Abstract
Background
Two features of alcohol addiction that have been widely studied in animal models are relapse drinking following periods of alcohol abstinence, and the escalation of alcohol consumption after chronic continuous or intermittent alcohol exposure. The genetic contribution to these phenotypes has not been systematically investigated.
Methods
HXB/BXH recombinant inbred (RI) rat strains were given access to alcohol sequentially as follows: alcohol (10%) as the only fluid for one week; alcohol (10%) and water in a two-bottle choice paradigm for 7 weeks (“pre-alcohol deprivation effect (ADE) alcohol consumption”); two weeks of access to water only (alcohol deprivation); two weeks of re-access to 10% alcohol and water (“post-ADE alcohol consumption”). The periods of deprivation and re-access to alcohol were repeated three times. The ADE was defined as the amount of alcohol consumed in the first 24 hr after deprivation minus the average daily amount of alcohol consumed in the week prior to deprivation. Heritability of the phenotypes was determined by ANOVA, and QTLs were identified.
Results
All strains showed increased alcohol consumption, compared to the predeprivation period, in the first 24 hr after each deprivation (ADE). Broad sense heritability of the ADEs was low (ADE1, 9.1%; ADE2, 26.2%; ADE3, 16.3%). Alcohol consumption levels were relatively stable over weeks 2 to 7. Post-ADE alcohol consumption levels consistently increased in some strains, and were decreased or unchanged in others. Heritability of pre- and post-ADE alcohol consumption was high and increased over time (week 2, 38.5%; week7, 51.1%; week 11, 56.8%; week 15, 63.9%). QTLs for pre- and post-ADE alcohol consumption were similar, but the strength of the QTL association with the phenotype decreased over time.
Conclusions
In the HXB/BXH RI rat strains, genotypic variance does not account for a large proportion of phenotypic variance in the ADE phenotype (low heritability), suggesting a role of environmental factors. In contrast, a large proportion of the variance across the RI strains in pre- and post-ADE alcohol consumption is due to genetically determined variance (high heritability).
Keywords: HXB/BXH recombinant inbred rat strains, alcohol consumption, alcohol deprivation effect, heritability, QTL
Introduction
A common feature of drug and alcohol addiction is relapse following a period of abstinence from drug use (Bossert et al., 2013; Koob and Le Moal, 1997; Le and Shaham, 2002; Martin-Fardon and Weiss, 2013). This return to compulsive alcohol seeking behavior is a key problem in the treatment of alcohol addiction (Anton et al., 2006; Heyser et al., 1997; O'Brien, 2005; Spanagel and Holter, 1999). In order to understand the neurobiological basis for relapse, which is thought to result from increased “craving” for the drug, and to develop medications to treat this aspect of addiction, numerous animal models have been developed (Bossert et al., 2013; Martin-Fardon and Weiss, 2013). These models include various forms of reinstatement of drug-seeking behavior (e.g., context-induced reinstatement, cue-induced reinstatement, stress-induced reinstatement), conditioned place preference and a procedure called the “alcohol deprivation effect” (ADE). The ADE is defined as a transient (usually one to several days) increase in alcohol intake following re-exposure to alcohol access after single or multiple periods of abstinence (McKinzie et al., 1996; Rodd-Henricks et al., 2000a; Salimov et al., 1993; Sinclair and Senter, 1967). The ADE has been suggested to represent a model for alcohol craving and relapse in rats and mice (Heyser et al., 1997; Sinclair and Li, 1989; Spanagel and Holter, 1999), and has been used in studies to assess the ability of pharmacological agents to modify relapse drinking (Sajja and Rahman, 2013; Spanagel and Holter, 1999; Suchankova et al., 2013).
Much of the work on the ADE has made use of rat lines that have been genetically selected for alcohol preference or avoidance (Bell et al., 2004; Rodd-Henricks et al., 2000a; Rodd-Henricks et al., 2000b). There are differences among these lines with respect to the occurrence and magnitude of the ADE. Some lines need repeated deprivation periods in order to display an ADE (O'Dell et al., 2004; Rodd-Henricks et al., 2000a), and some lines of alcohol-preferring rats do not display an ADE (Sinclair and Li, 1989; Vengeliene et al., 2003). These differences led to the suggestion that genetic background has a significant influence on the development and magnitude of the ADE (Bell et al., 2008; Vengeliene et al., 2003). However, to date there has not been a systematic study of the contribution of genotype and/or environment on the ADE.
While the ADE is a transient effect, both continuous chronic alcohol exposure and intermittent alcohol exposure (periods of alcohol exposure followed by periods of abstinence) have been reported to result in changes (usually an escalation) of “steady-state” (longer term) voluntary alcohol intake by mice and rats. We define this alcohol consumption, over a two-week period following the alcohol deprivation period, as “post-ADE alcohol consumption”. Such “post-ADE alcohol consumption” has been evaluated by others using numerous models that vary in the duration and route (e.g., alcohol vapor inhalation, alcohol injection) of the chronic or intermittent alcohol exposure periods, and the number and length of alcohol abstinence periods (Becker and Lopez, 2004; Camarini and Hodge, 2004; Cox et al., 2013; Gilpin et al., 2008; Lopez and Becker, 2005; O’Dell et al., 2004; Roberts et al., 2000). Many of these studies have used single inbred strains or lines of mice or rats that have been selected for high alcohol preference, but in a few instances several strains of mice or rats have been compared. Crabbe and his colleagues, comparing several different selected lines and inbred strains of mice, found that the degree of change in post-ADE alcohol consumption varied across genotypes (Crabbe et al., 2012; Rosenwasser et al., 2013). Differences were also found in comparisons of inbred rat strains and selected lines of high alcohol-preferring rats (Vengeliene et al., 2003). As for the ADE, these results suggest a genetic influence on changes in voluntary alcohol intake after chronic or intermittent exposure to alcohol, but no systematic analysis of the genetic influence on this phenotype has yet been performed.
We have now used a panel of recombinant inbred rat strains, the HXB/BXH rats (Printz et al., 2003), to perform a quantitative analysis of the role of genetics in determining responses to intermittent alcohol exposure. We have assessed both the ADE, and changes in prolonged alcohol consumption (post-ADE alcohol consumption patterns), following periods of alcohol consumption and deprivation in these rat strains. The design of this analysis allows us to measure the heritability of the phenotypes. Our results suggest a genetic contribution to levels of voluntary alcohol consumption both pre and post-ADE, but indicate that factors other than genetic background contribute significantly to the variations in the ADE.
Materials and Methods
Animals
Male rats from the HXB/BXH recombinant inbred (RI) strains were used for these studies (Printz et al., 2003). These rats were rederived and maintained by Dr. Morton Printz at the University of California San Diego (UCSD). The rats were originally developed from an intercross between two inbred strains, the Wistar origin spontaneously hypertensive rat (SHR/Ola) and a Brown Norway congenic (BN-Lx/Cub), by Dr. Michal Pravenec at Charles University in Prague, Czech Republic. The RI strains were bred in a gender reciprocal fashion, providing strains that differ in the source of mitochondrial DNA and the Y chromosome (HXB and BXH strains) (Printz et al., 2003). Complete DNA sequence information on both parental strains (http://phenogen.ucdenver.edu), an extensive marker set on RI strains (http://www.snp-star.edu), detailed brain transcriptome information for the parental strains (http://phenogen.ucdenver.edu/) and phenotypic QTLs (http://rgd.mcw.edu) are some of the available HXB/BXH RI panel resources.
Behavioral Phenotypes: Alcohol Consumption and Alcohol Deprivation Effect
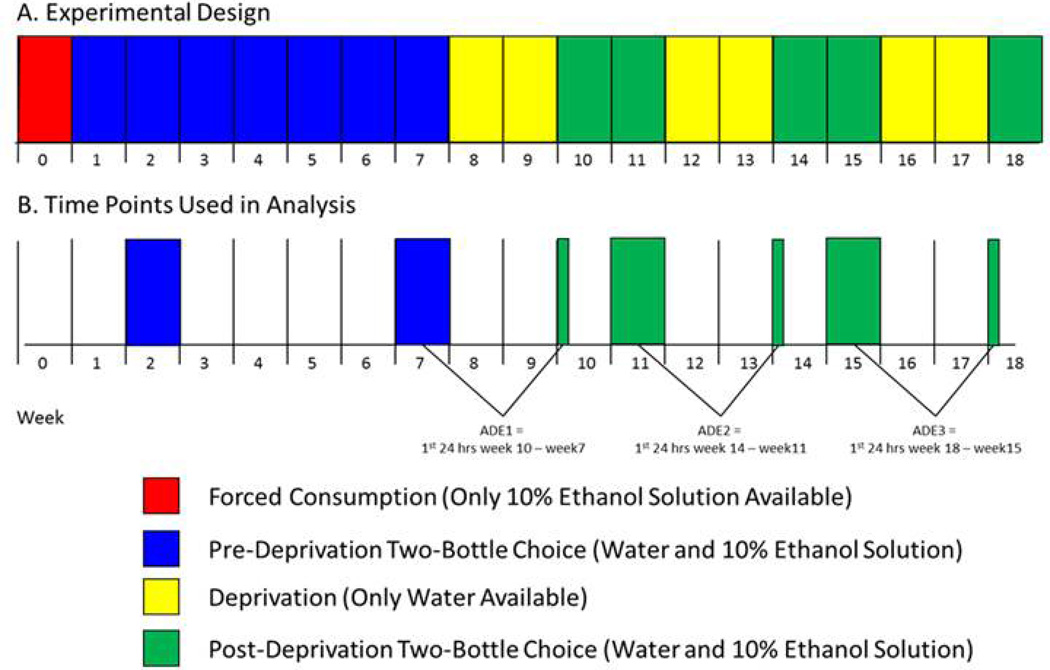
Male rats (70–100 days old at the start of the experiment; n=9–12 rats per strain) from 23 HXB/BXH RI strains, and the parental strains (BN-Lx/CubPrin, SHR/OlaIpcvPrin) were, for one week, kept in cages where ethanol solution was their only source of fluid (10% ethanol). Following this week, each rat was placed in a cage where water or a 10% ethanol solution were available in separate containers (“a two-bottle choice” paradigm (Tabakoff et al., 2009)) for a period of 7 weeks. This alcohol exposure was followed by a 2-week period where rats were only provided with water (“alcohol deprivation”), which was followed by a 2-week period of two-bottle choice alcohol exposure (water and 10% ethanol solution). After this, rats were subjected to a second 2-week alcohol deprivation period, followed by 2 weeks of 2-bottle choice alcohol exposure, a third 2-week alcohol deprivation period, and one week of 2-bottle choice alcohol exposure. The treatment schema is shown in Figure 1. For each rat, volumes of alcohol and water consumption were measured by weight on Monday, Wednesday and Friday of each week when alcohol solution in the two-bottle choice paradigm was offered (including alcohol consumption over the first 24 hr of re-access to alcohol following each deprivation period), and average daily alcohol consumption (g/kg) for each week was calculated. These studies were performed at UCSD, under a protocol approved by the UCSD IACUC.
Figure 1. Alcohol Consumption Paradigm for HXB/BXH RI Rat Strains.
A. During week 0, rats had access to 10% alcohol as their only fluid. During weeks 1 to 7 (pre-ADE alcohol consumption), rats were given alcohol access in a 2-bottle choice paradigm (water and 10% alcohol). There were then three 2-week periods of alcohol deprivation (water only): weeks 8 and 9, weeks 12 and 13, and weeks 16 and 17. Each deprivation period was followed by a period when rats again had access to water and 10% alcohol: weeks 10 and 11, weeks 14 and 15, and week 18. B. Average daily alcohol consumption levels during weeks 2 and 7 were used to assess “pre-ADE alcohol consumption”. The first ADE (ADE1) was determined as 24-hour alcohol consumption on the first day of re-exposure to alcohol after the first deprivation period, minus the average daily alcohol consumption from week 7. Weeks 12 and 13 are the second alcohol deprivation period, and ADE2 is 24-hour alcohol consumption at the second re-exposure to alcohol, minus average daily consumption during week 11. Weeks 16 and 17 are the third alcohol deprivation period, and ADE3 is defined as 24-hour alcohol consumption after the third re-exposure, minus average daily alcohol consumption during week 15. Average daily alcohol consumption during week 11 and during week 15 are defined as “post-ADE alcohol consumption”.
We defined the alcohol deprivation effect (ADE) as the amount of alcohol consumed (g/kg) during the first 24 hr after reintroduction of alcohol, minus the average daily amount (g/kg) consumed during the week prior to each deprivation period (i.e., average for weeks 7, 11 and 15 for calculation ADE1, ADE2 and ADE3, respectively). Two other phenotypes were investigated in the current study: average daily alcohol consumption levels for weeks 2 and 7 (defined as “pre-ADE alcohol consumption”), and “post-ADE alcohol consumption”, defined as the average daily alcohol consumption during week 11 or during week 15. Figure 1 and its legend illustrate the periods of measurement of each of these phenotypes.
Heritability
Broad-sense heritability (H2) of average daily alcohol consumption during weeks 2, 7, 11 and 15, as well as for each ADE, was calculated by a 1-way ANOVA using strain as the independent variable.
Quantitative Trait Loci (QTL)
Single Nucleotide Polymorphisms for HXB/BXH Panel
To obtain a marker set for QTL calculation in the HXB/BXH rat RI panel, we used a single nucleotide polymorphism (SNP) dataset available via the STAR Consortium (version rn3.4, obtained from http://www.snp-star.edu). SNP locations were updated to version rn5 of the rat genome by aligning the 40 bp genomic sequence surrounding each SNP (provided by STAR consortium) to the RGCS 5.0/rn5 assembly (Kent, 2002). Of the 20,283 original STAR markers, 19,401 could be definitively mapped to a physical location in the RGCS 5.0/rn5 assembly. Markers that did not differ across all genotyped HXB/BXH RI strains and several related inbred strains (54 strains in total) and markers that had a heterozygous genotype call in > 10% HXB/BXH RI panel and parental strains were deleted.
We also examined individual genotype calls based on the population structure. Initially, if a strain had an “unknown” call for a specific marker, a call was assigned to it if the marker was within an HXB/BXH panel haplotype block and it could be accurately imputed (498 unknown calls changed). Genotype calls that originally implied a double recombination event were assumed to be genotyping errors and were recoded as such (3,560 double recombinant calls changed). A double recombinant event within a strain is implied when a SNP genotype matches one parental strain, but is surrounded in close proximity by SNPs on either side with genotypes derived from the opposite parental strain. For this genotype to be correct, two recombinations of the DNA had to occur in the small stretch of DNA. This SNP dataset is available on the PhenoGen Website (http://phenogen.ucdenver.edu).
To simplify our QTL analysis, only SNPs that were fully informative among the RI strains were used. We then identified 1,486 strain distribution patterns (SDPs), i.e., “haplotype blocks”. An SDP was eliminated if the genotype distribution differed significantly from an even split between the two parental genotypes (nominal p-value<1.0×10−5). We also eliminated SDPs based on recombination rates within a given chromosome, using two methods: difference in maximum LOD scores (>2) as implemented in the checkAlleles function of the rqtl package of R (Broman et al., 2003), and the ratio of distance between SNPs in megabases to distance between SNPs in centimorgans (outliers within a chromosome based on 2 standard deviations). These additional QC criteria reduced the number of SDPs to 1,328. Finally, the set of SDPs was further reduced to 813 SDPs unique among the 21 HXB/BXH RI strains for which we have both genotype and phenotype data. Strain means were used to identify QTLs by performing a weighted marker regression QTL analysis (Carlborg et al., 2005). QTLs with empirical p-values less than 0.05 (based on permutation (Churchill and Doerge, 1994)) were considered significant, and markers with LOD scores ≥ 2.0 were considered “suggestive”, and were included as potential QTLs for alcohol consumption (Tabakoff et al., 2009). The 20 Mb region around each significant or suggestive marker is the QTL interval shown in Table 2.
Table 2.
QTLs for Pre-ADE and Post-ADE Alcohol Consumption.
| Week2 | Week7 | Week11 | Week15 |
|---|---|---|---|
| Chr 1:167.3 – 187.3 Mb 2.3 (0.740) |
|||
| Chr 1: 234.9 – 256.4 Mb 3.0 (0.280) |
Chr 1: 234.9 – 256.4Mb 2.7 (0.258) |
Chr 1: 234.9 – 256.4 Mb 2.2 (0.611) |
|
| Chr 2: 256.4 – 276.4 Mb 2.4 (0.667) |
Chr 2: 252.3 – 272.3Mb 2.0 (0.592) |
||
| Chr 3: 10.3 – 30.3 Mb 2.0 (0.739) |
|||
| Chr 6: 0 – 33.5 Mb 3.7 (0.038) |
Chr 6: 12.2 – 32.2 Mb 2.2 (0.606) |
Chr 6: 12.2 – 32.2 Mb 2.6 (0.224) |
|
| Chr 6: 43.4 – 63.4 Mb 2.2 (0.791) |
|||
| Chr 8: 94.8 – 114.8 Mb 2.0 (0.755) |
|||
| Chr 12: 31.0 – 52.4 Mb 2.7 (0.439) |
Chr 12: 31.0 – 51.0 Mb 2.1 (0.680) |
QTLs for alcohol consumption with LOD ≥ 2 (significant and “suggestive” QTLs) are shown. The top line in each box indicates the QTL location (Chr: Mb interval). The bottom line indicates the LOD score and p-value.
For each QTL, the estimated proportion of the genetic variance explained by the QTL in the RI panel was calculated by taking the R2 of a weighted 1-way ANOVA using strain means as the dependent variable and genotype for each strain as the independent variable. We also included all significant and suggestive markers in a multiple linear regression model (no interaction effects tested) to determine genetic variance explained using all markers.
Results
Alcohol Deprivation Effect
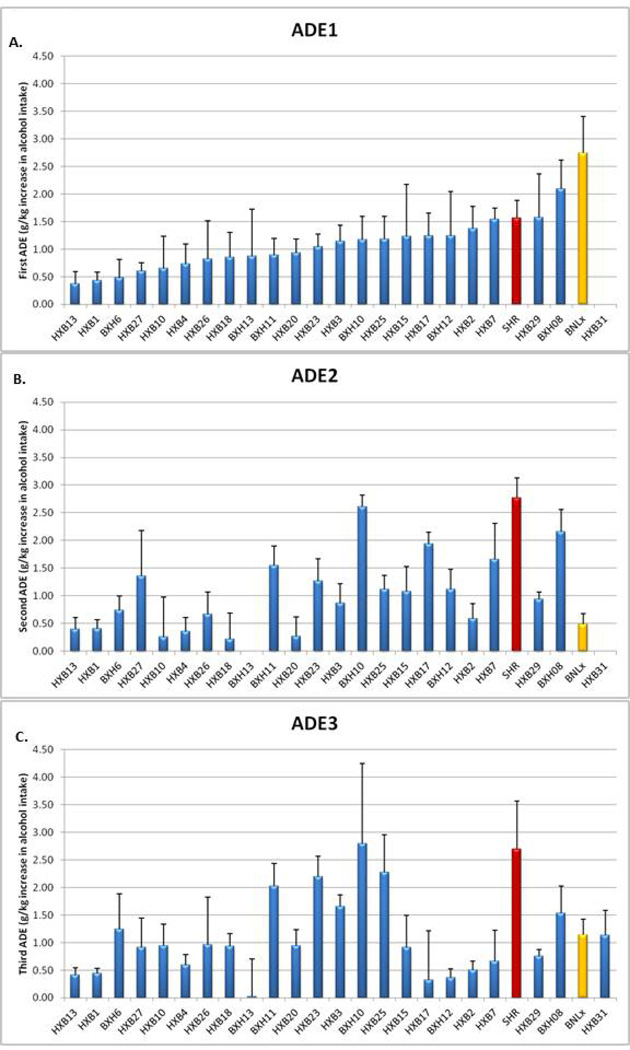
Alcohol deprivation effects (ADEs) were observed following each of the three periods of alcohol deprivation in the RI rat strains. As noted, we defined the ADE as the difference between the 24-hour alcohol intake on the first day of re-exposure to alcohol and the average daily alcohol consumption for the week prior to the deprivation period. The rank order of the ADEs by strain differ over the three deprivation periods, as shown in Figure 2. In some previous studies, when an ADE was not observed following a single deprivation period, the animals were subject to repeated periods of alcohol access and deprivation, and an ADE developed and/or was enhanced following multiple deprivation periods (e.g., Bell et al., 2004; Rodd-Henricks et al., 2000a). Our results demonstrate that, for some RI strains, the ADE does increase after a second and/or third deprivation period. However, for other strains, there is no change in the ADE with repeated periods of deprivation, and for some strains, the ADE decreases after the second or third deprivation period. Notably, there is little consistency of these effects with respect to rat strain (e.g., for HXB17, ADE2 is higher than ADE1, but ADE3 is lower than ADE1) (see Supplementary Figure 1). There was no significant correlation of the strain means for each ADE with alcohol consumption in the week preceding the deprivation period (for example, a strain with a high average level of pre-ADE alcohol consumption did not necessarily display a low ADE, arguing against the possibility of a “ceiling effect”) (Rodd-Henricks et al., 2000a). The heritability for the ADEs was quite low, although heritability increased to a level significantly different from 0 for the second and third ADEs (ADE1, 9.1%; ADE2, 26.2%; ADE3, 16.3%).
Figure 2. Strain Order for ADEs.
Three alcohol deprivation effects (ADE1, ADE2, ADE3) were measured in HXB/BXH recombinant inbred (RI) rat strains, as described in the text and Figure 1. In panel A (ADE1), RI strains are ordered according to the magnitude of ADE1, and this strain order is maintained for panels B (ADE2) and C (ADE3). Data are mean ± SEM. Data for SHR/OlaIpcvPrin rats are shown in red, and data for BN-Lx/CubPrin rats are shown in yellow.
Pre-ADE and Post-ADE Alcohol Consumption
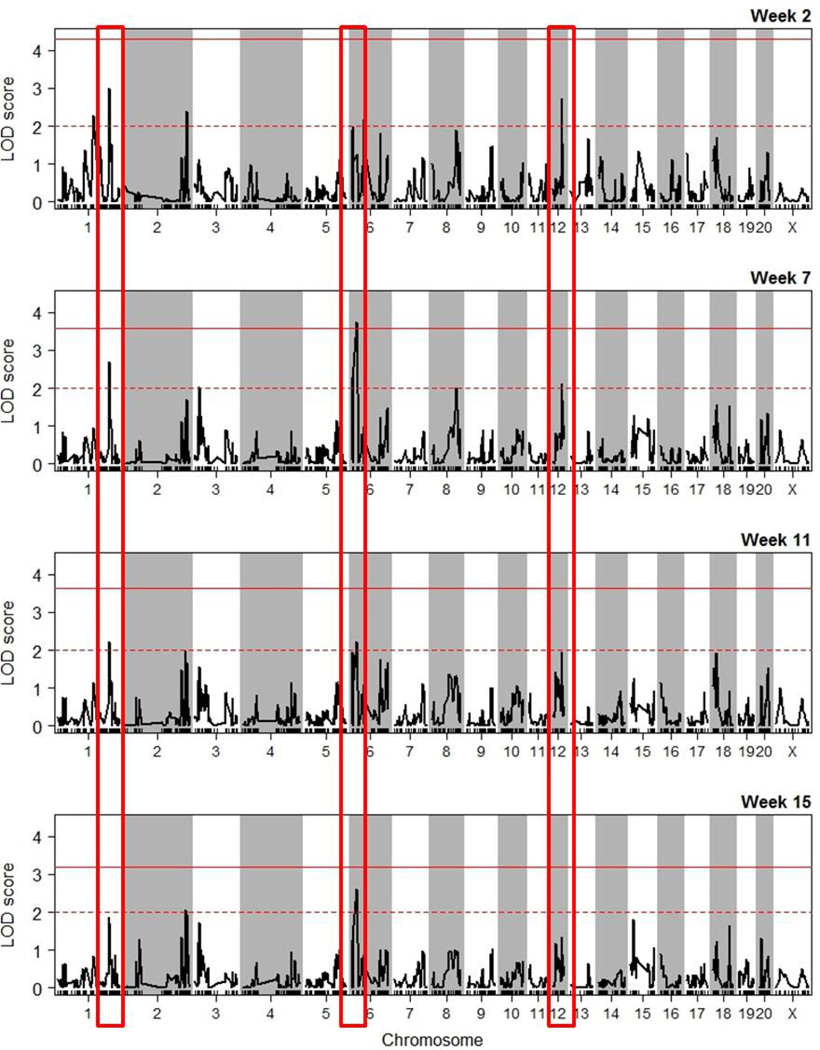
We previously reported that alcohol consumption levels by each RI strain were stable over the initial seven weeks of alcohol consumption (Tabakoff et al., 2009). The re-analysis of the data confirms that there are significant correlations of alcohol consumption across strains between weeks 2 and 7 (pre-ADE alcohol consumption), as well as between weeks of pre-ADE and post-ADE alcohol consumption (Table 1). Although there are some differences between weeks 2 and 7 of pre-ADE alcohol consumption, with some strains increasing consumption at week 7, compared to week 2, and other strains decreasing consumption, a one-way repeated measures ANOVA revealed that only 2 strains show significant differences in consumption between weeks 2 and 7 (unadjusted p value <0.05, Figure 3). To determine if periods of alcohol deprivation affect alcohol consumption levels (post-ADE alcohol consumption), we also compared drinking during weeks 11 (second week after ADE1) and 15 (second week after ADE2) to drinking during week 2. The one-way repeated measures ANOVA indicates that several strains showed significant changes during the post-ADE alcohol consumption period (Figure 3), with consistent changes for particular strains. Heritability of pre-ADE and post-ADE alcohol consumption is high, and tends to increase over time (week 2, 38.5%; week 7, 51.1%; week 11, 56.8%; week 15, 63.3%), indicating that increasing proportions of the phenotypic variance can be attributed to strain (genetic variance). Given the relatively high heritability for this phenotype, we determined QTLs for alcohol consumption at weeks 2, 7, 11 and 15. As shown in Table 2, the same suggestive QTL on chromosome 1 (234.9–256.4 Mb) was identified for alcohol consumption at weeks 2, 7 and 11; however, the LOD score decreased from 3.0 to 2.2, and this QTL no longer reached the suggestive threshold at week 15 (LOD < 2). Similarly, a suggestive QTL on chromosome 12 (31.0–52.4 Mb) had a LOD core of 2.7 at week 2, and this decreased to 2.1 at week 7. A significant QTL on chromosome 6 (0–33.5 Mb) was identified for week 7. The LOD score for this QTL also decreased over time. The LOD plots for each week of pre-ADE and post-ADE alcohol consumption (Figure 4) demonstrate a large number of small peaks, along with the decrease of significant and “suggestive” peaks over time. The data in Table 3 demonstrate that the identified significant and suggestive QTLs explain a substantial proportion of the genetic variance in alcohol consumption.
Table 1.
Correlation Matrix of Pre-ADE and Post-ADE Alcohol Consumption Levels.
| Week2 | Week7 | Week11 | Week15 | |
|---|---|---|---|---|
| Week2 | 0.70 | 0.76 | 0.59 | |
| (0.0002) | (<0.0001) | (0.0032) | ||
| Week7 | 0.70 | 0.94 | 0.93 | |
| (0.0002) | (<0.0001) | (<0.0001) | ||
| Week11 | 0.76 | 0.94 | 0.93 | |
| (<0.0001) | (<0.0001) | (<0.0001) | ||
| Week15 | 0.59 | 0.93 | 0.93 | |
| (0.0032) | (<0.0001) | (<0.0001) |
Data shown are Pearson correlation coefficient for alcohol consumption levels across the RI strains at weeks 2, 7, 11 and 15. P-values are in parentheses.
Figure 3. Strain Order for Pre-ADE and Post-ADE Alcohol Consumption Levels.
Alcohol consumption levels in a two-bottle choice paradigm were determined in HXB/BXH RI rat strains as described in the text and Figure 1. Panel A illustrates alcohol consumption levels at week 2 (pre-ADE alcohol consumption levels), ordered by strain according to the level of alcohol consumption. In Panels B, C and D, the data are presented in the same strain order as Panel A. Panel B: Difference in alcohol consumption between week 7 (pre-ADE alcohol consumption) and week 2. Panel C: Difference in alcohol consumption between week 11 (post-ADE alcohol consumption) and week 2. Panel C: Difference in alcohol consumption between week 15 (post-ADE alcohol consumption) and week 2. Data are mean ± SEM. Data for SHR/OlaIpcvPrin rats are shown in red; data for BN-Lx/CubPrin rats are shown in yellow. *Significant change, compared to week 2 (unadjusted p-value = 0.05, 1-way ANOVA with repeated measures).
Figure 4. QTLs for Each Weekly Drinking Measurement.
LOD plots for each week of pre-ADE and post-ADE alcohol consumption are shown. The dotted red line indicates the threshold for a “suggestive” QTL, and the solid red line indicates the threshold for a “significant” QTL. The red boxes highlight suggestive and significant QTLs on chromosome 1, 6 and 12 that display a reduced LOD score over time.
Table 3.
Estimates for Percent Genetic Variance Explained by QTLs.
| bQTL | Week2 | Week7 | Week11 | Week15 |
|---|---|---|---|---|
| Chr 1: (167.3 – 187.3)Mb |
39.2% | 18.7% | 21.9% | 16.9% |
| Chr 1: (234.9 – 256.4)Mb |
48.2% | 44.6% | 38.7% | 31.3% |
| Chr 2*: (252.3 – 272.3)Mb |
46.3% | 31.4% | 31.5% | 37.2% |
| Chr 3: (10.3 – 30.3) Mb |
19.5% | 35.9% | 28.9% | 31.3% |
| Chr 6: (0 – 33.5) Mb |
23.4% | 56.0% | 38.7% | 43.7% |
| Chr 6: (43.4 – 63.4) Mb |
37.9% | 19.4% | 21.2% | 17.3% |
| Chr 8: (94.8 – 114.8) Mb |
34.0% | 35.6% | 24.8% | 18.9% |
| Chr 12*: (31.0 – 52.4) Mb |
47.0% | 37.8% | 34.9% | 25.4% |
| ALL QTLs | 78.4% | 81.2% | 72.5% | 75.5% |
Estimates for percent genetic variance explained by each QTL, as well as the combination of all QTLs.
QTL intervals that contained different max LOD peak depending on the phenotype.
It should be pointed out that the data for week 2 of alcohol consumption are the same data that were used for previous analysis of the candidate genes associated with alcohol preference of these rat strains (Tabakoff et al., 2009), but in the present study, certain aspects of data analysis differed from the earlier study. In particular, in the current analysis, the definition of phenotypic outliers was altered (an outlier was defined as >3 within-strain SD from the strain mean, instead of 2 SD from the mean) in order to avoid overestimating the heritability (all data were used, as no values were >3 SD from the strain mean); for the QTL analysis, the parental strains were not used; markers were included only if there was no missing genotype for all of the RI strains; and SNP markers were realigned and filtered as described earlier. As a result of these differences, the calculated heritability for alcohol consumption at week 2 is slightly less than previously reported (Tabakoff et al., 2009), and the location and identity of week 2 QTLs are slightly different from the previous report (for week 2, same 4 QTLs on chromosomes 1, 6 and 12, but an additional QTL on chromosome 2).
Discussion
The heritability of a trait within a population is the proportion of phenotypic variance that is due to genetic factors. Based primarily on the heritability estimates for the phenotypes measured in the RI strains in this study, we conclude that there is little genetic contribution to the variation in the short-term (24 hr) increase in alcohol consumption that occurs immediately after periods of alcohol deprivation in rats (the alcohol deprivation effect, ADE), once this increase is adjusted for pre-ADE levels of alcohol drinking. On the other hand, both pre-ADE and post-ADE levels of alcohol consumption by rats have a substantial genetic contribution.
The alcohol deprivation effect (ADE) has been widely studied, and is suggested as a model of craving for alcohol and relapse alcohol drinking (Heyser et al., 1997; Li et al., 1993; Sinclair and Li, 1989; Spanagel and Holter, 1999). Although the definition of the ADE, as well as its duration and magnitude, differ in various studies, the phenomenon has been shown to occur in several selected lines and inbred strains of mice and rats (Bell et al., 2004; McKinzie et al., 1996; Rodd-Henricks et al., 2000a; Rodd-Henricks et al., 2000b; Sajja and Rahman, 2013; Salimov et al., 1993; Salimov et al., 1995; Vengeliene et al., 2003). P rats, that have been selectively bred to prefer alcohol, showed an ADE when they were given one month of continuous access to alcohol, followed by 12 hours or one to 8 weeks of deprivation (Rodd-Henricks et al., 2000b; Sinclair and Li, 1989). Using a voluntary alcohol consumption paradigm, with access to a single alcohol concentration, as in the present study, the magnitude of the ADE was not changed by repeated periods of deprivation, but the duration of the ADE was prolonged (Rodd-Henricks et al., 2000b). However, not all selectively-bred lines of alcohol-preferring rats show an ADE following a single alcohol deprivation period. A set of replicate lines that have been selected for high alcohol preference using a voluntary alcohol consumption paradigm, the HAD rats, when given access to a single alcohol concentration (10%), only displayed an ADE following relatively long deprivation periods or after two or three deprivation periods (Rodd-Henricks et al., 2000a). Similarly, rats selectively bred for low alcohol preference (NP and LAD rats) only showed an ADE when they were given concurrent access to multiple ethanol concentrations, and after several deprivation sessions (Bell et al., 2004). While in these studies, the magnitude of the alcohol deprivation effect either did not change or was increased following repeated deprivation periods, in our studies, the magnitude of the second and third ADEs increased in some rat strains and was unchanged or decreased in others, compared to the first ADE. It is notable that these changes in the ADE that occurred with repeated deprivation periods were not consistent for a particular RI strain.
The major finding of this study is that the heritability of the ADE phenotype was low, suggesting that there is modest influence of genetic variation across the RI rat strains on the variance in the ADE phenotype. Our finding of lower heritability for the ADE does not necessarily mean that genetics does not influence the ADE. Rather, it indicates that, in the HXB/BXH RI population, there is modest genetically determined variation for this particular trait. In terms of extrapolation of our results, Falconer and Mackay (Falconer and Mackay, 1996) state: “…whenever a value is stated for the heritability of a given character it must be understood to refer to a particular population under particular conditions. Values found in other populations under other circumstances will be more or less the same according to whether the structure of the population and the environmental conditions are more or less alike”. Therefore, depending on population and environmental conditions, our results may be interpreted to indicate that variations in the ADE may not be heavily influenced by genetic factors. Our findings may have important implications for translational alcoholism research. Although the ADE has become a model for alcohol relapse drinking, and is used to test medications for preventing relapse in humans, the current results should generate caution in investigators searching for genetic determinants of relapse drinking by humans.
Voluntary alcohol consumption, measured using a two-bottle choice paradigm, has been well studied, and is genetically-influenced in rats, as demonstrated by many selective breeding experiments, as well as the identification of QTLs and candidate genes that contribute to this phenotype (Colombo et al., 2006; Lumeng et al., 1977; McBride and Li, 1998; Murphy et al., 2002; Sommer et al., 2006; Spence et al., 2009; Tabakoff et al., 2009). The current results confirm the genetic influence that we previously described for the levels of alcohol consumption by the HXB/BXH RI strains during the second week of alcohol drinking (Tabakoff et al., 2009). In the present study, we also found that heritability increased from 2 to 7 weeks of alcohol consumption, and we identified consistent QTLs for alcohol consumption at both two weeks and seven weeks.
Chronic alcohol consumption over periods of several weeks to months, has been associated with an “escalation” of alcohol intake, and this escalation phenomenon occurs more rapidly when the chronic consumption period is punctuated by intermittent periods of forced abstinence (Becker and Lopez, 2004; Lopez and Becker, 2005; Spanagel and Holter, 1999). This type of escalation of alcohol intake has been suggested to reflect an increased motivation to consume alcohol, and to be indicative of the transition to an alcohol-dependent state (Gilpin et al., 2008; Roberts et al., 2000; Rodd et al., 2003; Spanagel and Holter, 1999). The genetic contribution to such changes in alcohol consumption has not been well characterized, although some strain and selected line differences have been reported (Crabbe et al., 2012; Rosenwasser et al., 2013; Vengeliene et al., 2003).
We noted consistent, RI strain-dependent increases or decreases in post-ADE alcohol drinking levels. In our study the genetic influence is evident from the finding that heritability of the alcohol consumption phenotype continues to increase over each period of post-ADE alcohol consumption. In the literature, there is sometimes a blurring of the distinction between the ADE and escalation of alcohol consumption after chronic alcohol exposure. For example, the ADE may be prolonged beyond 48 hr (lasting for several days) after repeated deprivation/re-exposure, compared to a single deprivation/re-exposure (Rodd-Henricks et al., 2000b). The present results, however, clearly distinguish, on a genetic basis, between the transient ADE which has low heritability, and levels of post-ADE chronic alcohol consumption, which are highly heritable.
QTLs are regions of the genome believed to harbor “candidate genes” that influence the associated phenotype. Recent evidence suggests that the genetic basis for complex traits may relate more to differences in gene expression than to polymorphisms that disrupt codons (Emilsson et al., 2008; Li and Burmeister, 2005; Schadt et al., 2003). QTL regions may thus represent genomic locations that control the expression levels of the candidate genes (eQTLs). We used eQTL and phenotypic QTL information in our previous analysis, which identified candidate genes and pathways associated with predisposition for alcohol consumption during the second week of exposure (Tabakoff et al., 2009). In the current work, we noted that even though heritability increased during the later post-ADE alcohol consumption periods, the magnitude of the QTL associations with phenotype decreased over these periods. An “uncoupling” of high heritability of transcript expression levels from the number of statistically detectable eQTLs was noted in studies of aging in C. elegans, and was attributed to the recruitment of more genetic loci with small effect sizes that determine the phenotype of transcript expression levels as the animals age (Vinuela et al., 2012). A similar process may be occurring with regard to the phenotype of post-ADE alcohol consumption, and may determine the decreased significance of any one QTL for the post-ADE alcohol consumption levels. That is, the repeated periods of deprivation and re-exposure to alcohol may induce changes in gene expression or function that contribute to altered alcohol consumption levels. The overall contribution of each locus to the additive effect of genotype on phenotype can change over time, demonstrating increased heritability, but diminished contribution of any one locus to the phenotype. Further analysis will be needed to identify the genetic loci and candidate genes associated with the changes that occur during the period of post-ADE alcohol consumption.
Because the ADE in our studies was found to have only a small heritable component, it is reasonable to consider the environmental factors that have been postulated to affect relapse drinking (ADE) by animals and humans. For the most part, research has focused on stress and alcohol-related cues as instigators of increased alcohol intake following periods of abstinence (withdrawal), particularly in individuals with a history of alcohol dependence (e.g., Breese et al., 2011). In these individuals, for example, stress may interact with alcohol- or other environmentally-induced neuroadaptations to produce excessive alcohol consumption. However, an ADE can be observed even in animals that do not show typical signs of alcohol physical dependence (Roberts et al., 2000), and “social” drinkers also report cue-induced drug craving (Carter and Tiffany, 1999).
Our data suggest that, in the HXB/BXH RI rat strains, the factors determining the consumption of ethanol differ in relationship to the immediacy of measurement after periods of deprivation in an “experienced drinker”. An early phase of drinking after deprivation (first 24 hours) is an event which has a strong environmental component. This initial phase of relapse is followed by a more stable and highly heritable level of consumption. The post-ADE drinking changes in a consistent manner in each RI strain across time after each deprivation period. We noted indications for recruitment of additional genetic determinants of post-ADE drinking over time, and for certain strains a significant and escalating increase in post-ADE drinking levels occurred after the first, second and third deprivation period. One can speculate whether it is the ADE drinking per se, or the steady escalation of chronic drinking in some strains of rats, that is the indicator of the genetic component of the trajectory to addiction. The answer to this speculation will profoundly impact the selection of targets for medications to block the development of and/or the treatment of alcoholism.
Supplementary Material
Acknowledgements
The authors thank Laura Breen and Joseph Gatewood for technical assistance with the behavioral data collection.
This work was supported in part by NIAAA/NIH (U01AA016649 INIA Project (PH); R24AA013162 (BT): AA006420 (GK); AAU01 Developmental Grant- INIA Project (MP)); NHLBI/NIH (HL035108 (MP)); Pearson Center for Alcoholism and Addiction Research (GK); Banbury Fund (BT)
References
- Anton RF, O'Malley SS, Ciraulo DA, Cisler RA, Couper D, Donovan DM, Gastfriend DR, Hosking JD, Johnson BA, LoCastro JS, Longabaugh R, Mason BJ, Mattson ME, Miller WR, Pettinati HM, Randall CL, Swift R, Weiss RD, Williams LD, Zweben A. Combined pharmacotherapies and behavioral interventions for alcohol dependence: the COMBINE study: a randomized controlled trial. Jama. 2006;295(17):2003–2017. doi: 10.1001/jama.295.17.2003. [DOI] [PubMed] [Google Scholar]
- Becker HC, Lopez MF. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 2004;28(12):1829–1838. doi: 10.1097/01.alc.0000149977.95306.3a. [DOI] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Boutwell CL, Hsu CC, Lumeng L, Murphy JM, Li TK, McBride WJ. Effects of long-term episodic access to ethanol on the expression of an alcohol deprivation effect in low alcohol-consuming rats. Alcohol Clin Exp Res. 2004;28(12):1867–1874. doi: 10.1097/01.alc.0000148101.20547.0a. [DOI] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Schultz JA, Peper CL, Lumeng L, Murphy JM, McBride WJ. Effects of short deprivation and re-exposure intervals on the ethanol drinking behavior of selectively bred high alcohol-consuming rats. Alcohol. 2008;42(5):407–416. doi: 10.1016/j.alcohol.2008.03.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Marchant NJ, Calu DJ, Shaham Y. The reinstatement model of drug relapse: recent neurobiological findings, emerging research topics, and translational research. Psychopharmacology (Berl) 2013;229(3):453–476. doi: 10.1007/s00213-013-3120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese GR, Sinha R, Heilig M. Chronic alcohol neuroadaptation and stress contribute to susceptibility for alcohol craving and relapse. Pharmacol Ther. 2011;129(2):149–171. doi: 10.1016/j.pharmthera.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Camarini R, Hodge CW. Ethanol preexposure increases ethanol self-administration in C57BL/6J and DBA/2J mice. Pharmacol Biochem Behav. 2004;79(4):623–632. doi: 10.1016/j.pbb.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Carlborg O, De Koning DJ, Manly KF, Chesler E, Williams RW, Haley CS. Methodological aspects of the genetic dissection of gene expression. Bioinformatics. 2005;21(10):2383–2393. doi: 10.1093/bioinformatics/bti241. [DOI] [PubMed] [Google Scholar]
- Carter BL, Tiffany ST. Meta-analysis of cue-reactivity in addiction research. Addiction. 1999;94(3):327–340. [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138(3):963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G, Lobina C, Carai MA, Gessa GL. Phenotypic characterization of genetically selected Sardinian alcohol-preferring (sP) and -non-preferring (sNP) rats. Addict Biol. 2006;11(3–4):324–338. doi: 10.1111/j.1369-1600.2006.00031.x. [DOI] [PubMed] [Google Scholar]
- Cox BR, Olney JJ, Lowery-Gionta EG, Sprow GM, Rinker JA, Navarro M, Kash TL, Thiele TE. Repeated cycles of binge-like ethanol (EtOH)-drinking in male C57BL/6J mice augments subsequent voluntary EtOH intake but not other dependence-like phenotypes. Alcohol Clin Exp Res. 2013;37(10):1688–1695. doi: 10.1111/acer.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Harkness JH, Spence SE, Huang LC, Metten P. Intermittent availability of ethanol does not always lead to elevated drinking in mice. Alcohol Alcohol. 2012;47(5):509–17. doi: 10.1093/alcalc/ags067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, Mouy M, Steinthorsdottir V, Eiriksdottir GH, Bjornsdottir G, Reynisdottir I, Gudbjartsson D, Helgadottir A, Jonasdottir A, Styrkarsdottir U, Gretarsdottir S, Magnusson KP, Stefansson H, Fossdal R, Kristjansson K, Gislason HG, Stefansson T, Leifsson BG, Thorsteinsdottir U, Lamb JR, Gulcher JR, Reitman ML, Kong A, Schadt EE, Stefansson K. Genetics of gene expression and its effect on disease. Nature. 2008;452(7186):423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th ed. Essex, England: Longman Group Ltd; 1996. p. 161. [Google Scholar]
- Gilpin NW, Richardson HN, Lumeng L, Koob GF. Dependence-induced alcohol drinking by alcohol-preferring (P) rats and outbred Wistar rats. Alcohol Clin Exp Res. 2008;32(9):1688–1696. doi: 10.1111/j.1530-0277.2008.00678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyser CJ, Schulteis G, Koob GF. Increased ethanol self-administration after a period of imposed ethanol deprivation in rats trained in a limited access paradigm. Alcohol Clin Exp Res. 1997;21(5):784–791. [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF, Le Moal M. Drug abuse: hedonic homeostatic dysregulation. Science. 1997;278(5335):52–58. doi: 10.1126/science.278.5335.52. [DOI] [PubMed] [Google Scholar]
- Le A, Shaham Y. Neurobiology of relapse to alcohol in rats. Pharmacol Ther. 2002;94(1–2):137–156. doi: 10.1016/s0163-7258(02)00200-0. [DOI] [PubMed] [Google Scholar]
- Li J, Burmeister M. Genetical genomics: combining genetics with gene expression analysis. Hum Mol Genet. 2005;14(Spec No. 2):R163–R169. doi: 10.1093/hmg/ddi267. [DOI] [PubMed] [Google Scholar]
- Li TK, Lumeng L, Doolittle DP. Selective breeding for alcohol preference and associated responses. Behav Genet. 1993;23(2):163–170. doi: 10.1007/BF01067421. [DOI] [PubMed] [Google Scholar]
- Lopez MF, Becker HC. Effect of pattern and number of chronic ethanol exposures on subsequent voluntary ethanol intake in C57BL/6J mice. Psychopharmacology (Berl) 2005;181(4):688–696. doi: 10.1007/s00213-005-0026-3. [DOI] [PubMed] [Google Scholar]
- Lumeng L, Hawkins DT, Li TK. New strains of rats with alcohol preference and nonpreference. In: Thurman RG, editor. Alcohol and Aldehyde Metabolizing Systems. Vol. 3. New York: Academic Press; 1977. pp. 537–544. [Google Scholar]
- Martin-Fardon R, Weiss F. Modeling relapse in animals. Curr Top Behav Neurosci. 2013;13:403–432. doi: 10.1007/7854_2012_202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Li TK. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit Rev Neurobiol. 1998;12(4):339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- McKinzie DL, Eha R, Murphy JM, McBride WJ, Lumeng L, Li TK. Effects of taste aversion training on the acquisition of alcohol drinking in adolescent P and HAD rat lines. Alcohol Clin Exp Res. 1996;20(4):682–687. doi: 10.1111/j.1530-0277.1996.tb01672.x. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li TK. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32(5):363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- O’Brien CP. Anticraving medications for relapse prevention: a possible new class of psychoactive medications. Am J Psychiatry. 2005;162(8):1423–1431. doi: 10.1176/appi.ajp.162.8.1423. [DOI] [PubMed] [Google Scholar]
- O’Dell LE, Roberts AJ, Smith RT, Koob GF. Enhanced alcohol self-administration after intermittent versus continuous alcohol vapor exposure. Alcohol Clin Exp Res. 2004;28(11):1676–1682. doi: 10.1097/01.alc.0000145781.11923.4e. [DOI] [PubMed] [Google Scholar]
- Printz MP, Jirout M, Jaworski R, Alemayehu A, Kren V. Genetic Models in Applied Physiology. HXB/BXH rat recombinant inbred strain platform: a newly enhanced tool for cardiovascular, behavioral, and developmental genetics and genomics. J Appl Physiol (1985) 2003;94(6):2510–2522. doi: 10.1152/japplphysiol.00064.2003. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Heyser CJ, Cole M, Griffin P, Koob GF. Excessive ethanol drinking following a history of dependence: animal model of allostasis. Neuropsychopharmacology. 2000;22(6):581–594. doi: 10.1016/S0893-133X(99)00167-0. [DOI] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, McKinzie DL, Murphy JM, McBride WJ, Lumeng L, Li TK. The expression of an alcohol deprivation effect in the high-alcohol-drinking replicate rat lines is dependent on repeated deprivations. Alcohol Clin Exp Res. 2000a;24(6):747–753. [PubMed] [Google Scholar]
- Rodd-Henricks ZA, McKinzie DL, Shaikh SR, Murphy JM, McBride WJ, Lumeng L, Li TK. Alcohol deprivation effect is prolonged in the alcohol preferring (P) rat after repeated deprivations. Alcohol Clin Exp Res. 2000b;24(1):8–16. [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Kuc KA, Murphy JM, Lumeng L, Li TK, McBride WJ. Effects of repeated alcohol deprivations on operant ethanol self-administration by alcohol-preferring (P) rats. Neuropsychopharmacology. 2003;28(9):1614–1621. doi: 10.1038/sj.npp.1300214. [DOI] [PubMed] [Google Scholar]
- Rosenwasser AM, Fixaris MC, Crabbe JC, Brooks PC, Ascheid S. Escalation of intake under intermittent ethanol access in diverse mouse genotypes. Addict Biol. 2013;18(3):496–507. doi: 10.1111/j.1369-1600.2012.00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajja RK, Rahman S. Nicotinic receptor partial agonists modulate alcohol deprivation effect in C57BL/6J mice. Pharmacol Biochem Behav. 2013;110:161–167. doi: 10.1016/j.pbb.2013.07.009. [DOI] [PubMed] [Google Scholar]
- Salimov R, Salimova N, Klodt P, Maisky A. Interaction between alcohol deprivation and morphine withdrawal in mice. Drug Alcohol Depend. 1993;34(1):59–66. doi: 10.1016/0376-8716(93)90046-s. [DOI] [PubMed] [Google Scholar]
- Salimov R, Salimova N, Ratkin A, Shvets L, Maisky A. Genetic control of alcohol deprivation effect in congenic mice. Alcohol. 1995;12(5):469–474. doi: 10.1016/0741-8329(95)00033-n. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, Linsley PS, Mao M, Stoughton RB, Friend SH. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422(6929):297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Sinclair JD, Li TK. Long and short alcohol deprivation: effects on AA and P alcohol-preferring rats. Alcohol. 1989;6(6):505–509. doi: 10.1016/0741-8329(89)90059-1. [DOI] [PubMed] [Google Scholar]
- Sinclair JD, Senter RJ. Increased preference for ethanol in rats following deprivation. Psychon Sci. 1967;8:11–12. [Google Scholar]
- Sommer W, Hyytia P, Kiianmaa K. The alcohol-preferring AA and alcohol-avoiding ANA rats: neurobiology of the regulation of alcohol drinking. Addict Biol. 2006;11(3–4):289–309. doi: 10.1111/j.1369-1600.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Holter SM. Long-term alcohol self-administration with repeated alcohol deprivation phases: an animal model of alcoholism? Alcohol Alcohol. 1999;34(2):231–243. doi: 10.1093/alcalc/34.2.231. [DOI] [PubMed] [Google Scholar]
- Spence JP, Liang T, Liu L, Johnson PL, Foroud T, Carr LG, Shekhar A. From QTL to candidate gene: a genetic approach to alcoholism research. Curr Drug Abuse Rev. 2009;2(2):127–134. doi: 10.2174/1874473710902020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchankova P, Steensland P, Fredriksson I, Engel JA, Jerlhag E. Ghrelin receptor (GHS-R1A) antagonism suppresses both alcohol consumption and the alcohol deprivation effect in rats following long-term voluntary alcohol consumption. PLoS One. 2013;8(8):e71284. doi: 10.1371/journal.pone.0071284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, Hubner N, Heinig M, Pravenec M, Mangion J, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders J, Grant B, Hoffman PL. Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengeliene V, Siegmund S, Singer MV, Sinclair JD, Li TK, Spanagel R. A comparative study on alcohol-preferring rat lines: effects of deprivation and stress phases on voluntary alcohol intake. Alcohol Clin Exp Res. 2003;27(7):1048–1054. doi: 10.1097/01.ALC.0000075829.81211.0C. [DOI] [PubMed] [Google Scholar]
- Vinuela A, Snoek LB, Riksen JA, Kammenga JE. Aging Uncouples Heritability and Expression-QTL in Caenorhabditis elegans. G3 (Bethesda) 2012;2(5):597–605. doi: 10.1534/g3.112.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.