Significance
HIV-1 infection and the associated disease AIDS are a major cause of human death worldwide with no vaccine or cure available. The assembly of virus particles involves numerous host and viral proteins that are potential therapeutic targets. We used high-resolution microscopy techniques to investigate how the virus hijacks cellular proteins to enable the release of virions from an infected cell. We show with high temporal and spatial resolution that components of the host endosomal sorting complex required for transport (ESCRT) machinery are recruited to the neck of the assembling virus to facilitate scission of the link between the virus and the cell.
Keywords: viral assembly, nanobody, TIR-FM, single molecule localization microscopy
Abstract
HIV-1 virions assemble at the plasma membrane of mammalian cells and recruit the endosomal sorting complex required for transport (ESCRT) machinery to enable particle release. However, little is known about the temporal and spatial organization of ESCRT protein recruitment. Using multiple-color live-cell total internal reflection fluorescence microscopy, we observed that the ESCRT-I protein Tsg101 is recruited together with Gag to the sites of HIV-1 assembly, whereas later-acting ESCRT proteins (Chmp4b and Vps4A) are recruited sequentially, once Gag assembly is completed. Chmp4b, a protein that is required to mediate particle scission, is recruited to HIV-1 assembly sites ∼10 s before the ATPase Vps4A. Using two-color superresolution imaging, we observed that the ESCRT machinery (Tsg101, Alix, and Chmp4b/c proteins) is positioned at the periphery of the nascent virions, with the Tsg101 assemblages positioned closer to the Gag assemblages than Alix, Chmp4b, or Chmp4c. These results are consistent with the notion that the ESCRT machinery is recruited transiently to the neck of the assembling particle and is thus present at the appropriate time and place to mediate fission between the nascent virus and the plasma membrane.
Live-cell fluorescence microscopy of assembling HIV-1 virions has established the temporal sequence in which various viral and host molecules are recruited to the assembly site (1–6). The HIV-1 genome is recruited first to the plasma membrane by a subdetectable number of molecules of the structural protein, Gag (3), and a steady accumulation of Gag ensues for 6–10 min (1, 2, 4). After Gag recruitment is completed, members of the endosomal sorting complex required for transport III (ESCRT-III) complex and the ATPase vacuolar protein sorting-associated protein 4A (Vps4A) are recruited transiently, for just a few minutes, to the site of assembly (4, 6). The ESCRT machinery functions during membrane fission in processes such as the formation of multivesicular bodies, the terminal stages of cytokinesis (7), and the budding of enveloped viruses such as HIV-1 (8, 9). These processes all have inverted topologies compared with the topology of endocytic events at the plasma membrane. HIV-1 hijacks the ESCRT machinery by recruiting its members through specific amino acid sequences, called late domains, in the major structural protein Gag. Specifically, the PTAP motif recruits tumor susceptibility gene 101 (Tsg101) and the LXXLF motif recruits ALG-2 interacting protein X (Alix), with PTAP being the functionally more important motif (10, 11). Biochemical and genetic assays have defined specific molecular interactions between ESCRT proteins that are recruited by Gag (8, 12–14), but a fine temporal and spatial mapping of the recruitment of viral and host components relative to each other is lacking. Here, using live-cell multiple-color total internal reflection fluorescence microscopy (TIR-FM), we demonstrate that the ESCRT protein Tsg101 is corecruited with Gag and accumulates progressively, whereas charged multivesicular body protein 4b (Chmp4b) and Vps4A are recruited sequentially and transiently to Gag assembly sites. Moreover, because diffraction-limited microscopy cannot resolve spatial differences the size of an HIV-1 virion (∼100 nm) (15–17), we determined the relative spatial positions of Gag and of several members of the ESCRT machinery in nascent virions using two-color superresolution imaging.
Results
Tsg101 Is Progressively Recruited to HIV Assembly Sites Along with Gag.
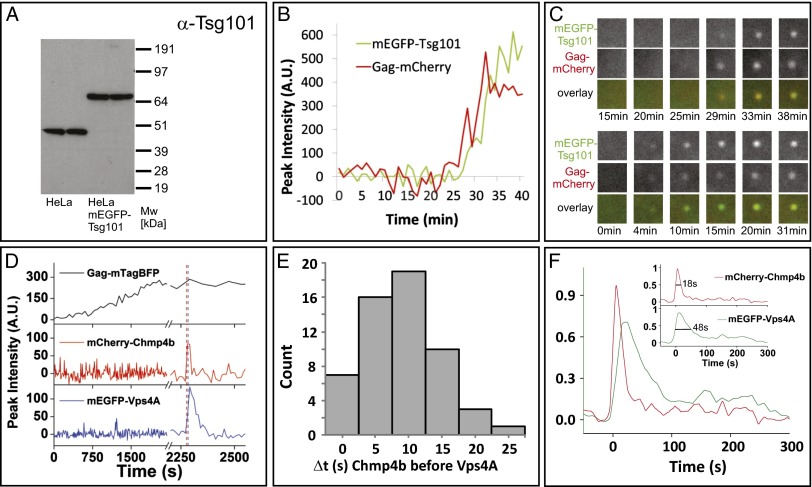
We first tested when Tsg101, a member of the ESCRT-I complex, is recruited to the site of Gag assembly (Fig. 1 A–C). We did not previously detect recruitment of this protein (4), which binds directly to HIV-1 Gag via the PTAP motif in the p6 domain of the polyprotein, likely due to high cellular background and a less sensitive detection system. To image Tsg101 recruitment, we generated a cell line stably expressing a version of Tsg101 tagged with monomeric EGFP (mEGFP) at endogenous levels (Fig. 1A). In this cell line, the expression of mEGFP–Tsg101 decreased the levels of the endogenous Tsg101 below the detection limit. The cell line showed no changes in growth rate, in numbers of multinucleated cells, or in shape (Fig. S1 A and B). These results imply that the tagged version is fully functional, because it substituted for the endogenous protein. We imaged the assembly of HIV-1 viral-like particles (VLPs) in this cell line following cotransfection of a 1:5 mix of Gag–mCherry and untagged Gag. Tsg101 was recruited together with Gag and remained at the site past the completion of Gag assembly (time course plotted in Fig. 1B and selected images shown in Fig. 1C with another example in the lower panels). Thus, it appears that Tsg101, an ESCRT protein that binds directly to Gag, is progressively recruited along with it to sites of VLP assembly and remains with the Gag VLP past the time frames when the signals for Chmp1b, Chmp4b, Chmp4c, and Vps4A returned to background levels (4). Using a comparable approach, we did not detect the recruitment of Alix to assembly sites.
Fig. 1.
Multiple-color live-cell fluorescence microscopy of the ESCRT proteins Tsg101 or Chmp4b with Vps4A during HIV-1 Gag assembly. (A) Expression levels of Tsg101 (45 kDa) and mEGFP–Tsg101 (72 kDa) in our stable cell line. (B) Example traces for Gag and Tsg101 recruitment to assembly sites. (C) The upper three rows show the puncta in the two channels and as an overlay for the traces in B. The lower three rows are from another assembly event.1.5 × 1.5 µm2. (D) An example from imaging HeLa cells stably expressing mEGFP–Vps4A and mCherry–Chmp4b transfected with plasmids expressing Gag:Gag–mTagBFP:Vpu (5:1:2). (E) Histogram depicting the time delay in seconds of the recruitment of Vps4A and Chmp4b to sites of Gag assembly as imaged with Gag–mTagBFP (56 events from six cells). (F) Averaged dwell times, relative to initial Chmp4b recruitment, for Chmp4b and Vps4A at Gag assembly sites (56 events from six cells). (Inset) Each of the fluorescent traces for Vps4A and Chmp4b normalized to its peak signal and resampled with 0.1-s time steps. The leading edge of each trace was aligned to time 0, and the amplitude was normalized to its peak. A cumulative dwell time graph was then produced for each protein, mCherry–Chmp4b and mEGFP–Vps4A, by averaging all traces (56 from six cells) at each time interval. Vps4A dwell time is 2–3 times longer than Chmp4b. The distribution width at half maximum was 48 s for Chmp4b and 18 s for Vps4A. Vps4A was often observed to have multiple peaks, represented by the low-level signal at long time points.
Chmp4b Is Recruited to HIV Assembly Sites Before Vps4A.
In previous studies, we found that both ESCRT-III proteins and Vps4A were recruited to assembling particles approximately coincident with the termination of Gag accumulation. However, a limitation of our earlier work was that the ESCRT proteins were imaged individually along with Gag, so the precise timing of the recruitment of each ESCRT protein relative to each other could not be determined (4). Therefore, we imaged mCherry–Chmp4b, an ESCRT-III protein essential for HIV-1 budding (8, 13, 14), and mEGFP–Vps4A, the ATPase believed to disassemble the ESCRT complexes to complete fission (8, 13, 14), during assembly of HIV-1 VLPs in cells stably expressing low levels of both fluorescently tagged host proteins (Fig. S1). VLP assembly was indicated by the increase of the fluorescent signal of Gag–mTagBFP until a plateau was reached (Fig. 1D) (1). Within a few minutes, recruitment of Chmp4b was observed. Notably, Vps4A recruitment was not simultaneous with Chmp4b, but followed shortly afterward (9 ± 6 s, ± SD; Fig. 1 D–F). As observed previously (4, 6), Chmp4b and Vps4A were transiently recruited to nascent VLPs (Fig. 1 D–F), but Vps4A remained associated with VLPs for more than twice as long (48 ± 5 s, the duration that the signal was more than half its maximum level) than Chmp4b (18 ± 5 s) (Fig. 1F). Although the Gag signal from the fully assembled VLP remained as close to its maximum value at the site of assembly, the Chmp4b and Vps4A signals return to background levels, implying that these members of the ESCRT machinery leave the site of assembly after the scission event (Fig. 1D).
Establishing Two-Color Superresolution Imaging with Nanobodies.
The above findings, and previous studies (4, 6), show that ESCRT-III proteins are transiently associated with nascent virions and the ESCRT-I protein Tsg101 is recruited at the same time as Gag. However, these studies lack the optical resolution needed to determine the location, relative to the assembling particle, where ESCRT proteins are recruited. To determine the spatial organization of ESCRT complexes during their recruitment to assembly sites, we developed a two-color superresolution technique similar to direct stochastic optical reconstruction microscopy (dSTORM) (Materials and Methods), where each dye for a particular color was driven to a transient dark state and then induced to blink with only one laser at high laser powers (488-nm laser for Atto488 or 561-nm laser for Atto594). The dyes were targeted to proteins of interest via nanobodies that recognized fusion proteins with photoactivatable (PA) variants of GFP and mCherry. The PA proteins were not switched to an emitting state, but used as targets for the nanobodies only. This approach allowed us to use a commercial microscope and commercially available probes and can be applied to a variety of other biological questions where functional fusions with PA fluorescent proteins are available but antibodies with high affinity and specificity to the proteins of interest are not. As a test of the sensitivity and specificity of our approach, we imaged Gag tagged with either PA–mGFP or PA–mCherry1 proteins detected with nanobodies (α-GFP–nanobody–Atto488 to detect Gag–PA–mGFP and α-RFP–nanobody–Atto594 to detect Gag–PA–mCherry1). In these experiments, we cotransfected Gag tagged with near-infrared fluorescent protein (iRFP) and used the iRFP signal to report the assembly of VLPs (Fig. 2A, right cell). The Gag–iRFP signal colocalized with assemblages of Gag–Atto488 (Fig. 2 B and E–H) and Gag–Atto594 (Fig. 2 C and E–H) imaged by superresolution microscopy. The superresolution images also showed smaller sporadic signals, which were also observed on the outside of cells and in cells that showed no Gag–iRFP signal above background (Fig. 2 A–E, left cell). These signals were ruled to be noise and were omitted from subsequent analysis. We performed a spatial clustering analysis (Bivariate Ripley’s K Test) and found that the relative distribution of Gag–Atto488 and Gag–Atto594 was not the consequence of random overlap (Fig. 2I), as evidenced by the signal (black line) rise outside the randomness confidence interval (red dotted lines). We repeated this analysis for a truncated version of Gag (GagΔp6), which fails to recruit the ESCRT machinery via Alix and Tsg101 (18–22), and found that GagΔp6–Atto488 and GagΔp6–Atto594 also colocalized (Fig. 2 J–L), which was supported by the clustering of these two signals in the Bivariate Ripley’s K Test (Fig. 2M).
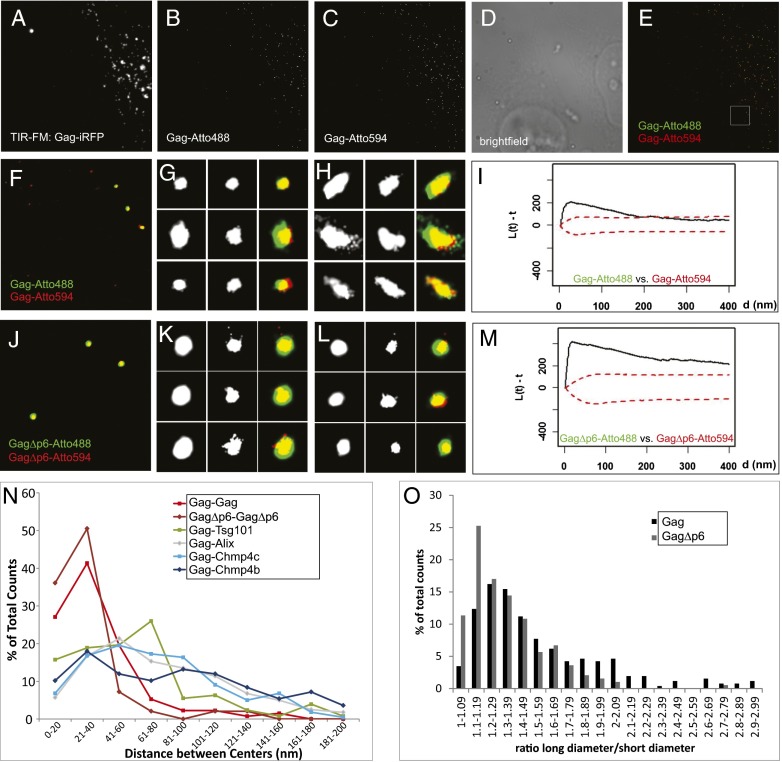
Fig. 2.
Establishing superresolution imaging in two colors. (A–H) HeLa cells transfected with Gag–iRFP, Gag–PA–mGFP, Gag–PA–mCherry1, and Gag (1:1:1:15) were imaged in TIR-FM to excite iRFP (A) and imaged in dSTORM-like mode to determine the location of Gag–PA–mGFP labeled with α-GFP–nanobody–Atto488 (B) and Gag–PA–mCherry1 labeled with α-RFP–nanobody–Atto594 (C). D shows the brightfield image for the ROI. The overlaid superresolution images (E, full ROI; F, enlarged section shown in white in E) show background signal from the untransfected cell and noncellular area and signal from three VLPs with Atto488 and Atto594 signal. Representative images for round (G) and elongated (H) Gag structures shown for Gag–Atto488 (Left), Gag–Atto594 (Center), and their overlay (Right). (I) Bivariate Ripley’s K Test for Gag–Atto488 and Gag–Atto594 signals showed that they cluster. (J–L) HeLa cells transfected with GagΔp6–iRFP, GagΔp6–PA–mGFP, GagΔp6–PA–mCherry1, and GagΔp6 (1:1:1:15) were also imaged in dSTORM-like mode to obtain localizations for Gag–PA–mGFP labeled with α-GFP–nanobody–Atto488 and for Gag–PA–mCherry1 labeled with α-RFP–nanobody–Atto594 (J). K and L show representative images for assemblages for GagΔp6–Atto488 (Left), GagΔp6–Atto594 (Center), and their overlay (Right). (M) Bivariate Ripley’s K Test for GagΔp6–Atto488 and GagΔp6–Atto594 signals showed that they cluster. (N) The distance between the centers of structures labeled with Atto488 or Atto594 varies depending on the proteins probed: shorter if both labels are on the same protein (for Gag, 37 ± 27 nm, or GagΔp6, 30 ± 23 nm) or longer if Gag is compared with Chmp4b (94 ± 74 nm), Chmp4c (80 ± 94 nm), or Alix (89 ± 61 nm). The mean distance between the centers of Gag and Tsg101 structures was significantly smaller (63 ± 46 nm) than for Chmp4b/c or Alix and Gag (P < 0.001). (O) Distributions of the ratios of long over short diameter for Gag structures (black bars) compared with GagΔp6 (gray bars). A–E, 40 × 40 µm2; F and J, 4 × 4 µm2; G, H, K, and L, 400 × 400 nm2.
Superresolution Imaging of Gag Assemblages.
The overall mean diameter of a Gag assemblage was 176 ± 84 nm (132 particles in two colors). The assemblages varied in shape, with a mean long diameter of 216 ± 95 nm and a short diameter of 136 ± 39 nm, giving an elongation ratio of 1.59 ± 0.51 (Fig. 2O, black bars). The majority of Gag assemblages appeared round (here defined as an elongation ratio less than 1.6; Fig. 2G), with a mean elongation of 1.29 ± 0.14 and a mean diameter of 155 ± 50 nm (Gag–Atto488) and 150 ± 48 nm (Gag–Atto594) (69 particles), consistent with the diameters found for HIV-1 particles by electron microscopy (18, 19). A minority (∼30%) of Gag assemblages was elongated, with a mean elongation ratio of 2.34 ± 0.60 (Fig. 2H, 29 particles). These assemblages were elongated in different radial directions (Fig. 2H), and thus the observed VLP elongation could not be explained by noncompensated drift, which usually occurs in one direction. Such elongated assemblages have been previously reported in superresolution microscopy experiments (20, 21), but it is unclear whether these represent two adjacent VLPs, morphologically abnormal individual VLPs, or perhaps assembly intermediates. Surprisingly, GagΔp6 formed fewer elongated assemblages (∼15%, elongation, 1.33 ± 0.25; Fig. 2O, gray bars) than full-length Gag, with a mean diameter of 134 ± 49 nm (97 particles) [Kolmogorov–Smirnov (KS) test, P < 0.05]. This experiment serves also as a control to rule out that elongated assemblages are not simply a distortion caused by assembling VLPs being trapped between cells and the glass surface. A more detailed discussion is given in SI Materials and Methods and Figs. S2 and S3.
Quantification of the distances between the centers of Gag–Atto488 and Gag–Atto594 assemblages (Fig. 2N, red line) showed a single population with a mean value of 37 ± 27 nm (132 particles). This distance is fourfold smaller than the mean diameter of Gag assemblages and the known diameter of individual HIV-1 particles. This strongly suggests that the signal from Gag–Atto488 and Gag–Atto594 originates from the same VLP. We observed a similar distribution of distances (Fig. 2N, brown line) between the truncated forms of Gag, GagΔp6–Atto488, and GagΔp6–Atto594, with a mean value of 30 ± 23 nm (97 particles). Therefore, our superresolution microscopy method is capable of determining the locations of fluorophores with subvirion resolution. Furthermore, we used these distances as standards representing completely overlapping structures, allowing us to determine the spatial relationship between the Gag and ESCRT assemblages.
ESCRT Proteins Are Recruited to the Vicinity of Gag Assemblages.
We imaged Gag along with ESCRT proteins that bind directly to Gag (Alix or Tsg101) or ESCRT proteins that are not thought to bind to Gag directly but act late in the ESCRT pathway to mediate particle scission (Chmp4b or Chmp4c). The relative localization of Gag and ESCRT proteins was determined in the presence of a dominant-negative mutant of Vps4A to trap the fission machinery at sites of virion assembly. This should only occur at the terminal stages of particle fission, as Vps4A is not recruited to the sites of assembly until ESCRT-III proteins are already at these sites, as determined in our three-color live-cell experiments (Fig. 1D). We used cell lines stably expressing low levels of tagged ESCRT proteins (Fig. S1 C–E and SI Materials and Methods). Cell lines expressing PA–mCherry1–Chmp4b (Fig. 3 A–E), PA–mCherry1–Chmp4c (Fig. 3 G–K), or PA–mCherry1–Alix (Fig. 3 M–Q) were transiently transfected with plasmids expressing Gag, Gag–PA–mGFP, Gag–iRFP, and Vps4A–K173Q and labeled with nanobodies (Gag–Atto488 with Chmp4b–Atto594, Chmp4c–Atto594, or Alix–Atto594). Parallel experiments were also done in HeLa cells transiently transfected with PA–mCherry1–Alix (Fig. S4 A–E) or PA–mCherry1–Tsg101 (Fig. 3 S–W). Gag assemblages often (20–44%) had ESCRT proteins in their direct vicinity (Fig. 3 and Fig. S4). The mean diameters for assemblages of ESCRT proteins were similar to each other (Chmp4b, 115 ± 46 nm, 179 particles; Chmp4c, 110 ± 41 nm, 231 particles; Alix, 101 ± 50 nm, 302 particles; and Tsg101, 100 ± 41 nm, 130 particles), but significantly smaller (P < 0.005) than corresponding Gag assemblages from the same cells (Gag for Chmp4b, 221 ± 117 nm; Gag for Chmp4c, 182 ± 59 nm; Gag for Alix, 198 ± 119 nm; Gag for Tsg101, 175 ± 52 nm) (Fig. 3).
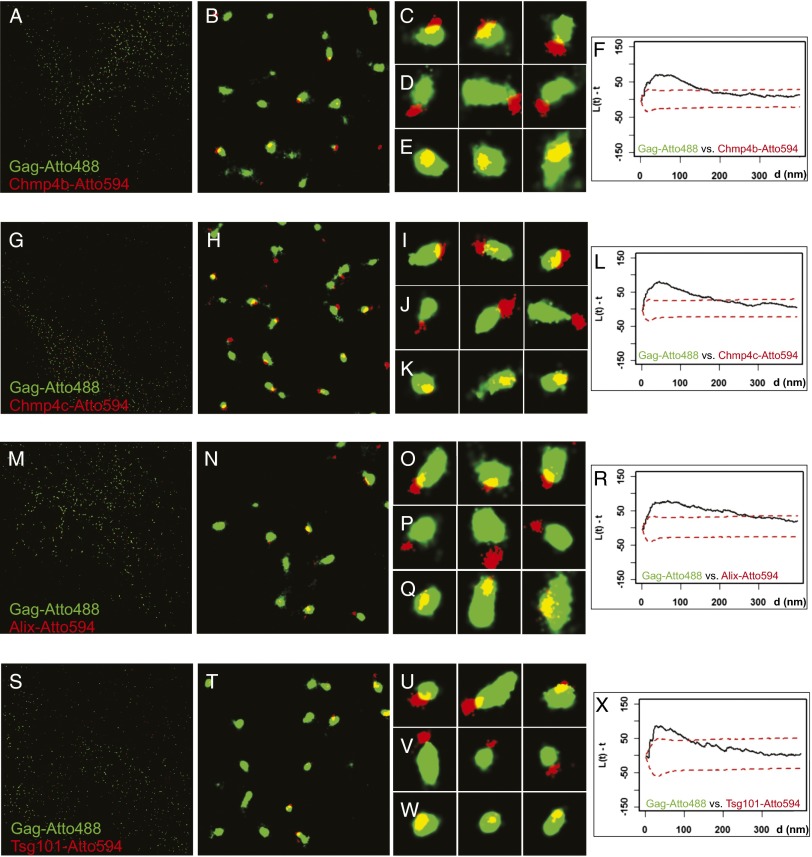
Fig. 3.
Superresolution imaging of Gag and ESCRT proteins. (A–E, G–K, M–Q, and S–W) HeLa cell line stably expressing PA-mCherry1-Chmp4b (A–E), PA–mCherry1–Chmp4c (G–K), or PA–mCherry1–Alix (M–Q) transiently transfected with Gag–PA–mGFP, Gag–iRFP, Gag and Vps4A–K173Q, or HeLa cells transiently transfected with PA–mCherry1–Tsg101, Gag–PA–mGFP, Gag–iRFP, Gag, and Vps4A–K173Q (S–W) were fixed and labeled with nanobodies targeted against GFP labeled with Atto488 (shown in green) or against RFP/mCherry labeled with Atto594 (shown in red). The mean diameters for structures of Chmp4b (115 ± 46 nm, 179 particles), Chmp4c (110 ± 41 nm, 231 particles), Alix (101 ± 50 nm, 302 particles), and Tsg101 (100 ± 41 nm, 130 particles) were smaller (P < 0.005) than the corresponding Gag structures (Chmp4b, 221 ± 117 nm; Chmp4c, 182 ± 59 nm; Alix, 198 ± 119 nm; Tsg101, 175 ± 52 nm). A, G, M, and S show the full ROI imaged and analyzed for the experiments at 40 × 40 µm2. B, H, N, and T show representative 4 × 4-µm2 areas from the large field, and C–E, I–K, O–Q, and U–W show representative 400 × 400-nm2 areas. (F, L, R, and X) Bivariate Ripley’s K Tests between assemblages labeled with Atto488 and Atto594. Gag–Atto488 assemblages clustered with Chmp4b–Atto594 (F), Chmp4c–Atto594 (L), Alix–Atto594 (R), and Tsg101–Atto594 (X).
The assemblages of Gag and ESCRT proteins either overlapped partially (Fig. 3 C, I, O, and U and Fig. S4C), were in direct contact without overlap (Fig. 3 D, J, P, and V and Fig. S4D), or overlapped completely (Fig. 3 E, K, Q, and W and Fig. S4E). When completely overlapping, the centers of the Gag and ESCRT assemblies were displaced from each other (Fig. 3 E, K, Q, and W and Fig. S4E). The association of Gag and ESCRT assemblies was significant and not the result of a random distribution, as determined by the clustering analysis (Chmp4b, Fig. 3F; Chmp4c, Fig. 3L; Alix, Fig. 3R and Fig. S4F; or Tsg101, Fig. 3X). As an independent test of the colocalization of Gag with Chmp4b, we imaged a Gag protein lacking p6 that cannot recruit Alix and Tsg101 (22–25). GagΔp6 assemblages did not localize with Chmp4b (Fig. S5A), and clustering analysis showed that there was no correlation between the position of assemblies of GagΔp6 and Chmp4b (Fig. S5B). As a control, we switched the fluorescent proteins on Gag and Chmp4b (Fig. S6). The degree of displacement of the ESCRT proteins from the center of Gag assemblages was variable (see also Fig. 2N), potentially reflecting variability in the size of the Gag and the ESCRT assemblages as well as variability in the angle of budding relative to the optical axis.
Distance analysis shows that Tsg101 is recruited to a position closer to the center of Gag assemblages than Alix and the ESCRT-III proteins. Each assemblage of ESCRT proteins was displaced relative to Gag, by a distance that was approximately equivalent to the mean radius of the Gag particle (Fig. 2N) and significantly greater than the distances measured for Gag–Gag. The mean distances between the centers of the Gag assemblages and those of Chmp4b, Chmp4c, or Alix were similar, but the distance between the centers of Gag and Tsg101 (Fig. 2N, green line) was smaller (P < 0.001). This difference is also observed in the cumulative distance plot (Fig. S7), which shows that the distances for Gag–Tsg101 were different from Gag–Gag and the other Gag–ESCRT pairs, whereas the distances for Gag–Gag and Gag–Chmp4b, Gag–Chmp4c, and Gag–Alix were not statistically significant (P < 0.001, Mann–Whitney and KS tests). Although Alix and Tsg101 recognize motifs that are physically near each other in the p6 domain of the Gag molecule, these results suggest that they may have physically distinct localizations relative to the assembling VLPs. This displacement of ESCRT protein assemblages relative to Gag assemblages was not a result of misalignment of the green and the red images, as it occurred at different directions within a field (Fig. 3).
Discussion
Our results reveal a specific temporal and spatial order to the recruitment of ESCRT proteins to the site of an assembling HIV-1 VLP. Tsg101, a component of ESCRT-I that binds directly to the p6 domain of Gag, is progressively recruited together with Gag during assembly. Once recruitment of Gag is complete, Chmp4b [a component of ESCRT-III required for HIV-1 budding thought to assemble into filaments (8, 13, 14) that drive particle fission] is transiently recruited to assembly sites. Ten seconds later, the ATPase Vps4A, which is thought to disassemble the filaments (8, 13, 14), is recruited. Both proteins subsequently depart completely from budding sites, with Vps4A lingering for over twice as long as Chmp4b. This finding is consistent with studies of cell division showing that Chmp4b is recruited first, although Chmp4b and Vps4B were imaged in separate cells and then the timing of their recruitment was synchronized to the timing of microtubule breakage (26).
Diffraction-limited microscopy cannot distinguish whether Tsg101, Chmp4b, or Vps4A is within the head of the VLP or at the neck connecting to the parental cell. Our results from superresolution imaging indicate that when the active state of the fission machinery is trapped by expressing a dominant-negative Vps4A, the Tsg101, Alix, Chmp4b, and Chmp4c molecules collect toward one side of the assembling Gag VLP, presumably at the membranous neck. Our superresolution imaging results, which reveal statistically significant displacements of ESCRT proteins relative to Gag, are inconsistent with the notion that ESCRT molecules are within the head of the VLPs. A recent paper based on superresolution imaging has proposed that the ESCRT proteins are recruited to the head of the VLPs, with ESCRT proteins acting as scaffolding (27). Although the analysis found similar-sized Gag and ESCRT assemblages as those that we identified and a similar overlap of Gag and ESCRT centroids (∼20%), in that report the Gag and ESCRT assemblages were separately averaged and then superimposed to be concentric. Differences may also arise from the overexpression of ESCRT proteins in that study. Furthermore, the scaffolding model is inconsistent with the live-cell imaging data of Chmp4b, which is recruited only transiently, and then leaves the virion (ref. 4 and this study).
Our data are consistent with the ESCRT proteins being recruited to the neck of the Gag VLPs during scission. We found that Tsg101 is significantly closer to the center of a Gag VLP than Alix and the ESCRT-III proteins. This was a surprising result, as Tsg101 and Alix can interact directly with the p6 domain of the structural protein Gag.
Although we observed that Tsg101 is recruited with Gag during VLP assembly, Tsg101 does not completely overlap spatially with Gag at late stages of assembly and its location is biased to the side of Gag assemblages. This suggests that Tsg101 preferentially binds to the Gag molecules closest to the neck of the nascent virion. This recruitment may be aided by the negative curvature of the membranous neck, causing the C-terminal p6 domains of Gag to spread farther apart. This arrangement of Gag molecules may ultimately result in the preferential recruitment of Tsg101 to the neck at late stages of VLP assembly. Coincident with the termination of Gag assembly, ESCRT-III proteins are then transiently recruited to polymerize at the neck, followed shortly afterward by Vps4A. Although we detected Tsg101 at sites of assembly at time frames when ESCRT-III and Vps4A signals returned to background, our live-cell imaging data do not allow us to determine whether Tsg101 remains with the VLP after scission or stays behind on the plasma membrane outside of the VLP. Studies by mass spectrometry found members of the ESCRT-I complex (such as Tsg101 and Vps28) and Alix with purified HIV-1 virions, but Chmp4b, Chmp4c, Vps4A, or other members of the ESCRT-III complex were not detected (28). Biochemical data demonstrated only a few molecules (3–12.5) of Alix in each HIV-1 VLP (13), three orders of magnitude lower than the number of Gag molecules per VLP (18). It remains to be determined if scission is contemporaneous with the recruitment of the ESCRT proteins, the Vps4A protein, or at another point. Independent of the ESCRT protein behavior during Gag assembly (coassembly with Tsg101or transient recruitment of Chmp4b, Chmp4c, and Vps4A), our results are consistent with the recruitment of all imaged ESCRT proteins to the membranous neck connecting the nascent virus and the plasma membrane, as suggested previously in various models for ESCRT protein function (8, 9).
Materials and Methods
This is a summary of the materials and methods used. The full description is provided in SI Materials and Methods.
Plasmid Construction.
All plasmids used in this study were constructed using standard molecular biological techniques following the manufacturers’ instructions. For Gag–mEGFP, the A206K mutation was first introduced in pEGFP-N1 to minimize EGFP dimerization (29); then Gag was PCR amplified and inserted via the BamHI and NotI restriction sites. The same restriction sites were used to replace mEGFP with other fluorescent proteins to generate the following plasmids: Gag–mTagBFP, Gag–mCherry, Gag–iRFP (30), Gag–PA–mGFP (PA GFP), and Gag–PA–mCherry1 (PA mCherry1) (31). To generate a plasmid expressing untagged Gag, mEGFP was deleted from the Gag–mEGFP plasmid using site-directed mutagenesis. Versions of these plasmids with the p6 domain of Gag missing were constructed by deleting the appropriate section from the plasmids with the Site-Directed Mutagenesis Lightning Kit (Agilent-Stratagene). Plasmids to make stably expressing cell lines were based in the pLNCX2 plasmid (Clontech) and monomeric fluorescent proteins. The following constructs were generated: pLNCX2–PA–mGFP–Chmp4b, pLNCX2–PA–mCherry1–Chmp4b, pLNCX2–PA–mCherry1–Chmp4c, pLNCX2–PA–mCherry1–Tsg101, pLNCX2–PA–mCherry1–Alix, pLNCX2–mEGFP–Vps4A, pLNCX2–mEGFP–Tsg101, and pLNCX2–mCherry–Chmp4b. The plasmid pCR3.1-Vpu was previously described (4).
Production of Clonal Cell Lines and Protein Detection via Western Blotting.
Stable cell lines expressing tagged versions of the ESCRT proteins were created by viral infection of HeLa cells using the appropriate pLNCX2 plasmids. Clones selected for this study exhibited growth rates and occurrence of multinucleated cells similar to the parental HeLa cell line (growth, cell numbers increased by a factor of 4.8–5.4 over 48 h for cell lines and by a factor of 5.2 for the parental HeLa cell line; multinucleated cells, 3.5–6.1% compared with 6.4% for HeLa cells; dividing cells were considered multinucleated; Fig. S1 A and B). Table S1 summarizes the cell lines created for this work. Protein expression of these cell lines was assayed via Western blotting with the following antibodies: α-Chmp4b (Santa Cruz sc-82556, rabbit polyclonal, 1:100), α-Alix (Santa Cruz sc-53540, mouse monoclonal, 1:1,000), α-Tsg101 (Santa Cruz sc-7964, mouse monoclonal, 1:200), α-Vps4A (abcam ab81697, rabbit polyclonal, 1:1,000), α-GFP [Clontech Living Colors 632381 (JL-8), mouse monoclonal, 1:1,000], α-dsRed (Clontech Living Colors, rabbit polyclonal 1:1,000), goat α-mouse–HRP (Sigma–Aldrich A-9917, 1:25,000), and goat α-rabbit–HRP (Sigma–Aldrich A-0545, 1:25,000). All blocking and incubations were done in 0.5% nonfat milk powder in Tris-buffered saline with Tween 20 except for mEGFP and mCherry detection, which were done in 5% (wt/vol) milk powder.
Cell Culture, Transfection, and Fixation Protocol for Superresolution Microscopy.
All cells were grown in DMEM (without pyruvate, Gibco) with 10% FBS at 37 °C and 5% CO2 using standard sterile techniques. The growth medium was supplemented with Geneticin at 0.4 µg/mL for maintenance of stable cell lines. For imaging, cells were seeded on 35 mm MatTek dishes with no. 1.5 coverslips coated with Fibronectin (Invitrogen) or poly-d-lysine (Cultrex). The medium was changed to one without phenol red. Cells were transfected with 1 µg DNA and 3 µL FuGENE 6 (Promega) in 100 µL Opti-Mem I according to manufacturer’s instructions. We verified that multiple expression plasmids were simultaneously expressed in cells and the various proteins incorporated into individual VLPs (Fig. S8). Twenty-four hours later, cells were washed, treated for exactly 1 min with 0.5% Saponin in PBS-T, fixed with 3.7% (wt/vol) PFA, washed, permeabilized with 0.5% Triton X-100, washed again, blocked with 4% (wt/vol) BSA, then incubated with α-GFP–nanobody–Atto488 and α-RFP–nanobody–Atto594 (Chromotek, cat. nos. gba488 and rba594, respectively) in 4% BSA for 1 h at room temperature, protected from light, according to the manufacturer’s instructions, and then washed and stored at 4 °C, protected from light. The α-RFP–nanobody–Atto594 does not cross-react with the fluorophore iRFP; the proteins were isolated from different species, and the iRFP is ∼80 amino acids longer than mCherry and shares no sequence similarity.
Two-color live-cell imaging of a stable cell line expressing mEGFP–Tsg101 was performed after transient transfection with 2 µg of total DNA (Gag-mCherry:Gag 1:5) and Lipofectamine 2000 (Life Technologies) in Opti-Mem I. For three-color live-cell imaging, a stable cell line expressing mEGFP–Vps4A and mCherry–Chmp4b was transiently transfected with 1 µg of total DNA (200 ng Vpu, 800 ng Gag–mTagBFP:Gag 1:5) and FuGENE6 in Opti-Mem I.
Superresolution Imaging.
For two-color superresolution imaging, the two colors (Atto488 and Atto594) were imaged sequentially. The dye molecules were driven from the ground state to a transient dark state with a thiol-containing buffer in combination with an enzyme mixture for oxygen scavenging (17, 32), whereas individual dye molecules were excited stochastically with high laser power at their excitation wavelength (488 nm for Atto488 or 561 nm for Atto594, respectively) to induce blinking on millisecond time scales. This approach allowed us to image each of these two dyes without cross-talk. Experiments were performed on a Nikon Eclipse Ti STORM microscope with a Nikon CFI Apo 100×/1.49 N.A. objective, an Agilent laser launch system, an Andor iXonEM+ camera, and an NSTORM Quad cube (Chroma). Cells in MatTek dishes were imaged in a buffer containing 50 mM Tris, pH 8, 10 mM NaCl, 10% (wt/vol) glucose, with the following chemicals added right before imaging (final concentration given): 500 µg/mL glucose oxidase, 143 mM β-mercaptoethanol, and 4 mg/mL catalase. We also added 1–2 µL of 100-nm Tetraspeck beads to aid in channel alignment. Once a suitable cell (excluding areas where cells had ripped off, Fig. S9) was selected via Gag–iRFP, Atto594 was imaged at the fastest frame rate for 20,000 frames using the 60-mW 561-nm laser set to 50% or higher to achieve blinking of the Atto594 dye molecules. Then, the proteins labeled with Atto488 were imaged at the fastest frame rate for 20,000 frames (or less, if no more signal was detected due to bleaching), with the 60-mW 488-nm laser set to a minimum of 67% to achieve blinking. The laser angles were set to achieve TIR to limit excitation to the plasma membrane and avoid structures from within the cell.
Image Reconstruction and Analysis.
The 14-bit superresolution images were fitted with a 2D Gaussian function and drift corrected using the Nikon NIS Elements software version 4.11. The signal from the 100-nm Tetraspeck beads was used to align the different channels. The images were opened in ImageJ (National Institutes of Health) as hyperstacks, thresholds (mean + 2 SD as minimum) applied, and then analyzed with the particle analysis menu, with the superresolution assemblages fitted to ellipses. The results from these fits were further analyzed using Microsoft Excel. All results are given as mean ± SD if not mentioned otherwise. For statistical analysis of clustering, regions of interest (ROIs) from superresolution images were selected to exclude cell boundaries and space not occupied by the cell. Centroids of structures for the analysis were input from the list given by the ImageJ ‘Analyze Particles’ routine on the ROIs. Bivariate Ripley’s K-based statistics were used to compare the distributions of the structures in two-color superresolution images to those from two overlapping spatially random distributions with the same area and density using the CellSpan program (33).
Fluorescence Microscopy.
All fluorescence microscopy experiments were performed either on a custom-built microscope based on an Olympus IX-70 equipped with an ApoN 60×/1.49 N.A. objective and a 405-nm, 488-nm, and 589-nm laser or an IX-81 with an UApoN 100×/1.49 N.A. objective and a 405-nm, 488-nm, 594-nm, and 647-nm laser. The cells were imaged under azimuthal TIR-FM (34) using a Cairn Opto-Split III with a cube and appropriate filters. Live-cell experiments were performed at 37 °C in cell imaging medium (CIM) (Hanks BBS, 10 mM Hepes, pH 7.4) with 1% FBS. For two-color live-cell experiments, Gag–mCherry and mEGFP–Tsg101 were imaged every minute simultaneously via an emission splitter from Cairn (OptoSplit III). For three-color live-cell experiments, Gag–mTagBFP was imaged every minute, with mEGFP–Vps4A and mCherry–Chmp4b imaged every 10 s. Both microscopes were equipped with a Hamamatsu Flash 4.0 camera. Timing of recruitment of mEGFP–Vps4A and mCherry–Chmp4b to VLP assembly sites was determined from the leading edge of each ESCRT protein’s recruitment peak. The baseline before recruitment and the maximum peak signal were compared with the calculation of a half-maximum signal. Interpolation between time points was then used to determine the leading edge time. This time was then used to compare the relative time between recruitment of Vps4A and Chmp4b (Fig. 1E) and to align all of the traces to construct the averaged recruitment dwell profile (Fig. 1F).
Supplementary Material
Acknowledgments
We thank C. Takacs from the S.M.S. laboratory for critical reading of our manuscript. The superresolution images were acquired on the Nikon STORM microscope system at the Bio-Imaging Resource Center at The Rockefeller University. This work was supported by National Institutes of Health Grants P50GM103297 (to M.B., D.S.J., and S.M.S.), 5R01AI089844 (to M.S.I. and S.M.S.), and R01AI50111 (to P.D.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321655111/-/DCSupplemental.
References
- 1.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454(7201):236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivanchenko S, et al. Dynamics of HIV-1 assembly and release. PLoS Pathog. 2009;5(11):e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci USA. 2009;106(45):19114–19119. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13(4):394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jouvenet N, Simon SM, Bieniasz PD. Visualizing HIV-1 assembly. J Mol Biol. 2011;410(4):501–511. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumgärtel V, et al. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13(4):469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- 7.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Sundquist WI, Kräusslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2(7):a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22(3):133–140. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344(1):55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 11.Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- 12.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100(21):12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114(6):701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 14.Morita E, et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9(3):235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313(5793):1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 16.Müller B, Heilemann M. Shedding new light on viruses: Super-resolution microscopy for studying human immunodeficiency virus. Trends Microbiol. 2013;21(10):522–533. doi: 10.1016/j.tim.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47(33):6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 18.Briggs JA, et al. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11(7):672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 19.Gross I, et al. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19(1):103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muranyi W, Malkusch S, Müller B, Heilemann M, Kräusslich HG. Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail. PLoS Pathog. 2013;9(2):e1003198. doi: 10.1371/journal.ppat.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunzenhäuser J, Olivier N, Pengo T, Manley S. Quantitative super-resolution imaging reveals protein stoichiometry and nanoscale morphology of assembling HIV-Gag virions. Nano Lett. 2012;12(9):4705–4710. doi: 10.1021/nl3021076. [DOI] [PubMed] [Google Scholar]
- 22.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7(12):1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 23.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 24.Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 25.Fisher RD, et al. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128(5):841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 26.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108(12):4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Engelenburg SB, et al. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014;343(6171):653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chertova E, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80(18):9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296(5569):913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 30.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29(8):757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subach FV, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6(2):153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8(12):1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Characterizing the topography of membrane receptors and signaling molecules from spatial patterns obtained using nanometer-scale electron-dense probes and electron microscopy. Micron. 2006;37(1):14–34. doi: 10.1016/j.micron.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Johnson DS, Jaiswal JK, Simon SM. Total internal reflection fluorescence (TIRF) microscopy illuminator for improved imaging of cell surface events. Curr Protoc Cytom. 2012;Chapter 12:Unit 12.29. doi: 10.1002/0471142956.cy1229s61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.