Significance
Several molecular systems with important adaptive roles have originated from the domestication of integrated phages (prophages). However, the evolutionary mechanisms and extent of prophage domestication remain poorly understood. In this work, we detected several hundred prophages originating from common integration events and described their dynamics of degradation within their hosts. Surprisingly, we observed strong conservation of the sequence of most vertically inherited prophages, including selection for genes encoding phage-specific functions. These results suggest pervasive domestication of parasites by the bacterial hosts. Because prophages account for a large fraction of bacterial genomes, phage domestication may drive bacterial adaptation.
Keywords: prokaryotes, viruses
Abstract
Integrated phages (prophages) are major contributors to the diversity of bacterial gene repertoires. Domestication of their components is thought to have endowed bacteria with molecular systems involved in secretion, defense, warfare, and gene transfer. However, the rates and mechanisms of domestication remain unknown. We used comparative genomics to study the evolution of prophages within the bacterial genome. We identified over 300 vertically inherited prophages within enterobacterial genomes. Some of these elements are very old and might predate the split between Escherichia coli and Salmonella enterica. The size distribution of prophage elements is bimodal, suggestive of rapid prophage inactivation followed by much slower genetic degradation. Accordingly, we observed a pervasive pattern of systematic counterselection of nonsynonymous mutations in prophage genes. Importantly, such patterns of purifying selection are observed not only on accessory regions but also in core phage genes, such as those encoding structural and lysis components. This suggests that bacterial hosts select for phage-associated functions. Several of these conserved prophages have gene repertoires compatible with described functions of adaptive prophage-derived elements such as bacteriocins, killer particles, gene transfer agents, or satellite prophages. We suggest that bacteria frequently domesticate their prophages. Most such domesticated elements end up deleted from the bacterial genome because they are replaced by analogous functions carried by new prophages. This puts the bacterial genome in a state of continuous flux of acquisition and loss of phage-derived adaptive genes.
The ubiquity and abundance of bacteriophages (or phages) makes them key actors in bacterial population dynamics (1). Although all phages are able to propagate horizontally between cells, temperate phages also propagate vertically in bacterial (lysogenic) lineages, typically by integrating into the bacterial chromosome as prophages. Very few genes are expressed in the prophage, which replicates with the bacterial chromosome (2). The evolutionary interests of integrated phages (prophages) are partly aligned with those of the host chromosome because rapid proliferation of the latter effectively increases prophage population. Accordingly, prophages protect the host against further phage infection (3), from phagocytosis (4), and provide bacterial pathogens with virulence factors (5). Temperate phages also encode accessory genes that increase host fitness under certain conditions, such as increased growth under nutrient limitation (6), biofilm formation (7), and antibiotic tolerance (8). Some prophages provide bacteria with regulatory switches (9). Functional prophages might also be used as biological weapons by lysogens, because their induction can counteract or delay colonization by nonlysogens (10–12). High diversity and high turnover of temperate phages result in a constant input of new genes in the host genome (13, 14). For example, Escherichia coli prophages contribute to more than 35% of the gene diversity (pangenome) of the species (15). Most temperate phages integrate into a few very specific and conserved integration hot spots in chromosomes and their sequences are adapted to the local frequency of DNA motifs, suggesting adaptation of the phage sequence to the requirements of the prophage state (15).
Independently of occasional contributions to bacterial fitness, intact prophages are molecular time bombs that kill their hosts upon activation of the lytic cycle (2). It has been shown that bacterial pseudogenes are under selection for rapid deletion from bacterial genomes (16, 17). Prophage inactivation should be under even stronger selection because these elements can kill the cell. One might thus expect rapid genetic degradation of prophages: either they activate the lytic cycle and kill the cell before accumulating inactivating mutations or they are irreversibly degraded and deleted from the host genome. Bacterial chromosomes have numerous cryptic (defective) prophages and other prophage-derived elements that might result from this evolutionary dynamics (13, 14). Accordingly, functional studies of the full repertoire of prophages of bacterial genomes suggest that the majority of prophages are defective at some level: excision, virion formation, lysis, or infective ability (18, 19).
Bacterial genomes encode many molecular systems presumably derived from defective prophages. These include gene transfer agents (GTAs) that transfer random pieces of chromosomal DNA to other cells (20), and bacteriocins and type 6 secretion systems (T6SSs) that are involved in bacterial antagonistic associations (21, 22). Model phage-derived elements, like GTAs or T6SSs, are streamlined and genetically very stable. However, genomes contain a number of elements derived from prophages that fit less neatly in the above categories and perform a number of functions with diverse degrees of efficiency: they parasite other phages, kill other bacteria, or transfer host DNA (23). Finally, prophage-derived structures are also involved in complex animal–bacteria associations (24, 25). These different elements blur the distinction between stable phage-derived elements and prophages ongoing genetic degradation, suggesting that some defective prophages provide adaptive functions to bacteria.
Temperate phages and their hosts develop complex antagonistic and mutualistic interactions: depending on the circumstances, prophages can either kill bacteria or increase their fitness. There are no systematic studies of which trend dominates the evolutionary dynamics of prophage–bacteria interactions. Here, we bring to the fore a key related question: how do prophages evolve within the bacterial genome? To answer it, we identified the repertoire of vertically inherited prophages of Escherichia coli and Salmonella enterica. The analysis of these prophages revealed unexpected evolutionary patterns suggesting widespread contribution of prophages to bacterial fitness.
Results
Prophages Display Signs of Degradation.
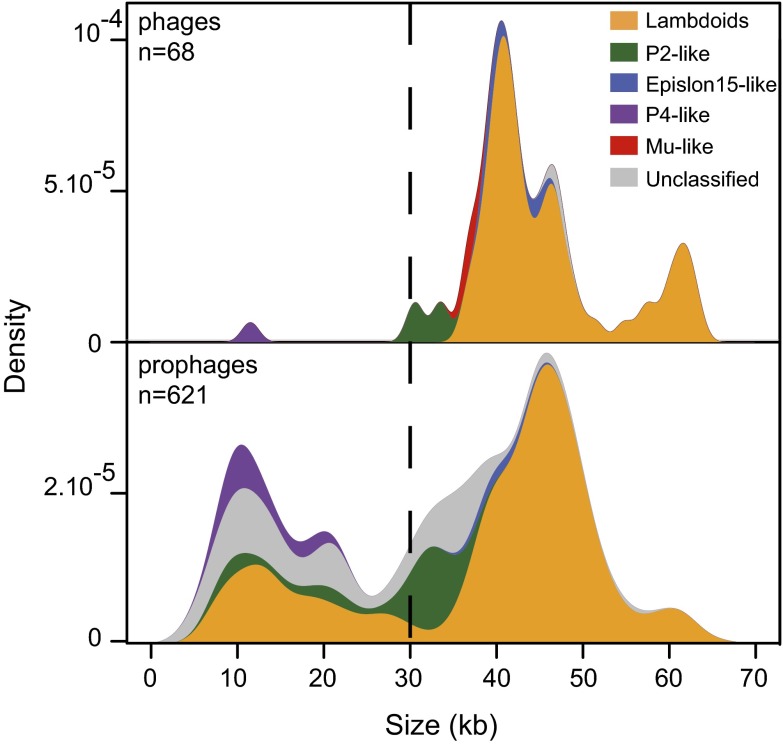
In this study, we analyzed a dataset of phages and prophages of E. coli and S. enterica that we have previously identified (15), to which we added prophages from recently published genomes. Prophages were identified using several tools and their precise limits were determined by comparative genomics and expert curation (Materials and Methods). We identified 624 prophages among 58 and 27 fully assembled genomes of E. coli (474 prophages) and S. enterica (150 prophages), respectively. Because intact prophages are likely to kill the cell upon induction of the lytic cycle, there should be strong selection for mutations leading to prophage inactivation. We therefore expected to find few large prophages, corresponding to recent integrations, and then a gradient of smaller and smaller prophages having endured diverse levels of genome degradation. Surprisingly, the distribution of the genome size of prophages is clearly bimodal with a class of small and another of large elements (Fig. 1). A near complete separation between the two classes occurs for prophage genome size of ∼30 kb, the size of the smallest autonomous dsDNA phage infecting enterobacteria in GenBank (Fig. 1). Many prophages (37%) are smaller than 30 kb, even though we excluded from the analysis the very small prophages difficult to distinguish from other mobile genetic elements (Materials and Methods). This suggests either the presence of two different populations of prophages or rapid degradation of large prophages and then stabilization of the resulting elements in the genome.
Fig. 1.
Probability distributions of the genome size of the 68 dsDNA temperate caudophages infecting enterobacteria (Upper) and of the Caudovirales prophages (Lower). The taxonomic groups are indicated on the Right of the figure.
The bimodal distribution of prophages could be due to differences in taxonomic groups, e.g., small prophages and large prophages could derive from different types of phages. To test this hypothesis, we classified phages and prophages in taxa using a genome similarity score that includes information on the patterns of gene presence/absence and sequence similarity (following refs. 15 and 26). Most of the small prophages (74%) could be assigned to known taxa. These prophages are systematically smaller than the phages of the same taxa in GenBank, a clear indication that they have endured some genetic degradation (P < 0.0001, Wilcoxon test). Moreover, 48% of the small prophages could be classified as lambdoid or P2-like phages (Fig. 1), for which all known representatives are larger than 30 kb. Few small prophages are clearly distinct from autonomous dsDNA phages (13%): we identified 3 ssDNA Inoviruses and 28 P4-like satellite prophages. The remaining small prophages lack homologs of the characteristic proteins of P4-like or SaPI satellite phages (27, 28): Sid, Pif, CpmA, and CpmB (blastp, e value > 0.001) (29, 30). To study the gene repertoires of small prophages, and their differences, we built protein families for the whole dataset of prophages and temperate phages of enterobacteria (Materials and Methods). Small prophages are significantly enriched in tail genes compared with temperate caudophages of enterobacteria (P < 0.0001, χ2 test) (Fig. S1A). Hence, small prophages rarely encode characteristic satellite phage proteins, they are often phylogenetically close to large prophages, and they often encode structural proteins. This shows that most small prophages are not satellite phages, but prophages resulting from the genetic degradation of larger elements.
The observed bimodality of prophage size could result from systematic large neutral deletions of genetic material within the prophage. The deletion spectrum in the chromosome of Salmonella is not consistent with this pattern, showing a clear predominance of small deletions (31). Nevertheless, we tested this hypothesis by simulating genetic deletions of different sizes within prophages while requiring conservation of the flanking bacterial core genes. Our results show that a pattern dominated by large deletions leads to unbalanced deletion of genes in the prophage: genes encoded in the central part of the element (like lysis or packaging genes) are much more frequently deleted than genes at the edges (integrases and cargo genes) (Fig. S2). On the contrary, small deletions (and the experimental deletion spectrum) lead to a much weaker dependency of the probability of deletion with the position in the prophage. The comparison of the results of the simulations with the functions encoded in small lambdoid prophages relative to known lambdoid phages does not show significant differences in most functional classes (Fig. S1B). It is thus not compatible with the predominance of large neutral deletions in prophage evolution.
Many Prophages Are Vertically Inherited.
Our dataset includes an average of 8.2 and 5.6 prophages per genome for E. coli and S. enterica, respectively. Prophages at similar loci in different genomes can derive from a single ancestral prophage (orthologous prophages) or from multiple independent integrations at the same loci (15). Hence, prophages in a given chromosomal locus are a mixture of orthologous and nonorthologous prophages. We used a conservative set of four criteria to distinguish them (SI Materials and Methods and Fig. S3). First, two orthologous prophages must be integrated into the same chromosomal locus (flanked by the same bacterial core genes). Second, orthologous prophages must have a high genomic similarity score (R ≥ 0.7). Third, orthologous prophages are replicated like any part of the bacterial chromosome and should thus exhibit similar neutral substitution rates. Synonymous positions are under weak selection, and we require prophages to have average synonymous substitution rates similar to the genes of the core genome. In a few cases, the inferred ancestral prophage genome was much larger than expected given the distribution of genome lengths of temperate phages infecting enterobacteria. This suggested that we needed a fourth criterion: as orthologous families derive from a single ancestral prophage, the gene diversity (pangenome) of a given family of orthologous prophages must not be much larger than the number of genes present in the prophage encoding the highest number of genes (Materials and Methods). With these strict filters, our step-by-step method eliminated nonorthologous prophages and removed or split families into multiple smaller families. In the end, we identified 100 families of orthologous prophages (71 in E. coli and 29 in S. enterica). The majority of the integration events (72%) was observed in one single strain, presumably because they occurred very recently. Around 28% of the inferred integration events involved a prophage that has left remnants in more than one strain, i.e., produced a family of orthologous prophages. These families include 372 prophages (60% of the total) and contain from 2 to 15 orthologous elements (Fig. S4). This suggests that many prophages in a species are derived from a single ancestral integration event despite frequent prophage loss. The elements of a given family of orthologous prophages have remained in the bacterial chromosome the same number of years and should exhibit comparable levels of genetic degradation. Indeed, nearly all families of orthologous prophages (90%) include elements of either the large or the small classes of prophage size, but not both.
The groups of prophages with and without orthologous elements do not have significantly different taxonomic distributions (P = 0.2, χ2 test; Fig. S5), suggesting no particular taxonomic bias in the prophages that reside in bacterial chromosomes for longer periods of time (Fig. S5). As prophages in general, the orthologous prophages include mostly groups of lambdoids (59%) and a smaller number of P2-like (13%) elements. As expected, orthologous prophages are found in more closely related bacterial strains compared with nonorthologous prophages integrated into the same loci (P < 0.0001, Wilcoxon test). Prophages with orthologous elements are also shorter than prophages lacking orthologs (30.9 vs. 36.7 kb on average; P < 0.0001, Wilcoxon test), which likely results from their longer residence time. The number of genes lost in all elements of a family cannot be precisely quantified because we ignore the genome of the ancestral phage. However, the comparison of pairs of orthologous prophages allows the quantification of the patterns of differential gene loss in the prophage family, i.e., losses that did not take place in all elements. Prophages have endured an average of 7.8 such gene losses per pair (5.6 kb on average; Materials and Methods). The median deletion is 500 nt long, and only 40% of pairs of orthologous prophages do not display any indels.
Spontaneous excision rates of complete prophages are of the order of 10−6 per cell division for Lambda under nonstressful conditions and are otherwise orders of magnitude higher (32). Therefore, fully functional prophages are unlikely to remain in the chromosome for a long time. To assess how many of the prophage families might constitute functional prophages, we analyzed the only strain in our dataset for which the function of all prophages has been experimentally studied (O157:H7 Sakaï) (18). We found orthologous elements for 15 of the 16 defective prophages in this genome. We found no single orthologous prophage for the only fully functional phage. This restricted analysis supports the claim that prophages endure rapid genetic degradation.
Vertically Inherited Prophages Are Under Purifying Selection.
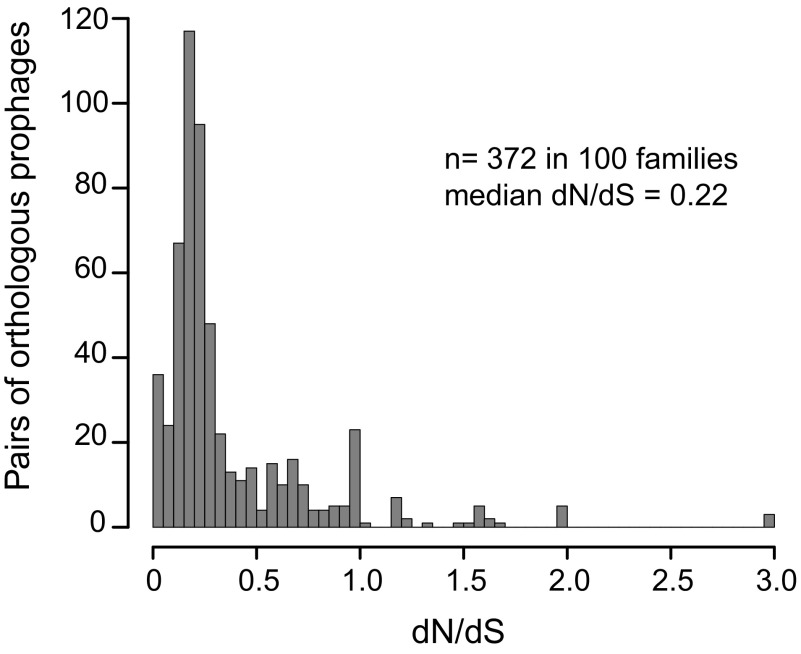
To investigate the action of natural selection on the genes of prophages, we computed the ratio of nonsynonymous over synonymous substitution rates (dN/dS) for the pools of orthologous genes within bacterial core genes and within orthologous prophages. As expected, bacterial core genomes display very low dN/dS values (median dN/dS = 0.06; P < 0.0001, Wilcoxon test). Prophage genes display higher dN/dS ratios. However, and very surprisingly, most orthologous prophages display a dN/dS ratio much lower than 1 (median dN/dS = 0.22; P < 0.0001, Mann–Whitney test). The preferential purge of nonsynonymous mutations by natural selection suggests selection for maintaining the function of the genes encoded in prophages (Fig. 2). A similar dN/dS distribution was observed for a subset of prophages for which orthology was defined using even more stringent criteria (Fig. S6; Materials and Methods). Very few pairs of orthologous prophages (6%) show dN/dS values consistent with neutral, positive, or diversifying selection (dN/dS ≥ 1). The low dN/dS values are not an artifact associated with the small number of genes or the low density of SNPs in the dataset, because dN/dS values are similar for the most divergent genomes where the signal is the strongest (Fig. S7). In fact, dN/dS is constant along the range of dS values, consistent with the rapid imprint of purifying selection in dN/dS in E. coli (33). Small prophages are as constrained by purifying selection as the large prophages (median dN/dS = 0.23 and 0.22, respectively; P = 0.09, Wilcoxon test).
Fig. 2.
Histogram of the ratio of nonsynonymous to synonymous substitutions (dN/dS) between orthologous prophages.
Recombination with incoming phages can imprint a signal of purifying selection on prophages by introducing an overabundance of synonymous polymorphisms resulting from purifying selection on phages. To test whether recombination caused our unexpected observations, we detected recombinant genes among orthologous prophages using seven different methods and a combination of them. In each analysis, we removed the gene families showing significant evidence of recombination or phylogenetic incongruence. This led to the rejection of between 8% and 32% of the genes (Table S1). The joint analysis using pairwise homoplasy index (PHI)/neighbor similarity score (NSS)/maximum χ2 (MaxChi), Prunier, and MaxChi on concatenates rejected 16% of the genes (34–36). In all of the eight variants of the analysis, we observed dN/dS values for the nonrecombining genes very significantly smaller than 1 (Table S1). These previous analyses cannot account for the nearly complete replacement of the prophage by homologous recombination. They also cannot detect cases of independent integration of very similar phages in different genomes. Such events should lead to incongruence between the prophage and the host phylogenetic trees. Our analysis shows that only 1 of the 18 prophage families with five or more taxa have phylogenetic trees significantly different from the hosts (P < 0.05, both for Shimodeira–Hasegawa and Approximately Unbiased tests followed by Bonferroni correction). Removing this family makes no significant difference in the dN/dS distribution, which remains significantly smaller than 1 (dN/dS = 0.16; P < 0.0001, Mann–Whitney test). Hence, although our results confirm the existence of recombination between prophages and phages (or other prophages), as previously shown (37), this effect is not the major cause of the observed low dN/dS values.
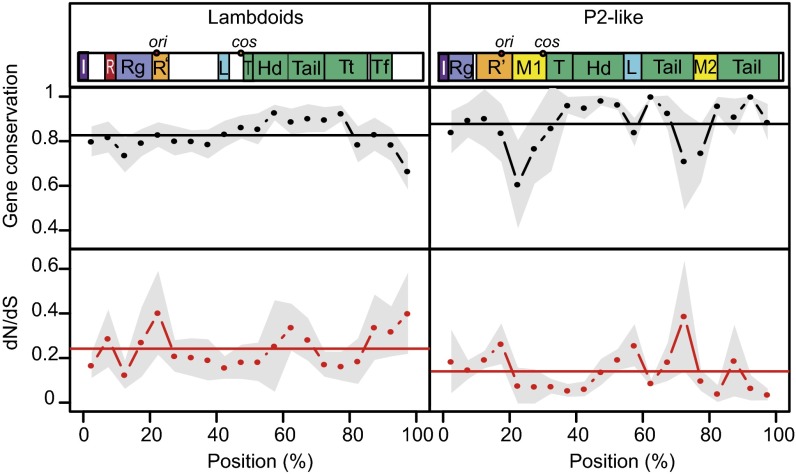
To investigate the patterns of substitution rates and gene loss in prophages, we analyzed each gene in function of its position in the prophage genome. Gene positions are highly conserved in lambdoids and in P2-like phages (38, 39). We restricted our attention to lambdoid and P2-like large prophages (55% of the dataset) because they can be mapped accurately in this genetic organization. Overall, there is a nearly constant high degree of gene conservation in orthologous prophages (Fig. 3). Comparison with Fig. S2 confirms our previous observation that prophage degradation in our dataset does not result from random neutral large deletions. The high degree of gene conservation might partly result from the strict rules used to define prophage orthology. In lambdoids, the region encoding the tail proteins is slightly more conserved and the cargo region (extreme end after the tail genes) is less conserved. P2-like prophages are also well conserved along the genome except for two regions (“morons” 1 and 2), which typically contain fast evolving accessory genes (39). The regions of the prophages where gene loss is less frequent are also those where genes have lower dN/dS values (except P2-like moron region 1, which has low dN/dS values) (Fig. 3). This is consistent with purifying selection on these genes. Regions encoding infection-related functions, such as replication or capsid proteins, are very conserved in prophages. These results were confirmed independently by the analysis of prophage gene families in terms of functional categories, which are not limited to large P2-like and lambdoid prophages (Fig. S8). As a whole, the data suggest pervasive positive contribution of prophage genes to bacterial fitness.
Fig. 3.
Average gene conservation and nonsynonymous to synonymous substitution ratios (dN/dS) along prophage genomes. (Left) Lambdoids. (Right) P2-like prophages. Only large (≥30 kb) prophages were considered in this analysis because small prophages cannot be confidently represented on the normalized genome map. Schematic representations of lambdoid and P2-like genomes are given on top and are based on Lambda (Left) and P2 (Right) genome architectures. Hd, head; I, integration; L, lysis; M1, moron region 1; M2, moron region 2; R, recombination; R', replication; Rg, regulation; T, terminase(s); Tf, tail fiber; Tt, tail tip. The gray contours represent the 95% confidence intervals of gene conservation and dN/dS ratios. The horizontal lines indicate median values of gene conservation and dN/dS ratios.
Discussion
We found that orthologous prophages are numerous, representative of the diversity of enterobacterial phages and have endured genetic degradation since the ancestral integration into the genome. This has presumably rendered them defective. Surprisingly, most orthologous prophages show strong signs of purifying selection. Previous studies have shown that many recently acquired genes in bacteria are under purifying selection (40). We have mentioned above that many prophages carry accessory genes that are adaptive to bacteria (6–8). Our results differ from previous works in one essential aspect: in our study, many of the prophage genes under stronger purifying selection encode core phage-related functions, like tail and lysis proteins. This suggests that prophage functions are under selection in the bacterial chromosome.
We observed a strongly bimodal distribution of prophage genome size, which is neither caused by phage taxonomic biases nor by large neutral deletions of genetic material. Bimodality could result from rapid inactivating gene losses followed by much slower genetic degradation of the remaining genes (14). The slowdown of genetic inactivation could result from purifying selection on certain genes as observed in the dN/dS analysis. This raises the question of why bacteria are not accumulating even larger numbers of prophage genes. We suggest that analog/homolog gene replacements may lead to frequent gene loss. Genomes of enterobacteria are constantly acquiring prophages of a relatively small number of taxonomic groups. Extant prophage genes may thus suddenly become under relaxed selection when homologous or analogous genes arrive in the host. This may lead to the replacement of prophage genes by others performing similar functions.
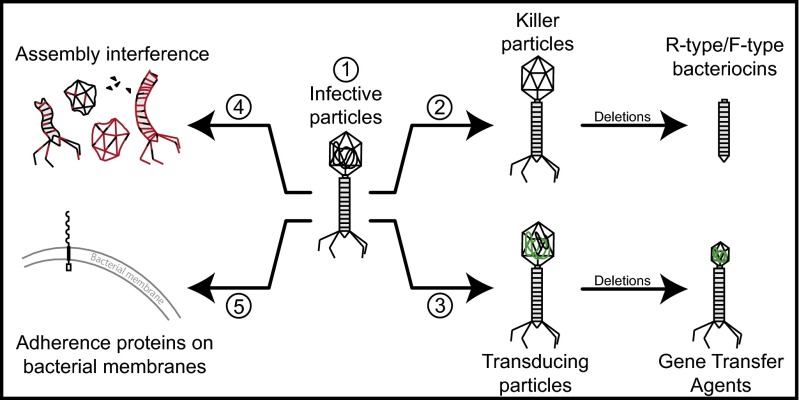
Our results show that many phage-derived functions are under purifying selection. We suggest this is because they are adaptive for their host. Previous studies have shown that prophages unable to produce viable virions upon infection can protect from superinfection, excise, package DNA, and even infect other cells (8, 18, 19). Therefore, partly degraded prophage elements can have a number of adaptive functions, as described below (Fig. 4).
Fig. 4.
Putative functions of orthologous prophages conserved in their hosts. (1) Functional prophages can be used as molecular weapons to kill nonlysogens through the production of infective particles. (2) Defective prophages can produce noninfective particles (phage killer particles and R/F-type bacteriocins) that kill sensitive cells. (3) Prophages can form transducing particles and gene transfer agents (GTAs) that promote host DNA exchange (displayed in green). (4) Degraded prophages might interfere with the assembly of other phages (represented in red) leading to the formation of defective particles. (5) Prophage structural proteins often display Ig-like domains that might be used by their hosts for adherence in niche colonization.
Functional prophages allow populations of lysogens to kill nonlysogen competitors (11). However, this advantage rapidly disappears by the creation of lysogens in the susceptible populations (12) and comes at the cost of cell lysis in a fraction of the population. Our results indicating that intact prophages are rapidly lost suggest that this strategy might be very short-lived or rarely used.
Phage-derived bacteriocins kill cells whose genomes lack their cognate immunity genes (21, 41). Contrary to fully functional phages, they do not reinfect other cells nor do they produce lysogens, preventing the creation of immunity in other populations. R-type and F-type bacteriocins are typically composed of domesticated tail and lysis genes from myophages and siphophages, respectively. This fits the observed gene repertoires of some families of small prophages in our dataset. Notably, the largest orthologous prophage family has a phylogenetic tree that mirrors that of the bacterial host, lacks integrases, and seems to have been stabilized in a large number of strains of Salmonella (Fig. S9). This element is related to P2-like phages (Myoviridae) and could therefore correspond to a R-type bacteriocin. The putative domestication of this prophage might even predate the split between Escherichia and Salmonella, because we identified a very similar small prophage also missing an integrase at the same position in two E. coli strains (Fig. S9). Most prophage families do not fit so well the description of R-type or F-type bacteriocins but could correspond to phage killer particles. These elements behave like bacteriocins but are very diverse genetically, presenting characteristics ranging from streamlined elements like R-type and F-type bacteriocins to nearly complete phages (23, 42–44). Defective phages can easily give rise to phage killer particles. For example, PBSX and other noninfectious defective phage particles were termed phage killer particles or protophages and act de facto as bacteriocins (42, 44). Some of these systems are conserved among different isolates and might be widespread among bacterial species (23, 43, 45). Altered particles of T-even phages also display a bacteriocin activity (46). It is possible that many of the orthologous prophages correspond to R/F-type bacteriocins or phage killer particles.
GTAs are found in diverse prokaryotic clades and are thought to have originated from domestication of general transducing phages (47). They typically encode structural genes, lack integrases, and evolve under purifying selection. General transducing phages P22 and P1 also produce and transfer phage particles that contain exclusively bacterial DNA (48, 49). An increased rate of general transduction by these phages can be obtained by a small number of different mutations altering the head morphogenesis process (50, 51). Therefore, a defective phage may lead to a transducing agent in very few mutational steps. Accordingly, several prophage families in our dataset encode a nearly complete set of structural and lysis genes while lacking many functions associated with regulation, replication, and integration/excision. Also, the size of GTAs, between 14 and 30 kb (20), is in close agreement with the lower peak of prophage size distribution observed in our dataset (Fig. 1). GTAs package unspecific DNA and this can be easily achieved by pac-based phage terminases but not by cos-based terminases (52). Lambdoids use either pac or cos systems (53), and the former might act as GTAs. However, P2-like phages use cos packaging systems (54) and are unlikely to become GTAs. Nevertheless, the mechanism by which GTAs package random fragments of DNA is not well understood and it remains possible that transencoded packaging systems fill phage particles with host DNA.
Our results suggest that bacteria select for the conservation of components of virion particles. Virions of lambdoids are composed of over 500 protein units, and the architecture of the phage particle depends on their precise interaction (55). If the assembly of different phage particles in the cell results in protein interactions between components of the different virions, this might lead to the production of defective phage particles. If a phage infects a cell containing prophages, their expression may interfere with the incoming phage and diminish its viable progeny. This would result in higher fitness for bacterial populations carrying prophages. This idea is supported by the observation that double coinfections by lambdoids lead to fewer virions compared with single infections (56). A similar type of molecular interference has been proposed to explain why proteins forming large complexes are less prone to horizontal gene transfer and duplication (57, 58). Given the high complexity of phage particles (providing ample potential for interference) and the abundance of phage infections in natural environments (providing strong fitness gains for bacteria avoiding phage infections), we speculate that selection for the maintenance of interfering prophages might be advantageous for the host. This advantage might, however, disappear in the long term due to rapid phage diversification.
There may be many other uses of phage proteins that for the moment are purely speculative. For example, about 25% of caudophages encode proteins with Ig-like domains (59). These phages are abundant in mucosal surfaces of Metazoans, protecting them from bacterial infections and allowing phages to have a constant supply of hosts (60). Conversely, expressing prophage structural proteins at the surface of bacterial cells could aid bacterial infection or niche colonization. For example, prophage tail proteins expressed after treatment with mitomycin C or UV light mediate Streptococcus mitis and Enterococcus faecalis platelet binding, favoring infective endocarditis (19, 61). Prophage structural proteins might thus favor physical interactions of bacteria with their environment.
The functions encoded by some prophages are compatible with selection for the use of prophages in antagonistic relations with other bacteria (as biological weapons, phage-derived bacteriocins, or killer particles), for horizontal transfer (GTAs), for protection against other phages, or for bacterial colonization. Several cases have been described in the literature of degraded phages performing such functions. However, no single type of phage-derived element fits the gene repertoires of all orthologous prophages that we have identified. This suggests that prophages provide several different functions. These prophage-derived functions may be very generic. Because closely related phages are constantly arriving at the bacterial genome, prophage-derived genes are likely to be frequently superseded by other incoming prophage-derived genes. As mentioned above, this should lead to frequent analogous/homologous gene replacements. Occasionally prophage-derived elements may evolve toward a new highly specialized function. This will lead to their enduring domestication.
Materials and Methods
Data.
We downloaded 58 and 27 complete genomes of E. coli and S. enterica, respectively, from the Bacterial section of National Center for Biotechnology Information (NCBI) RefSeq (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/; last accessed February 2013), and 68 temperate caudophage genomes infecting enterobacteria from the Virus section of NCBI RefSeq (ftp://ftp.ncbi.nih.gov/genomes/Viruses/; last accessed February 2013).
Prophage Detection and Classification.
Prophages were detected as in ref. 15, except that prophages with no match to core phage genes were discarded. This resembles the implementation of the stringent option of Phage Finder (62) but it does not exclude a priori the smallest prophages. Prophages detected at rearrangement breakpoints were removed from the analysis because their positions could not be confidently defined in all genomes. To avoid gene loss overprediction due to imprecise delimitation of prophages, we built the families of homologous proteins found in all genomes between the same two flanking core genes. Each protein family was then considered as phage-related or host-related but not both. Information about the 624 prophages is detailed in Dataset S1.
The resulting 624 prophages were classified by comparing their gene repertoires with those of phages infecting enterobacteria (15). Gene repertoire relatedness (R) between pairs of (pro)phages was defined as follows: , where S(Ai,Bi) is the similarity score of the pair i of homologous proteins between (pro)phage A and (pro)phage B (varying from 0.4 to 1), M is the total number of homologs shared by (pro)phages A and B, and nA and nB are the total number of proteins of (pro)phages A and B, respectively.
Other Materials and Methods.
Details on the computation of core genomes and bacterial distances, the definition of orthologous prophages, the functional assignment of prophage proteins, deletion simulations, estimation of recombination, and the computation of synonymous and nonsynonymous substitution rates are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jean-François Gout for helpful comments on an earlier version of this manuscript. This work was supported by European Research Council starting Grant EVOMOBILOME 281605 (to E.P.C.R.) and a grant from the Ministère de l'Enseignement Supérieur et de la Recherche (to L.-M.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405336111/-/DCSupplemental.
References
- 1.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399(6736):541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 2.Ptashne M. Genetic Switch: Phage Lambda and Higher Organisms. 2nd Ed. Cambridge, MA: Blackwell; 1992. [Google Scholar]
- 3.Campbell AM. Bacteriophages. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: ASM; 1996. pp. 2325–2338. [Google Scholar]
- 4.Steinberg KM, Levin BR. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc Biol Sci. 2007;274(1621):1921–1929. doi: 10.1098/rspb.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waldor MK, Friedman DI. Phage regulatory circuits and virulence gene expression. Curr Opin Microbiol. 2005;8(4):459–465. doi: 10.1016/j.mib.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Edlin G, Lin L, Bitner R. Reproductive fitness of P1, P2, and Mu lysogens of Escherichia coli. J Virol. 1977;21(2):560–564. doi: 10.1128/jvi.21.2.560-564.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gödeke J, Paul K, Lassak J, Thormann KM. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J. 2011;5(4):613–626. doi: 10.1038/ismej.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, et al. Cryptic prophages help bacteria cope with adverse environments. Nat Commun. 2010;1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabinovich L, Sigal N, Borovok I, Nir-Paz R, Herskovits AA. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. 2012;150(4):792–802. doi: 10.1016/j.cell.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Bossi L, Fuentes JA, Mora G, Figueroa-Bossi N. Prophage contribution to bacterial population dynamics. J Bacteriol. 2003;185(21):6467–6471. doi: 10.1128/JB.185.21.6467-6471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown SP, Le Chat L, De Paepe M, Taddei F. Ecology of microbial invasions: Amplification allows virus carriers to invade more rapidly when rare. Curr Biol. 2006;16(20):2048–2052. doi: 10.1016/j.cub.2006.08.089. [DOI] [PubMed] [Google Scholar]
- 12.Gama JA, et al. Temperate bacterial viruses as double-edged swords in bacterial warfare. PLoS One. 2013;8(3):e59043. doi: 10.1371/journal.pone.0059043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canchaya C, Fournous G, Brüssow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53(1):9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- 14.Casjens S. Prophages and bacterial genomics: What have we learned so far? Mol Microbiol. 2003;49(2):277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 15.Bobay LM, Rocha EP, Touchon M. The adaptation of temperate bacteriophages to their host genomes. Mol Biol Evol. 2013;30(4):737–751. doi: 10.1093/molbev/mss279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence JG, Hendrix RW, Casjens S. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 2001;9(11):535–540. doi: 10.1016/s0966-842x(01)02198-9. [DOI] [PubMed] [Google Scholar]
- 17.Kuo CH, Ochman H. The extinction dynamics of bacterial pseudogenes. PLoS Genet. 2010;6(8):e1001050. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asadulghani M, et al. The defective prophage pool of Escherichia coli O157: Prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009;5(5):e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matos RC, et al. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 2013;9(6):e1003539. doi: 10.1371/journal.pgen.1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang AS, Zhaxybayeva O, Beatty JT. Gene transfer agents: Phage-like elements of genetic exchange. Nat Rev Microbiol. 2012;10(7):472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84(5-6):499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 22.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106(11):4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell A. Defective bacteriophages and incomplete prophages. In: Fraenkel-Conrat H, Wagner RR, editors. Regulation and Genetics. Vol 8. New York: Springer; 1977. pp. 259–328. [Google Scholar]
- 24.Hurst MR, Glare TR, Jackson TA. Cloning Serratia entomophila antifeeding genes—a putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol. 2004;186(15):5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shikuma NJ, et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343(6170):529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohwer F, Edwards R. The Phage Proteomic Tree: A genome-based taxonomy for phage. J Bacteriol. 2002;184(16):4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick RP, Subedi A. The SaPIs: Mobile pathogenicity islands of Staphylococcus. Chem Immunol Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- 28.Christie GE, Dokland T. Pirates of the Caudovirales. Virology. 2012;434(2):210–221. doi: 10.1016/j.virol.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ubeda C, et al. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol Microbiol. 2009;72(1):98–108. doi: 10.1111/j.1365-2958.2009.06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damle PK, et al. The roles of SaPI1 proteins gp7 (CpmA) and gp6 (CpmB) in capsid size determination and helper phage interference. Virology. 2012;432(2):277–282. doi: 10.1016/j.virol.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S, Ke R, Hughes D, Nilsson M, Andersson DI. Genome-wide detection of spontaneous chromosomal rearrangements in bacteria. PLoS One. 2012;7(8):e42639. doi: 10.1371/journal.pone.0042639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottesman ME, Yarmolinsky MB. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- 33.Rocha EPC, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239(2):226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 34.Abby SS, Tannier E, Gouy M, Daubin V. Detecting lateral gene transfers by statistical reconciliation of phylogenetic forests. BMC Bioinformatics. 2010;11:324. doi: 10.1186/1471-2105-11-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172(4):2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin DP, et al. RDP3: A flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26(19):2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Paepe M, et al. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: The role of Rad52-like recombinases. PLoS Genet. 2014;10(3):e1004181. doi: 10.1371/journal.pgen.1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell A, Botstein D. Evolution of the lambdoid phages. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1983. pp. 365–380. [Google Scholar]
- 39.Nilsson A, Haggard Ljungquist E. The P2-like bacteriophages. In: Calendar RL, Abedon ST, editors. The Bacteriophages. 2nd Ed. Vol 1. New York: Oxford Univ Press; 2006. pp. 365–390. [Google Scholar]
- 40.Daubin V, Ochman H. Bacterial genomes as new gene homes: The genealogy of ORFans in E. coli. Genome Res. 2004;14(6):1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy KG. Colicinogeny and related phenomena. Bacteriol Rev. 1975;39(4):464–515. doi: 10.1128/br.39.4.464-515.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradley DE. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967;31(4):230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garro AJ, Marmur J. Defective bacteriophages. J Cell Physiol. 1970;76(3):253–263. doi: 10.1002/jcp.1040760305. [DOI] [PubMed] [Google Scholar]
- 44.Wood HE, Dawson MT, Devine KM, McConnell DJ. Characterization of PBSX, a defective prophage of Bacillus subtilis. J Bacteriol. 1990;172(5):2667–2674. doi: 10.1128/jb.172.5.2667-2674.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerdes JC, Romig WR. Complete and defective bacteriophages of classical Vibrio cholerae: Relationship to the kappa type bacteriophage. J Virol. 1975;15(5):1231–1238. doi: 10.1128/jvi.15.5.1231-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duckworth DH. The metabolism of T4 phage ghost-infected cells. I. Macromolecular synthesis and ransport of nucleic acid and protein precursors. Virology. 1970;40(3):673–684. doi: 10.1016/0042-6822(70)90212-6. [DOI] [PubMed] [Google Scholar]
- 47.Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15(2):54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119(1):75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 49.Wall JD, Harriman PD. Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology. 1974;59(2):532–544. doi: 10.1016/0042-6822(74)90463-2. [DOI] [PubMed] [Google Scholar]
- 50.Casjens S, et al. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J Mol Biol. 1992;227(4):1086–1099. doi: 10.1016/0022-2836(92)90523-m. [DOI] [PubMed] [Google Scholar]
- 51.Iida S, Hiestand-Nauer R, Sandmeier H, Lehnherr H, Arber W. Accessory genes in the darA operon of bacteriophage P1 affect antirestriction function, generalized transduction, head morphogenesis, and host cell lysis. Virology. 1998;251(1):49–58. doi: 10.1006/viro.1998.9405. [DOI] [PubMed] [Google Scholar]
- 52.Ebel-Tsipis J, Botstein D, Fox MS. Generalized transduction by phage P22 in Salmonella typhimurium. I. Molecular origin of transducing DNA. J Mol Biol. 1972;71(2):433–448. doi: 10.1016/0022-2836(72)90361-0. [DOI] [PubMed] [Google Scholar]
- 53.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9(9):647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 54.Nilsson AS, Haggård-Ljungquist E. Evolution of P2-like phages and their impact on bacterial evolution. Res Microbiol. 2007;158(4):311–317. doi: 10.1016/j.resmic.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 55.Häuser R, et al. Bacteriophage protein-protein interactions. Adv Virus Res. 2012;83:219–298. doi: 10.1016/B978-0-12-394438-2.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Refardt D. Within-host competition determines reproductive success of temperate bacteriophages. ISME J. 2011;5(9):1451–1460. doi: 10.1038/ismej.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci USA. 1999;96(7):3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker CR, Hanson-Smith V, Johnson AD. Following gene duplication, paralog interference constrains transcriptional circuit evolution. Science. 2013;342(6154):104–108. doi: 10.1126/science.1240810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fraser JS, Yu Z, Maxwell KL, Davidson AR. Ig-like domains on bacteriophages: A tale of promiscuity and deceit. J Mol Biol. 2006;359(2):496–507. doi: 10.1016/j.jmb.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 60.Barr JJ, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110(26):10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bensing BA, Siboo IR, Sullam PM. Proteins PblA and PblB of Streptococcus mitis, which promote binding to human platelets, are encoded within a lysogenic bacteriophage. Infect Immun. 2001;69(10):6186–6192. doi: 10.1128/IAI.69.10.6186-6192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fouts DE. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006;34(20):5839–5851. doi: 10.1093/nar/gkl732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.