Significance
Polychlorinated biphenyls (PCBs) as persistent organic pollutants are widespread in the sediments of lakes, rivers, and harbors. Although the PCB detoxification through microbial reductive dechlorination has been extensively studied for more than 20 y, the difficulty in cultivating PCB dechlorinators in pure culture impedes further characterization, optimization, and application in in situ bioremediation. By combining traditional culture techniques with next-generation sequencing technology, this study reports the successful cultivation and characterization of three PCB-respiring Dehalococcoides mccartyi strains in pure culture and identification of their key functional genes, which advances the PCB bioremediation and our understanding of organohalide respiration of PCBs.
Abstract
Fastidious anaerobic bacteria play critical roles in environmental bioremediation of halogenated compounds. However, their characterization and application have been largely impeded by difficulties in growing them in pure culture. Thus far, no pure culture has been reported to respire on the notorious polychlorinated biphenyls (PCBs), and functional genes responsible for PCB detoxification remain unknown due to the extremely slow growth of PCB-respiring bacteria. Here we report the successful isolation and characterization of three Dehalococcoides mccartyi strains that respire on commercial PCBs. Using high-throughput metagenomic analysis, combined with traditional culture techniques, tetrachloroethene (PCE) was identified as a feasible alternative to PCBs to isolate PCB-respiring Dehalococcoides from PCB-enriched cultures. With PCE as an alternative electron acceptor, the PCB-respiring Dehalococcoides were boosted to a higher cell density (1.2 × 108 to 1.3 × 108 cells per mL on PCE vs. 5.9 × 106 to 10.4 × 106 cells per mL on PCBs) with a shorter culturing time (30 d on PCE vs. 150 d on PCBs). The transcriptomic profiles illustrated that the distinct PCB dechlorination profile of each strain was predominantly mediated by a single, novel reductive dehalogenase (RDase) catalyzing chlorine removal from both PCBs and PCE. The transcription levels of PCB-RDase genes are 5–60 times higher than the genome-wide average. The cultivation of PCB-respiring Dehalococcoides in pure culture and the identification of PCB-RDase genes deepen our understanding of organohalide respiration of PCBs and shed light on in situ PCB bioremediation.
Polychlorinated biphenyls (PCBs) as priority persistent organic pollutants (1) are ranked fifth on the US Environmental Protection Agency Superfund Priority List of Hazardous Compounds (2). PCBs were massively produced and sold as complex mixtures (e.g., Aroclor 1260) for industrial uses, resulting in their widespread distribution in sediments of lakes, rivers, and harbors (2). Although the production of PCBs was banned in most countries by the late 1970s, their persistence in nature and bioaccumulation in food chains continue to pose a significant health risk for humans (3). The pitfalls of the most commonly used chemical methods for PCB remediation via dredging include risk of leaking contaminants, identifying suitable disposal methods for large quantities of contaminated soil, and the invasive and disruptive impact on the surrounding ecosystem (4).
In as early as 1987, detoxification of PCBs through reductive dechlorination by indigenous anaerobic bacteria was reported at contaminated sites (5) and confirmed in laboratory studies (6), opening up the possibility of an environmentally attractive in situ microbial detoxification strategy. However, progress in this direction has been slow due to the challenges involved in cultivation of PCB-respiring bacteria. Correspondingly, to date, only three bacterial strains showed PCB dechlorination activity, Dehalobium chlorocoercia DF-1 (7), Dehalococcoides mccartyi 195 (8), and D. mccartyi CBDB1 (9), none of which have been shown to be capable of respiring on the commercial PCBs as is needed for in situ PCB bioremediation and for identification of key functional genes (10). On the other hand, several bacterial genera have been implicated in PCB dechlorination, including Dehalococcoides, Dehalogenimonas, and Dehalobacter (11–14), and these serve as a rich environmental resource for isolation and comparative study of the genes and processes underlying organohalide respiration.
In this study, we report the successful isolation and characterization of three D. mccartyi strains (CG1, CG4, and CG5) that metabolically dechlorinate the complex commercial PCB mixture Aroclor 1260. This was made possible by the synergy of high-throughput sequencing-based metagenomic and metatranscriptomic profiling with traditional culture techniques, to establish the viability of an alternate electron acceptor for bacterial isolation from PCB-enriched cultures (12). Further genomic, transcriptomic, functional, and biochemical characterization of these isolates helped to identify and confirm novel genes encoding reductive dehalogenases (RDases) for catalyzing the PCB dechlorination.
Results
Metagenomic Profiling Indicates the Feasibility of Using Tetrachloroethene as an Alternative Electron Acceptor to Isolate PCB Dechlorinators.
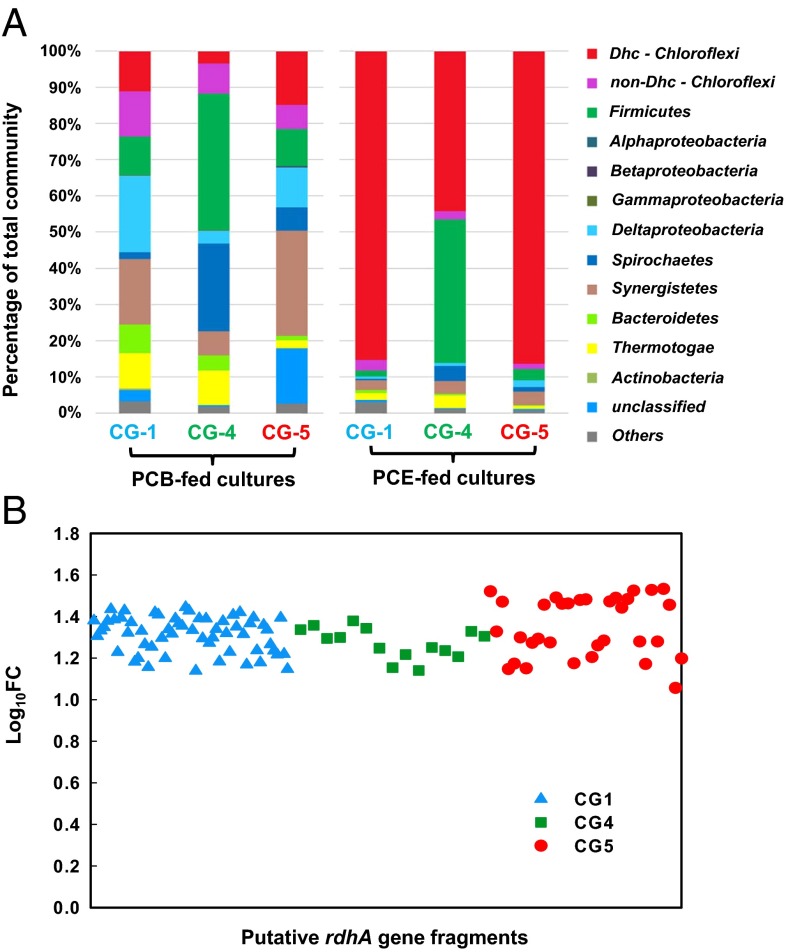
Consistent with the challenges faced by other researchers, despite our enrichment of three PCB-dechlorinating microbial communities (CG-1, CG-4, and CG-5) (12), repeated attempts to enrich dechlorinating bacteria as the dominant taxa through sequential transfers in defined mineral medium amended with lactate and Aroclor 1260 were unsuccessful. Profiling of the bacterial communities in the enrichments revealed that the relative abundance of putative PCB dechlorinators (Dehalococcoides of Chloroflexi phylum) only reached 11.1% (CG-1), 3.4% (CG-4), and 14.8% (CG-5) (Fig. 1A), suggesting that further transfers were unlikely to be useful for enriching them to dominance as is required for the subsequent isolation process. Chloroethenes are known to support the growth of Dehalococcoides (13) and so can potentially serve as an alternative substrate for their enrichment. However, this process also has drawbacks, with the possibility of the obtained strain losing PCB dechlorination activity during the enrichment process (13). Therefore, monitoring the transfer process through genomic approaches can help to guide the enrichment process and increase the odds. Kinetic characterization of the three microbial communities, CG-1, CG-4, and CG-5, on different halogenated compounds (chloroethenes and chlorophenols) indicated that all three cultures dechlorinate tetrachloroethene (PCE) to trichloroethene (TCE), cis-dichloroethene (cis-DCE), and trans-DCE without a lag phase (Fig. S1A), suggesting PCE as a potential alternative electron acceptor. It turned out that after a single culture transfer on PCE, the Dehalococcoides in cultures CG-1, CG-4, and CG-5 became dominant with relative abundances of 85.2%, 44.2%, and 86.3%, respectively (Fig. 1A and Fig. S1B). Polymorphism analysis of shotgun metagenomic sequencing data from PCE- and PCB-fed mixed cultures mapped to the genome of reference D. mccartyi 195 provided strong evidence that a single D. mccartyi strain was dominant and retained during the transfers of CG-4 and CG-5 (Fig. S1C; also seen in 16S rRNA gene sequencing). In contrast, Fig. S1C showed evidence of two strains in equal abundance in PCB-fed culture CG-1 (indistinguishable by 16S rRNA gene sequencing), of which only one was enriched in the PCE-fed culture. This was later confirmed through polymorphism analysis via mapping to the genomes of the respective Dehalococcoides isolate (Fig. S1D). Metabolic pathway analysis and characterization of the family of RDase genes confirmed the retention of key metabolic pathways and RDase genes during culture transfer on PCE for CG-1, CG-4, and CG-5 (Fig. 1B and Dataset S1A). Taken together, these results supported the feasibility of using PCE as an alternative electron acceptor for enriching PCB-dechlorinating Dehalococcoides. Indeed, after 10 serial transfers on PCE, the relative abundance of Dehalococcoides accounted for 96.3%, 97.0%, and 91.0% of the total microbial communities in culture CG-1, CG-4, and CG-5, respectively (Dataset S1B). Subsequently, the cultures were subjected to serial dilution-to-extinctions in defined mineral medium amended with peptidoglycan-attacking antibiotic, ampicillin (which Dehalococcoides are resistant to), and with acetate as a carbon source and H2 as an electron donor. This resulted in successful isolation of three D. mccartyi strains, CG1, CG4, and CG5, and the purity of each culture was confirmed by multilocus sequence typing (MLST) targeting four housekeeping genes (16S rRNA, adk, atpD, and rpoB) (Fig. S1E).
Fig. 1.
Metagenomic profiling of the enrichment process in medium with lactate as the sole carbon source. (A) Enrichment of Dehalococcoides (Dhc, in red) from PCB dechlorinating cultures via a single transfer with PCE as an alternative electron acceptor. Stacked bar graph depicts microbial community compositions (obtained by Illumina sequencing of 16S rRNA genes) at the phylum level. See Fig. S1B for corresponding graphs at the genus level. (B) Retention of potential rdhA gene fragments when changing the electron acceptor from PCBs to PCE in culture CG-1 (58 fragments), CG-4 (15 fragments), and CG-5 (33 fragments). FC, fold change of normalized metagenomic read abundance between PCB-fed and PCE-fed cultures.
Distinct Dechlorination Pathways and PCB-Dependent Growth in Pure Cultures.
The isolates, strains CG1, CG4, and CG5, were confirmed to have retained the same PCB dechlorination activity as their respective parent enrichment cultures (12), for which abiotic controls showed no obvious difference from the original Aroclor 1260 after 6 mo of incubation (Fig. 2A). To further characterize their dechlorination specificities, the cultures were inoculated into medium amended with eight PCB congeners (234-234-CB, 234-245-CB, 236-245-CB, 245-245-CB, 2345-234-CB, 2345-236-CB, 2345-245-CB, and 2356-245-CB) representing 54.8 mol % of PCBs in Aroclor 1260. The three strains remove chlorines predominantly from meta- and para-positions but with markedly distinct specificities (Fig. S2 A and B). Strain CG1 mainly removes 33′-meta-chlorines from 23456-, 2345-, 2346-, and 234-chlorophenyl rings (Fig. S2 A and B) of PCB congeners in Aroclor 1260, resulting in the accumulation of 245-245-CB, 236-245-CB, 2356-24-CB, 245-24-CB, 236-24-CB, 246-24-CB, and 245-25-CB (Fig. 2A). A limited amount of chlorine removal was observed at 55′-meta- and 44′-para-positions. In contrast, the chlorine-attacking preference for strain CG4 was found to be 44′-para > 55′-meta > 33′-meta (Fig. S2A), removing flanked para-chlorines from 2345-, 2346-, and 245-chlorophenyl rings and flanked meta-chlorines from 23456-, 2345-, 245-, and 234-chlorophenyl rings (Fig. 2A). Finally, strain CG5 showed the most extensive PCB dechlorination activity (Fig. S2 A and B), removing chlorines primarily from the 33′- and 55′-meta-positions of 2345-, 234-, 235-, 236-, and 245-chlorophenyl rings (Fig. 2A) and flanked para-chlorines from 2345-, 2346-, and 245-chlorophenyl rings.
Fig. 2.

Reductive dechlorination of Aroclor 1260 in pure cultures after 6 mo of incubation and PCB- and PCE-dependent growth of the Dehalococcoides isolates. (A) Absolute difference in the congener distribution of Aroclor 1260 residues between the abiotic control and pure cultures CG1, CG4, and CG5. Negative mol % indicates the amount of PCBs being dechlorinated; positive numbers represent produced PCB congeners. Growth of the Dehalococcoides isolates is coupled with reductive dechlorination of (B) Aroclor 1260 and (C) PCE. Controls are cultures without PCB and PCE amendment. Error bars represent SDs of triplicate cultures. See Fig. S2C for PCB and PCE dechlorination and cell growth supported by chlorine removal.
To investigate whether PCBs and PCE support the growth of strain CG1, CG4, and CG5, cell numbers were quantified along with the reductive dechlorination of Aroclor 1260 (Fig. 2B) and PCE (Fig. 2C) using quantitative real-time PCR (qPCR). After 150 d of incubation, the cell density of strains CG1, CG4, and CG5 increased 8.2-, 12.0-, and 16.2-fold to 5.9 × 106, 6.8 × 106, and 10.4 × 106 cells per mL, respectively, with 23.7, 27.7, and 83.6 µM chlorine removal from PCBs (Fig. S2C). Comparatively, the increase of cell numbers in PCE-fed pure cultures was much higher, i.e., 37.9-, 31.4-, and 28.0-fold to 1.3 × 108, 1.2 × 108, and 1.3 × 108 cells per mL with 1.1, 1.4, and 1.4 mM chlorine removed in cultures CG1, CG4, and CG5, respectively. The average cell growth of the three isolates on PCE was 1.0 × 1014 cells per mole of chlorine removed. This is comparable with that for PCBs, i.e., 2.1 × 1014 cells per mole of chlorine removed. No growth was observed in the control bottles without PCBs/PCE amendment (Fig. 2 B and C).
Genomic and Transcriptomic Characterization of the Isolates.
Whole-genome shotgun sequencing and targeted finishing experiments were used to obtain closed genome sequences for all three D. mccartyi strains, to aid in their further characterization. Although CG4 and CG5 have similar sized genomes (1.38 Mb and 1.36 Mb) and number of predicted protein-coding ORFs (1,421 vs. 1,413), CG1 has a larger genome (1.47 Mb) and correspondingly more ORFs (1,557) (Fig. 3A). Phylogenetic analysis showed that strains CG1, CG4, and CG5 clustered into distinct D. mccartyi subgroups (Victoria, Cornell, and Pinellas) represented by previously sequenced strains VS (15), 195 (16), and CBDB1 (17), respectively (Fig. 3B). Overall, the three genomes were organized in a fashion similar to other sequenced D. mccartyi strains but with significant pockets of differences concentrated in specific genomic regions (Fig. 3C and Fig. S3A). Some of these were in addition to the two high-plasticity regions (HPRs) (15) flanking both sides of the Ori that were observed in all three genomes (Fig. 3C). Strikingly, the majority of the genes in these strain-specific regions coded for proteins with unknown function (Dataset S1C), whereas the HPRs were enriched in RDases (Fig. 3C).
Fig. 3.
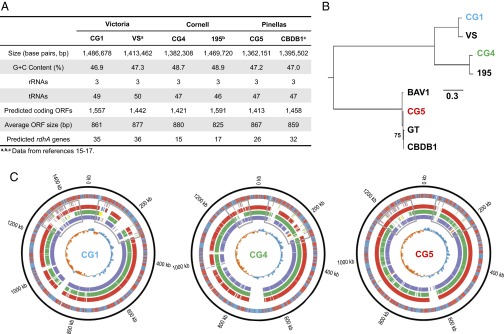
Comparative genomic characterization of D. mccartyi isolates. (A) Genome overview of closed genomes for novel strains (CG1, CG4, and CG5) and previously sequenced isolates (VS, 195, and CBDB1) belonging to the three Dehalococcoides subgroups. (B) Maximum-likelihood tree based on whole-genome SNP analysis. The three isolated Dehalococcoides strains were clustered into different respective subgroups. The numbers marked at the branches are confidence values based on 100 bootstrap replications, and only values less than 100 are noted. (C) Circos plot representation of genes and synteny for the three Dehalococcoides isolates. Tracks from inside to outside: track 1, GC skew plot; tracks 2–4, nucleotide alignment to reference strain VS (purple, forward strand; orange, reverse strand), strain 195 (green, forward strand; yellow, reverse strand), and strain CBDB1 (red, forward strand; blue, reverse strand); track 5, predicted rdhAs and rdhBs (green, rdhA on forward strand; purple, rdhA on reverse strand; yellow, rdhB on forward strand; orange, rdhB on reverse strand); and track 6, ORFs (blue, forward strand; red, reverse strand). The high-plasticity regions (HPRs) are enclosed in light gray boxes (HPR1, right side of the Ori; HPR2, left side of the Ori).
Characterization of full-length RDase homologous (rdhA) genes in the strain genomes revealed 35, 15, and 26 rdhA genes in CG1, CG4, and CG5, respectively, and all except for 3 of these have a corresponding rdhB gene encoding putative RDase membrane anchor proteins (within 70 bp of the 3′ end). Phylogenetic analysis of all 192 rdhA genes present in the three isolates and in D. mccartyi strain 195, CBDB1, VS, GT, and BAV1 (Fig. S3B) showed the presence of divergent clades of rdhA genes within each strain, suggesting that they may have been acquired from a diverse pool of available rdhA genes or originated from gene fragment recombinations rather than arising from duplication and mutagenesis events after a single acquisition. Furthermore, a higher tendency for Dehalococcoides of the same subgroup to share similar rdhA genes was observed (Fig. S3B), which may imply a shared evolutionary relationship between subtype specification and rdhA gene acquisition/divergence.
To identify the genes responsible for PCB and PCE dechlorination, transcriptomic (PCE-fed pure cultures) and metatranscriptomic (PCB-fed mixed cultures) analyses as well as qPCR (PCE- and PCB-fed pure cultures) using specific primers for each rdhA gene were used. Surprisingly, we found that the most highly transcribed rdhA genes were identical when cultures were fed with either PCBs or PCE, i.e., RD17 for CG1 (RD-CG1-17, 1,306,532–1,308,046 on the genome), RD1 for CG4 (RD-CG4-1, 64,056–65,504 on the genome), and RD1 for CG5 (RD-CG5-1, 64,583–66,031 on the genome), suggesting that the same dominant rdhAs were involved in chlorine removal from both PCBs and PCE (Fig. 4 A and B). The second most highly transcribed RD-CG5-20 (1,240,912–1,242,399 on the genome) gene in the PCE-fed pure culture CG5 shared over 93% similarity (in a 1.5-kbp region) with the pceA gene of D. mccartyi 195, indicating that RD-CG5-20 may also be involved in PCE dechlorination. Transcriptomic and metatranscriptomic analyses (with the exception of PCB-fed mixed culture CG-1) are largely in concordance with the qPCR results (Fig. S3C). Transcriptomic analysis for PCB-fed mixed culture CG-1 was complicated by the presence of two similar Dehalococcoides strains in the culture and the inability to deconvolute their respective contributions to the sequence data. Overall, the transcription levels of RD-CG1-17, RD-CG4-1, and RD-CG5-1 genes were much higher than the genome-wide average (5–60 times) (Fig. S3C) and among the top 10 highly transcribed genes (Dataset S2) in nearly all cultures, confirming the active metabolic role of dechlorination in these cultures.
Fig. 4.
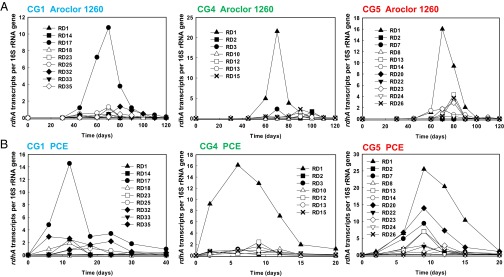
Transcription profiles of potential PCB-/PCE-RDase genes in pure cultures. The cultures are amended with (A) Aroclor 1260 or (B) PCE. Transcription changes (obtained via qPCR by using gene-specific primers) at various time points are shown as fold change compared with their respective 16S rRNA gene copies. Note that transcripts corresponding to RD17 gene of strain CG1 (RD-CG1-17), RD1 gene of strain CG4 (RD-CG4-1), and RD1 gene of strain CG5 (RD-CG5-1) are preferentially transcribed in both PCB- and PCE-fed cultures.
To further confirm that dechlorination of PCBs and PCE was catalyzed by the same RDases, in vitro assays were conducted with crude cell lysates from PCE-fed pure cultures that completely dechlorinated PCE to TCE and DCEs. The crude cell lysates in bottles spiked with Aroclor 1260 yielded the same dechlorination profiles as those observed in the active cultures (Fig. S4A). In the in vitro assay bottle amended with PCE or 2345-245-CB, the rate of chlorine removal from PCE was two times the rate for the PCB congener (Fig. S4B). Purified RD-CG1-17, RD-CG4-1, and RD-CG5-1 from native PAGE gels were also shown to be capable of removing chlorines from both 2345-245-CB and PCE in in-gel dechlorination assays (Fig. S4C), confirming their role in both PCB and PCE dechlorination. These three RDases were then designated pcbA1 (RD-CG1-17), pcbA4 (RD-CG4-1), and pcbA5 (RD-CG5-1) with pcbA4 and pcbA5 sharing 97% amino acid identity (Fig. S3B). These RDase genes can serve as potential biomarkers for assessing PCB dechlorination activities.
Discussion
Studies pertaining to characterization of PCB-respiring bacteria and subsequent identification of RDase genes have been limited due to difficulties in isolating these fastidious bacteria and challenges in generating sufficient biomass for characterizing RDase genes. Here we used high-throughput genome and transcriptome sequencing and integrated analyses to establish that PCB-dechlorinating Dehalococcoides could be enriched and isolated by using PCE as an alternative electron acceptor, providing an opportunity for further characterization of the genomes and RDase genes of the isolates. Dechlorinating bacteria rarely grow on agar plates, and isolation is normally achieved by enriching the bacteria to dominance in a mixed culture and then via serial dilution-to-extinctions (18–20). This strategy has been ineffective for isolating PCB-dechlorinating bacteria because their relative abundance in PCB-fed enrichment cultures remained at a low level (11, 14). The dechlorinating bacteria failed in growth competition with other bacteria in PCB-fed cultures due to their low cell densities resulting from the extremely low bioavailability of PCBs (10, 14). To circumvent this problem, we previously used TCE and vinyl chloride instead of PCBs to enrich and isolate PCB-dechlorinating bacteria from culture AD14 (13). Even though we obtained two Dehalococcoides isolates from the culture, the two strains did not show PCB dechlorination activity, possibly as a result of loss of PCB-dechlorinators or PCB RDase genes during the enrichment process. The technical difficulties in differentiating highly similar bacterial strains present in the same culture further confounds the challenge in isolation of PCB-dechlorinating bacteria, e.g., as observed in PCB-fed mixed culture CG-1 where the two D. mccartyi strains have identical 16S rRNA gene sequence (12). The use of high-throughput genomic approaches in this study was key to confirming that PCE is an appropriate alternative to PCBs, to enrich PCB-dechlorinating bacteria without activity loss. Capitalizing on those advantages, three PCB-dechlorinating D. mccartyi strains were successfully isolated via a five-step procedure: (i) transfer of the culture on Aroclor 1260 to enrich PCB-dechlorinating bacteria and to phase out other halorespiring microbes incapable of growing on PCBs, (ii) substrate screening to identify PCE as an alternative to PCBs and metagenomic profiling to check whether PCB-dechlorinating bacteria had been highly enriched after a single transfer on PCE, (iii) continuous culture transfers on PCE to enrich the PCB-dechlorinating bacteria to become dominant, (iv) metagenomic analyses to confirm the persistence of PCB-dechlorinating bacteria and rdhA genes during culture transfers before proceeding with isolation process, and (v) verification of PCB dechlorination activity in pure cultures and further genomic and transcriptomic characterization. The overall procedure shows the synergy of genomic approaches with traditional culture techniques for isolation of PCB-dechlorinating Dehalococcoides and may also be applicable to other hard-to-isolate bacteria.
The capability of bacteria to grow on the halogenated compounds is critical for their in situ bioremediation. For example, the biostimulation and bioaugmentation of chlorinated ethenes at contaminated sites have been successfully implemented by using the obligate organohalide respiring bacteria (21). However, among the identified PCB-dechlorinating bacteria of Dehalococcoides, Dehalobium, Dehalogenimonas, and Dehalobacter genera, no microorganisms that metabolically dechlorinate commercial PCBs have been obtained in pure culture. To our knowledge, this study is the first documentation that Dehalococcoides in pure culture couple their growth with extensive dechlorination of complex commercial PCBs. The meta- and para-dechlorination specificities of these isolates could help to relieve dioxin-like toxic effects of PCBs in the environment. The less-chlorinated products are susceptible to complete degradation by other bacteria that attack the nonchlorinated sites using monooxygenases or dioxygenases (10). The three characterized PCB-respiring isolates belonging to the three subgroups of Dehalococcoides may form a group of bacterial models to promote PCB bioremediation and to advance our understanding of organohalide respiration of PCBs.
Having a diverse pool of distinct rdhA genes within the Dehalococcoides genome confers evolutionary benefits because it enables wider substrate specificity, higher dehalogenation potential, and, consequently, swifter adaption to potential environmental changes. The RDases that have been characterized to date have been shown to be highly substrate-specific for chlorine removal, including the five rdhAs (pceA, mbrA, tceA, vcrA, and bvcA) identified from Dehalococcoides for catalyzing the four-step dechlorination of PCE to ethene (22–26). For the isolates in this study, however, a single dominant rdhA gene (pcbA1, pcbA4, or pcbA5) was highly transcribed in each pure culture amended with aliphatic PCE or aromatic Aroclor 1260 composed of more than 30 PCB congeners. These findings highlight a previously unknown diversity in the substrate specificity range of RDases and add to the current diversity of PCE RDases (i.e., pceA and mbrA) identified in Dehalococcoides. Additionally, the identified rdhA genes could potentially be used as biomarkers for monitoring PCB dechlorination activity in bioremediation applications. In all, cultivation of PCB-respiring Dehalococcoides in pure culture and identification of PCB-RDase genes deepen our understanding of organohalide respiration of PCBs and shed light on in situ PCB bioremediation.
Materials and Methods
Full protocols are available in SI Materials and Methods.
Cultivation and Isolation.
Completely defined, anaerobic mineral salts medium for bacterial enrichment and isolation was prepared as described (12, 13, 20). Enrichment of PCB-dechlorinating Dehalococcoides was achieved by sequential transfers in 100 mL medium amended with sodium lactate (10 mM) and the chlorinated substrate, 30 ppm (80.65 µM) Aroclor 1260 or 0.7 mM PCE. Further isolation of Dehalococcoides strains was conducted by serial dilutions in 20-mL vials filled with 10 mL of medium amended with 10 mM sodium acetate, hydrogen (5 × 104 Pa), 0.7 mM PCE, and 50 ppm ampicillin. In pure cultures, 8 PCB congeners (5 ppm each) were added individually to verify PCB dechlorination pathways.
Analytical Techniques.
PCBs were extracted with isooctane and quantified by gas chromatography (GC) equipped with an electron capture detector and a DB-5 capillary column as described (12, 13). Chlorinated ethenes were analyzed by GC using flame ionization detector and a GS-GasPro column (13).
DNA/RNA Extraction, qPCR, Sequencing, and Bioinformatic Analysis.
Total community DNA was extracted from PCB- or PCE-fed mixed cultures for metagenomic sequencing. DNA for genome sequencing and RNA for transcriptomic analyses of PCE dechlorination were extracted from PCE-fed pure cultures. Repeated attempts to extract RNA from PCB-fed pure cultures failed to generate enough RNA for building RNA-seq libraries. Therefore, PCB-fed mixed cultures were used for RNA extraction for metatranscriptomic analyses of PCB dechlorination. qPCR enumeration of Dehalococcoides cells and RDase genes with gene-specific primers (Dataset S3) was performed with QuantiTect SYBR Green PCR kit (26). Details of cell harvest, library preparation and sequencing, genome assembly, ORF prediction and annotation, metagenomic SNP (Single Nucleotide Polymorphism)-based analyses, phylogenetic and transcriptional regulation analyses, and data deposition information are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by the Science and Engineering Research Council, Agency for Science, Technology and Research under the Industrial Microbiology Genome Informatics platform and Project 102 101 0025 and by the National Research Foundation, Prime Minister’s Office, Singapore, under its Competitive Research Programme (CRP Award NRF-CRP 5-2009-05).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw sequencing reads in this paper have been deposited in National Center for Biotechnology Information’s short read archive (accession nos. SRR1050472, SRR1050378, SRR1050473, SRR1055187, SRR1055188, SRR1055189, SRR1050524, SRR1050525, SRR1050519, SRR1050523, SRR1050527, SRR1050528, SRR1051208, SRR1051209, and SRR1051210). Closed genomes reported in this paper have been deposited in the GenBank database (accession nos. CP006949, CP006950, and CP006951).
See Commentary on page 11919.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404845111/-/DCSupplemental.
References
- 1.Stockholm Convention on Persistent Organic Pollutants 1971. The 12 POPs Under the Stockholm Convention (Stockholm Conv on Persistent Org Pollut, Stockholm)
- 2.Agency for Toxic Substances and Disease Registry . CERCLA Priority List of Hazardous Compounds. Washington, DC: Agency for Toxic Subst and Dis Regist; 2006. [Google Scholar]
- 3.Falandysz J, Rose M, Fernandes AR. Mixed poly-brominated/chlorinated biphenyls (PXBs): Widespread food and environmental contaminants. Environ Int. 2012;44:118–127. doi: 10.1016/j.envint.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Rahuman M, Pistone L, Trifiro F, Miertus S. 2000. Destruction technologies for polychlorinated biphenyls (PCBs). Proceedings of Expert Group Meetings on POPs and Pesticides Contamination (Int Cent for Sci and High Technol, UN Ind Dev Organ, Vienna), pp 10–12.
- 5.Brown JF, Jr, et al. Polychlorinated biphenyl dechlorination in aquatic sediments. Science. 1987;236(4802):709–712. doi: 10.1126/science.236.4802.709. [DOI] [PubMed] [Google Scholar]
- 6.Quensen JF, 3rd, Tiedje JM, Boyd SA. Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediments. Science. 1988;242(4879):752–754. doi: 10.1126/science.242.4879.752. [DOI] [PubMed] [Google Scholar]
- 7.May HD, Miller GS, Kjellerup BV, Sowers KR. Dehalorespiration with polychlorinated biphenyls by an anaerobic ultramicrobacterium. Appl Environ Microbiol. 2008;74(7):2089–2094. doi: 10.1128/AEM.01450-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fennell DE, Nijenhuis I, Wilson SF, Zinder SH, Häggblom MM. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol. 2004;38(7):2075–2081. doi: 10.1021/es034989b. [DOI] [PubMed] [Google Scholar]
- 9.Adrian L, Dudková V, Demnerová K, Bedard DL. “Dehalococcoides” sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl Environ Microbiol. 2009;75(13):4516–4524. doi: 10.1128/AEM.00102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sowers KR, May HD. In situ treatment of PCBs by anaerobic microbial dechlorination in aquatic sediment: Are we there yet? Curr Opin Biotechnol. 2013;24(3):482–488. doi: 10.1016/j.copbio.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bedard DL, Ritalahti KM, Löffler FE. The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl Environ Microbiol. 2007;73(8):2513–2521. doi: 10.1128/AEM.02909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, He J. Phylogenetically distinct bacteria involve extensive dechlorination of aroclor 1260 in sediment-free cultures. PLoS ONE. 2013;8(3):e59178. doi: 10.1371/journal.pone.0059178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, He J. Dechlorination of commercial PCBs and other multiple halogenated compounds by a sediment-free culture containing Dehalococcoides and Dehalobacter. Environ Sci Technol. 2013;47(18):10526–10534. doi: 10.1021/es4017624. [DOI] [PubMed] [Google Scholar]
- 14.Bedard DL. A case study for microbial biodegradation: Anaerobic bacterial reductive dechlorination of polychlorinated biphenyls-from sediment to defined medium. Annu Rev Microbiol. 2008;62:253–270. doi: 10.1146/annurev.micro.62.081307.162733. [DOI] [PubMed] [Google Scholar]
- 15.McMurdie PJ, et al. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 2009;5(11):e1000714. doi: 10.1371/journal.pgen.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seshadri R, et al. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science. 2005;307(5706):105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- 17.Kube M, et al. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol. 2005;23(10):1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- 18.Maymó-Gatell X, Chien Y, Gossett JM, Zinder SH. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276(5318):1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 19.Adrian L, Szewzyk U, Wecke J, Görisch H. Bacterial dehalorespiration with chlorinated benzenes. Nature. 2000;408(6812):580–583. doi: 10.1038/35046063. [DOI] [PubMed] [Google Scholar]
- 20.He J, Ritalahti KM, Yang KL, Koenigsberg SS, Löffler FE. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature. 2003;424(6944):62–65. doi: 10.1038/nature01717. [DOI] [PubMed] [Google Scholar]
- 21.Löffler FE, Edwards EA. Harnessing microbial activities for environmental cleanup. Curr Opin Biotechnol. 2006;17(3):274–284. doi: 10.1016/j.copbio.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Magnuson JK, Stern RV, Gossett JM, Zinder SH, Burris DR. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Appl Environ Microbiol. 1998;64(4):1270–1275. doi: 10.1128/aem.64.4.1270-1275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnuson JK, Romine MF, Burris DR, Kingsley MT. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: Sequence of tceA and substrate range characterization. Appl Environ Microbiol. 2000;66(12):5141–5147. doi: 10.1128/aem.66.12.5141-5147.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller JA, et al. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl Environ Microbiol. 2004;70(8):4880–4888. doi: 10.1128/AEM.70.8.4880-4888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krajmalnik-Brown R, et al. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl Environ Microbiol. 2004;70(10):6347–6351. doi: 10.1128/AEM.70.10.6347-6351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chow WL, Cheng D, Wang S, He J. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J. 2010;4(8):1020–1030. doi: 10.1038/ismej.2010.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.