Abstract
Sexual antagonism occurs when an allele is beneficial in one sex but costly in the other. Parental antagonism occurs when an allele is beneficial when inherited from one sex but costly when inherited from the other because of fitness interactions among kin. Sexual and parental antagonisms together define four genetic niches within the genome that favor different patterns of gene expression. Natural selection generates linkage disequilibrium among sexually and parentally antagonistic loci with male-beneficial alleles coupled to alleles that are beneficial when inherited from males and female-beneficial alleles coupled to alleles that are beneficial when inherited from females. Linkage disequilibrium also develops between sexually and parentally antagonistic loci and loci that influence sex determination. Genes evolve sex-specific expression to resolve sexual antagonism and evolve imprinted expression to resolve parental antagonism. Sex-specific chromosomes allow a gene to specialize in a single niche.
Natural selection acts differently on genes in female and male bodies and on genes of maternal and paternal origin. In these “environments,” genes may exhibit sex-specific or imprinted expression or may influence sex determination.
Every diploid individual of a sexually reproducing population is derived from an egg fertilized by a sperm. Therefore, males considered collectively have the same reproductive value as females because half the genes of the next generation will be derived from males and half from females (Kokko et al. 2006). This fundamental symmetry is independent of sex ratio and mating system and applies also to hermaphrodites in male and female roles. Despite their equal stakes in posterity, males and females have evolved distinct morphologies and reproductive strategies as indirect consequences, often very indirect, of the ancient dichotomy between production of larger gametes by one sex and smaller gametes by the other (Queller 1997). Vive la différence!
Not only are half the genes of the next generation present in females of the current generation, but half the genes of the current generation were inherited from females. Genes of maternal and paternal origin (hereafter matrigenes and patrigenes) are each transmitted to 50% of the next generation of gametes and resulting offspring. Therefore, matrigenic and patrigenic alleles benefit equally from an individual’s survival and reproduction. This symmetry is broken when organisms interact with kin to whom they are unequally related via their mother and father (Haig 1997; Úbeda and Gardner 2010). Adaptive responses to such asymmetries of relatedness can favor gene expression that is conditional on parental origin. “Imprinted” expression can cause matrigenes and patrigenes to have opposing effects on disputed phenotypes (Haig 2000a; Holman and Kokko 2014).
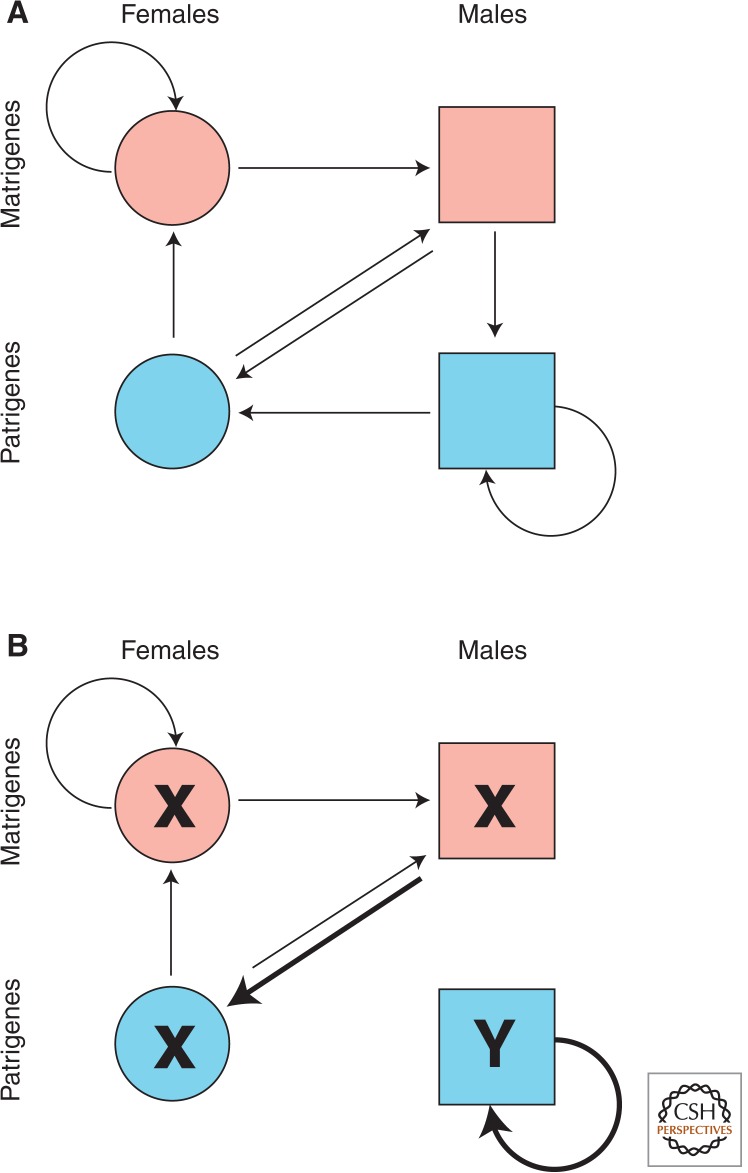
Anatomical, physiological, and behavioral sex differences are thus associated with two selective asymmetries, one obvious (natural selection acts differently on genes in female and male bodies) and the other less obvious (natural selection acts differently on genes of maternal and paternal origin). These asymmetries define orthogonal partitions of the gene pool into pairs of “environments” in which there is niche-specific selection (Fig. 1A). Differential selection in male and female niches reinforces sexual dimorphism by processes of sex-specific adaptation. Differential selection in matrigenic and patrigenic niches can result in imprinted gene expression and molecular adaptations acting at cross-purposes within organisms.
Figure 1.
Sex-related partitions of the gene pool. The gene pool is independently subdivided into female (circles) and male (squares) niches and into matrigenic (pink) and patrigenic (blue) niches. Gene flow between the environments is represented by arrows. Thin arrows represent 50% gene flow in each generation. Thick arrows represent 100% gene flow. (A) Autosomal alleles experience all four environments. (B) Y-linked genes experience only the patrigenic male environment. X-linked genes are excluded from this environment.
PHENOTYPIC CONFLICT AND ALLELIC COMPETITION
“Sexual conflict,” as the term is currently used in the literature, encompasses a set of loosely connected phenomena. It sometimes refers to conflictual relations between males and females and sometimes to antagonistic pleiotropy of a gene’s effects in male and female bodies. Rather than review how terms have been used and misused, this section will define how terms will be used in this article. We will use conflict to refer to interactions in which different phenotypic agents have contradictory goals; competition to refer to processes by which alleles at a locus compete for representation in the gene pool; and antagonism to refer to fitness trade-offs in competition among alleles (for similar distinctions between conflict and competition, see Cosmides and Tooby 1981; Haig 1997).
Sexual conflict will refer to phenotypic interactions in which males and females “prefer” different outcomes. The classic example is when a male attempts to mate with a female who attempts to avoid his mating attempt (Parker 1979). Parental conflict will refer to phenotypic interactions in which matrigenes and patrigenes “prefer” different outcomes. The classic example is when patrigenes and matrigenes have opposing effects on fetal growth (Haig and Graham 1991). Sexual antagonism occurs when an allele has a relative advantage in one sex but relative disadvantage in the other (Rice 1987). By analogy, parental antagonism occurs when an allele has a relative advantage when inherited from one sex but relative disadvantage when inherited from the other (Haig 1997).
Sexual and parental conflict both occur within a generation: the first between male and female organisms and the second between patrigenic and matrigenic suborganisms. In contrast, sexual and parental antagonism involve pleiotropic effects of a single gene in different generations. Sexual antagonism occurs because an allele finds itself sometimes in males and sometimes in females; parental antagonism occurs because an allele is inherited sometimes from males and sometimes from females.
“Parental” is not the ideal label for conflict between the haploid contributions of parents because parental conflict is easily misinterpreted as referring to conflict between the diploid parents (unfortunately, we have found no better adjective). Genes of mothers and matrigenes of offspring have distinct evolutionary “interests,” as do genes of fathers and patrigenes of offspring (Haig 1992; Burt and Trivers 1998; Wilkins and Haig 2002; Queller 2003). Parental conflict between matrigenes and patrigenes is not the same as sexual conflict between mothers and fathers.
Sexual conflict and sexual antagonism are distinct. Neither implies the other. Sexual conflict involves antagonistic adaptations that are mutually reinforcing. Males possess adaptations to overcome resistance because females resist; females possess adaptations to resist coercion because males coerce. The genes responsible for resistance are maintained because of the genes responsible for coercion. They justify each other’s existence. Sexual antagonism, in contrast, is a relation between competing alleles in which one allele’s gain is another allele’s loss. It may involve traits that do not involve any direct interaction between the sexes. Our distinction, between sexual conflict and sexual antagonism, corresponds to the distinction of Chapman et al. (2003) between interlocus and intralocus sexual conflict. We prefer our terminology because lumping together these disparate notions as “sexual conflict” is a recipe for conceptual confusion.
Consider two illustrative hypotheses: (1) Seminal proteins benefit a male’s sperm in competition with sperm of other males but reduce the longevity of females with whom he mates (Chapman et al. 1995). Sexual conflict is present because a trait that increases male fitness reduces the fitness of females, but sexual antagonism is absent because genes that encode male-specific proteins are never expressed in females. (2) Tyrosine at position 282 of the hemachromatosis protein (HFE) causes greater retention of iron and is costly in men because of iron overload but beneficial in women because of periodic loss of blood at menstruation and childbirth (Little 1996). Sexual antagonism is present because the HFE allele that encodes Tyr282 is beneficial in females but costly in males. Sexual conflict is absent because the allele's expression in females does not reduce the fitness of males.
Parental conflict and parental antagonism are also distinct. Consider an extended example. Igf2 encodes a growth enhancer (IGF-II) and Igf2r a growth inhibitor (IGF2R). In the absence of imprinting, patrigenes at both loci benefit from greater fetal growth than matrigenes, but allelic competition favors a compromise between levels of expression that are optimal in matrigenic and patrigenic niches (because unimprinted alleles are constrained to the same expression in both niches). At the optimal compromise, an Igf2 allele with increased expression would have a fitness benefit in the patrigenic niche outweighed by a fitness cost in the matrigenic niche, whereas an Igf2r allele with increased expression would have a fitness cost in the patrigenic niche outweighed by a fitness benefit in the matrigenic niche. Thus, expression at both loci is subject to parental antagonism. However, matrigenes and patrigenes have identical expression and there is no phenotypic conflict. IGF-II and IGF2R cooperate to produce the level of growth that is optimal for unimprinted genes.
Imprinted alleles are expressed differently in matrigenic and patrigenic niches and thereby create the possibility of phenotypic conflict. An Igf2 allele with decreased expression as a matrigene but unchanged expression as a patrigene can outcompete an unimprinted allele, as can an Igf2r allele with decreased expression as a patrigene but unchanged expression as a matrigene. Patrigenes and matrigenes belong to different suborganisms with different optimal levels of growth. Parental conflict between the suborganisms is expressed by IGF-II pushing growth toward the patrigenic optimum and IGF2R pushing growth toward the matrigenic optimum (Haig and Graham 1991).
In a model of Wilkins and Haig (2001), parentally antagonistic selection favored progressively lower matrigenic expression of Igf2 and progressively higher patrigenic expression, with the opposite at Igf2r. At both loci, imprinted alleles outcompeted unimprinted alleles and highly expressed imprinted alleles outcompeted more lowly expressed imprinted alleles. In the domain of allelic competition, imprinted alleles with increased expression of Igf2 and Igf2r were evolutionary allies because each justified the other’s existence in competition with alternative imprinted and unimprinted alleles at their respective loci. At the evolutionary equilibrium, there was all or none parent-specific expression at both loci. Parental antagonism was resolved because successful alleles could do no better in either matrigenic or patrigenic niches. However, in the domain of parental conflict, patrigenic expression of Igf2 and matrigenic expression of Igf2r had opposing effects on growth.
SEXUAL ANTAGONISM
Sexual antagonism occurs when natural selection favors different alleles in different environments, in this case male and female bodies. Each generation, alleles that have survived selection in one sex are mixed with alleles that have survived selection in the other sex and are randomly assigned to male and female bodies of the next generation. Single-locus models of antagonistic selection with frequent gene mixing can maintain polymorphism but only under restrictive conditions. The usual outcome is fixation of whichever allele does best, on average, across the ensemble of selective environments (Hedrick 1999; Prout 2000).
Stable polymorphism requires that heterozygotes have higher average fitness than both homozygotes. In the case of sexual antagonism, selection in males and females occurs in parallel with changes in zygotic allele frequencies determined by the harmonic means of sex-specific fitnesses (Levene 1953). For the same arithmetic mean, the harmonic mean decreases with larger differences between the sex-specific fitnesses. Therefore, conditions for stable polymorphism become less restrictive for stronger sexual antagonism under the assumption that fitness differences between the sexes are greater for homozygotes than for heterozygotes (Prout 2000).
Sexual antagonism can occur with intragenomic conflict. Meiotic drive is usually sex-specific because male and female meiosis are mechanistically distinct (Úbeda and Haig 2004, 2005). If a driving allele is to be maintained at a polymorphic equilibrium, then its “unfair” advantage in spermatogenesis or oogenesis must be balanced by viability or fertility costs elsewhere in the life cycle. If these costs are shared by the sexes, rather than restricted to the sex in which drive occurs, then the polymorphism will be sexually antagonistic. In other words, the driving allele will be more fit than the alternative allele in the sex in which it drives with the relative allelic fitnesses reversed in the other sex.
PARENTAL ANTAGONISM
Kin are significant actors in the lives of many organisms. Sex differences of survival, dispersal, mating behavior, or reproductive role cause organisms to interact differently with their mothers and fathers or with their mothers’ and fathers’ kin. As a result, matrigenes and patrigenes experience different relatedness to social partners and are subject to antagonistic selection when phenotypic outcomes that increase matrigenic inclusive fitness reduce patrigenic inclusive fitness, or vice versa (Haig 2000b; Brandvain 2010; Úbeda and Gardner 2010, 2011, 2012; Van Cleve et al. 2010; Brandvain et al. 2011). Matrigenic and patrigenic niches may be further subdivided by sex if males and females experience different kinship environments (Fig. 1A).
Optimal outcomes differ for an allele in matrigenic and patrigenic niches when its locus influences a trade-off between the fitness of self (ego) and of “asymmetric kin.” An individual is asymmetric kin of ego if the individual has different probabilities of carrying copies of ego’s matrigenes and patrigenes (Haig 1997). Thus, mothers are asymmetric kin of their offspring but offspring are symmetric kin of their mothers. Paternal half-siblings (patrisibs) and maternal half-siblings (matrisibs) are asymmetric kin but full-siblings are symmetric kin. Aunts, uncles, and cousins are usually asymmetric kin.
A necessary condition for parental antagonism is that genotypes with the same pair of alleles inherited from opposite parents make unequal contributions to the next generation either because the reciprocal heterozygotes have unequal fitness or are produced in unequal numbers. We will adopt the convention that Aa heterozygotes inherit A from their mother, whereas aA heterozygotes inherit A from their father. Two simple scenarios will illustrate how interactions with asymmetric kin can cause Aa and aA to experience different social environments.
Individual males of the greater spear-nosed bat, Phyllostomus hastatus, defend harems of unrelated females and father most of a harem’s offspring. Females of a harem give birth synchronously, each to a single pup. Pups huddle together in their mothers’ absence (McCracken and Bradbury 1981; Porter and Wilkinson 2001; Haig 2010). Suppose that A is a rare allele causing pups to divert a portion of their milk intake from individual growth to thermogenesis. An Aa pup will huddle solely with aa patrisibs because only its mother carries A, but an aA pup will huddle with aa and aA patrisibs because their mutual father is heterozygous for A. Therefore, aA pups will be warmer than Aa pups for the same expenditure of fuel and will have higher survival than Aa pups.
A longer interval until the birth of a younger sibling increases a child's chances of survival but reduces its mother’s fecundity, such that the child has fewer younger siblings. Suppose that each of a mother’s children has a different father and A is a rare allele that increases the interbirth interval, perhaps by promoting more intense suckling and prolonging lactational amenorrhea of the mother. Aa and aA sucklings cause the same prolongation of the subsequent birth interval, relative to aa, and therefore enjoy the same increment in fitness, but aA sucklings occur in families in which aA patrisibs are each cared for by different mothers whereas Aa sucklings occur in families in which Aa matrisibs are cared for by the same mother. As a consequence of A’s effects on maternal fecundity, there will be more aA patrisibs than Aa matrisibs and A will make a greater contribution to the next generation as a patrigene than as a matrigene.
The balance of selective forces shifts with allele frequencies and the changing genetic composition of families. The above scenarios considered the simple situation when A was rare. As A increases in frequency, it begins to occur in families in which both parents are Aa or aA and then in families in which one or both parents are AA. Thus, the genetic environments of A as a matrigene and as a patrigene will depend on allelic frequency, as will the genotypic fitnesses of aa, Aa, aA, and AA.
MULTILOCUS INTERACTIONS
Consider a gene pool divided between two habitats, marsh (M) and fen (F), with limited migration between M and F. Habitat-specific selection would result in marsh-adapted alleles attaining high frequency in M and fen-adapted alleles attaining high frequency in F. Marsh-adapted alleles would be associated with marsh-adapted alleles, and fen-adapted alleles with fen-adapted alleles, at all loci subject to habitat-specific selection. Now suppose that M and F represent males and females. In this case, there is complete hybridization between sexual “habitats” in every generation, followed by random assignment of progeny to habitats in the next generation. Can nonrandom allelic associations be maintained under these conditions?
Linkage disequilibrium (LD) is generated between two sexually antagonistic loci with independent effects on sex-specific fitness (Patten et al. 2010; Úbeda et al. 2011). This LD arises from recurrent admixture of gene pools subject to selection in male and female niches. Sperm are enriched for male-beneficial alleles at both loci and eggs for female-beneficial alleles. Therefore, the next generation of zygotes is enriched for haplotypes that couple male-beneficial alleles and for haplotypes that couple female-beneficial alleles. LD is also generated between two parentally antagonistic loci with independent effects on parent-specific fitness (Patten et al. 2013). Favored haplotypes couple patrigenically beneficial alleles or couple matrigenically beneficial alleles. Nonrandom associations can be intuitively understood as arising because alleles that do well in the same niche, and occur on the same chromosome, tend to be transmitted together to the next generation. The strength of these associations increases with the tightness of linkage between the loci.
Sexually antagonistic alleles have a relative advantage in one sex and are thus preferentially inherited from that sex. Therefore, alleles at a parentally antagonistic locus in the patrigenic niche will tend to occur on haplotypes with male-beneficial alleles but on haplotypes with female-beneficial alleles in the matrigenic niche. Therefore, selection favors haplotypes that couple male-beneficial alleles with patrigenically beneficial alleles and haplotypes that couple female-beneficial alleles with matrigenically beneficial alleles (Patten et al. 2013). The process resembles mutual hitchhiking in which alleles at the two loci are carried along in each other’s draft.
Local adaptation can create habitat-specialist islands within a habitat-generalist genome (Nosil et al. 2009) and select for chromosomal inversions that bind together coadapted clusters of alleles (Kirkpatrick and Barton 2006). Sex-specific selection can be seen as a special case of divergent selection in different habitats. By analogy to models of ecological divergence, sexual antagonism can create complementary haplotypes with sexually antagonistic effects and favor reduced recombination between the haplotypes.
SEX DETERMINATION
Sexually antagonistic haplotypes can be viewed as differentially adapted for distinct sexual “habitats.” From this perspective, sex determination is the process by which alleles occupy habitats, and sex-biasing alleles influence habitat choice. Sex-biasing alleles also influence the choice of matrigenic or patrigenic habitats in the generation after they bias sex determination. Therefore, a male-beneficial/patrigenically beneficial haplotype would “prefer” to be present in males whereas a female-beneficial/matrigenically beneficial haplotype would “prefer” to be present in females. Thus, the former class of haplotypes will become preferentially associated with male-biasing alleles and the latter with female-biasing alleles. The ability to match genotype to habitat facilitates ecological specialization in models of sympatric speciation (Ravigné et al. 2004, 2009). By analogy, associations of sexually and parentally antagonistic loci with sex-biasing loci facilitate sexual and parental specialization in the genome. For simplicity of exposition, the discussion that follows will focus on the interaction between sexually antagonistic loci and sex-biasing loci.
Consider a baseline scenario in which sex chromosomes and sex-specific meiotic drive are absent. All chromosome segments benefit from an individual developing as whichever sex has higher expected fitness, given environmental conditions and the individual’s genotype at sexually antagonistic loci. Different segments will have spent different amounts of time in male and female bodies because of their associations with sexually antagonistic and sex-biasing alleles.
Natural selection favors haplotypes that couple male-beneficial alleles and complementary haplotypes that couple female-beneficial alleles, with the resulting LD accentuating the local strength of sexually antagonistic selection (Patten et al. 2010). Natural selection will also generate LD that associates male-beneficial haplotypes with male-biasing alleles and female-beneficial haplotypes with female-biasing alleles because such associations ensure that haplotypes are preferentially present in the sex in which they have a relative advantage. Sex-biasing alleles, in effect, influence the choice between male and female habitats and become associated with genes that benefit from the choice. They create genomic neighborhoods that spend more than the average time in one sex and therefore accumulate sexually antagonistic polymorphisms (Jordan and Charlesworth 2011).
A system in which sex-biasing haplotypes are scattered throughout the genome is inherently unstable (Rice 1986) because local conditions for sexually antagonistic polymorphism are progressively relaxed as a genomic region gains influence in sex determination, whereas conditions for sexually antagonistic polymorphism become more restrictive in regions that lose influence. The “victor” of competition between unlinked sex-biasing loci for control of sex determination will generally be the locus that is associated with greater sexually antagonistic variance in fitness (van Doorn and Kirkpatrick 2007). This dynamic process tends to favor control of sex determination being concentrated in a single genomic region with a high hurdle of sexual antagonism to be overcome for the region’s replacement by a new sex-determining locus. The generation of complementary haplotypes—one male-biasing and male-beneficial, the other female-biasing and female-beneficial—favors the suppression of recombination between the haplotypes which, in turn, favors further accumulation of sexually antagonistic differences between the nascent sex chromosomes (Rice 1984; Charlesworth 1991).
Sex-specific meiotic drive sows discord within the genome with respect to sex determination because driving haplotypes will accumulate alleles that promote development as the sex in which drive occurs, even if this is not the sex that maximizes fitness for loci segregating independently of drive (Úbeda and Haig 2004, 2005; Burt and Trivers 2006). At the same time, haplotypes that are driven against will accumulate sex-biasing alleles that promote development of the nondriving sex. Genomic segments that segregate independently of drive should also accumulate sex-biasing alleles to counter drive-associated departures from the optimal sex ratio.
Sex-specific drive could thus explain some cases of replacement of polygenic sex determination by sex chromosomes or of one system of sex chromosomes by another (van Doorn and Kirkpatrick 2007; Yoshida and Kitano 2012). Although distortions of sex ratio are expected during transitions of one system of sex determination to another, the genome is expected to eventually return to consensus sex ratios via the parliament of genes and the suppression of drive. In some cases, the driving chromosome that precipitated a change will be lost in the process, and, in others, it will be retained but with its drive suppressed (Kozielska et al. 2010). Systems of meiotic drive are usually associated with inversions (Charlesworth and Hartl 1978; Burt and Trivers 2006) and these inversions would facilitate the recruitment of sex-biasing and sexually antagonistic alleles onto driving haplotypes.
SEX CHROMOSOMES
Autosomal genes experience all four niches defined by partition of the gene pool by sex and sex of origin (Fig. 1A), whereas sex-linked genes experience some niches but not others (Fig. 1B). Specifically, Y-linked genes are restricted to the patrigenic–male niche and X-linked genes are excluded from it. As a consequence, sexual and parental antagonism are experienced differently by autosomal, X-linked, and Y-linked loci.
Y-linked genes are always patrigenes in males and are therefore not subject to parentally or sexually antagonistic selection. They should be strongly biased to promote male fitness and patrigenic interests (Hurst 1994).
The situation for X-linked genes is more complex for two reasons. First, an X-linked allele is present in female bodies twice as often as in male bodies. Second, it shares female bodies with another X-linked allele but is on its own in male bodies. These factors cancel for X-linked alleles with additive effects on a sexually antagonistic phenotype because each allele has twice the influence on phenotype in males as in females. Dominant effects of rare X-linked alleles are expressed twice as often in females as in males, whereas rare recessive effects are experienced predominantly by males. Therefore, X chromosomes will tend to accumulate polymorphisms in which female-beneficial alleles have dominant effects and male-beneficial alleles have recessive effects (Rice 1984). A population genetic model found a slight bias toward fixing male-beneficial alleles on the X chromosome (Patten and Haig 2009).
With respect to parental antagonism, an X-linked gene is a patrigene only in females but a matrigene in both sexes. Matrigenic effects are unopposed by a patrigenic allele in males whereas X-linked matrigenes and patrigenes have equal selective weight in females. Therefore, parentally antagonistic selection is predicted to be biased toward matrigenic interests at X-linked loci relative to autosomal loci (Patten and Haig 2009).
“RESOLUTION” OF PARENTAL AND SEXUAL ANTAGONISM
Sexual antagonism is resolved (with respect to standing allelic variation) by the loss of sexually antagonistic polymorphism. It is resolved (with respect to mutation) by the fixation of an allele that is unbeatable in both sexes. Similarly, parental antagonism is resolved in the short term by loss of polymorphism and in the long term by the fixation of an allele that is unbeatable in both matrigenic and patrigenic niches.
Sexual antagonism could be resolved by a gene duplication in which the two paralogs become specialized for different sexes (Connallon and Clarke 2011, 2013; Gallach and Betrán 2011). In theory, one could imagine duplications at each and every locus subject to sexual antagonism with the genome composed of specialist teams, one on the field in male bodies but on the bench in female bodies, the other with roles reversed. This arrangement would resemble the game called football in the United States in which different teams take the field on offense and defense. With respect to sexual specialization, the genome resembles more the game called football in the rest of the world in which compromises between offensive and defensive prowess are ubiquitous because players must perform in both roles.
The formal possibility of male-specific and female-specific subgenomes poses the question why the sexes are so similar. Gonochorism (separate sexes) has ancient origins on the tree of life, yet males and females are often immediately recognizable as members of the same species, presumably because of constraints on sexual divergence imposed by a shared genome. Extreme sexual dimorphism, as shown by scale insects or barnacles with dwarf males, is the exception rather than the rule, perhaps analogous to the limited global appeal of American football.
Two consequences of duplication and sexual subfunctionalization may contribute to the long-term maintenance of sexual resemblance. First, substitutions at one locus that would be beneficial in both sexes are not readily transferrable between loci. Taken in isolation, this effect is cancelled by duplicate genes providing twice the target for mutation. Second, each duplicate will be subject to selection in only one sex. Therefore the strength of selection relative to drift and mutation is weakened for substitutions that would have benefited both sexes. Species in which most genes are sexual specialists may have a long-term evolutionary disadvantage because the two sexes must evolve separately to meet shared environmental challenges.
Sexual and parental antagonism can occur not only with respect to the amino acid sequence of proteins but also with respect to the strength and tissue specificity of promoters. Sexual and parental antagonism over levels of expression are resolved in different ways because expression in male and female niches occurs in different bodies but expression in matrigenic and patrigenic niches occurs in shared bodies, indeed in shared nuclei. If optimal expression differs between the sexes, then sexual antagonism can be resolved by the evolution of sex-specific expression with the female optimum achieved in female bodies and the male optimum in male bodies (Fig. 2A).
Figure 2.
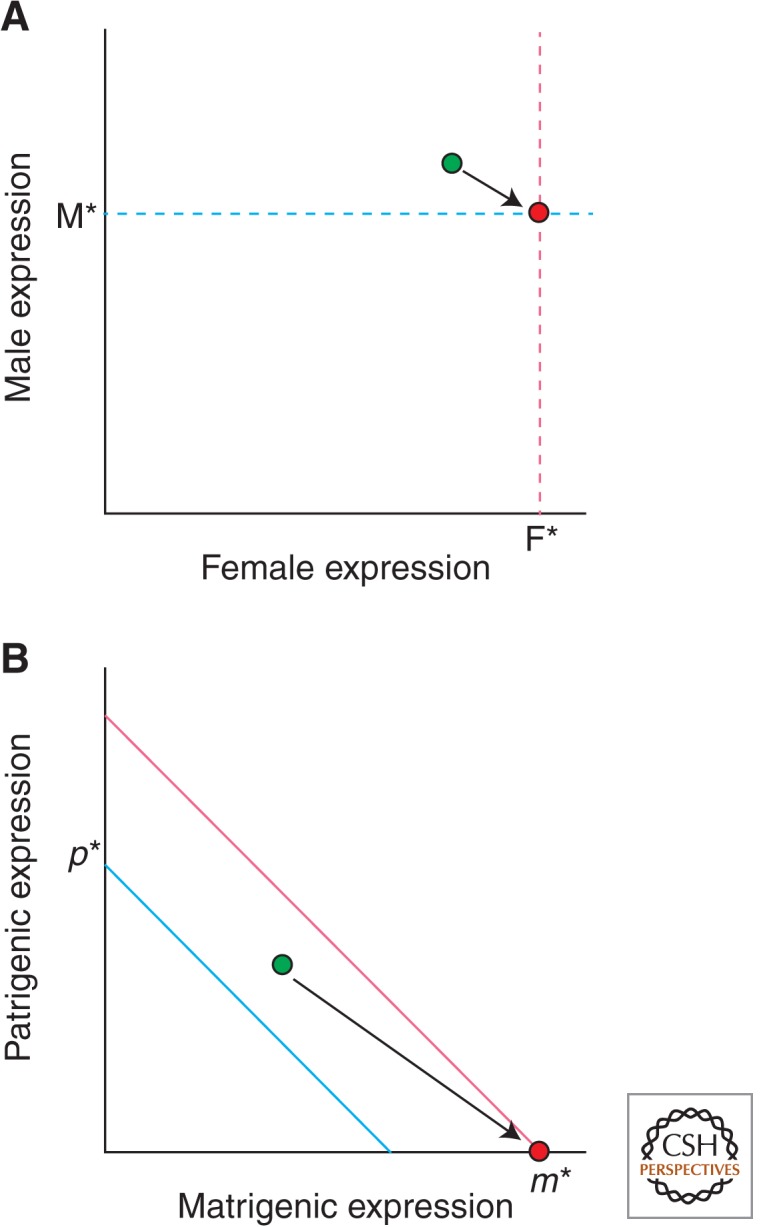
Resolution of sexual and parental antagonism. (A) The optimal level of expression in males is M* and in females F*. Sex expression is initially the same in both sexes at a compromise between their respective optima (green dot). Sex-specific expression allows M* to be achieved in males and F* in females (red dot). (B) The optimal level of expression for patrigenes is p* and for matrigenes m* (in this example, m* > p*). The blue line represents all combinations of matrigenic and patrigenic expression that sum to p* and the reddish line represents all combinations that sum to m*. Gene expression is initially unimprinted at a compromise between m* and p* (green dot). Imprinted expression allows silencing of the patrigenic allele and expression of m* by the matrigenic allele (red dot).
In contrast, the level of expression within a nucleus is determined by the sum of matrigenic and patrigenic expression, and this sum cannot simultaneously maximize matrigenic and patrigenic inclusive fitness when there is parental antagonism (Fig. 2B). Silencing of an allele when it occupies the niche that favors the lesser sum, but its expression at the optimal level in the other niche, is the evolutionarily stable outcome of conflict between matrigenes and patrigenes over level of transcription (Haig and Westoby 1989; Úbeda and Haig 2003). All-or-none imprinting is evolutionarily stable because the amount of mRNA is optimal in the niche in which an allele is expressed but the allele cannot reduce its expression below zero in the other niche. This resolution of parental antagonism has been called “the loudest voice prevails” (Wilkins and Haig 2003).
Quantitative biases of parent-specific expression, as distinct from all-or-none imprinting, have been reported (Khatib 2007). Expression at these loci is most easily explained as not being subject to antagonistic selection because biased expression is not predicted to be evolutionarily stable in the presence of parental antagonism (although see Greenwood-Lee et al. 2001). Alleles with biased expression could be selectively neutral variants at dosage-insensitive loci or could represent asymmetric “divisions of labor” between matrigenic and patrigenic roles at dosage-sensitive loci. The optimal level of expression at a dosage-sensitive locus not subject to parental antagonism is the same for matrigenes and patrigenes. Any combination of matrigenic and patrigenic expression that sums to this optimal amount is evolutionarily stable (Haig 1997, 2000a). Unbiased expression, with each allele producing half the optimal amount, has been considered the most plausible way of achieving optimal expression because matrigenes and patrigenes have been presumed to be equivalent in the absence of reasons to believe otherwise. This default assumption can be questioned within gene clusters that contain loci subject to all-or-none imprinting because matrigenic and patrigenic haplotypes are demonstrably nonequivalent.
CONCLUDING REMARKS
A gene is typically uncertain of whether it will occupy a male or female habitat in the next generation and, by extension, of whether it will occupy a matrigenic or patrigenic habitat in the generation after that. Differential selection in each pair of habitats gives rise to sexual and parental antagonism. Two kinds of resolutions are possible. In the first, a gene evolves different expression in different habitats: sexually dimorphic expression in response to sexual antagonism or imprinted expression in response to parental antagonism. Second, a gene might “choose” its habitat—either by influencing sex determination directly or by linkage to a sex-determining locus. A gene that occupies only one habitat can specialize. Not every antagonism can be resolved by tampering with expression levels nor can every gene choose its sex. Therefore, the sexual ecology of the genome will involve interactions among habitat specialists and habitat generalists.
Footnotes
Editors: William R. Rice and Sergey Gavrilets
Additional Perspectives on The Genetics and Biology of Sexual Conflict available at www.cshperspectives.org
REFERENCES
- Brandvain Y 2010. Matrisibs, patrisibs, and the evolution of genomic imprinting on autosomes and sex chromosomes. Am Nat 176: 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandvain Y, Van Cleve J, Úbeda F, Wilkin JF 2011. Demography, kinship, and the evolving theory of genomic imprinting. Trends Genet 27: 251–257 [DOI] [PubMed] [Google Scholar]
- Burt A, Trivers R 1998. Genetic conflicts in genomic imprinting. Proc R Soc B 265: 2393–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Trivers R 2006. Genes in conflict. Harvard University Press, Cambridge, MA [Google Scholar]
- Chapman T, Liddle LF, Kalb MF, Partridge L 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373: 241–244 [DOI] [PubMed] [Google Scholar]
- Chapman T, Arnqvist G, Bangham J, Rowe L 2003. Sexual conflict. Trends Ecol Evol 18: 41–47 [Google Scholar]
- Charlesworth B 1991. The evolution of sex chromosomes. Science 251: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Hartl DL 1978. Population dynamics of the segregation distorter polymorphism of Drosophila melanogaster. Genetics 89: 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clarke AG 2011. The resolution of sexual antagonism by gene duplication. Genetics 187: 919–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connallon T, Clarke AG 2013. Antagonistic versus nonantagonistic models of balancing selection: Characterizing the relative timescales and hitchhiking effects of partial selective sweeps. Evolution 67: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmides LM, Tooby J 1981. Cytoplasmic inheritance and intragenomic conflict. J Theor Biol 89: 83–129 [DOI] [PubMed] [Google Scholar]
- Gallach M, Betrán E 2011. Intralocus sexual conflict resolved through gene duplication. Trends Ecol Evol 26: 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood-Lee JM, Taylor PD, Haig D 2001. The inclusive fitness dynamics of genomic imprinting. Selection 2: 101–116 [Google Scholar]
- Haig D 1992. Genomic imprinting and the theory of parent–offspring conflict. Sem Devel Biol 3: 153–160 [Google Scholar]
- Haig D 1997. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proc R Soc B 264: 1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D 2000a. The kinship theory of genomic imprinting. Annu Rev Ecol Syst 31: 9–32 [Google Scholar]
- Haig D 2000b. Genomic imprinting, sex-biased dispersal, and social behavior. Ann NY Acad Sci 907: 149–163 [DOI] [PubMed] [Google Scholar]
- Haig D 2010. The huddler's dilemma: A cold shoulder or a warm inner glow. In Social behaviour: Genes, ecology and evolution (ed. Székely T, Moore AJ, Komdeur J), pp. 107–109 Cambridge University Press, Cambridge [Google Scholar]
- Haig D, Graham C 1991. Genomic imprinting and the strange case of the insulin-like growth factor-II receptor. Cell 64: 1045–1046 [DOI] [PubMed] [Google Scholar]
- Haig D, Westoby M 1989. Parent-specific gene expression and the triploid endosperm. Am Nat 134: 147–155 [Google Scholar]
- Hedrick PW 1999. Antagonistic pleiotropy and genetic polymorphism: A perspective. Heredity 82: 126–133 [Google Scholar]
- Holman L, Kokko H 2014. The evolution of genomic imprinting: Costs, benefits and long-term consequences. Biol Rev Camb Philos Soc 10.1111/brv.12069 [DOI] [PubMed] [Google Scholar]
- Hurst LD 1994. Embryonic growth and the evolution of the mammalian Y chromosome: I. The Y as an attractor for selfish growth factors. Heredity 73: 223–232 [DOI] [PubMed] [Google Scholar]
- Jordan CY, Charlesworth D 2011. The potential for sexually antagonistic polymorphism in different genome regions. Evolution 66: 505–516 [DOI] [PubMed] [Google Scholar]
- Khatib H 2007. Is it genomic imprinting or preferential expression? Bioessays 29: 1022–1028 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton N 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokko H, Jennions MD, Brooks R 2006. Unifying and testing models of sexual selection. Annu Rev Ecol Syst 37: 43–66 [Google Scholar]
- Kozielska M, Weissing FJ, Beukeboom LW, Pen I 2010. Segregation distortion and the evolution of sex-determining mechanisms. Heredity 104: 100–112 [DOI] [PubMed] [Google Scholar]
- Levene H 1953. Genetic equilibrium when more than one ecological niche is available. Am Nat 87: 331–333 [Google Scholar]
- Little P 1996. Woman’s meat, a man’s poison. Nature 382: 494–495 [DOI] [PubMed] [Google Scholar]
- McCracken GF, Bradbury JW 1981. Social organization and kinship in the polygynous bat Phyllostomus hastatus. Behav Ecol Sociobiol 8: 11–34 [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D 2009. Divergent selection and heterogenous genomic divergence. Mol Ecol 18: 375–402 [DOI] [PubMed] [Google Scholar]
- Parker GA 1979. Sexual selection and sexual conflict. In Sexual selection and reproductive competition in insects (ed. Blum MS, Blum NA), pp. 123–166 Academic, New York [Google Scholar]
- Patten MM, Haig D 2009. Maintenance or loss of genetic variation under sexual and parental antagonism at a sex-linked locus. Evolution 63: 2888–2895 [DOI] [PubMed] [Google Scholar]
- Patten MM, Haig D, Úbeda F 2010. Fitness variation due to sexual antagonism and linkage disequilibrium. Evolution 64: 3638–3642 [DOI] [PubMed] [Google Scholar]
- Patten MM, Úbeda F, Haig D 2013. Sexual and parental antagonism shape genomic architecture. Proc R Soc B 280: 20131795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter TA, Wilkinson GS 2001. Birth synchrony in greater spear-nosed bats, Phyllostomus hastatus. J Zool 253: 383–390 [Google Scholar]
- Prout T 2000. How well does opposing selection maintain variation? In: Evolutionary genetics: From molecules to morphology (ed. Singh RS, Krimbas CB), pp. 157–181 Cambridge University Press, Cambridge [Google Scholar]
- Queller DC 1997. Why do females care more than males? Proc R Soc B 264: 1555–1557 [Google Scholar]
- Queller DC 2003. Theory of genomic imprinting conflict in social insects. BMC Evol Biol 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravigné V, Olivieri I, Dieckmann U 2004. Implications of habitat choice for protected polymorphisms. Evol Ecol Res 6: 125–145 [Google Scholar]
- Ravigné V, Dieckmann U, Olivieri I 2009. Live where you thrive: Joint evolution of habitat choice and local adaptation facilitates specialization and promotes diversity. Am Nat 174: E141–E169 [DOI] [PubMed] [Google Scholar]
- Rice WR 1984. Sex chromosomes and the evolution of sexual dimorphism. Evolution 38: 735–742 [DOI] [PubMed] [Google Scholar]
- Rice WR 1986. On the instability of polygenic sex determination: The effect of sex-specific selection. Evolution 40: 633–639 [DOI] [PubMed] [Google Scholar]
- Rice WR 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41: 911–914 [DOI] [PubMed] [Google Scholar]
- Úbeda F, Gardner A 2010. A model for genomic imprinting in the social brain: Juveniles. Evolution 64: 2587–2600 [DOI] [PubMed] [Google Scholar]
- Úbeda F, Gardner A 2011. A model for genomic imprinting in the social brain: Adults. Evolution 65: 462–475 [DOI] [PubMed] [Google Scholar]
- Úbeda F, Gardner A 2012. A model for genomic imprinting in the social brain: Elders. Evolution 66: 1567–1581 [DOI] [PubMed] [Google Scholar]
- Úbeda F, Haig D 2003. Dividing the child: Genomic imprinting and evolutionary games. Genetica 117: 103–110 [DOI] [PubMed] [Google Scholar]
- Úbeda F, Haig D 2004. Sex-specific meiotic drive and selection at an imprinted locus. Genetics 167: 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Úbeda F, Haig D 2005. On the evolutionary stability of Mendelian segregation. Genetics 170: 1345–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Úbeda F, Haig D, Patten MM 2011. Stable linkage disequilibrium owing to sexual antagonism. Proc R Soc B 278: 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cleve J, Feldman MW, Lehmann L 2010. How demography, life history, and kinship shape the evolution of genomic imprinting. Am Nat 176: 440–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449: 909–912 [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D 2001. Genomic imprinting of two antagonistic loci. Proc R Soc B 268: 1861–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D 2002. Parental modifiers, antisense transcripts and loss of imprinting. Proc R Soc B 269: 1841–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JF, Haig D 2003. What good is genomic imprinting: The function of parent-specific gene expression. Nat Rev Genet 4: 359–368 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Kitano J 2012. The contribution of female meiotic drive to the evolution of neo-sex chromosomes. Evolution 66: 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]