Significance
Hundreds of microRNAs become dramatically elevated in the plasma or serum of acetaminophen (APAP) overdose patients and then recover back toward normal during successful treatment with the APAP antidote, N-acetyl cysteine (NAC). Importantly, the elevation of these circulating miRNAs can precede the rise in the standard biomarker, alanine aminotransferase (ALT), and the recovery of these miRNAs during NAC treatment is more rapid than ALT. We identify a set of 11 miRNAs whose profiles and dynamics in circulation during NAC treatment can discriminate APAP hepatotoxicity from another common hepatotoxic condition, ischemic hepatitis. These findings suggest that miRNAs are sensitive diagnostic and prognostic biomarkers for liver injury with broad potential for use in monitoring drug-induced liver injury in clinical and research contexts.
Keywords: hsa-miR-122-5p, hsa-miR-3646-3p, hsa-miR-412
Abstract
We have identified, by quantitative real-time PCR, hundreds of miRNAs that are dramatically elevated in the plasma or serum of acetaminophen (APAP) overdose patients. Most of these circulating microRNAs decrease toward normal levels during treatment with N-acetyl cysteine (NAC). We identified a set of 11 miRNAs whose profiles and dynamics in the circulation during NAC treatment can discriminate APAP hepatotoxicity from ischemic hepatitis. The elevation of certain miRNAs can precede the dramatic rise in the standard biomarker, alanine aminotransferase (ALT), and these miRNAs also respond more rapidly than ALT to successful treatment. Our results suggest that miRNAs can serve as sensitive diagnostic and prognostic clinical tools for severe liver injury and could be useful for monitoring drug-induced liver injury during drug discovery.
Acetaminophen (APAP) overdose (OD) is the major cause of acute liver failure and death due to analgesics in the developed world (1–3), with over 500 deaths per year in the United States (3, 4). APAP misuse results in over 78,000 emergency department (ED) visits annually, at a cost of more than $86 million/year (5).
Patients who present with a report of APAP ingestion are assessed for treatment with N-acetyl cysteine (NAC) by measuring serum APAP concentrations and liver function tests, including alanine aminotransferase (ALT) activity (4). There are limits to the utility of the current biomarker, ALT, in assessing the status of potentially APAP-poisoned patients. For example, ALT may take greater than 72 h to reach peak activity in blood (6), and elevated ALT activity is not specific for APAP hepatotoxicity. New biomarkers could serve as more sensitive and specific signatures to predict hepatotoxicity following APAP overdose and to distinguish APAP hepatotoxicity from other causes of liver disease. Ideal biomarkers should identify liver damage at an early stage, accelerating appropriate treatment.
MicroRNAs (miRNAs) are short (∼22 nt in length) regulatory RNAs that control gene expression posttranscriptionally (7, 8). The human genome contains more than 1,000 genes encoding distinct miRNAs whose levels in a biological sample can be quantified with great sensitivity and precision using quantitative real-time PCR (qRT-PCR) (9). Many miRNAs are expressed tissue-specifically or enriched in certain cell types, with the expression pattern providing signatures for the physiological or pathological status of specific cells and tissues (10, 11). Importantly, miRNAs can be exported from cells and are detectable in stable complexes extracellularly, in blood, serum, or plasma (12).
There is ample evidence that circulating extracellular miRNAs in blood can serve as biomarkers for internal organ physiology and pathology (11, 13, 14). Increased levels of miR-122 and miR-192 were found in the plasma of APAP-overdosed (300 mg/kg) mice (15). A number of additional miRNAs (miR-135a, miR-466g, miR-574-5p, and miR-1196) were elevated in the plasma of mice with higher APAP overdoses (500 mg/kg) (13). Starkey et al. (16) assayed the levels of four miRNAs in human plasma and found that miR-122 and miR-192 were elevated in APAP hepatotoxicity.
We evaluated the use of circulating extracellular miRNAs as biomarkers for diagnosing APAP poisoning and in discriminating APAP hepatotoxicity from ischemic hepatitis, a common form of dramatic liver injury caused by reduced blood flow to the liver. We profiled the levels of 372 extracellular miRNAs in plasma of non–APAP-exposed, healthy individuals and in serum or plasma of patients with APAP poisoning or ischemic hepatitis and used a computational approach to study the effect of liver injury on circulating miRNA profiles. We identified many miRNAs that change dramatically in hepatotoxic conditions, including a set of 11 circulating miRNAs whose profiles and dynamics in patients undergoing NAC treatment discriminated APAP hepatotoxicity from ischemic hepatitis. We also show that the elevation of certain miRNA biomarkers for APAP toxicity can precede a detectable rise in the standard protein biomarker, ALT, as reported previously in mice (15). Our results suggest that miRNAs can serve as sensitive diagnostic and prognostic clinical tools for APAP overdose and, possibly, other hepatotoxic conditions.
Results and Discussion
Circulating miRNA Profiles Provide a Measure of Liver Toxicity in Humans.
In this study we clustered 49 patients (Dataset S1, Tab S1) based solely on their miRNA profiles (for 221 miRNAs) from the first available blood draw, disregarding their ALT activity, type of injury (APAP or ischemic hepatitis), and patient history. Both serum and plasma were used in this analysis. Minimal difference in using either of these sources has been shown (17). In our hands, profiles of miRNA from serum or plasma from the same patient showed high correlation (Fig. S1), indicating that they can be compared directly. Of note, the rigorous centrifugation process (Materials and Methods) reduces contamination by miRNA-rich platelets (18).
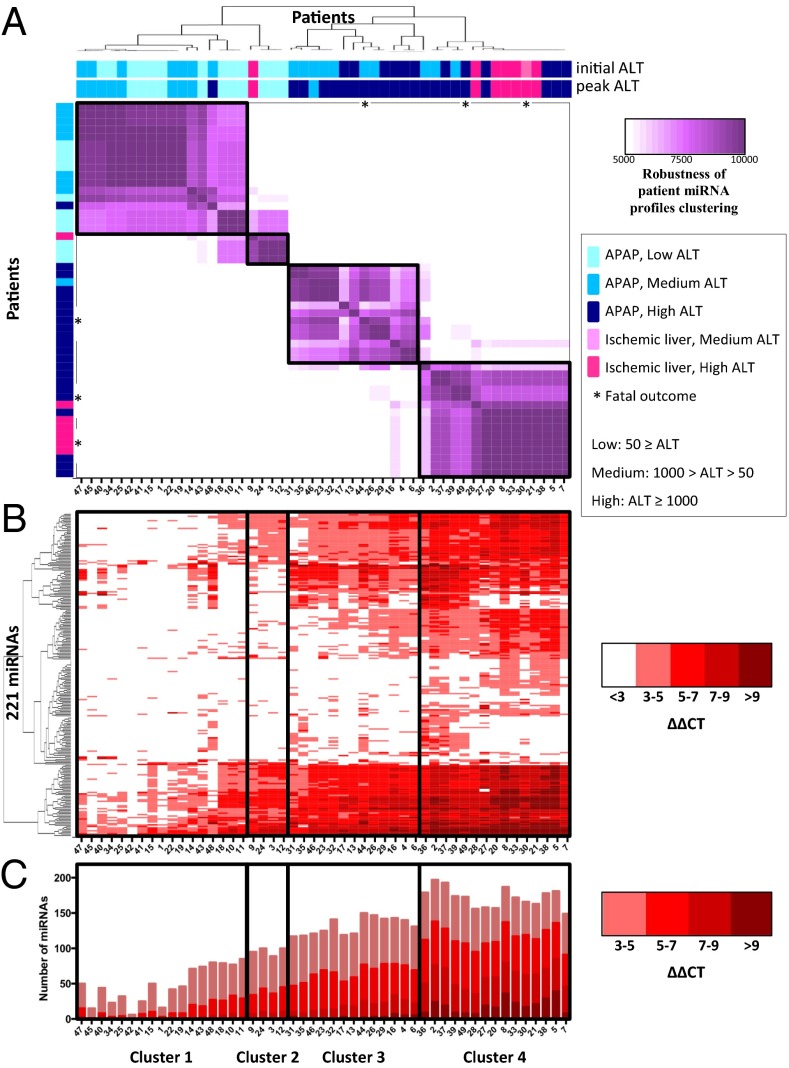
Despite the heterogeneity of the patient population and the variance in the timing of sample collection, we were able to apply a two-level clustering approach (Materials and Methods) that provides a high level of confidence in the consistency of the clusters obtained. Fig. 1A shows the results of this approach, where we identified four clusters. The source of the sample (i.e., plasma or serum) did not bias the clustering (Dataset S1, Tab S1). Although the clustering was based only on the miRNA profiles, we observed that these clusters, interestingly, correlate with peak ALT activity: patients in clusters 1 and 2 have low (ALT ≤ 50) and medium (50 < ALT < 1,000) peak ALT activity, and patients in clusters 3 and 4 have high (ALT ≥ 1,000) peak ALT activity. The last three patients in cluster 1 showed high similarity to patients in cluster 2.
Fig. 1.
Patient clustering based on earliest miRNA profiles. (A) A clustering of 49 patients (designated by patient ID, are ordered in rows and columns, from left to right, top to bottom) was generated based on miRNA profiles from the earliest available sample (within 2 d of hospital admittance). Each patient is designated according to APAP overdose or ischemic hepatitis, initial ALT, peak ALT during hospital stay, and outcome. Each heat map cell corresponds to the number of times that the patient’s miRNA profiles were coclustered out of 10,000 runs of the random k-means algorithm (see details in Materials and Methods). The resulting heat map defines four robust clusters (designated by black boxes, across A, B, and C), denoted as clusters 1–4. (B) The heat map of 221 miRNAs displaying the ΔΔCT = CTSample – Mean CTHealthy. Columns correspond to patients, and rows correspond to unique miRNAs. (C) Plots representing the total number of miRNAs elevated in each patient compared with healthy controls. The colors of the stacked bars correspond to ΔΔCT = CTSample – Mean CTHealthy.
Elevations in miRNA profiles, compared with healthy controls, are represented in Fig. 1 B and C. Patients in cluster 2 showed a mild elevation in miRNA profiles compared with patients in cluster 1. The clinical significance of why this group (cluster 2) clustered separately from the other three clusters is currently unclear, and additional patients’ samples may address this question. Although the initial ALT activity of some patients in cluster 3 were more similar to cluster 1, their initial miRNA profiles were more similar to the miRNA profiles of cluster 4, and moreover, the eventual peak ALT activity of cluster 3 patients were more like cluster 4 (Fig. 1A).
Cluster 4 consists of patients with high peak ALT, and, notably, includes six out of the seven ischemic-hepatitis patients. This cluster shows the highest increase in miRNA levels (Fig. 1 B and C). The extent of miRNA elevation tends to increase across clusters 1 through 4 (Fig. 1 B and C).
When we focused only on the APAP overdose patients in clusters 1–4, we found a set of miRNAs that are increased (ΔΔCT ≥ 3, equivalent to ≥ eightfold change), with an upward trend of ΔΔCT across the clusters (Dataset S1, Tabs S6 and S7). This upward trend correlates with increasing ALT activity and a global increase in miRNA profiles (Fig. 1C, and last row in Dataset S1, Tab S7). In addition, we identified miRNAs that increase above the ΔΔCT ≥ 3 threshold only in patients with high ALT activity found in clusters 3 and 4 (Dataset S1, Tab S8). This correlation suggests that these miRNAs could be used in conjunction with, or in place of, ALT as markers to approximate the severity of APAP induced liver injury.
The number of miRNAs that we observed elevated in hepatotoxic patients, and the extent of elevation for many of those miRNAs, is striking. Approximately half of these patients exhibited at least eightfold (ΔΔCT ≥ 3) elevation of more than 100 miRNAs (Fig. 1C). In many of these patients, dozens of miRNAs were elevated more than 100-fold. Because we performed a more comprehensive set of assays, we describe significantly more elevated miRNAs in the serum of patients with APAP-induced liver injury than has been reported (13–16). Elevated miRNAs in our study include the previously identified miR-122-5p (13, 15, 16), but the previously implicated miR-192 (16) showed inconsistent elevation in our hands. There were reductions in some circulating miRNAs compared with healthy controls. However, these reductions did not display any consistent patterns across the patients (Fig. S2).
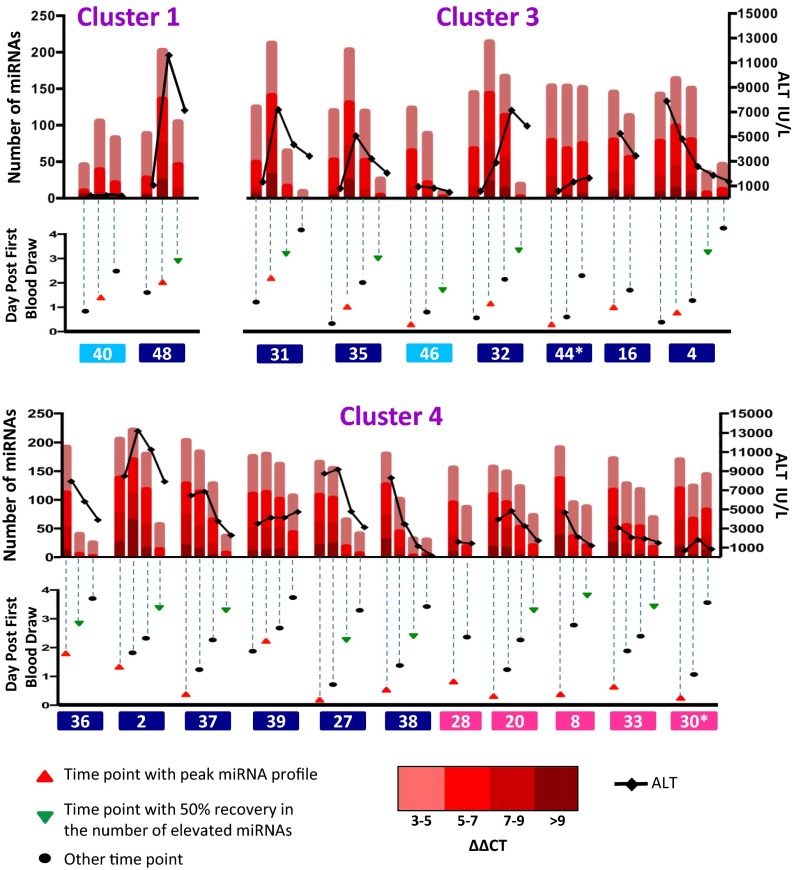
Interestingly, miRNA levels of patients no. 40 and no. 48 (from cluster 1) and patients no. 31, no. 32, and no. 35 (from cluster 3) continued to rise after admittance (Fig. 3). To determine whether this increase resulted in an miRNA profile similar to patients in cluster 4, we performed the clustering again, this time replacing the initial miRNA sample for the above five patients with a subsequent sample that showed the greatest increase in miRNA profiles (see red triangle in Fig. 3). The resulting clustering is shown in Fig. S3. Indeed, the peak samples of four out of these five patients clustered with samples from cluster 4.
Fig. 3.
Dynamics of miRNA profiles during NAC treatment. Profiles of ALT (black line), miRNA changes (red bars), and sample time point (triangle or oval) are shown for patients with at least two available samples within 4 d of hospital admittance, for which at least 100 miRNAs are elevated (ΔΔCT ≥ 3) during this time window. Other patients are shown in Fig. S4. Time points with peaked miRNA profiles are designated with a red triangle. A subsequent time point where the number of elevated miRNAs has decreased by at least 50% is designated with a green triangle. Additional time points analyzed are designated by black ovals. Patients are labeled with patient ID and color-coded according to their peak ALT as in Fig. 1.
Circulating miRNA Profiles Can Distinguish Between APAP Overdose and Liver Pathology of Ischemic-Hepatitis Origin.
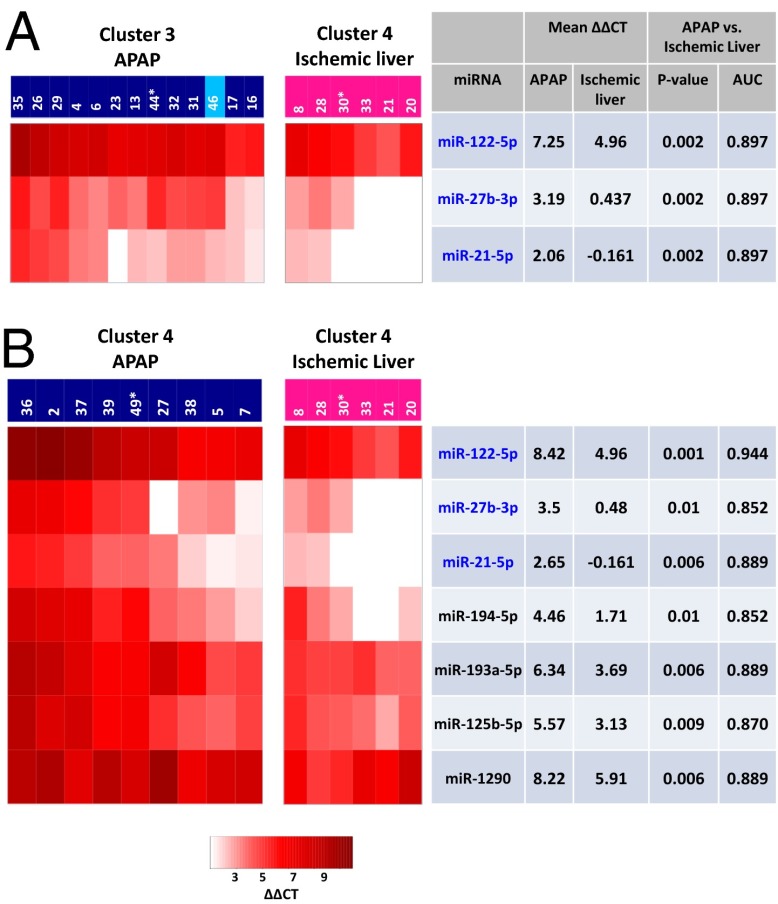
We investigated whether miRNA expression profiles in the earliest available blood sample could distinguish between APAP-overdosed and ischemic-hepatitis patients. The most stringent comparison would be between patients with similar elevations in miRNA profiles. Accordingly, we compared the miRNA profiles of the APAP patients in cluster 3 and cluster 4, respectively, with that of ischemic-hepatitis patients in cluster 4. Three miRNAs (miR-122-5p, miR-27b-3p, and miR-21-5p) emerged as differentially increased between ischemic-hepatitis patients and APAP patients in both cluster 3 and cluster 4 (Fig. 2 A and B). Moreover, among cluster 4 patients, additional miRNAs (miR-194-5p, miR-193a-5p, miR-125b-5p, and miR-1290) contributed to distinguishing APAP from ischemic-hepatitis damage (Fig. 2B). Receiver operating characteristic (ROC) curve analysis was used to validate the discrimination between the groups, as can be seen by the favorable area under the curve (AUC) values (AUC > 0.85 in Fig. 2). The differences in these miRNAs could be helpful in distinguishing between APAP and ischemic-hepatitis patients upon hospital admission. Elevated circulating miRNAs that overlap between the APAP and ischemic-hepatitis patients are probably associated with nonspecific liver injury (Dataset S1, Tab S6). These miRNAs could be valuable as a tool in the drug discovery and development process to potentially predict drug-induced liver injury (DILI), the major cause of preclinical and aftermarket withdrawal.
Fig. 2.
Unique miRNAs can differentiate between APAP and ischemic-hepatitis patients. Separate heat maps comparing APAP patients in cluster 3 (A) and cluster 4 (B) to ischemic-hepatitis patients in cluster 4. Each column corresponds to an individual patient; each row corresponds to a specific miRNA. Mean ΔΔCTs, comparing both APAP and ischemic hepatitis to healthy controls are displayed. P value results show the significance of the comparison between APAP and ischemic-hepatitis patients. The area under the ROC curve (AUC), as a measure of discrimination between APAP and ischemic hepatitis, is also shown. Patients are labeled with patient ID and color-coded according to their peak ALT as in Fig. 1. The miRNA selection criteria included those in which (i) the meanΔΔCTAPAP ≥ 2 (for cluster 3) or meanΔΔCTAPAP ≥ 3 (for cluster 4), and ΔΔCTAPAP – ΔΔCTIschemic hepatitis ≥ 2, and (ii) P value ≤ 0.01. Filter 1 resulted in 3 and 19 miRNAs in clusters 3 and 4, respectively. Filter 2 reduced the number of miRNAs to 3 and 7 in these clusters.
Evolution of miRNA Profiles in Individual Patients During NAC Treatment.
We analyzed miRNA profiles and ALT activity in sequential time points over a 4-d time period from the day of hospital admittance. For this analysis, we included those patients for whom at least two time points were available and that displayed at least 100 miRNAs elevated (ΔΔCT ≥ 3) during this time window (Fig. 3). Despite the dramatic elevation in circulating miRNA levels in the earliest clinical sample obtained from our APAP-overdose patients and ischemic-hepatitis patients, many of these miRNAs recovered to near normal levels as the patients underwent i.v. NAC therapy (Fig. 3). The exceptions included APAP-overdose patient no. 44 and ischemic-hepatitis patient no. 30, both of whom died, and APAP patients no. 39 and no. 16, who showed only slight recovery in miRNA profiles and persistently high ALT activity within the 4-d window but exhibited miRNA recovery after the 4-d window.
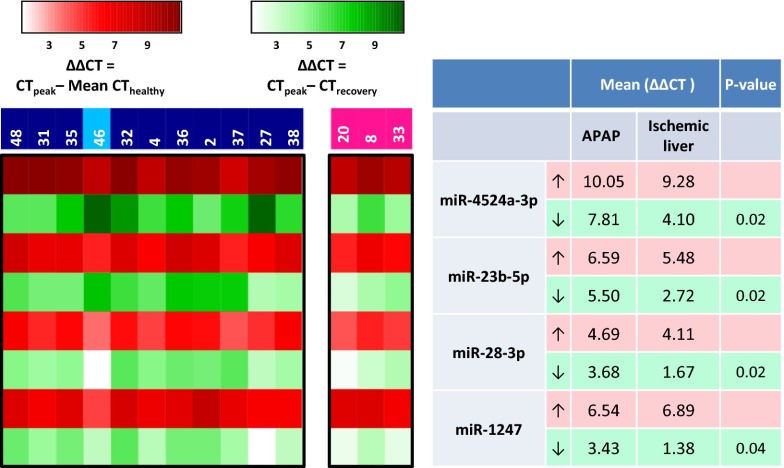
In general, for the APAP-overdose patients, the pace and degree of recovery, as measured by circulating miRNA profiles, was more substantial than for the ischemic-hepatitis patients. For example, greater than 50% recovery occurred in APAP-overdosed patients no. 27 and no. 38 within 2 d whereas it required 3.5 d for ischemic-hepatitis patient no. 8 to reach that level. To define a specific miRNA recovery profile that distinguishes APAP from ischemic liver, we first identified the miRNAs that showed similar elevations in all patients at the peak of their miRNA profile (red triangle in Fig. 3). We then examined the levels of those miRNAs at the point where the number of miRNAs elevated had decreased at least 50% (green triangle in Fig. 3). Patients who did not show recovery to 50% were excluded. We then compared the recovery of the similarly elevated miRNAs in APAP patients to those of ischemic-hepatitis patients. This analysis yielded four miRNAs (miR-4524a-3p, miR-23b-5p, miR-28-3p, and miR-1247-5p) that peaked to similar levels in all patients, yet showed significantly more robust recovery in APAP OD patients compared with ischemic-hepatitis patients (Fig. 4 and Table 1). We propose that these miRNAs can be used to help distinguish between APAP and ischemic-hepatitis patients based on their differences in response to NAC treatment.
Fig. 4.
miRNAs that distinguish recovery in APAP vs. ischemic-hepatitis patients. The heat map reveals miRNAs that increase to similar levels in both APAP and ischemic-hepatitis patients compared with healthy controls as shown in red. The extent of miRNA recovery from the peak to recovery is depicted in green. Only recovered APAP and ischemic-hepatitis patients were used for this analysis.
Table 1.
Summary of APAP-associated circulating miRNAs
| microRNA | Elevated in all clusters, ranked by cluster 4* | Highest elevated in circulation† | Most abundant in healthy liver‡ | Initial response§ | Rebound¶ | APAP vs. ischemic first sample|| | APAP vs. ischemic recovery** |
| miR-3646-3p | 1 | 2 | 1 | ||||
| miR-412 | 2 | 1 | 1 | ||||
| miR-2467-3p | 3 | 3 | 2 | 6 | |||
| miR-1207-5p | 4 | 5 | 8 | ||||
| miR-138-1-3p | 5 | 6 | 9 | ||||
| miR-605 | 6 | 4 | 5 | ||||
| miR-4258 | 7 | 8 | 6 | ||||
| miR-372 | 8 | 9 | 13 | ||||
| miR-4524a-3p | 9 | 7 | 1 | ||||
| miR-631 | 10 | 14 | |||||
| miR-19b-1-5p | 10 | 4 | |||||
| miR-122-5p | 11 | 1 | 1 | ||||
| miR-483-5p | 12 | ||||||
| miR-1260a-5p | 2 | ||||||
| miR-4454-5p | 3 | ||||||
| miR-4286-5p | 4 | ||||||
| miR-451a-5p | 5 | ||||||
| miR-4516 | 6 | 11 | |||||
| miR-21-5p | 7 | 3 | |||||
| miR-29c-3p | 8 | ||||||
| miR-22-3p | 9 | ||||||
| miR-126-3p | 10 | ||||||
| miR-194-5p | 11 | 4 | |||||
| miR-16-5p | 12 | ||||||
| miR-373-5p | 3 | ||||||
| miR-1203 | 7 | ||||||
| miR-542-5p | 10 | ||||||
| mir-551a | 12 | ||||||
| miR-1286 | 15 | ||||||
| miR-337-5p | 16 | ||||||
| let7i-3p | 17 | ||||||
| miR-5095 | 2 | ||||||
| miR-3191-3p | 3 | ||||||
| miR-1910 | 4 | ||||||
| miR-202-3p | 5 | ||||||
| miR-27b-3p | 2 | ||||||
| miR-193a-5p | 5 | ||||||
| miR-125b-5p | 6 | ||||||
| miR-1290 | 7 | ||||||
| miR-23b-5p | 2 | ||||||
| miR-28-3p | 3 | ||||||
| miR-1247 | 4 |
n = no. of patients.
Ten miRNAs that are elevated with ΔΔCT ≥ 3 in all four clusters ranked by highest elevation in cluster 4 (n = 49).
Top 12 miRNAs most elevated in APAP toxicity patients in clusters 3 and 4 (n = 23).
Top 12 miRNAs found in liver tissue (based on three biopsied).
Seventeen miRNAs elevated with ΔΔCT ≥ 3 within 12 h of APAP overdose (n = 5).
Six miRNAs that show ΔΔCT ≥ 3 after APAP overdose, recover ΔΔCT ≥ 3 after NAC treatment, then rise again with ΔΔCT ≥ 1.5 after NAC ends (n = 7).
Seven miRNAs with distinct differences in response in the earliest blood draw between APAP toxicity and ischemic liver (n = 28).
Four miRNAs that do not recover as well with NAC treatment in ischemic liver vs. APAP toxicity (n = 14).
NAC is the treatment of choice for APAP overdose because of its efficacy in replacing APAP-depleted glutathione stores in the liver, but it is also used in other instances of acute liver failure that do not necessarily impact the glutathione pathway. It is interesting that patients with ischemic liver injury also displayed some recovery in miRNA profile with NAC treatment. Several recent reviews question the benefit of NAC in non–APAP-induced acute liver failure (NAI-ALF) (19). Our data suggest that NAC is effective at decreasing the elevated levels of circulating miRNAs seen in ischemic hepatitis and may be useful in other hepatotoxic conditions.
Notably, although most elevated circulating miRNAs recover in conjunction with successful NAC treatment, miR-1290 (Fig. 5) remained elevated for at least 2 d after other miRNAs began to decrease toward normal levels. More extended longitudinal studies of APAP overdose patients may reveal whether slower recovering miRNAs eventually return to normal and perhaps provide insight into the longer-term process of liver recovery from drug-induced injury.
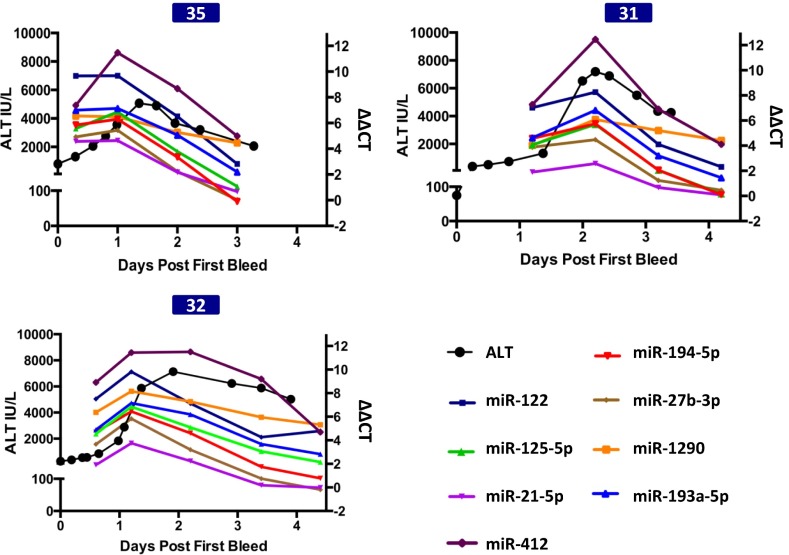
Fig. 5.
miRNAs precede ALT, both in initial elevation and in recovery during treatment. Three patients had ALT activity (black line) that rose above 1,000 IU/L after many of the miRNAs showed significant elevation. Shown are profiles of ALT and APAP distinguishing miRNAs (identified in Fig. 2), miR-1290 (orange line), which recovered slowly, and miR-412 (purple line), which is the most elevated miRNA.
Earliest Effects of APAP on Circulating miRNAs.
Patients no. 10, no. 12, no. 15, no. 41, and no. 43 (clusters 1 and 2) were admitted relatively quickly, within 12 h after reported APAP overdose. All had high APAP blood levels and low ALT. Because of their timely treatment, these patients’ ALT activity never rose above normal. Seventeen miRNAs were identified as elevated (ΔΔCT ≥ 3) in the earliest blood sample from these patients (Table 1). Further study is needed to determine whether this set of early responding miRNAs signifies an inflammatory response to APAP before major liver damage begins (20).
miRNA Profiles in APAP OD Patients Can Rebound After Cessation of NAC Treatment.
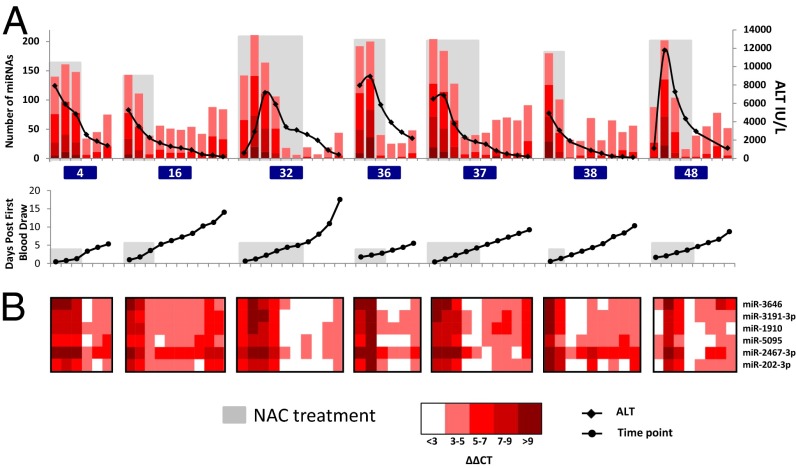
In our study, 13 APAP-overdosed patients had high peak ALT and multiple time points available to assay for circulating miRNAs (Fig. 3). Seven of these patients (no. 4, no. 16, no. 32, no. 36, no. 37, no. 38, and no. 48) were hospitalized for extended periods of time (greater than 5 d). These patients had apparently recovered, as monitored by continued reduction of ALT, and also showed recovery in their miRNA profiles during NAC treatment. However, their toxicity-associated miRNA profile partially rebounded after cessation of NAC (Fig. 6A). Interestingly, more than 90% of the miRNAs that are elevated in these patients at the point of highest rebound were also elevated at the earlier peak point. Fig. 6B shows the dynamics of six miRNAs that exhibited rebound in all seven patients.
Fig. 6.
Certain patients show a rebound trend in a subset of miRNAs after stopping NAC treatment. Profiles of ALT, miRNA changes, and sample time point are shown for seven patients that show rebound in miRNA profiles after stopping treatment with NAC. (A) NAC treatment period is shaded with gray. Patients are labeled with patient ID and color-coded according to their peak ALT as in Fig. 1. (B) Heatmaps for six miRNAs that showed a ΔΔCT ≥ 3 after APAP overdose, recovered by ΔΔCT ≥ 3 after NAC treatment, and then rose again by ΔΔCT ≥ 1.5 after NAC treatment, in at least five of the seven patients.
These rebounding miRNAs could reflect lingering APAP pathology that was not completely ameliorated by NAC treatment. Alternatively, they could be a consequence of NAC itself, or of regenerative liver processes. Further longitudinal study of more patients after NAC treatment could determine whether circulating miRNAs can be used to monitor the long-term status of patients with APAP-overdose hepatotoxicity. It has been reported that some patients who received prolonged i.v. NAC treatment, within 8 h of overdose ingestion, nevertheless required liver transplantation (21). It is possible that standard NAC treatment protocols may not be aggressive enough, in all cases, to allow for sustained recovery from hepatotoxicity, as is reflected in the rebound of elevated miRNAs.
Changes in Circulating miRNA Profiles Associated with Liver Injury Precede Changes in ALT Activity.
Of the 13 APAP-overdosed patients with high peak ALT and multiple available time points, 6 had presented during or after the ALT had peaked (no. 16, no. 4, no. 36, no. 37, no. 27, and no. 38) and 7 had presented before the ALT had peaked (no. 48, no. 31, no. 35, no. 32, no. 44, no. 2, and no. 39) (Fig. 3). In 3 of the 7 patients (no. 31, no. 32, and no. 35), time points where the ALT was still below 1,000 IU/L were available for the analysis of circulating miRNAs. Interestingly, many of the miRNAs identified above as indicative of APAP poisoning (miR-122-5p, miR-27b-3p, miR-21-5p, miR-125b-5p, miR-194-5p, miR-193a-5p, and miR-1290) (Fig. 2) exhibited dramatically elevated levels before a rise in ALT to above 1,000 IU/L (Fig. 5). In the remaining 4 patients that presented early, time points for the analysis of circulating miRNAs before rise in ALT were unavailable; nevertheless, miRNAs were observed to be elevated in these patients as well, before the time of peak ALT (Fig. 3). Overall, the results are promising; however, further studies with additional patients are needed to validate the signature of circulating miRNAs in APAP-induced liver toxicity.
Origins and Significance of Circulating miRNAs in APAP Overdose.
The source of the increased levels of circulating miRNAs due to APAP hepatotoxicity is not known. The miRNAs associated with hemolysis or red blood cell contamination (22) do not overlap the APAP-associated circulating miRNAs identified in this study. Because APAP overdose can be toxic to the liver (3), it is reasonable to suppose that the miRNAs elevated in circulation are a result of hepatocyte lysis. In that case, the miRNAs elevated in circulation would be those that are highly expressed in liver tissues. However, interestingly, the profile of circulating miRNAs that are most elevated in APAP-overdose patients does not match the expression profile of miRNAs in healthy human liver tissue (Table 1). Among the top 12 miRNAs in healthy liver, only one (miR-122-5p) was also among the top 12 miRNAs elevated in APAP-overdose patients’ circulation. Moreover, although miR-122-5p is the most abundant miRNA in healthy liver, it is only the 11th most elevated miRNA in the circulation of APAP-overdosed patients.
Although the miRNA profile in healthy liver tissue from our qRT-PCR data is similar to that of previously published deep sequencing of liver small RNAs, certain miRNAs with high signals, particularly miR-4454-5p, miR-4516, and miR-4286-5p, were not detected by small RNA sequencing (23). Previous studies have shown that miRNA profiles by deep sequencing can vary substantially from qRT-PCR profiles from the same samples (24). It is also possible that these three sequences may represent another class of RNA because the miRBase (9) curation is somewhat uncertain. Regardless, miR-4454-5p, miR-4516, and miR-4286-5p signals were not elevated in APAP samples compared with healthy samples and are not included in our signatures.
These observations suggest that the circulating miRNA signature of APAP toxicity likely reflects processes more complex than simple hepatocyte lysis. It is possible that many of the miRNAs that we detected in circulation could be selectively exported from intact (although stressed) liver cells (25, 26). Perhaps the elevated miRNAs are much more stable in circulation than the other liver miRNAs. It is also possible that other tissues could export these miRNAs into circulation as indirect responses to systemic effects of liver injury.
Conclusions
An important element of our findings is that circulating miRNAs are potentially powerful biomarkers for early diagnosis of APAP-induced liver injury in human patients and for the timely and accurate monitoring of treatment. We believe that these circulating miRNAs can be substantially more informative than the current biomarker, ALT. Low ALT activity can signify mild or absent toxicity, or conversely, can reflect a late stage in the disease progression where the liver is severely compromised, depleted of ALT (Fig. 3, patients no. 30 and no. 44). Elevated ALT is not a quantitative measure of the severity of the injury or prognosis. By contrast, the broader dynamic range and quick responsiveness of the miRNAs to hepatotoxic agents and to treatment with NAC can add valuable information beyond that provided by ALT. Importantly, our results indicate that, if clinical samples of plasma are available soon after APAP overdose, an elevated APAP-diagnostic miRNA signature could be detected in circulation before an increase in ALT. Moreover, our data also show that lack of recovery of circulating miRNA levels may be indicative of adverse patient outcomes. A larger patient cohort is required to confirm whether a lack of miRNA recovery in the early phases of NAC therapy could be used as a predictor of potentially fatal hepatotoxicity and as candidacy for liver transplantation. To apply miRNA profiling in this fashion in the clinical setting, it will be necessary to develop practical clinical assays for circulating miRNA levels with appropriately rapid turnaround time for data acquisition and analysis.
Materials and Methods
Patient Parameters.
The parameters of patients in this study are described in SI Materials and Methods.
RNA Extraction, Reverse Transcription, Preamplification, and Real-Time PCR.
Plasma was isolated, RNA was extracted, and qRT-PCR assays were performed as described in SI Materials and Methods.
Data Analysis.
CT values were calculated using the Viia 7 software (Threshold, 0.04; Baseline, Automatic). Samples with low reverse transcription efficiency, defined by a miRTC CT value greater than 19, were excluded from subsequent analysis. Mean CT value, SD, and a score (mean × SD) were calculated for each miRNA across all samples. miRNAs that had a CT value greater than 32 in more than 20 samples (not including healthy controls) were removed (two miRNAs, 1277-3p and 2355-5p). miRNAs were then ranked based on their score, and the mean and SD of all of the scores were determined. The set of miRNAs whose scores were lower than mean −1.5 SD (of all scores) were chosen as the least variable set for normalization (20 miRNAs). All CT values were normalized to the mean CT value of this least variable set (by subtraction), multiplied by −1, and shifted to a positive scale. For each miRNA, fold changes (in log scale) relative to healthy controls were calculated as ΔΔCT = CTsample – mean CTHealthy. (Raw values, miRNAs in the normalization set, the normalized values, and ΔΔCTs are found in Dataset S1, Tabs S2–S5). A set of 221 miRNAs that showed a ΔΔCT ≥ 3 in at least five patients was chosen for further analysis.
Data from the earliest time point for each of the 49 patients was used in a two-level clustering analysis (27). The profiles of the above 221 miRNAs were first clustered using the random k-means algorithm implemented in the R environment (with k = 3) (28). This algorithm randomly selects k-centers and computes the partition of the samples into clusters. Because this algorithm depends on the initial centers, it does not necessarily provide the same solution each time. To compensate for this sensitivity to initial conditions, we ran the algorithm 10,000 independent times and counted, for each pair of miRNA profiles, the number of times they co-occur in the same cluster (referred to as an accumulation matrix in the literature) (27). These coclustering frequencies were then used to cluster the samples by hierarchical clustering, and the results were visualized using a heat map (gplots in R). This approach assumes that profiles that colocalize to a cluster multiple times, under different initial k-centers, are genuinely similar. Note that this two-level clustering approach allows a number of clusters greater than k to emerge. Values for k were tested (2–4), and a k value of 3 was chosen for good separation on the one hand, while also allowing a sufficient number of patients in each cluster for statistical tests. All reported P values were calculated using the Wilcoxon test in R. For receiver operating characteristic (ROC) curve analysis, we used the ROCR package in R (29). The ΔΔCT values were used as the input for ROC analysis and to calculate the area under the curve (AUC).
Supplementary Material
Acknowledgments
Julie Meyer and Jonathan Schaffer of QIAGEN provided technical support and pilot reagents for this study. Members of the University of Massachusetts Memorial Emergency Department donated control plasma. Catherine Sterling (V.R.A. laboratory) read and commented on the manuscript. This work was supported financially by the McNeil Consumer Healthcare Division of McNeil PPC, Inc., the Silverman Chair in Natural Sciences, and a University of Kansas Medical Center Liver Center Pilot Grant.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412608111/-/DCSupplemental.
References
- 1.Craig DGN, Lee A, Hayes PC, Simpson KJ. Review article: The current management of acute liver failure. Aliment Pharmacol Ther. 2010;31(3):345–358. doi: 10.1111/j.1365-2036.2009.04175.x. [DOI] [PubMed] [Google Scholar]
- 2.Bronstein AC, et al. 2008 Annual report of the American Association of Poison Control Centers’ National Poison Data System (NPDS): 26th annual report. Clin Toxicol (Phila) 2009;47(10):911–1084. doi: 10.3109/15563650903438566. [DOI] [PubMed] [Google Scholar]
- 3.Larson AM, et al. Acute Liver Failure Study Group Acetaminophen-induced acute liver failure: Results of a United States multicenter, prospective study. Hepatology. 2005;42(6):1364–1372. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 4.Budnitz DS, Lovegrove MC, Crosby AE. Emergency department visits for overdoses of acetaminophen-containing products. Am J Prev Med. 2011;40(6):585–592. doi: 10.1016/j.amepre.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 5.Bond GR, Novak JE. The human and economic cost of paracetamol (acetaminophen) overdose. Pharmacoeconomics. 1995;8(3):177–181. doi: 10.2165/00019053-199508030-00001. [DOI] [PubMed] [Google Scholar]
- 6.Green TJ, et al. When do the aminotransferases rise after acute acetaminophen overdose? Clin Toxicol (Phila) 2010;48(8):787–792. doi: 10.3109/15563650.2010.523828. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. microRNAs: Tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 8.Siow RCM, Clough GF. Spotlight issue: MicroRNAs in the microcirculation—from cellular mechanisms to clinical markers. Microcirculation. 2012;19(3):193–195. doi: 10.1111/j.1549-8719.2012.00166.x. [DOI] [PubMed] [Google Scholar]
- 9.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ji S, et al. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62(5):716–726. doi: 10.1136/gutjnl-2011-301083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu R, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58(3):610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 12.Ward JA, et al. Circulating cell and plasma microRNA profiles differ between non-ST-segment and ST-segment-elevation myocardial infarction. Fam Med Med Sci Res. 2013;2(2):108. doi: 10.4172/2327-4972.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward J, Bala S, Petrasek J, Szabo G. Plasma microRNA profiles distinguish lethal injury in acetaminophen toxicity: A research study. World J Gastroenterol. 2012;18(22):2798–2804. doi: 10.3748/wjg.v18.i22.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laterza OF, et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55(11):1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 15.Wang K, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106(11):4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starkey Lewis PJ, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54(5):1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, et al. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE. 2012;7(7):e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willeit P, et al. 265 Plasma microRNAa as biomarkers for platelet inhibition. Heart. 2013;99:A139–A140. [Google Scholar]
- 19.Sales I, Dzierba AL, Smithburger PL, Rowe D, Kane-Gill SL. Use of acetylcysteine for non-acetaminophen-induced acute liver failure. Ann Hepatol. 2013;12(1):6–10. [PubMed] [Google Scholar]
- 20.Jaeschke H, Williams CD, Ramachandran A, Bajt ML. Acetaminophen hepatotoxicity and repair: The role of sterile inflammation and innate immunity. Liver Int. 2012;32(1):8–20. doi: 10.1111/j.1478-3231.2011.02501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyon S, Klein-Schwartz W. Hepatotoxicity despite early administration of intravenous N-acetylcysteine for acute acetaminophen overdose. Acad Emerg Med. 2009;16(1):34–39. doi: 10.1111/j.1553-2712.2008.00296.x. [DOI] [PubMed] [Google Scholar]
- 22.Pritchard CC, et al. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev Res (Phila) 2012;5(3):492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schee K, et al. Deep sequencing the microRNA transcriptome in colorectal cancer. PLoS ONE. 2013;8(6):e66165. doi: 10.1371/journal.pone.0066165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 26.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fred ALN, Jain AK. Proceedings of the 16th International Conference on Pattern Recognition. Vol 4. New York: IEEE; 2002. Data clustering using evidence accumulation; pp. 276–280. [Google Scholar]
- 28.Hartigan JA, Wong MA. Algorithm AS 136: A K-Means Clustering Algorithm. Appl Stat. 1979;28:100. [Google Scholar]
- 29.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: Visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.