 2002; Hirota et al. 2008).
DISCOVERIES THAT MERGE MPF WITH CDK
The Rao–Johnson studies showed that nuclear–cytoplasmic interactions are important for regulating mitosis. Masui and Markert (1971) published a paper entitled “Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes.” They hypothesized the presence of a cytoplasmic factor called MPF in the embryogenesis of frog (Rana pipiens) eggs treated with progesterone. They could identify cytoplasmic activities by injecting cytoplasm from progesterone-treated oocytes at various stages of maturation. The most effective cytoplasm peaked in parallel with maturation. The responsible substance was unidentified at the time, but it was cytoplasmic, as the production of MPF was not affected by removal of the nucleus. MPF was later established to cause germinal vesicle breakdown when injected into the frog (Xenopus oocytes) and to induce mitotic metaphase in a cell-free system.
Seventeen years after the initial discovery, Lohka et al. (1988) reported the purification of MPF from egg extract using ammonium sulfate precipitation and six chromatographic steps. Two protein bands were present in the most highly purified preparations. This material contained an intense protein kinase activity that phosphorylated histone H1. That same year, Maller and colleagues (Gautier et al. 1988) reported that one of the two components in the purified MPF was actually the homolog of the fission yeast Cdc2 kinase (a homolog of budding yeast Cdc28). The other band was later identified as mitotic cyclin (Hunt 2004), the level of which underwent cell-cycle stage-specific degradation by ubiquitin-mediated proteolysis that led to the inactivation of the Cdc2–cyclin complex. This finding resolved several enigmas regarding M-phase transitions in the cell cycle. Exciting discoveries were reported almost simultaneously from several laboratories, which caused a coalescence of mitosis research that previously had consisted of people working in disparate disciplines with different organisms, such as fission yeast, budding yeast, flies, clams, frog oocytes, and mammalian cells. The unification of cell-cycle research thus occurred through the discovery of MPF and CDK as the conserved cell-cycle regulators from yeast to higher eukaryotes (Nurse 1990). Molecular biology of cancer research was also enormously influenced by the discovery of the basic mechanism of cell-division cycle control, symbolized by CDK.
CYCLIN AND SECURIN DESTRUCTION IN YEAST AND MAMMALIAN MITOSIS
Using clam oocyte extracts, Hershko and colleagues identified the large complex called a cyclosome (Sudakin et al. 1995) (alternatively called anaphase-promoting complex [APC]) (Peters et al. 1996; Hershko 1999) that contains cyclin-B-selective ubiquitin ligase activity. This APC/cyclosome modifies mitotic cyclin by polyubiquitination, promoting its destruction by 26S proteasomes. The same complex was found to be necessary for ubiquitin-mediated degradation of fission yeast Cut2, budding yeast Pds1, and human hPTTG1 (collectively called “securin”) that are essential for chromosome segregation (Funabiki et al. 1996; Yamamoto et al. 1996; Jallepalli et al. 2001). (Securin/PTTG is not essential in the mouse; separase can be regulated by Cdk phosphorylation.) Securin destruction is needed for proper chromosome segregation and the signal sequence for destruction, called the destruction box, can be swapped between fission yeast mitotic cyclin Cdc13 and securin Cut2 so that the timing of destruction should be under the same signal pathway. The role of the APC/cyclosome is thus to coordinate mitotic control with chromosome segregation. The actual role of securin is to physically associate with a protease called separin/separase, essential for cleavage of the cohesin subunit. Securin acts as a chaperone/inhibitor of separin/separase. The loss of securin promotes activation of separase activity.
IDENTIFICATION OF CENTROMERES IN BUDDING AND FISSION YEASTS
Eukaryotic gene cloning and sequencing studies first flourished using S. cerevisiae as the model eukaryote because this organism contained a 2-µm plasmid that was exploited as a source of extrachromosomal genes. Any gene inserted into that plasmid (or integrated into the genomic DNA) with marker DNA could be isolated and sequenced, leading to identification of the gene product. Transformation, which changes the properties of yeast cells by introducing new or altered genes, was most powerful for elucidating the properties of endogenous genes. Basically, the same transformation method was used in many other organisms, including mammals, to identify and manipulate genes of interest.
For example, Hinnen et al. (1978) transformed a leucine-requiring yeast strain, leu2, using a plasmid that carried the yeast LEU2 gene. Resulting Leu+ transformants contained a plasmid carrying either the LEU2 gene or chromosomally integrated gene dependent on the presence or absence of the replication origin in plasmid DNA. The plasmid DNA sequence integrated into the yeast genomic DNA behaved like the yeast genomic DNA in mitosis and meiosis. A significant breakthrough was made when Clarke and Carbon (1980a,b) identified a functional yeast centromere. They first isolated the CDC10 gene, which is close to the centromere of chromosome III, by transformation of ts cdc10 mutants isolated in the laboratory of Lehland Hartwell. They were able to establish the directionality of a cloned piece of DNA with respect to the genetic map. In the second stage of their investigation, Clarke and Carbon successfully isolated a short piece of a functional centromere. When present on a plasmid carrying a yeast chromosomal DNA replicator, this DNA (designated CEN3) enabled the plasmid to function as a minichromosome, both mitotically and meiotically. These circular minichromosomes are stable in mitosis and segregate as yeast chromosomes in the first and second meiotic divisions. Indeed, the functional centromere of budding yeast was rather short, only several hundred base pairs. In other organisms, such as fission yeast, flies, and mammals, however, the centromeres are far larger, requiring different approaches for identification and isolation.
Analyses of artificially constructed mini-chromosomes by pulsed-field gel electrophoresis showed that the size of the S. pombe centromere is 30- to 130-kb long, much larger than that of S. cerevisiae, which is on the order of 0.1 kb (Chikashige et al. 1989; Hahnenberger et al. 1991; Takahashi et al. 1992). Linear mini-chromosomes were obtained by double truncation followed by the addition of telomeric sequences. The circular minichromosomes were isolated by the gap-repair method (Hahnenberger et al. 1989; Niwa et al. 1989). These 30- to 160-kb minichromosomes were useful to define the S. pombe functional centromere regions and also to determine the entire repetitious centromere sequence (Niwa et al. 1989; Takahashi et al 1992). The pericentromeric repeats are heterochromatic because histone H3 is methylated and heterochromatin protein 1 (HP-1), which affects accurate chromosome segregation like Swi6, is abundant. The central centromere region associates with histone H3-like CENP-A and Mis12, which form the base of the kinetochore and are essential for equal chromosome segregation. Pericentromeric repeats are actually transcribed and directed by RNA interference, which flanks the central centromere region. The central centromere region tethers CENP-A-like, Cnp1-containing nucleosomes and promotes kinetochore assembly in mitosis. A number of proteins bound to central centromere regions are all conserved in higher eukaryotes. Although the DNA sequence organization of centromeres differs greatly among organisms, proteins bound to the centromere and pericentromere regions are highly conserved, allowing the conservation of segregation mechanisms that require centromere-binding proteins.
LINEAR DNA ENDS IN THE LARGE NUCLEUS OF Tetrahymena HELPED ISOLATE TELOMERES
Ciliates are a group of protozoans characterized by the presence of hair-like cilia. Unlike most other eukaryotes, these organisms, such as Tetrahymena thermophile, possess two different nuclei. A micronucleus, containing ordinary chromosomes, serves as the germ line nucleus, but does not express its genes, whereas the large nucleus is generated from the micronucleus by amplification of gene-sized DNAs. Free ribosomal RNA genes were discovered by Gall (1974) in the macronucleus. Blackburn and Gall (1978) studied sequences occurring at their termini in the extrachromosomal recombinant DNA (rDNA) genes and found a tandemly repeated hexanucleotide C4A2 sequence 5′ (C-C-C-C-A-A)n 3′, in which the parameter n ranges from 20 to 70. Thus, Tetrahymena was an ideal organism for identification of the linear DNA end sequence.
Szostak and Blackburn (1982) then shifted to the use of yeast and constructed a linear yeast plasmid by joining fragments from the termini of Tetrahymena rDNA to a yeast vector. Thus, yeast can use DNA ends from distantly related organisms, suggesting that structural features required for telomere replication might be highly conserved in evolution. The linear plasmid was then used as a vector to clone telomeres from yeast. One Tetrahymena linear end was removed, and yeast fragments that functioned as an end on a linear plasmid were selected. Szostak and Blackburn (1982) successfully isolated yeast telomeres and suggested that all yeast chromosomes appeared to have a common telomere sequence. Yeast telomeres appear to be similar in structure to the rDNA of Tetrahymena, regarding the presence of specific nicks or gaps within a simple repeated sequence.
Telomeres are protected from fusion, degradation, or recombination, common properties of DNA damaged by γ-irradiation, which induces double-stranded breakage. An enigmatic question was what kind of structure at the ends of linear DNA allows their complete replication. Shampay et al. (1984) showed that yeast chromosomal telomeres terminate in a DNA sequence consisting of tandem irregular repeats of the general form C1-3A. The same repeat units could be added to the ends of Tetrahymena telomeres, in an apparently non-template-directed manner, during their replication on linear plasmids in yeast.
Greider and Blackburn (1985) then found a novel activity in Tetrahymena cell-free extracts that adds tandem TTGGGG repeats onto synthetic telomere primers. The single-stranded DNA oligonucleotides (TTGGGG)4 and TGTGTGGGTGTGTGGGTGTGTGGG, containing the Tetrahymena and yeast telomeric sequences, respectively, each functioned as primers for elongation, whereas nontelomeric DNA oligomers did not. A novel telomere terminal transferase, later identified as the ribonucleoprotein enzyme called telomerase, is involved in the addition of telomeric repeats necessary for the replication of chromosome ends in eukaryotes. Telomere shortening was later shown to be related to senescence in all eukaryotes.
DIVERSE MITOTIC MUTANT PHENOTYPES BROADENED OUR UNDERSTANDING OF MITOSIS
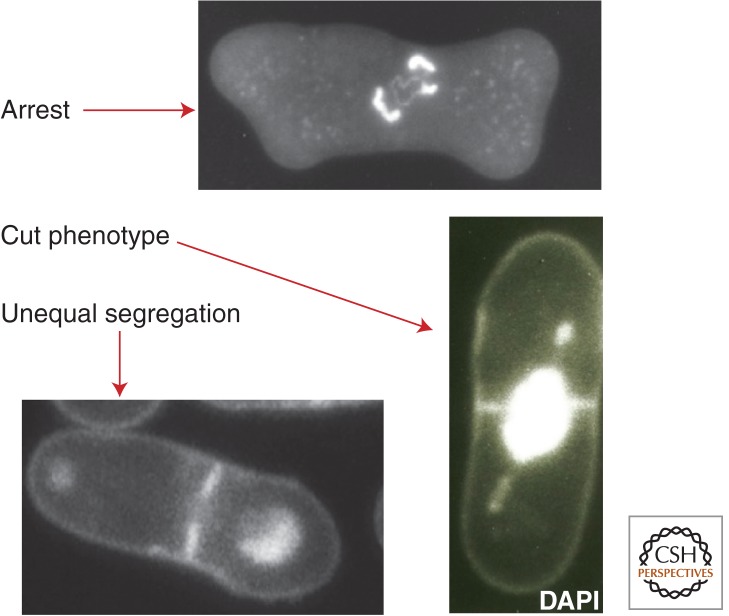
Mutants defective in mitosis have been isolated from various model organisms from fungi to mammals, with the aim of understanding gene functions essential for mitosis. Mammalian culture cell ts mutants have also been isolated (Nishimoto et al. 1978). Here, two model organisms, fission yeast and fruit flies, are considered as examples. Systematic screening of fission yeast mitotic mutants (ts and cold-sensitive [cs]) was performed. A number of genes were identified and their products were characterized (reviewed in Yanagida 2005). Three principal chromosome segregation defects (arrest, cut, and unequal) were found for this organism at the restrictive temperature (Fig. 4). For example, β-tubulin nda3 cs mutant was mitotically arrested at 22°C because of the absence of the spindle, resulting in the activation of SAC (Hiraoka et al. 1984), whereas DNA topoisomerase II (top2) ts mutants displayed the drastic cut phenotype. In these cells, cytokinesis bisected the nucleus that had failed to divide during anaphase because of a DNA topoisomerase II (Top2) mutation (Uemura and Yanagida 1984). Centromere-associating protein mutants such as mis6 produced unequal chromosome segregation, resulting in cells with large and small daughter nuclei (Hayashi et al. 2004).
Figure 4.
Three types of chromosome segregation defects in fission yeast mutants cultured at the restrictive temperature. Arrest, β-tubulin nda3 mutant; cut phenotype, top2 mutant; unequal segregation, centromere-binding protein mis6 mutant. DNA is stained with DAPI.
Various aspects of chromosome segregation could be understood through gene functions essential for mitosis. Some of them are closely related to cell-cycle control, including stage-specific protein modification and proteolysis. SAC-related APC/cyclosome ubiquitin ligase and ubiquitin-mediated anaphase proteolysis lead to the destruction of mitotic cyclin and securin, whereas protein phosphatases (PP1, PP2A) and protein kinases (PKA, aurora), other than CDK, are required for controlling different stages of mitosis. Assembly and proper functioning of the mitotic kinetochore and spindle apparatus are highly complex, requiring multiple gene functions. Key players in chromosome segregation are cohesin, condensin, the securin–separase complex, Top2, and kinetochore microtubule destabilizers. They function not only in mitosis, but also in interphase displaying distinct functions. Most mitotic genes identified are conserved in higher eukaryotes, so that the basic mechanism of eukaryotic chromosome segregation in mitosis should, likewise, be largely conserved. There are some curious exceptions, however. For example, fission yeast centromere protein Mis18, required for priming centromeres to load CENP-A/cenH3 protein Cnp1, is conserved in vertebrates, but not in nonvertebrates, such as fruit flies (Fujita et al. 2007). Concerning this essential centromere protein, vertebrates are more similar to fungi.
Wang's discovery of DNA topoisomerase (Wang 1991) brought relief to many biologists who worked on various aspects of DNA metabolism, because it was apparent that DNA winding, unwinding, catenation, and decatenation had to occur for DNA to be properly used and archived. However, no one had any idea how those functions were performed. The answer was that many specific DNA topoisomerases control different aspects of the topological problems of DNA (Wang 1991). To elucidate the role of topoisomerase in forming mitotic chromosomes, a biochemical genetic approach was used. S. pombe mutants defective in Top2 activity were isolated by assaying a great number of mutant extracts. With ts and cs top2 mutants, it was established that Top2 is required for both chromosome segregation and condensation (Uemura and Yanagida 1984; Uemura et al. 1987). In the laboratories of Wang and Botstein, Top2 was shown to be essential for budding yeast mitosis (Holm et al. 1986). Type 1 topoisomerase partly overlaps in function (relaxing activity) with Top2, so that the double mutant produced a nonmitotic phenotype in which the cell-cycle block occurred during interphase and the nucleolus was destroyed (Uemura and Yanagida 1984).
The curious cut (cell untimely torn) phenotype, which bisected the nucleus during cytokinesis (Fig. 4), was used as a cytological marker to isolate other mutants with similar phenotypes. A number of mutants producing cut-like phenotypes were isolated and their gene products were identified. All cut-like mutants turned out to be defective in important mitotic steps, such as chromosome condensation, segregation, activation of separase, control of ubiquitin-mediated proteolysis, spindle formation, spindle elongation, cytokinesis, etc. (Yanagida 2005). Among them, mutants in top2, separase cut1, and condensin cut3 produced highly similar phenotypes. They may be implicated in the removal of interphase components from chromosomes before chromosome segregation (Yanagida 2009; Akai et al. 2011). Chromosome condensation may be actually visualized as a result of the removal of proteins and RNAs from chromosomes before segregation.
Drosophila is an attractive organism in which to study both the rapid rounds of mitosis typical of embryonic development and the longer cell cycles of diploid tissues later in development (Glover 1989). Powerful molecular biological studies of fruit fly mitosis became possible after the discovery of Drosophila mutants and use of reagents, pioneered by Nuesslein-Volhard (St Johnston and Nuesslein-Volhard 1992). Glover and associates discovered polo and aurora, protein kinases with important roles in the progression from early to late mitosis (Sunkel and Glover 1988; Glover et al. 1995). The discovery of these kinases inaugurated a new mechanistic approach to understand how the entire mitotic process, including chromosome segregation and subsequent cytokinesis, is regulated. Although CDK1 was inactivated during the transition from metaphase to anaphase, polo and aurora remain active until telophase because they also regulate cytokinesis. Drosophila bearing mutant forms of polo and aurora revealed severe defects in mitosis and cytokinesis with pleiotropic defects in chromosome segregation. Polo and aurora are present from fungi to higher eukaryotes (Lane and Nigg 1996). These are profoundly important in orchestrating mitotic events, such as CDK regulation, spindle formation, maintenance of centrosome structural integrity, centrosome activation, SAC, chromosome cohesion and condensation, and progression of cytokinesis (Earnshaw and Cooke 1991; Golsteyn et al. 1994; Kumagai and Dunphy 1996; Biggins and Murray 2001).
The concept of chromatid cohesion was initially developed in fruit fly genetics. In PubMed, the terminology of sister chromatid cohesion started with the papers of Orr-Weaver (Kerrebrock et al. 1992; (Miyazaki and Orr-Weaver 1994) that clarified the phenotype of the fruit fly mei-S332 mutation that displayed a defect in chromatid cohesion. Mei-S332/Shugoshin is now known to protect cohesion at centromeres in coordination with type 2A phosphatase (Kitajima et al. 2004). The mutant mei-S332, which stands for “meiotic from Salaria 332” (isolated in Salaria, Rome, Italy), was originally described in 1968 (Sandler et al. 1968). The mutant showed chromosome nondisjunction defects in homozygotes of both sexes, hinting that this gene is required in a common meiotic process for separating sister chromatids. A subsequent paper (Goldstein 1980) suggested that mei-S332 mutants are defective in sister chromatid cohesiveness, which was felt to be an important factor in chromosome segregation. Holloway et al. (1993) later investigated this issue using vertebrate cells and showed that the inactivation of Cdk1 was not required for sister chromatid separation, but proteolysis might be needed to dissolve the linkage (“glue”) between them. However, their hypothesis that ubiquitin-mediated proteolysis was required turned out to be indirect. There exists a protease that directly cleaves the protein responsible for sister chromatid cohesion. Studies on budding yeast showed that the protein complex, called cohesin, was responsible for sister chromatid cohesion and that cohesion was disrupted in mitosis (Guacci et al. 1997; Uhlmann et al. 1999, 2000). A thiol protease, separase, cleaves the cohesin subunit Scc1/Rad21, dissociating the ring-like cohesin from chromosomes and allowing disjunction of sister chromatids.
FUTURE PROSPECTS FOR MITOSIS
Although mitosis was first discovered and described in detail commencing in the 19th century, molecular biological approaches assumed importance only after the onset of recombinant DNA technology, that is, gene cloning and sequencing. Whereas a large number of essential proteins and their functional complexes during mitosis are now known and their number is still increasing, very few are understood mechanistically at the atomic level. In the near future, biochemistry and structural biology may become the dominant methods for solving basic questions in mitosis.
Thereafter, physiological, medical, and evolutionary fields related to mitosis will flourish. For example, little is understood about the alteration of mitosis under various nutritional and environmental stresses. Patterns of mitosis may change greatly under different physiological, nutritional, and senescent conditions. Comparative studies of mitosis in stem and nonstem cells are of interest, as mitosis is a plausible stage for the origin of asymmetric properties between daughter cells, which are essential features of cell differentiation. In the end, the importance of evolutionary variations in mitosis must be stressed, as changes in mitosis are known to be the rich resources of diversification of organisms.
ACKNOWLEDGMENTS
The author is greatly indebted to past and present laboratory members in both Kyoto University and Okinawa Institute of Science and Technology Graduate University (OIST) for lively and stimulating discussions and a productive research environment. The present article is imperfect in citing important discoveries and research topics, as the field of mitosis is now so broad and deep. The author apologizes for the lack of citations of the work of many people who contributed significantly to the development of the field. The author thanks Norihiko Nakazawa for preparing illustrations in Figures 1 and 2. The author is greatly indebted to Steven D. Aird (OIST) for editing the manuscript.
Footnotes
Editors: Mitsuhiro Yanagida, Anthony A. Hyman, and Jonathon Pines
Additional Perspectives on Mitosis available at www.cshperspectives.org
REFERENCES
- Akai Y, Kurokawa Y, Nakazawa N, Tonami-Murakami Y, Suzuki Y, Yoshimura SH, Iwasaki H, Shiroiwa Y, Nakamura T, Shibata E, et al. 2011. Opposing role of condensin hinge against replication protein A in mitosis and interphase through promoting DNA annealing. Open Biol 1: 110023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery OT, Macleod CM, McCarty M 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med 79: 137–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltzer F 1964. Theodor Boveri. Science 144: 809–815 [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Biggins S, Murray AW 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev 15: 3118–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Gall JG 1978. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120: 33–53 [DOI] [PubMed] [Google Scholar]
- Boveri T 2008. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 121: 1–84 [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M 1989. Composite motifs and repeat symmetry in S. pombe centromeres: Direct analysis by integration of NotI restriction sites. Cell 57: 739–751 [DOI] [PubMed] [Google Scholar]
- Clarke L, Carbon J 1980a. Isolation of the centromere-linked CDC10 gene by complementation in yeast. Proc Natl Acad Sci 77: 2173–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J 1980b. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287: 504–509 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Cooke CA 1991. Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of a pathway of structural changes in the chromosomes during metaphase and early events in cleavage furrow formation. J Cell Sci 98: 443–461 [DOI] [PubMed] [Google Scholar]
- Fantes P, Nurse P 1977. Control of cell size at division in fission yeast by a growth-modulated size control over nuclear division. Exp Cell Res 107: 377–386 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M 2007. Priming of centromere for CENP-A recruitment by human hMis18α, hMis18β, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M 1996. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature 381: 438–441 [DOI] [PubMed] [Google Scholar]
- Gall JG 1974. Free ribosomal RNA genes in the macronucleus of Tetrahymena. Proc Natl Acad Sci 71: 3078–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J, Norbury C, Lohka M, Nurse P, Maller J 1988. Purified maturation-promoting factor contains the product of a Xenopus homolog of the fission yeast cell cycle control gene cdc2+. Cell 54: 433–439 [DOI] [PubMed] [Google Scholar]
- Glover DM 1989. Mitosis in Drosophila. J Cell Sci 92: 137–146 [DOI] [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H 1995. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81: 95–105 [DOI] [PubMed] [Google Scholar]
- Goldstein LS 1980. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster. Chromosoma 78: 79–111 [DOI] [PubMed] [Google Scholar]
- Golsteyn RM, Schultz SJ, Bartek J, Ziemiecki A, Ried T, Nigg EA 1994. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J Cell Sci 107: 1509–1517 [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43: 405–413 [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger KM, Baum MP, Polizzi CM, Carbon J, Clarke L 1989. Construction of functional artificial minichromosomes in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci 86: 577–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnenberger KM, Carbon J, Clarke L 1991. Identification of DNA regions required for mitotic and meiotic functions within the centromere of Schizosaccharomyces pombe chromosome I. Mol Cell Biol 11: 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH 1971. Genetic control of the cell division cycle in yeast: IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res 69: 265–276 [DOI] [PubMed] [Google Scholar]
- Hartwell LH 1991. Twenty-five years of cell cycle genetics. Genetics 129: 975–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Reid B 1970. Genetic control of the cell-division cycle in yeast: I. Detection of mutants. Proc Natl Acad Sci 66: 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M 1973. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics 74: 267–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ 1974. Genetic control of the cell division cycle in yeast. Science 183: 46–51 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M 2004. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- Hershko A 1999. Mechanisms and regulation of the degradation of cyclin B. Philos Trans R Soc Lond B Biol Sci 354: 1571–1575; discussion 1575–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A, Hicks JB, Fink GR 1978. Transformation of yeast. Proc Natl Acad Sci 75: 1929–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto Y 1956. Cell division without mitotic apparatus in sea urchin eggs. Exp Cell Res 11: 630–636 [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M 1984. The NDA3 gene of fission yeast encodes β-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell 39: 349–358 [DOI] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134 [DOI] [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW 1993. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell 73: 1393–1402 [DOI] [PubMed] [Google Scholar]
- Holm C, Meeks-Wagner DW, Fangman WL, Botstein D 1986. A rapid, efficient method for isolating DNA from yeast. Gene 42: 169–173 [DOI] [PubMed] [Google Scholar]
- Hunt T 2004. The discovery of cyclin (I). Cell 116 (2 Suppl): S63–S64, 1 p following S65 [DOI] [PubMed] [Google Scholar]
- Inoue S 1953. Polarization optical studies of the mitotic spindle: I. The demonstration of spindle fibers in living cells. Chromosoma 5: 487–500 [DOI] [PubMed] [Google Scholar]
- Inoue S, Dan K 1951. Birefringence of the dividing cell. J Morphol 89: 423–455 [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C 2001. Securin is required for chromosomal stability in human cells. Cell 105: 445–457 [DOI] [PubMed] [Google Scholar]
- Johnson RT, Rao PN 1970. Mammalian cell fusion: Induction of premature chromosome condensation in interphase nuclei. Nature 226: 717–722 [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Miyazaki WY, Birnby D, Orr-Weaver TL 1992. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics 130: 827–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510–517 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 273: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Lane HA, Nigg EA 1996. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol 135: 1701–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold U 1958. Studies on recombination in Schizosaccharomyces pombe. Cold Spring Harb Symp Quant Biol 23: 161–170 [DOI] [PubMed] [Google Scholar]
- Li R, Havel C, Watson JA, Murray AW 1993. The mitotic feedback control gene MAD2 encodes the α-subunit of a prenyltransferase. Nature 366: 82–84 [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Hayes MK, Maller JL 1988. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc Natl Acad Sci 85: 3009–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi I 1973. A myosin-like protein in the cortical layer of the sea urchin egg. J Cell Biol 59: 542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui Y, Markert CL 1971. Cytoplasmic control of nuclear behavior during meiotic maturation of frog oocytes. J Exp Zool 177: 129–145 [DOI] [PubMed] [Google Scholar]
- Matsuura H 1951. Chromosome studies on Trillium kamtschaticum Pall. and its allies: XXIV. The association of kinetochores of non-homologous chromosomes at meiosis. Chromosoma 4: 273–283 [DOI] [PubMed] [Google Scholar]
- Mazia D, Dan K 1952. The isolation and biochemical characterization of the mitotic apparatus of dividing cells. Proc Natl Acad Sci 38: 826–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1929. A cytological and genetical study of triploid maize. Genetics 14: 180–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1931. The order of the genes C, Sh and Wx in Zea mays with reference to a cytologically known point in the chromosome. Proc Natl Acad Sci 17: 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1932. A correlation of ring-shaped chromosomes with variegation in Zea mays. Proc Natl Acad Sci 18: 677–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1941. The stability of broken ends of chromosomes in Zea mays. Genetics 26: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1950. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci 36: 344–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1984. The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- McClintock B, Hill HE 1931. The cytological identification of the chromosome associated with the R-G linkage group in Zea mays. Genetics 16: 175–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM 1957. The growth of single cells: 1. Schizosaccharomyces pombe. Exp Cell Res 13: 244–262 [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL 1994. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet 28: 167–187 [DOI] [PubMed] [Google Scholar]
- Nishimoto T, Eilen E, Basilico C 1978. Premature of chromosome condensation in a ts DNA-mutant of BHK cells. Cell 15: 475–483 [DOI] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M 1989. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J 8: 3045–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P 1990. Universal control mechanism regulating onset of M-phase. Nature 344: 503–508 [DOI] [PubMed] [Google Scholar]
- Paweletz N 2001. Walther Flemming: Pioneer of mitosis research. Nat Rev Mol Cell Biol 2: 72–75 [DOI] [PubMed] [Google Scholar]
- Peters JM, King RW, Hoog C, Kirschner MW 1996. Identification of BIME as a subunit of the anaphase-promoting complex. Science 274: 1199–1201 [DOI] [PubMed] [Google Scholar]
- Rao PN, Johnson RT 1970. Mammalian cell fusion: Studies on regulation of DNA synthesis and mitosis. Nature 225: 159–164 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP 1990. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol 110: 81–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Hard R 1990. Newt lung epithelial cells: Cultivation, use, and advantages for biomedical research. Int Rev Cytol 122: 153–220 [DOI] [PubMed] [Google Scholar]
- Robinow CF, Marak J 1966. A fiber apparatus in the nucleus of the yeast cell. J Cell Biol 29: 129–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L, Lindsley DL, Nicoletti B, Trippa G 1968. Mutants affecting meiosis in natural populations of Drosophila melanogaster. Genetics 60: 525–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J, Szostak JW, Blackburn EH 1984. DNA sequences of telomeres maintained in yeast. Nature 310: 154–157 [DOI] [PubMed] [Google Scholar]
- St Johnston D, Nusslein-Volhard C 1992. The origin of pattern and polarity in the Drosophila embryo. Cell 68: 201–219 [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell 6: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel CE, Glover DM 1988. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci 89: 25–38 [DOI] [PubMed] [Google Scholar]
- Szostak JW, Blackburn EH 1982. Cloning yeast telomeres on linear plasmid vectors. Cell 29: 245–255 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell 3: 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Yamamoto M, Yanagida M 1981. Sequential alterations in the nuclear chromatin region during mitosis of the fission yeast Schizosaccharomyces pombe: Video fluorescence microscopy of synchronously growing wild-type and cold-sensitive cdc mutants by using a DNA-binding fluorescent probe. J Cell Sci 52: 271–287 [DOI] [PubMed] [Google Scholar]
- Uemura T, Yanagida M 1984. Isolation of type I and II DNA topoisomerase mutants from fission yeast: Single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J 3: 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M 1987. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50: 917–925 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K 1999. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400: 37–42 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103: 375–386 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wang JC 1991. DNA topoisomerases: Why so many? J Biol Chem 266: 6659–6662 [PubMed] [Google Scholar]
- Yamamoto A, Guacci V, Koshland D 1996. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s). J Cell Biol 133: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M 2005. Basic mechanism of eukaryotic chromosome segregation. Philos Trans R Soc Lond B Biol Sci 360: 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida M 2009. Clearing the way for mitosis: Is cohesin a target? Nat Rev Mol Cell Biol 10: 489–496 [DOI] [PubMed] [Google Scholar]