Significance
Respiratory syncytial virus (RSV) causes major disease in pediatric and elderly patients, urging the development of efficacious therapeutics. This study establishes a recombinant RSV reporter strain for drug discovery and identifies an entry inhibitor class targeting the viral fusion (F) protein. Biochemical, structural, and functional characterization of the inhibitor spotlights two microdomains governing the conformational stability of prefusion F. Mutations in these domains cause broad cross-resistance against the compound and RSV entry inhibitors in preclinical and clinical development, without mandatory loss of in vivo pathogenicity, challenging the possible clinical benefit of current RSV entry inhibitor classes. Anti-RSV campaigns should better target postentry steps or proactively circumvent resistance to entry inhibition. The resistant RSV reporter strain developed here establishes a strategy toward this goal.
Keywords: antiviral drugs, drug resistance
Abstract
Respiratory syncytial virus (RSV) is a leading pediatric pathogen that is responsible for a majority of infant hospitalizations due to viral disease. Despite its clinical importance, no vaccine prophylaxis against RSV disease or effective antiviral therapeutic is available. In this study, we established a robust high-throughput drug screening protocol by using a recombinant RSV reporter virus to expand the pool of RSV inhibitor candidates. Mechanistic characterization revealed that a potent newly identified inhibitor class blocks viral entry through specific targeting of the RSV fusion (F) protein. Resistance against this class was induced and revealed overlapping hotspots with diverse, previously identified RSV entry blockers at different stages of preclinical and clinical development. A structural and biochemical assessment of the mechanism of unique, broad RSV cross-resistance against structurally distinct entry inhibitors demonstrated that individual escape hotspots are located in immediate physical proximity in the metastable conformation of RSV F and that the resistance mutations lower the barrier for prefusion F triggering, resulting in an accelerated RSV entry kinetics. One resistant RSV recombinant remained fully pathogenic in a mouse model of RSV infection. By identifying molecular determinants governing the RSV entry machinery, this study spotlights a molecular mechanism of broad RSV resistance against entry inhibition that may affect the impact of diverse viral entry inhibitors presently considered for clinical use and outlines a proactive design for future RSV drug discovery campaigns.
Respiratory syncytial virus (RSV) is a member of the paramyxovirus family, which consists of mostly highly contagious nonsegmented, negative polarity RNA viruses that spread through the respiratory route. RSV disease is the leading cause of virus infection-induced death among children less than 1 year of age (1) and can be life-threatening to the elderly and the immunocompromised (2, 3). Reinfection with RSV can occur throughout life (4), but infants born prematurely, or with bronchopulmonary dysplasia or a congenital heart defect, are at highest risk of developing severe disease (5). In a typical case, initial RSV infection of airway epithelia cells is followed by rapid spread from the nasopharynx to the lower airways that can affect respiratory function through excessive mucus, necrotic epithelial debris, and inflammatory cells obstructing the airways (6, 7).
Attempts to develop an effective RSV vaccine have been fruitless thus far, because the virus is poorly immunogenic overall and neutralizing antibody titers wane quickly after infection (4). Although ribavirin was approved for RSV treatment, clinical use is minimal because of efficacy and toxicity issues (8, 9). The humanized neutralizing antibody palivizumab is used for immunoprophylaxis of high-risk pediatric patients, but high costs prohibit broad-scale implementation (5, 10–13).
Clinical disease associated with infection by several paramyxoviruses such as mumps virus or measles virus (MeV) originate predominantly from immunopathogenic effects, which makes therapeutic treatment challenging, because viral replication is typically immune-controlled and titers decline when symptoms become manifest (14, 15). In the case of RSV infection, however, several studies have suggested that pathogenesis is not the result of host immunopathology alone. Rather, higher viral loads were recognized as a predictor for severe lower respiratory infection in infants (16), and RSV titers on day three of hospitalization were indicative for increased need for intensive care in hospitalized children less than 2 y old (17). These observations suggest that efficacious therapeutics given early to hospitalized children may improve downstream morbidity and reduce immunopathology, opening a window for improved disease management and making RSV a premier target for drug discovery campaigns.
RSV particles are wrapped by a lipid envelope that contains two integral membrane glycoproteins, the attachment (G) and F proteins (4). Although G acts as a major virulence factor (18), the F protein alone is capable of mediating particle docking to target cells and membrane fusion (19, 20). Consistent with this central role of F in initiating RSV infection, the protein emerged as a major target of small-molecule RSV inhibitors, since recent open screens identified several structurally distinct classes of RSV entry blockers (21–24). Spotlighted by the benefit of the peptidic HIV entry inhibitor Fuzeon, interference with viral entry represents a clinically viable antiviral strategy (25).
High-resolution structures of RSV F trimers were solved in both the metastable prefusion (26) and final postfusion (27, 28) conformations, and show the hallmarks of class I viral fusion proteins: Each F monomer features two helical heptad repeat (HR) domains, one (HR-A) located adjacent to a membrane attack domain, the fusion peptide, and the other (HR-B) flanking the transmembrane domain. Although the HR-A is broken up into multiple distinct segments in the prefusion structure, triggering of the entry machinery leads to the assembly of an elongated HR-A triple-helix, insertion of the fusion peptide into the target membrane, and folding of the HR-B helices into the groves of the HR-A coiled coil. In the resulting thermodynamically highly stable six-helix bundle (6HB) structure, the fusion peptides and transmembrane domains and, thus, the target and donor membranes, are brought into close proximity (29). Based on the appearance of resistance mutations in F protein HR domains and biochemical evidence, some of the most advanced RSV entry inhibitors were suggested to prevent the creation of fusion pores through competitive interference with 6HB closure (21, 30).
Here, we describe a previously unidentified structural class of small-molecule RSV entry inhibitors through the development and implementation of an automated drug-screening assay. Resistance mutations located to two hotspots in the RSV F protein, setting the stage for a structural and mechanistic characterization of viral escape through molecular modeling, biochemical assays, and the generation of genetically controlled resistant RSV recombinants. Our results highlight molecular determinants that control triggering of the RSV membrane fusion machinery and outline a cross-resistance mechanism for broad RSV escape from structurally diverse entry inhibitors. We assessed the effect of resistance on viral pathogenesis in a small animal model of RSV infection to determine whether this escape mechanism may jeopardize the therapeutic potential of RSV drug candidates considered for clinical use (31).
Results
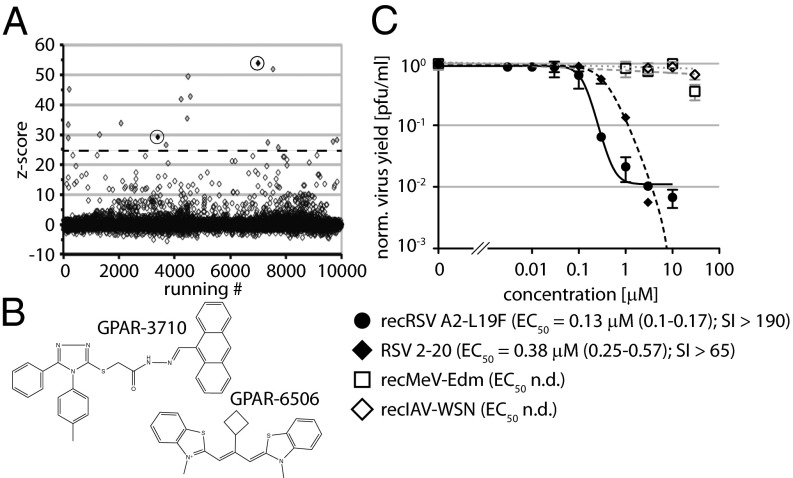
To identify novel anti-RSV drug candidates, we screened a 10,000-entry small-molecule library against a previously generated recombinant (rec) RSV strain harboring an additional transcription unit encoding for renilla luciferase (32, 33). Applying recently established assay conditions for automated antiparamyxovirus drug screens (33), this exercise returned a hit candidate pool of 17 compounds, each with a primary screening score exceeding 10 times the overall SD of the assay. (Fig. 1A).
Fig. 1.
Discovery of a previously unidentified small-molecule class of RSV inhibitors. (A) Results of a test screen of recRSV A2-L19F-renilla against a 10,000-entry compound library. Data for each compound were normalized, scaled, and are plotted by screening scores (z score). The dashed line marks the hit selection cutoff (10 assay − SD); circled hits maintained >90% RSV inhibition in 0.5 μM concentration counterscreens. (B) Structures of compounds circled in A. (C) Dose–response curves for sourced hit GPAR-3710 against paramyxoviruses and orthomyxoviruses. In addition to the recRSV A2-L19F screening strain, RSV clinical isolate 2–20 (40) was tested. Values are means of three experiments ± SD. Where applicable, EC50 concentrations were determined through four-parameter variable slope regression modeling; values in parentheses denote 95% confidence intervals. CC50 concentrations are based on host metabolic activity after 24-h exposure; highest concentration assessed 25 μM. n.d., not determined; SI, CC50/EC50.
Two-concentration counterscreens and cytotoxicity testing yielded two candidates that showed >90% RSV inhibition at one-10th (0.5 μM) of the original screening concentration (Fig. 1B). Based on structural considerations and cytotoxicity profiles, of these, GPAR-3710 was sourced for structure-integrity verification. The sourced compound combined low cytotoxicity with target-specific and dose-dependent inhibition of different RSV strains with active concentrations in the nanomolar to low micromolar range (Fig. 1C and Fig. S1).
A Previously Unidentified Chemical Class of Small-Molecule RSV Entry Inhibitors.
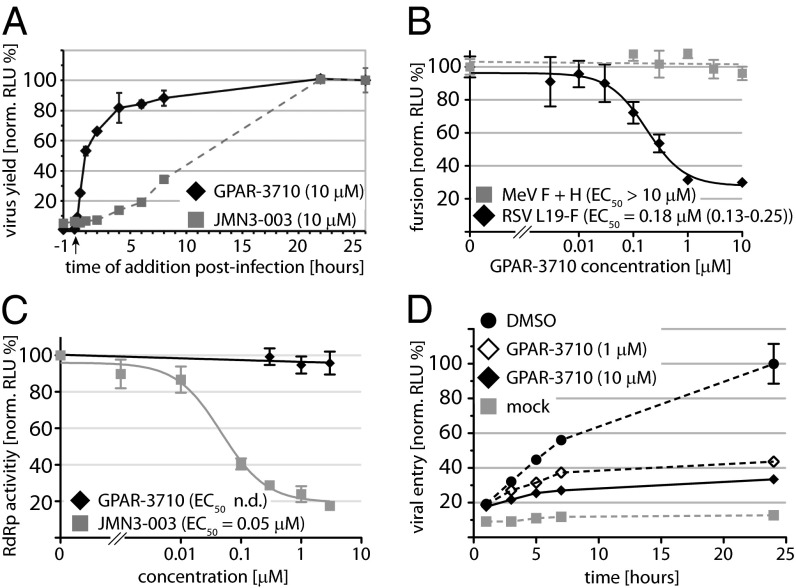
For mechanistic characterization, we first subjected GPAR-3710 to a time-of-addition study to narrow the step in the viral life cycle blocked by the compound. Maximal inhibition of virus replication was observed only when we added the compound at the time of infection, whereas essentially all antiviral activity was lost when GPAR-3710 was administered later than 4 h after infection (Fig. 2A). By comparison, a pan inhibitor of myxovirus polymerase function previously developed by our group, JMN3-003 (34), remained potently inhibitory even when added 8 h after infection. This time-of-addition profile points toward inhibition of virus attachment or cell entry by the compound. For cross-examination, we tested GPAR-3710 in two plasmid-based reporter assays that specifically measure bioactivity of the viral entry (35) and polymerase (36) machinery, respectively. RSV F protein-mediated membrane fusion activity was specifically and potently inhibited by the compound in these assays (Fig. 2B), whereas activity of the viral RNA-dependent RNA-polymerase (RdRp) complex remained unaffected (Fig. 2C). The perceived higher potency of the compound against live virus than transiently expressed F protein (99% inhibition in Fig. 1C vs. 80% inhibition in Fig. 2B) most likely originates from the potentiating effect of repeat opportunities for compound interference presented during multiple-step virus replication.
Fig. 2.
Mechanistic characterization of GPAR-3710. (A) Time-of-addition study of GPAR-3710 in comparison with the broad spectrum RdRp inhibitor JMN3-003. Compound was added at the indicated time points. The arrow marks the time of infection with recRSV A2-L19F-renilla. All samples were harvested 26 h p.i., and progeny titers were determined by TCID50 titration using renilla luciferase activity as the readout. Values are means of three experiments ± SD. (B) Quantitative dose–response cell-to-cell fusion assay using the DSP-chimeric reporter proteins and ViviRen renilla luciferase substrate. MeV F and H glycoprotein expression constructs are included for specificity control. Values were normalized for vehicle (DMSO)-treated samples and represent means of at least three experiments ± SD. Regression modeling is as in Fig. 1C. (C) Transient RSV luciferase replicon reporter assay to determine viral RdRp activity in the presence of GPAR-3710. The previously identified RdRp inhibitor JMN3-003 was used for specificity control. Values were normalized for vehicle (DMSO)-treated samples and represent means of three experiments ± SD; n.d., not determined. (D) Kinetic virus-to-cell fusion assay using the DSP reporter proteins and EnduRen live cell luciferase substrate. Cells were spin-inoculated with recRSV A2-L19F and shifted from 4° to 37 °C at 0 h. Values represent means of three experiments ± SD. Mock denotes cell mixtures that remained uninfected.
To directly monitor the effect of the inhibitor on the rate of viral entry, we established a quantitative RSV entry assay that monitors virus-to-cell fusion in near real time (Fig. 2D). RSV particles were spin-inoculated on a monolayer of cells expressing either the amino- or carboxyl-terminal halves of an eGFP-renilla luciferase chimeric protein (37). Simultaneous fusion of the incoming viral particles with two adjacent target cells results in cell content mixing, restoring eGFP fluorescence, and renilla luciferase activity. When executed in the presence of increasing GPAR-3710 concentrations, this assay revealed a significant, dose-dependent reduction of the RSV entry rate by the compound (Fig. 2D). Taken together, these observations characterize the GPAR-3710 scaffold as a previously unidentified class of small-molecule RSV entry inhibitors.
Escape Mutations Locate to the RSV F Protein.
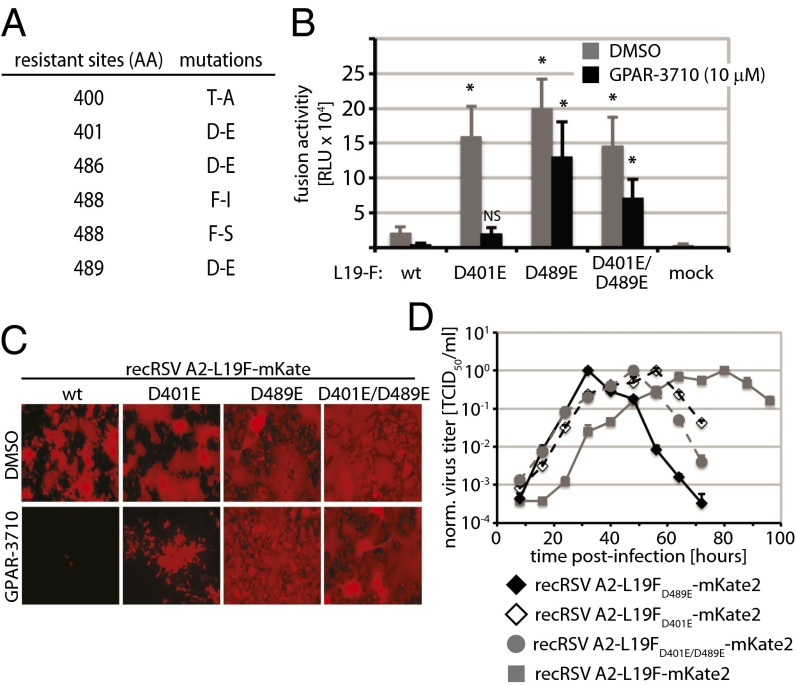
As a hallmark for pathogen-directed antiviral compounds, the experimental induction of viral escape from inhibition is typically straightforward, and resistance mutations usually locate to the viral protein physically targeted by the compound. We provoked RSV escape from GPAR-3710 inhibition through gradual adaptation to growth in the presence of increasing compound concentrations. Robust resistance—defined by viral growth in the presence of 30 μM (>200× EC50 concentration) of the compound—reliably appeared within a 30-d adaptation period. We concentrated on the viral entry machinery in search for the molecular basis for escape and determined the F protein sequences of six independently adapted RSV strains. Candidate mutations were rebuilt in an expression plasmid encoding the RSV line19 (L19) F protein (38) through directed mutagenesis, followed by first-pass resistance testing in transient cell-to-cell fusion assays carried out in the presence of the compound (Fig. S2).
In each of the six strains, we identified a single point mutation in the F protein that contributed to the phenotype (Fig. 3A). The mutations clustered in two linear microdomains (400 and 489) of RSV F, spanning residues 400–401 and 486–489, respectively. Mutations D401E and D489E were selected for transient cell-to-cell fusion assays in the presence and absence of the compound. In addition, we generated and analyzed an FD401E/D489E-double mutant construct. All mutant F were hyperfusogenic compared with the standard L19-F protein (Fig. 3B). Particularly robust resistance was observed when the D489E mutation was present.
Fig. 3.
Resistance profile of GPAR-3710. (A) Resistance mutations identified in the RSV F protein through viral adaptation. (B) Resistance quantification using transiently expressed RSV L19-F mutants, the DSP-based quantitative cell-to-cell fusion assay, and ViviRen luciferase substrate. Values represent means of six experiments ± SD. Datasets were subjected to one-way ANOVA and Bonferroni’s multiple comparison posttest; asterisks indicate statistically significant differences of values obtained for individual mutants compared with equally treated, unmodified L19-F (P < 0.05; NS, not significant). Mock denotes cells transfected with vector DNA instead of F expression plasmid. (C) Fluorescence microphotographs of recovered RSV recombinants expressing mKate2 and harboring F mutants instead of standard F. Photographs were taken 44 h p.i. after incubation in the presence of 10 μM GPAR-3710 or vehicle (DMSO). (D) Growth curves of the recovered RSV recombinants at 37 °C. Cell-associated viral titers were determined through TCID50 titration by using mKate2-derived fluorescence as readout. Values are means of three experiments ± SD.
To verify the role in escape in the context of viral infection, we transferred the mutant F constructs into the genetically controlled cDNA background of a recombinant RSV A2, harboring the L19-F protein (32). In addition, we generated a cDNA construct containing the FD401E/D489E double-mutant in the place of parental L19-F. All three mutant recRSVs were recovered successfully and showed resistance to GPAR-3710, based on efficient spread through cell monolayers in the presence of the compound (Fig. 3C). They all also showed accelerated growth rates compared with standard recRSV A2-L19F (Fig. 3D).
Structural Basis for Cross-Resistance Against Diverse RSV F Inhibitors.
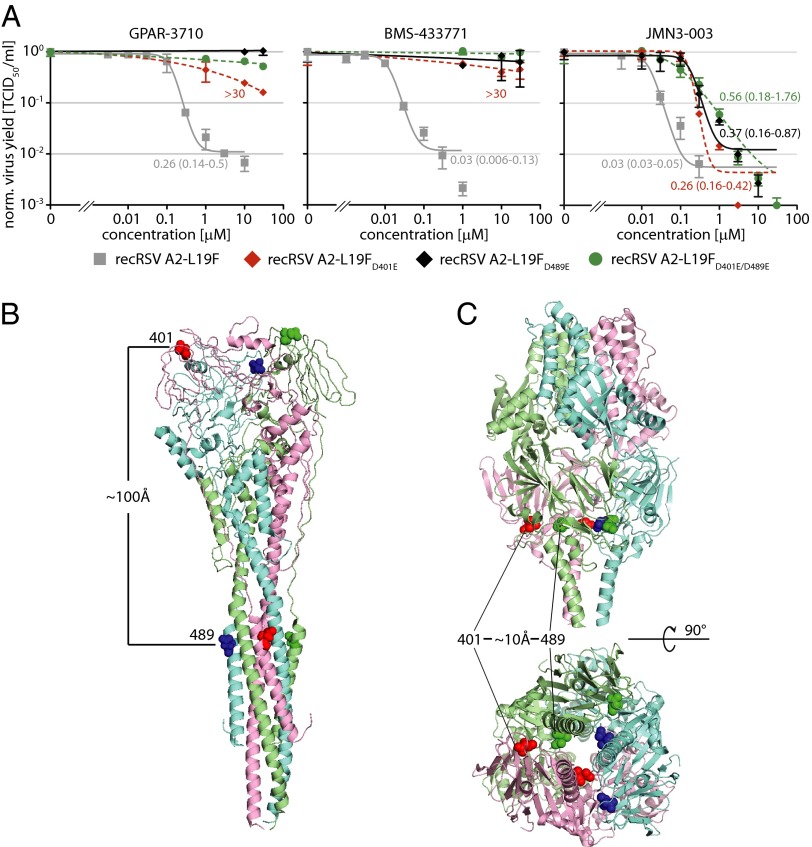
Previous anti-RSV drug discovery campaigns have yielded several structurally distinct, highly potent small-molecule entry inhibitor classes that reportedly likewise induced escape mutations in the F 392–401 and/or 486–489 microdomains (Table S1). Lead analogs of some of these inhibitor classes are at different stages of preclinical and clinical development. To quantify viral resistance, we generated dose–response curves for GPAR-3710 and, for comparison, a developmentally advanced RSV entry inhibitor, BMS-433771, against the three RSV recombinants (31). Mutations in either microdomain resulted in a more than 100-fold increased extrapolated EC90 concentration for either compound (Fig. 4A), confirming robust resistance.
Fig. 4.
Structural mapping of cross-resistance hot spots in prefusion and postfusion RSV F. (A) Dose–response curves of the four recovered RSV recombinants against GPAR-3710 and, for comparison, the clinically advanced RSV entry inhibitor BMS-433771 and broad spectrum RdRp blocker JMN3-003. Values are mean normalized cell-associated viral titers of three experiments ± SD; EC90 concentrations were calculated as in Fig. 1C when applicable. Highest concentration assessed, 30 μM. (B and C) Ribbon representations of RSV F in the postfusion (B; PDB ID code 3RRT) and prefusion (C; PDB ID code 4JHW) conformation, colored by monomer. Solid spheres represent for each monomer amino acid side chains at positions 401 and 489, respectively. Side views and, for prefusion F, a view from the viral envelope up, are shown.
Based on a biochemical target analysis, it was proposed that BMS-433771 populates a hydrophobic pocket in the HR-A triple helix that contains residue 489, preventing assembly of the 6HB fusion core during F refolding into its postfusion conformation (30). Surprisingly, however, both resistance domains map to opposing ends of the rod-like postfusion F structure, separated by ∼100 Å from each other (Fig. 4B). Recently, the structure of RSV F was solved also in the in the metastable prefusion state (26). When we localized the hotspots in this structure, we found residues 401 and 489 to be positioned in close proximity of each other (<10 Å) at the base of the head domain in prefusion F (Fig. 4C). The close proximity of individual resistance hotspots in prefusion F was not limited to escape mutants specifically identified through viral adaptation to GPAR-3710, because both the F 392–394 and 399–401 microdomains are likewise located in immediate proximity to residues 486–489 in prefusion F (Fig. S3).
Resistance Mutations Alter the Rate of F-Mediated Membrane Fusion.
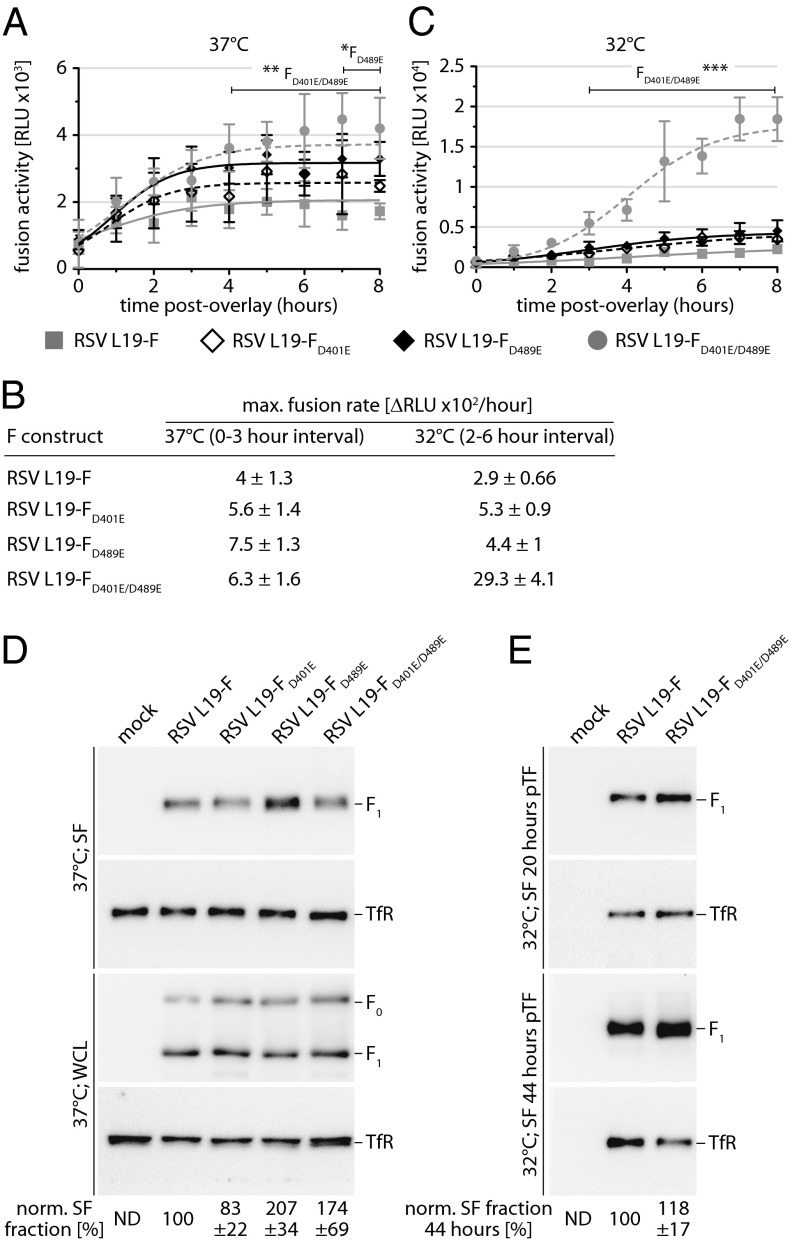
Prompted by proximity of both resistance hotspots in prefusion F and the accelerated growth rates of the RSV recombinants, we asked whether mutations in either microdomain affect prefusion F refolding rates. Using the kinetic cell-to-cell fusion assay, we assessed the rates of fusion pore formation mediated by the different F mutants. At physiological temperature, maximal fusion rates of all three F mutants were increased compared with that of standard F (Fig. 5 A and B). To fully appreciate the altered refolding kinetics of the mutated F variants, we determined fusion rates under reduced energy conditions (32 °C incubation temperature). Then, none of the individual mutations showed a statistically significant accelerating effect on fusion kinetics. In contrast, the FD401E/D489E double mutation significantly boosted the fusion rate at 32 °C, indicating a temperature-sensitive phenotype (Fig. 5 B and C).
Fig. 5.
Resistance mutations alter the F fusion kinetic. (A and C) Kinetic cell-to-cell fusion assays with transiently expressed F mutants, DSP-based luciferase reporter, and EnduRen live cells substrate. Fusion was followed at 37 °C (A) or 32 °C (C) by monitoring reconstituted DSP renilla luciferase activity in 30-min time intervals. Values are means of four replicates ± SD; *P < 0.05, **P < 0.01, ***P < 0.001. (B) Straight-line nonlinear fit regression modeling to calculate maximal F-induced fusion rates from datasets shown in A and C. Models are based on time intervals showing in first approximation linear signal increases, and numbers show best-fit slopes ± SEM. (D) Cell surface expression (SF) and whole-cell steady-state levels (WCL) of transiently expressed RSV F mutants after incubation of cells at 37 °C. Blots were probed with specific antibodies directed against RSV F (precursor F0 and cleaved F1 material is marked) or cellular transferrin receptor (TfR). Numbers denote mean densitometry quantitations of four experiments ± SD, all normalized for TfR and expressed relative to standard L19-F. Mock denotes cells transfected with vector DNA instead of F expression plasmid. (E) Cell surface expression of the L19-FD401E/D489E double mutant after incubation of cells at 32 °C. Proteins were harvested after 20 h and at steady state (44 h) after transfection. Blot development and densitometry quantifications as specified in A.
A densitometric analysis of whole-cell lysates and cell surface-expressed F material and immunoblotting demonstrated enhanced cell surface steady-state levels of the FD489E mutant compared with standard RSV L19-F (Fig. 5D). However, levels of the FD401E/D489E double mutant were slightly lower than those of FD489E, and intracellular transport rates of the double mutant and standard F remained essentially identical when cells were incubated at 32 °C (Fig. 5E).
These results suggest that higher bioactivity of the double mutant does not result from increased surface expression but indicate a synergistic effect of changes in each resistance hotspot on F bioactivity. To assess the broader applicability of this resistance model, we inversed the inhibitor/escape mutant combination by generating F-K394R and F-D489Y substitutions (reportedly mediating resistance to BMS-433771; Table S1) and testing resistance of these F constructs to inhibition by GPAR-3710. Each of these mutations caused a hyperfusogenic phenotype and mediated cross-resistance against GPAR-3710 (Fig. S4 A–D). Remarkably, also an F-F140I substitution increased F cell-to-cell fusion activity and resulted in resistance to GPAR-3710 (Fig. S4A). This mutation is located in the F fusion peptide region and was previously reported only in conjunction with resistance to BMS-433771. Our results suggest that these fusion peptide mutations phenocopy the effect of substitutions in the F 392–401 and 486–489 microdomains, resulting in cross-resistance to diverse entry inhibitors.
Effect of Resistance Mutations on Viral Pathogenicity.
To test whether a reduced structural stability of the mutated prefusion F constitutes the underlying mechanism for resistance, we adapted to RSV F a recently established class I viral envelope protein fusion-core assay that biochemically monitors the formation of the thermodynamically stable 6HB fusion core (39), which is indicative of fusion protein refolding into the postfusion conformation. Intact F trimers were natively extracted from gradient-purified viral particles, followed by gel fractionation under mildly denaturing, nonreducing conditions. Presence of the stable 6HB core in postfusion F complexes should be reflected by predominant migration of the extracted material as homotrimers, whereas metastable prefusion F trimers have a higher propensity to disintegrate. Thermal refolding of standard prefusion F into the postfusion conformation through brief heat shock of particles at increasing temperatures before extraction resulted in a gradual surge in trimeric material (Fig. S5). When we examined standard F and the three drug resistant mutants in this assay, the mutant trimers predominantly migrated as stable trimers, whereas standard F was mostly monomeric (Fig. 6A).
Fig. 6.
Stability and in vivo pathogenesis of resistant RSV recombinants. (A) Fusion core assay. F complexes were natively extracted from purified viral particles and fractionated through nonreducing TA-PAGE under mildly denaturing conditions. Immunoblots (IB) were probed with specific antibodies directed against the RSV F protein. The migration pattern of F monomers and fusion core-stabilized trimers is indicated: wt, standard L19F. Numbers below the graph show the relative F trimer:F monomer ratio based on densitometric quantification of signal intensities. (B) Thermal stability of resistant RSV virions. Recombinants were incubated at different temperatures for 24 h in the absence of target cells, followed by TCID50 titration of remaining infectivity. Values were normalized for aliquots immediately stored at –80 °C for 24 h, and represent means of three experiments ± SD. Asterisks denote statistical analysis of differences between test groups and standard recRSV A2-L19F by one-way ANOVA and Bonferroni’s multiple comparison posttest; *P < 0.05. (C) BALB/cJ mice were infected intranasally with 1 × 105 pfu of the indicated virus, lungs harvested 4 d p.i., and viral titers were determined through immunoplaque assays. Symbols represent individual animals of each group (n = 5), lines show means ± SEM. Cross bars denote statistical analysis of differences between test groups and standard recRSV A2-L19F by one-way ANOVA and Bonferroni’s multiple comparison posttest; *P < 0.05, NS, not significant. (D) Lungs of BALB/cJ mice infected as in C were harvested 8 d p.i. and processed for PAS staining. Representative airways are shown; mock denotes lungs of uninfected animals and bars (Scale bar: 100 μm.)
These findings spotlight that the resistance mutations reduce the structural stability of prefusion F complexes. To test whether this phenotype is mirrored by increased sensitivity of the recombinant virions to thermal inactivation, we subjected virus preparations to a 24-h incubation step at different temperatures in the absence of target cells. The individual mutant strains showed an intermediate but significant reduction in titers compared with standard recRSV after incubation at 32–39 °C (recRSV A2-L19FD401E) or 39 °C (recRSV A2-L19FD489E), respectively (Fig. 6B). Moreover, temperature sensitivity was most pronounced in the case of the recRSV A2-L19FD401E/D489E double mutant, because titers of this strain were significantly lower over the whole temperature range assessed. Recombinant RSV harboring the resistance mutations specifically reported for BMS-433771 (recRSV A2-L19FK394R and recRSV A2-L19FD489Y) showed analogous phenotypes when subjected to these assays (Fig. S6 A and B), underscoring the broader applicability of this escape mechanism from entry inhibitors.
Heightened thermosensitivity may coincide with lowered viral fitness in vivo, which could render drug-resistant variants clinically insignificant. We used an established mouse model for RSV infection (40) to assess pathogenicity of the mutant viruses. Only the two recombinants expressing single-mutant F variants that had emerged spontaneously during adaptation were subjected to this study. Lung titers of BALB/cJ mice infected with recRSV A2-L19FD489E were slightly reduced compared with animals exposed to standard recRSV (Fig. 6C). However, viral loads of animals infected with recRSV A2-L19FD401E remained unchanged.
The induction of extensive mucus production is one of the key features associated with RSV pathogenesis (41) and serves as an indicator for the severity of RSV disease in the mouse model (38, 42). When we infected animals with the two mutant recombinants and standard recRSV, the recRSV A2-L9FD489E mutant was only slightly mucogenic compared with mock-infected mice (Fig. 6D). In contrast, the recRSV A2-L19FD401E recombinant showed strong mucus induction at a level at least equivalent to that seen in lungs of animals infected with parental recRSV A2-L19F. Taken together, these results demonstrate that the individual F mutations, which each mediate robust resistance of recombinant RSV to diverse entry inhibitors, are not mandatorily associated with reduced viral pathogenesis in vivo.
Discussion
As a leading cause for infant hospitalization from viral respiratory disease, RSV has emerged in the past decade as a major target for the development of novel vaccines and therapeutics. Although formalin-inactivated RSV vaccines and subunit vaccines based on the G and F viral glycoproteins were associated in several past trials with disease enhancement (43–46), live attenuated vaccine candidates tested to date were—although safe in infants—poorly immunogenic, reflected by insufficient neutralizing antibody titers (47). Antiviral therapeutics may ameliorate disease in high-risk pediatric patients and possibly the elderly, especially because viral load in the early phase of clinical disease was found predictive for the severity of disease progression and the risk of life-threatening complications (16, 17).
Large-scale screening campaigns to identify novel therapeutic candidates against RSV were compromised thus far by the lack of appropriate reporter strains that were developed for robust automated drug discovery assays. Our study demonstrates the value of the recombinant RSV strain expressing renilla luciferase that we recently generated (32) for this task. Major advantages over conventional RSV-based assays explored for high-throughput campaigns are the broad dynamic range of the luciferase reporter; the availability of a full set of subinfection assays for MOA characterization that are genetically matched to the screening strain; the option to readily assess resistance in genetically controlled viral recombinants by using an efficient reverse genetics system; and the high pathogenicity of the reporter strain in the mouse model compared with standard laboratory RSV strains (40), opening a straightforward path toward small-animal efficacy testing of lead candidates.
By design, the high-throughput assay developed for our screen has a higher propensity to identify early and intermediate stage inhibitors of the viral life cycle (i.e., inhibitors of viral attachment, fusion, and viral polymerase activity) than blockers of viral assembly and egress, because the latter would act downstream of luciferase reporter expression. Consistent with this assumption, the protocol yielded as the most prominent hit candidate a previously unidentified RSV-specific virus entry inhibitor class when tested in a 10,000-compound proof-of-concept campaign. Characterization of the hit compound in infection and subinfection reporter assays and tracing of resistance to point mutations in the viral F protein confirmed interference with an F-mediated membrane merger as the underlying mechanism of anti-RSV activity. The RSV entry machinery emerges as highly susceptible to inhibition by small-molecule inhibitors, because structurally distinct lead compounds identified in several independent drug discovery campaigns all block membrane fusion (21–24). Possible reasons for this prevalence may include that entry inhibition poses lower demands on the compound than other inhibition strategies, because it does not mandate plasma membrane permeability of the inhibitor. In addition, paramyxovirus F proteins are comparably large in size and undergo—like other class I viral fusion proteins (29)—complex conformational rearrangements to mediate membrane merger. Combined, the result is the presentation of multiple druggable targets for small-molecule interference. In contrast to the F proteins of most members of the paramyxovirus family (48), RSV F is capable of mediating not only fusion but also viral attachment (19, 20), which may further broaden the spectrum of possible drug targets.
Five of the previously identified RSV entry inhibitors were synthetically developed to therapeutic candidate level and are at different stages of preclinical and clinical assessment (21–24, 31). Photoaffinity labeling assays implied physical binding of compounds representing two different inhibitor classes to F residue Y198 (21, 30), which is located in the HR-A domain and was proposed to reside in the immediate vicinity of HR-B residue D489 in a hydrophobic pocket in the final 6HB fusion core (illustrated in Fig. S7). In addition to these two classes, resistance hotspots were determined also for the remaining three scaffolds and, in analogy to our characterization of GPAR-3710, in all cases included F residues in the 400 and/or 489 microdomains (22, 24). Although recognized as surprising that distinct chemical inhibitor classes supposedly target the same F microdomain with high affinity (30), previous studies concluded that all of these diverse compounds prevent RSV entry through docking into the same hydrophobic cavity around residue Y198 in the central HR-A triple helix (21, 30), which emerges transiently during assembly of the 6HB structure. Curiously, none of the resistance mutations reported for any of these compounds maps to HR-A residues that surround Y198 and are implicated in forming the proposed target cavity, or any other position in the HR-A domain. However, this hypothesis was developed before the prefusion RSV F structure was solved, and previous work did not consider possible effects of resistance mutations on the conformational stability of prefusion F or the kinetics of viral entry.
Although biochemical data reveal direct binding of two inhibitor classes to F residue Y198 (21, 30), we propose based on three major lines of evidence—structural insight, biochemical characterization, and functional data—that unprecedented broad cross-resistance of RSV against multiple structurally diverse entry inhibitors is based on indirect escape. First, compound docking into postfusion RSV F structures failed to provide a mechanistic explanation for the hotspot around F residues 392–401 in resistance (30). We demonstrate that the 400 and 489 microdomains are located at opposing ends of the postfusion F structure, but are posited in close proximity to each other at the intersection of stalk and head domain in prefusion F. Interestingly, several studies investigating related paramyxovirus F proteins have identified this network of noncovalent interactions between prefusion F stalk and head as a major determinant for controlling the conformational stability of the trimer (49, 50). We have furthermore demonstrated that point mutations in this region confer resistance against a small-molecule entry inhibitor of MeV that we have developed (51). Second, point mutations in either of the two resistance hotspots reduced the structural stability of the prefusion RSV F trimer in biochemical assays and resulted in enhanced spontaneous viral inactivation rates in the absence of target cells. Third, membrane fusion rates of resistant F proteins were enhanced compared with those of the parent trimer, indicating accelerated refolding of the complex from the prefusion into the stable postfusion conformation.
Taken together, these observations spotlight an effective mechanism of secondary RSV resistance, in which escape mutations accumulate in F microdomains that govern the structural stability of the prefusion complex. Refolding rates of these conformationally destabilized mutant F trimers are enhanced, resulting in a hyperfusogenic phenotype and, possibly, a narrowed window of opportunity for small-molecule docking and interference with F trimer rearrangements leading to fusion pore formation. Different escape pathways were also identified for HIV resistance to the peptidic entry inhibitor Fuzeon (52, 53). However, Fuzeon escape did not coincide with resistance to other entry inhibitors (54). Broad cross-resistance against a structurally highly diverse panel of entry inhibitors appears unique to RSV F and may amount to a substantial obstacle in the clinic.
Despite the reduction in prefusion F conformational stability, fitness of one of the resistant recRSVs was unchanged compared with the standard recRSV in a mouse pathogenesis model, whereas the other was appreciably attenuated. Recently established, this mouse model using the recRSV A2-L19F strain exhibits higher lung viral loads, more airway mucus, and more severe respiratory distress than both the parental A2 and line 19 strains (38, 42), recapitulating key clinical parameters of infant RSV bronchiolitis (38, 40, 55). Although it is not determined yet whether pathogenicity of a resistant RSV-FD401E would be equally unchanged in the human host, efficient replication in particular of this recombinant in the mouse model raises concern that resistance mutations in the hotspot around F residue 400 could become prevalent in circulating RSV strains should any of these entry inhibitor classes experience broad clinical use.
Our data thus indicate that RSV entry inhibitors currently considered for clinical use are at risk to lose therapeutic benefit in the clinic because of rapidly emerging viral resistance. We propose that future RSV drug discovery campaigns are better directed at inhibiting postentry steps of viral replication or be proactively designed to conceptually circumvent broad cross-resistance against entry inhibitors. For instance, using the resistant recRSV A2-L19FD489E-renilla virus described in this study as the screening agent should have a high propensity to yield hit candidates that either act postentry or, if mechanistically possible, block viral entry without being compromised by cross-resistance.
Materials and Methods
Cell Culture, Transfection, and Virus Stocks.
All cell lines were maintained in DMEM supplemented with 7.5% (vol/vol) FBS at 37 °C and 5% CO2. Baby hamster kidney (BHK21) cells stably expressing T7 polymerase (BSR-T7/5 cells) were incubated at every third passage in the presence of 500 μg/mL G-418 (Geneticin). Cell transfections were carried out by using Lipofectamine 2000 (Invitrogen) or GeneJuice reagent (EMD Millipore). Standard RSV virus stocks were prepared by infecting HEp-2 cells (ATCC HB-8065) at a multiplicity of infection (MOI) of 0.01 pfu per cell at 37 °C, followed by incubation at 32 °C for 7–9 d. Cell-associated progeny virus was released through one freeze/thaw cycle, and titers were determined by TCID50 titration or immunoplaque assay as described (56). RecRSV-ren stocks were purified through ultracentrifugation through a 20%/60% (wt/vol) one-step sucrose gradient (90 min at 100,000 × g, 4 °C). The virus-containing fraction was diluted in 1 mM Tris at pH 7.2, 100 mM NaCl, and 10 mM EDTA (TNE buffer), pelleted at 60,000 × g for 30 min at 4 °C, and resuspended in TNE buffer.
Generation and Recovery of Recombinant RSV.
Point mutations were introduced through directed mutagenesis into a shuttle vector containing the RSV L19-F ORF, followed by transfer of the modified SacII/SalI line19 F fragment into pSynkRSV A2-L19F-renilla or pSynkRSV A2-L19F-mKate2 (32). Recombinants were recovered as described (32) and subjected to RT-PCR and cDNA sequencing for confirmation of specific point mutations.
Compounds.
All compounds were dissolved in dimethyl sulfoxide (DMSO) and stored at −80 °C. Sourced compounds were obtained form Ambienter (previously described pan-myxovirus inhibitor 09167; ref. 57) and Vitas-M Laboratory or MolPort (GPAR-3710 stocks). The screening library was obtained from ChemDiv. For screening, 2 × 104 (96-well plate format) or 6 × 103 (384-well plate format) HEp-2 cells per well were seeded into solid-wall microtiter plates. Test articles dissolved in DMSO were added at 5 μM final concentration (final DMSO content was below 0.1% vol/vol). As internal reference, four wells each on each plate were treated with the pan-myxovirus inhibitor JMN3-003 (final concentration 1 μM) or vehicle (DMSO) only. Cells were infected with recRSV A2-L19F-ren (MOI = 0.2 pfu per cell), and renilla luciferase activities were quantified in a Synergy H1 (BioTek) multimode microplate reader after 44- to 48-h incubation.
HTS Data Analysis.
Raw data sets were automatically reformatted and imported into the cellHTS2 application package (58, 59). Data were analyzed according to the plate median method: Each value was normalized to the median value for all compound wells of the plate, and normalized values were scaled to the median absolute deviation of the plate. The SciFinder database package (American Chemical Society) was used to query chemical databases with hit candidate structures to evaluate known bioactivities of analogs, commercial availability, and free intellectual property space.
Dose–Response Curves, Efficacy, and Cytotoxicity.
Cells infected (MOI = 0.05 pfu per cell) with recRSV A2-L19F, recRSV A2-L19F-ren, recRSV A2-L19F-mKate2, or GPAR-3710-resistant variants thereof were incubated in the presence of serial dilutions of compound for 44 h, followed by titration of cell-associated progeny particles or quantification of reporter expression as specified. If possible, 50 or 90 percent effective concentrations (EC50 or EC90 values, respectively) were calculated based on four-parameter variable-slope nonlinear regression modeling of mean values of at least three experiments. To quantify the effect of compound on cell metabolic activity, cells were incubated in the presence of serial compound dilutions (30 μM highest) for 44 h, then subjected to a nonradioactive cytotoxicity assay (CytoTox 96; Promega) according to the manufacturer’s instructions. Assay values were normalized to vehicle (DMSO) controls according to % toxicity = 100 – 100 × (sample − reference)/(vehicle − reference).
Time-of-Addition Assays.
HEp-2 cells were spin-inoculated (1,000 × g; 30 min; 4 °C; MOI = 10 pfu/mL) with purified recRSV A2-L19F-ren. Compound was added at the specified times preinfection or postinfection, and luciferase activities determined 26 h after infection. Reference samples received volume equivalents of vehicle (DMSO).
Minireplicon Reporter Assay.
Based on a described pT7-RSV-luciferase minigenome reporter (36), an RSV minigenome construct was generated under the control of the constitutive RNA pol I promoter (pHH-RSV-repl-firefly). Huh-7 cells were cotransfected with this plasmid and plasmids pRSV-L, pRSV-M2-1, pRSV-N, and pRSV-P under CMV promoter control. Compounds GPAR-3710 or JMN3-003 were added in serial dilutions, luciferase reporter activities were determined 40 h after transfection, and EC50 concentrations were calculated as above if possible.
End-Point Cell-to-Cell Fusion Assay.
A dual split-protein cell content mixing assay was used to quantify the extent of cell-to-cell fusion mediated by RSV F. For transfection, 293T cells received plasmid DNA encoding eGFP-renilla luciferase dual-split fusion proteins DSP1–7 or DSP8–11 (37), respectively. One cell population also received plasmid DNA encoding RSV L19F or, for control, MeV F and H proteins (35). Cell populations were mixed at an equal ratio 4 h after transfection and incubated in the presence of the specified amounts of GPAR-3710 for 26 h. Activity of reconstituted luciferase were quantified after loading of cells with 10 μM ViviRen (Promega) for 30 min.
Virus Entry Kinetics Assay.
To determine infection kinetics, 293T cells transfected with the plasmids encoding the DSP1–7 or DSP8–11 (37), respectively, were mixed at equal ratios, preloaded with EnduRen life cell substrate as described (60), and spin-inoculated with recRSV A2-L19F (1,000 × g; 30 min; 4 °C; MOI = 6 pfu per cell) in the presence of GPAR-3710 or DMSO. Activity of reconstituted luciferase was recorded at the specified time points.
Kinetic Cell-to-Cell Fusion Assay.
The DSP1-7 and DSP8-11 expression plasmids were transfected into 293T cells, and transfected cells were detached and reseeded in an equal ratio. Cells were then transfected with standard or mutant RSV L19-F-encoding plasmids, loaded with EnduRen luciferase substrate as above, and incubated at 32 °C or 37 °C. Luciferase activity was recorded at the specified time points.
Microscopy.
Fluorescence microphotographs were taken on a Zeiss Axio Observer D.1 inverted microscope at a magnification of 200×. For phase-contrast microphotographs, a Nikon Diaphot 200 inverted microscope was used at a magnification of 200×.
Virus Adaptation.
HEp-2 cells were infected with recRSV A2-L19F-mKate2 at an MOI of 0.1 pfu per cell and incubated in the presence 0.1 μM GPAR-3710. When extensive red fluorescence emerged, fresh cell monolayers were reinfected with 10-fold diluted cell-associated virions in the presence of increasing compound concentrations. Total RNA was extracted (RNeasy purification kit; Qiagen) from individually adapted clones when GPAR-3710 concentrations of 30 μM were tolerated, cDNAs were generated by using random hexamer primers, and the F-encoding ORF were amplified and subjected to DNA sequencing. Candidate mutations were rebuilt in RSV-L19F expression plasmids and subjected to cell-to-cell fusion assays in the presence of compound. Selected confirmed mutations were rebuilt in the pSynkRSV A2-L19F-mKate2 plasmid background and the corresponding recombinants were recovered.
Surface Biotinylation, SDS/PAGE, and Immunoblotting.
Protein surface expression was determined as described (61) with the following modifications: 293T cells (8× 105 per well in a six-well plate format) were transfected with 2 μg of plasmid DNA encoding the specified RSV F construct. Washed cells were biotinylated with 0.5 mg/mL sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate (Pierce), quenched, and subjected to precipitation by using immobilized streptavidin (GE Healthcare) after lysis in RIPA buffer (1% sodium deoxycholate, 1% Nonidet P-40, 150 mM NaCl, 50 mM Tris⋅Cl at pH 7.2, 10 mM EDTA, 50 mM sodiumfluoride, and protease inhibitors). Washed precipitates were fractioned by SDS/PAGE, blotted onto PVDF membranes (GE Healthcare), and F protein material was immunostained by using the motavizumab monoclonal antibody. Immunoblots were developed by using a ChemiDoc digital imaging system (Bio-Rad) and subjected to densitometry quantification by using the Image Lab software package (Bio-Rad).
Fusion Core Assay.
Standard and mutant recRSV A2-L19F were grown at 32 °C. Cell-free viral particles were harvested, pelleted, resuspended, and purified by ultracentrifugation through a 20%/60% (wt/vol) one-step sucrose gradient, and subjected to cold extraction of native plasma membrane proteins by using Native Sample Buffer [100 mM Tris⋅Cl at pH 8.6, 10% (vol/vol) glycerol, 0.0025% Bromophenol Blue, 0.1% digitonin, and 25 mM iodoacetamide], and clearance centrifugation (20,000 × g; 15 min; 4 °C). Extracts were mixed with Laemmli sample buffer with 0.5% SDS and fractionated on 3–8% (wt/vol) NuPAGE Tris-Acetate gradient gels (Life Technologies), followed by immunoblotting as above.
Temperature Sensitivity Assay.
Standard and resistant recRSV A2-L19F strains as specified were divided into equal aliquots, which either were frozen at −80 °C or incubated at the indicated temperature for 24 h followed by freezing, and remaining virus titers were determined by TCID50 titration.
In Vivo Infection.
BALB/cJ mice (Jackson Laboratories) were anesthetized by i.m. injection of a ketamine-xylazine solution and infected intranasally with 1 × 105 pfu of recRSV A2-L19F, recRSV A2-L19FD401E, or recRSV A2-L19FD489E. All animal procedures were performed according to the guidelines of the Emory University Institutional Animal Care and Use Committee.
Lung Titers.
Mice were euthanized day 4 after infection (p.i.), and the left lung lobe was extracted, weighed, and homogenized by using a BeadBeater (Biospec Products). Homogenates were serially diluted, transferred to HEp-2 cells, and cells were overlaid 1 h p.i. with minimum essential medium (MEM) containing 10% (vol/vol) FBS, penicillin G, streptomycin sulfate, amphotericin B solution, and 0.75% methylcellulose. Six days p.i., cells were fixed with methanol and plaques were visualized by immunodetection as described (40, 42).
Mucin Expression.
Mice were euthanized with fatal-plus 8 d p.i. (40, 42), and heart-lung tissue was harvested and fixed in 10% formalin. Lung tissue sections embedded in paraffin blocks were stained with periodic acid-Schiff (PAS) stain to visualize mucin expression. PAS-stained slides were digitally scanned by using a Zeiss Mirax Midi microscope (Carl Zeiss Microimaging).
Statistical Analysis.
To determine active concentrations from dose–response curves, four parameter variable slope regression modeling was performed by using the Prism (GraphPad) software package. Results were expressed as 50% or 90% inhibitory concentrations with 95% asymmetrical confidence intervals. Statistical significance of differences between sample groups were assessed by one-way or two-way analysis of variance (ANOVA) in combination with Bonferroni multiple comparison posttests as specified in the figure legends. Experimental uncertainties are identified by error bars, representing SD or SEM as specified.
Supplementary Material
Acknowledgments
We thank N. Kondo and Z. Matsuda for the DSP cell lines and DSP expression plasmids, J. Crowe for the motavizumab monoclonal antibodies, C. Shoffeitt for assistance with histology, and A. L. Hammond for critical reading of the manuscript. This work was supported, in part, by Public Health Service Grants AI087798 and AI095227 (to M.L.M.), and AI071002 and HD079327 from the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases and NIH/National Institute of Child Health and Human Development, respectively (to R.K.P.).
Footnotes
Conflict of interest statement: R.K.P. and M.L.M. are inventors on a disclosure filing describing the use of recRSV A2-L19FD489E-luciferase (renilla or firefly) for drug discovery. Distribution of these RSV strains and the pHH-RSV-repl-firefly reporter plasmid is regulated by a material transfer agreement from Georgia State University. M.L.M. and Emory University are entitled to licensing fees derived from various agreements Emory University has entered into related to products used in the research described in this paper. This study could affect his personal financial status. The terms of this agreement have been reviewed and approved by Emory University in accordance with its conflict of interest policies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405198111/-/DCSupplemental.
References
- 1.Thompson WW, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 2.Elliot AJ, Fleming DM. Influenza and respiratory syncytial virus in the elderly. Expert Rev Vaccines. 2008;7(2):249–258. doi: 10.1586/14760584.7.2.249. [DOI] [PubMed] [Google Scholar]
- 3.Ebbert JO, Limper AH. Respiratory syncytial virus pneumonitis in immunocompromised adults: Clinical features and outcome. Respiration. 2005;72(3):263–269. doi: 10.1159/000085367. [DOI] [PubMed] [Google Scholar]
- 4.Collins PL, Crowe JE., Jr 2007. Respiratory syncytial virus and metapneumoviruses. Fields Virology, eds Knipe DM, Howley PM (Lippincott Williams & Wilkins, Philadelphia), 5th Ed, Vol 2, pp 1601–1645.
- 5.Mahadevia PJ, Masaquel AS, Polak MJ, Weiner LB. Cost utility of palivizumab prophylaxis among pre-term infants in the United States: A national policy perspective. J Med Econ. 2012;15(5):987–996. doi: 10.3111/13696998.2012.690013. [DOI] [PubMed] [Google Scholar]
- 6.Aherne W, Bird T, Court SD, Gardner PS, McQuillin J. Pathological changes in virus infections of the lower respiratory tract in children. J Clin Pathol. 1970;23(1):7–18. doi: 10.1136/jcp.23.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lugo RA, Nahata MC. Pathogenesis and treatment of bronchiolitis. Clin Pharm. 1993;12(2):95–116. [PubMed] [Google Scholar]
- 8.Anderson LJ, Parker RA, Strikas RL. Association between respiratory syncytial virus outbreaks and lower respiratory tract deaths of infants and young children. J Infect Dis. 1990;161(4):640–646. doi: 10.1093/infdis/161.4.640. [DOI] [PubMed] [Google Scholar]
- 9.Hall CB, et al. American Academy of Pediatrics Committee on Infectious Diseases: Use of ribavirin in the treatment of respiratory syncytial virus infection. Pediatrics. 1993;92(3):501–504. [PubMed] [Google Scholar]
- 10.Broor S, et al. A prospective three-year cohort study of the epidemiology and virology of acute respiratory infections of children in rural India. PLoS ONE. 2007;2(6):e491. doi: 10.1371/journal.pone.0000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahadevia PJ, Makari D, Masaquel A. Methodological concerns regarding cost-effectiveness analysis of palivizumab in Florida Medicaid. Arch Pediatr Adolesc Med. 2012;166(10):968–969, author reply 969–970. doi: 10.1001/archpediatrics.2012.1591. [DOI] [PubMed] [Google Scholar]
- 12.Kamal-Bahl S, Doshi J, Campbell J. Economic analyses of respiratory syncytial virus immunoprophylaxis in high-risk infants: A systematic review. Arch Pediatr Adolesc Med. 2002;156(10):1034–1041. doi: 10.1001/archpedi.156.10.1034. [DOI] [PubMed] [Google Scholar]
- 13.Hampp C, Kauf TL, Saidi AS, Winterstein AG. Cost-effectiveness of respiratory syncytial virus prophylaxis in various indications. Arch Pediatr Adolesc Med. 2011;165(6):498–505. doi: 10.1001/archpediatrics.2010.298. [DOI] [PubMed] [Google Scholar]
- 14.de Vries RD, Mesman AW, Geijtenbeek TB, Duprex WP, de Swart RL. The pathogenesis of measles. Curr Opin Virol. 2012;2(3):248–255. doi: 10.1016/j.coviro.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Griffin DE. Measles virus-induced suppression of immune responses. Immunol Rev. 2010;236:176–189. doi: 10.1111/j.1600-065X.2010.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeVincenzo JP, El Saleeby CM, Bush AJ. Respiratory syncytial virus load predicts disease severity in previously healthy infants. J Infect Dis. 2005;191(11):1861–1868. doi: 10.1086/430008. [DOI] [PubMed] [Google Scholar]
- 17.El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis. 2011;204(7):996–1002. doi: 10.1093/infdis/jir494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teng MN, Whitehead SS, Collins PL. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology. 2001;289(2):283–296. doi: 10.1006/viro.2001.1138. [DOI] [PubMed] [Google Scholar]
- 19.Techaarpornkul S, Barretto N, Peeples ME. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol. 2001;75(15):6825–6834. doi: 10.1128/JVI.75.15.6825-6834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Techaarpornkul S, Collins PL, Peeples ME. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology. 2002;294(2):296–304. doi: 10.1006/viro.2001.1340. [DOI] [PubMed] [Google Scholar]
- 21.Roymans D, et al. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc Natl Acad Sci USA. 2010;107(1):308–313. doi: 10.1073/pnas.0910108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas JL, et al. Small molecules VP-14637 and JNJ-2408068 inhibit respiratory syncytial virus fusion by similar mechanisms. Antimicrob Agents Chemother. 2005;49(6):2460–2466. doi: 10.1128/AAC.49.6.2460-2466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cianci C, et al. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob Agents Chemother. 2004;48(2):413–422. doi: 10.1128/AAC.48.2.413-422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton CJ, et al. Structural characterization of respiratory syncytial virus fusion inhibitor escape mutants: Homology model of the F protein and a syncytium formation assay. Virology. 2003;311(2):275–288. doi: 10.1016/s0042-6822(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 25.Berkhout B, Eggink D, Sanders RW. Is there a future for antiviral fusion inhibitors? Curr Opin Virol. 2012;2(1):50–59. doi: 10.1016/j.coviro.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 26.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342(6158):592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85(15):7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson KA, et al. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci USA. 2011;108(23):9619–9624. doi: 10.1073/pnas.1106536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plemper RK. Cell entry of enveloped viruses. Curr Opin Virol. 2011;1(2):92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cianci C, et al. Targeting a binding pocket within the trimer-of-hairpins: Small-molecule inhibition of viral fusion. Proc Natl Acad Sci USA. 2004;101(42):15046–15051. doi: 10.1073/pnas.0406696101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roymans D, Koul A. Treatment of respiratory syncytial virus infection: Past, present and future. In: Resch B, editor. Human Respiratory Syncytial Virus Infection. Croatia: InTech, Rijeka; 2011. pp. 197–215. [Google Scholar]
- 32.Hotard AL, et al. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology. 2012;434(1):129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan D, et al. Dual myxovirus screen identifies a small-molecule agonist of the host antiviral response. J Virol. 2013;87(20):11076–11087. doi: 10.1128/JVI.01425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumm SA, et al. Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS ONE. 2011;6(5):e20069. doi: 10.1371/journal.pone.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brindley MA, Takeda M, Plattet P, Plemper RK. Triggering the measles virus membrane fusion machinery. Proc Natl Acad Sci USA. 2012;109(44):E3018–E3027. doi: 10.1073/pnas.1210925109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dochow M, Krumm SA, Crowe JE, Jr, Moore ML, Plemper RK. Independent structural domains in paramyxovirus polymerase protein. J Biol Chem. 2012;287(9):6878–6891. doi: 10.1074/jbc.M111.325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondo N, Miyauchi K, Matsuda Z. Monitoring viral-mediated membrane fusion using fluorescent reporter methods. Curr Protoc Cell Biol. 2011;Chapter 26:Unit 26.29. doi: 10.1002/0471143030.cb2609s50. [DOI] [PubMed] [Google Scholar]
- 38.Moore ML, et al. A chimeric A2 strain of respiratory syncytial virus (RSV) with the fusion protein of RSV strain line 19 exhibits enhanced viral load, mucus, and airway dysfunction. J Virol. 2009;83(9):4185–4194. doi: 10.1128/JVI.01853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Côté M, Zheng YM, Liu SL. Receptor binding and low pH coactivate oncogenic retrovirus envelope-mediated fusion. J Virol. 2009;83(22):11447–11455. doi: 10.1128/JVI.00748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stokes KL, et al. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol. 2011;85(12):5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JE, Gonzales RA, Olson SJ, Wright PF, Graham BS. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod Pathol. 2007;20(1):108–119. doi: 10.1038/modpathol.3800725. [DOI] [PubMed] [Google Scholar]
- 42.Lee S, et al. Vaccine-elicited CD8+ T cells protect against respiratory syncytial virus strain A2-line19F-induced pathogenesis in BALB/c mice. J Virol. 2012;86(23):13016–13024. doi: 10.1128/JVI.01770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancock GE, et al. Generation of atypical pulmonary inflammatory responses in BALB/c mice after immunization with the native attachment (G) glycoprotein of respiratory syncytial virus. J Virol. 1996;70(11):7783–7791. doi: 10.1128/jvi.70.11.7783-7791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy BR, Sotnikov AV, Lawrence LA, Banks SM, Prince GA. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3-6 months after immunization. Vaccine. 1990;8(5):497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
- 45.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol. 1969;89(4):405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 46.Kim HW, et al. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89(4):422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 47.Collins PL, Melero JA. Progress in understanding and controlling respiratory syncytial virus: Still crazy after all these years. Virus Res. 2011;162(1–2):80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamb RA, Parks GD. 2007. Paramyxoviridae: The viruses and their replication. Fields Virology, eds Knipe DM, Howley PM (Lippincott Williams & Wilkins, Philadelphia), 5th Ed, Vol 1, pp 1449–1496.
- 49.Lee JK, Prussia A, Snyder JP, Plemper RK. Reversible inhibition of the fusion activity of measles virus F protein by an engineered intersubunit disulfide bridge. J Virol. 2007;81(16):8821–8826. doi: 10.1128/JVI.00754-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439(7072):38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doyle J, et al. Two domains that control prefusion stability and transport competence of the measles virus fusion protein. J Virol. 2006;80(3):1524–1536. doi: 10.1128/JVI.80.3.1524-1536.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eggink D, et al. Detailed mechanistic insights into HIV-1 sensitivity to three generations of fusion inhibitors. J Biol Chem. 2009;284(39):26941–26950. doi: 10.1074/jbc.M109.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baldwin CE, et al. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J Virol. 2004;78(22):12428–12437. doi: 10.1128/JVI.78.22.12428-12437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reeves JD, et al. Enfuvirtide resistance mutations: Impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J Virol. 2005;79(8):4991–4999. doi: 10.1128/JVI.79.8.4991-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lukacs NW, et al. Differential immune responses and pulmonary pathophysiology are induced by two different strains of respiratory syncytial virus. Am J Pathol. 2006;169(3):977–986. doi: 10.2353/ajpath.2006.051055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radecke F, et al. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14(23):5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore TW, et al. Synthesis and metabolic studies of host directed inhibitors for anti viral therapy. ACS Med Chem Lett. 2013;4(8):762–767. doi: 10.1021/ml400166b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boutros M, Brás LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7(7):R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelz O, Gilsdorf M, Boutros M. web cellHTS2: A web-application for the analysis of high-throughput screening data. BMC Bioinformatics. 2010;11:185. doi: 10.1186/1471-2105-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brindley MA, et al. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J Virol. 2013;87(21):11693–11703. doi: 10.1128/JVI.01945-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plemper RK, Compans RW. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J Virol. 2003;77(7):4181–4190. doi: 10.1128/JVI.77.7.4181-4190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.