Significance
Mycoviruses are viruses that infect fungi and replicate in fungi. Previously, no mycoviruses had been discovered with negative-stranded (−)ssRNA genomes. Here, we characterize a (−)ssRNA mycovirus that infects a fungal plant pathogen. Although its genome and organization are significantly different from those of current mononegaviruses, this virus is closely related to viruses in families Nyamiviridae and Bornaviridae that infect animals. This discovery may provide insights into the global ecology and evolution of (−)ssRNA viruses. Furthermore, since many (−)ssRNA viruses are serious human pathogens, this system is likely to provide a less hazardous way to study replication of a (−)ssRNA virus and could be useful in establishing a system to screen antiviral compounds against (−)ssRNA viruses.
Keywords: mononegavirus, fungal plant pathogen, rapeseed stem rot, biological control
Abstract
Mycoviruses are widespread in nature and often occur with dsRNA and positive-stranded RNA genomes. Recently, strong evidence from RNA sequencing analysis suggested that negative-stranded (−)ssRNA viruses could infect fungi. Here we describe a (−)ssRNA virus, Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1), isolated from a hypovirulent strain of Sclerotinia sclerotiorum. The complete genome of SsNSRV-1 is 10,002 nt with six ORFs that are nonoverlapping and linearly arranged. Conserved gene-junction sequences that occur widely in mononegaviruses, (A/U)(U/A/C)UAUU(U/A)AA(U/G)AAAACUUAGG(A/U)(G/U), were identified between these ORFs. The analyses 5′ and 3′ rapid amplification of cDNA ends showed that all genes can be transcribed independently. ORF V encodes the largest protein that contains a conserved mononegaviral RNA-dependent RNA polymerase (RdRp) domain. Putative enveloped virion-like structures with filamentous morphology similar to members of Filoviridae were observed both in virion preparation samples and in ultrathin hyphal sections. The nucleocapsids are long, flexible, and helical; and are 22 nm in diameter and 200–2,000 nm in length. SDS/PAGE showed that the nucleocapsid possibly contains two nucleoproteins with different molecular masses, ∼43 kDa (p43) and ∼41 kDa (p41), and both are translated from ORF II. Purified SsNSRV-1 virions successfully transfected a virus-free strain of S. sclerotiorum and conferred hypovirulence. Phylogenetic analysis based on RdRp showed that SsNSRV-1 is clustered with viruses of Nyamiviridae and Bornaviridae. Moreover, SsNSRV-1 is widely distributed, as it has been detected in different regions of China. Our findings demonstrate that a (−)ssRNA virus can occur naturally in fungi and enhance our understanding of the ecology and evolution of (−)ssRNA viruses.
Mycoviruses (or fungal viruses) are widespread among the major taxonomic groups of fungi (1). Although many mycoviruses are associated with latent infections, some hold great promise for exploitation as biological agents to control fungal diseases. For example, the hypovirus that infects the chestnut blight fungal pathogen Cryphonectria parasitica was used successfully to control chestnut blight in Europe (2). Likewise, two other mycoviruses, namely Rosellinia necatrix megabirnavirus 1 and Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1, were shown to have the potential to control fungal diseases (3–5). Mycoviruses have also been used to enhance the understanding of the basic principles of the viral life cycle, such as viral replication and proliferation and interaction with their fungal hosts (6).
Most mycoviruses are composed of either double-stranded (ds) or positive single-stranded (+)ssRNA genomes, and a few have a DNA genome (4). At present, mycoviruses with RNA genomes are classified into 12 families, of which 4 contain dsRNA viruses, 6 comprise (+)ssRNA viruses, and 2 families (Metaviridae and Pseudoviridae) accommodate RNA reverse-transcribing genomes (1). However, an increasing number of mycoviruses that differ from those classified have been found (7–9). Recently, Kondo et al. (10) identified L protein-like gene sequences of a putative (−)ssRNA virus in the genome of Erysiphe pisi, a fungal plant pathogen, and they also assembled incomplete genomic sequences of (−)ssRNA viruses in transcriptome shotgun assembly libraries of another fungal pathogen, Sclerotinia homoeocarpa, and suggested that (−)ssRNA viruses are most likely to exist in fungi (10). However, to date it is not known whether (−)ssRNA viruses do in fact occur in fungi and their properties also remain as yet unknown.
Mononegaviruses are members of the order Mononegavirales with nonsegmented (−)ssRNA genomes and are 8.9–19 kb in size. Generally, the virions of mononegaviruses have large enveloped structures with variable morphologies, and individual particles frequently exhibit pleomorphism. Mononegaviruses are grouped into five families: Bornaviridae, Filoviridae, Paramyxoviridae, Rhabdoviridae, and Nyamiviridae. Some mononegaviruses are notorious human viral pathogens, such as the Ebola, human respiratory syncytial, measles, Nipah, and rabies viruses. Many mononegaviruses are found in vertebrates, whereas only a few of the mononegaviruses have been reported to infect plants and invertebrates (nematode) (11, 12).
Sclerotinia sclerotiorum is a devastating fungal plant pathogen with a wide host range including more than 400 plant species (13). Due to a lack of resistant cultivars and the fact that large-scale application of fungicides could result in a potential threat to the environment, diseases caused by S. sclerotiorum cannot be controlled efficiently. Previously, we reported that a hypovirulence-associated DNA mycovirus could be exploited as a potential biocontrol agent of diseases caused by S. sclerotiorum (4, 5). This finding encouraged us to screen more viruses that confer hypovirulence to S. sclerotiorum. In the present study we found that a hypovirulent strain AH98 (Fig. S1) was coinfected by two viruses: One is a hypovirus, highly similar to Sclerotinia sclerotiorum hypovirus 1 (SsHV-1) (14); and the other is a (−)ssRNA virus that is related to mononegaviruses, and was designated S. sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1). Here, we report the properties of this unique mycomononegavirus, including genome organization, viral morphology, evolution, and distribution in China.
Results
Properties of SsNSRV-1 Virus Particles.
The virions of SsNSRV-1 purified from mycelia of strain AH98 are filamentous, 25–50 nm in diameter, ∼1,000 nm in length (Fig. 1A), and similar in filamentous virion shape to the Ebola virus (15). However, unlike the case of the Ebola virus, no spikes were clearly observed by transmission electron microscopy (TEM) to be protruding from the outer layers of the SsNSRV-1 virions. When the virions were broken, helical and flexible substructures were observed (Fig. 1A). These helical substructures had an optimum absorption spectrum at 260 nm, indicating the presence of nucleic acid. Such substructures, known as nucleocapsids, are ubiquitous in mononegviruses, such as in the Marburg, rabies, and Newcastle disease viruses; simian virus 5; Sendai virus 1; and others (16–21).
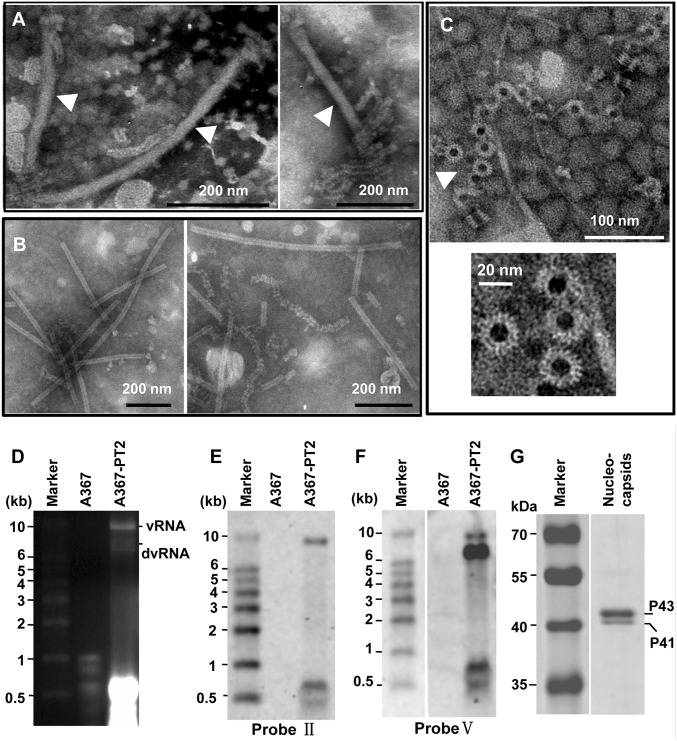
Fig. 1.
Morphology and structure of SsNSRV-1 particles and RNPs. Particles and RNPs were purified from mycelia of strains AH98 and Ep-1PNA367-PT2 and negatively stained with 2% PTA [(wt/vol), pH 7.4]. (A) Filamentous, possibly enveloped virions (marked by arrowheads), and RNPs. (B) Purified tight or loose coils of RNPs. (C) Rings that make up the coils and NP monomers (marked by arrowhead). (D) Agarose gel electrophoresis of viral RNAs extracted from virions purified from the mycelia of strain Ep-1PNA367-PT2. This gel contained 1.2% agarose and 1% formaldehyde (vol/vol) and the RNA sample was run in 1× Mops buffer (pH 7.0). The marker was RL10000 (TaKaRa). (E) Northern blot analysis of RNAs isolated from SsNSRV-1 particles using probe II. Probe II amplified from the N gene showed a single band with a size of 10 kb, identical to the size of the SsNSRV-1 genome. (F) Northern blot analysis of RNAs isolated from SsNSRV-1 particles using probe V. Probe V amplified from the L gene showed two bands of sizes of 10 and 7 kb. The smaller band was the mixture of defective RNAs of SsNSRV-1 genome. (G) SDS/PAGE analysis of SsNSRV-1 NPs. Samples collected from sucrose gradient fractions corresponding to 25% sucrose were subjected to SDS/PAGE. The sizes of the Coomassie blue-stained proteins were estimated by comparison with protein weight markers.
SsNSRV-1 nucleocapsids are single, left-handed, helical structures which, when tightly coiled, have a diameter of 20–22 nm and a length of 200–2,000 nm (Fig. 1B). The conformation of SsNSRV-1 nucleocapsids showed wide variations when observed by TEM (Fig. 1 B and C). They were observed to have tightly stacked turns and an overall appearance of relatively stiff rods and also as loosely coiled helices that were extended to varying degrees. Furthermore, nucleoprotein (NP) monomers that make up the coils were observed in some images (Fig. 1C). Such dense or loose coils have often been observed in mononegavirus preparations as RNA–nucleoprotein complexes (RNPs) (21).
The nuclocapsids and tubular structures were also observed in ultrathin fungal hyphal section by TEM. The outer layer of the tubular structures is apparently a membrane, whereas the inner part comprises nucleocapsids (Fig. S2). These tubular structures are similar to the purified virions. Thus, we suggest that the SsNSRV-1 virions may be enveloped by a membrane, and called “enveloped virion-like structures” (EVLSs).
That the viral genome of SsNSRV-1 is ssRNA in nature was convincingly demonstrated using a combined method of nuclease digestion analysis and RT-PCR amplification. Moreover, the RNPs of SsNSRV-1 can be used directly as a template for inverse transcription, which is also a unique feature of mononegaviruses (21).
Complete Sequence and Organization of SsNSRV-1 Genome.
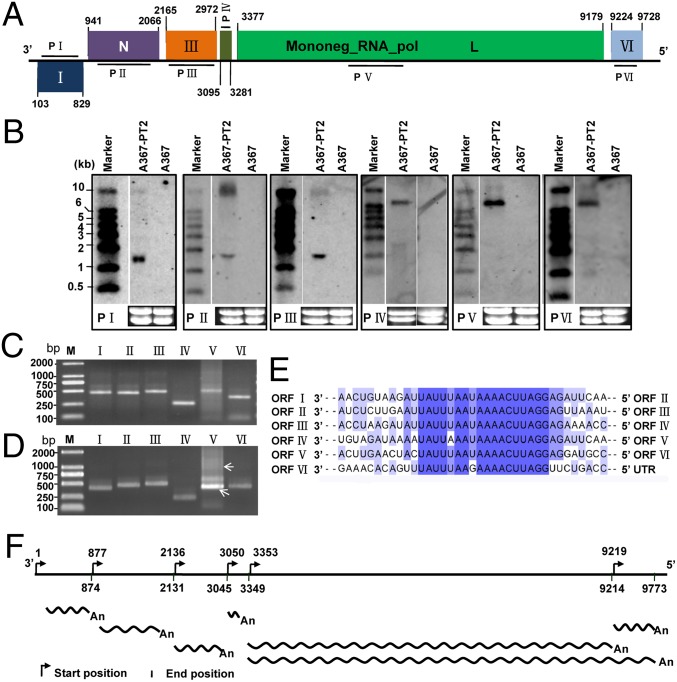
The assembled RNA genome sequence of SsNSRV-1 is 10,002 nt with at least fivefold coverage of every site and was confirmed by Northern blot analysis (Fig. 1 E and F). The viral RNA lacks a poly(A) tail at 3′ terminus and is devoid of a capped 5′ terminus because this terminus could be ligated to the 3′ terminus or be linked directly to an oligonucleotide by T4 RNA ligase, and then successfully RT-PCR amplified (Fig. S3). The GC content of the whole genome is 38.8%, and the 5′ and 3′ terminal regions do not exhibit obvious complementarity. The full length of the SsNSRV-1 genome is predicted to have six major ORFs (ORFs I–VI), and these nonoverlapping ORFs are arranged linearly in the viral genome (Fig. 2A). In addition, conserved noncoding sequences [3′-(A/U)(U/A/C)UAUU(U/A)AA(U/G)AAAACUUAGG(A/U)(G/U)-5′] were identified downstream of each ORF (Fig. 2E); the consensus sequences are underlined. These gene-junction sequences in the viral genome are ubiquitous and are a characteristic feature in mononegaviruses (22). Interestingly, downstream of ORF V, there is an additional ORF (ORF VI) which has not been found in currently known mononegaviruses. Further sequencing analysis showed that there is a stop codon between ORFs V and VI, and a conserved noncoding sequence is also present between them (Fig. 2E).
Fig. 2.
Genomic organization of SsNSRV-1. (A) Genome size and organization. ORFs II–VI are in the same frame, whereas ORF I is in another frame. A conserved domain in the L protein is shown. Boxes on the genome indicate the position and size of each ORF, which are labeled with Roman numerals, except for two ORFs, N and L, which encode the nucleoprotein (N) and RNA-dependent RNA polymerase (RdRp) protein (L). (B) Northern blot analysis of SsNSRV-1 ORFs I–VI. Total RNAs were extracted from mycelia of Ep-1PNA367 (shown as A367), which was virus-free; and Ep-1PNA367-PT2 (shown as A367-PT2), which was transfected by SsNSRV-1. Internal gene-specific DNA fragments were PCR amplified and used to make probes P I through P VI. Primer pairs were designed based on the sequence of each ORF (Table S3). DIG-labeled λ DNA was used as a probe to detect the RNA weight marker RL10000 (TaKaRa). In comparison with the SsNSRV-1-free Ep-1PNA367, all the mRNAs of SsNSRV-1 ORFs I–VI were specifically detected in Ep-PNA367-PT2. (C) Agarose gel electrophoresis of the 5′ RACE of ORFs I–VI on 1.5% agarose gel. (D) Agarose gel electrophoresis of the 3′ RACE of ORFs I–VI on 1.5% agarose gel. Arrows show the positions that contain the 3′ RACE product from ORF V. The primers used are listed in Table S3. (E) Comparison of putative gene-junction regions between the ORFs in SsNSRV-1. Alignment of the putative gene-junction sequences is shown in a 3′-to-5′ orientation. Conserved sequences are highlighted in purple. Different shades indicate the levels of conserved sequence, and the darkest color shade shows the most conserved sequence. (F) The possible transcript map of SsNSRV-1 based on the 5′ and 3′ RACE.
Northern blot showed that all six ORFs of SsNSRV-1 were indeed expressed using corresponding probes I–VI (Fig. 2B). The predicted ORFs were further confirmed by RT-PCR analysis. The 5′ and 3′ rapid amplification of cDNA ends (RACE) analysis results suggest that all six genes could be transcribed independently (Fig. 2 C and D), whereas ORF V could be cotranscribed with ORF VI because two types of 3′ RACE products were amplified (Fig. 2D and Table S1). All sequences of 5′ and 3′ RACE are shown in Table S1. Based on the 5′ and 3′ RACE results, a transcript map of SsNSRV-1 is shown (Fig. 2F).
Subviral RNA species could be also isolated from virion preparations (Fig. 1D and Fig. S4), and these RNAs could be hybridized to a probe derived from ORF V, but could not be detected with a probe derived from ORF II (Fig. 1 E and F). These RNA species were poly(A) tailed at the 3′ end and circularized with T4 RNA ligase, and then subjected to cDNA synthesis and terminal sequence determination (Fig. S5A). The results showed that these RNA species are defective viral RNA whose 5′ and 3′ termini are incomplete, where ORFs I and II are almost fully deleted (Fig. S5B). Our SsNSRV-1 genome sequence has been deposited in GenBank under accession no. KJ186782.
Analysis of Putative Polypeptides of SsNSRV-1.
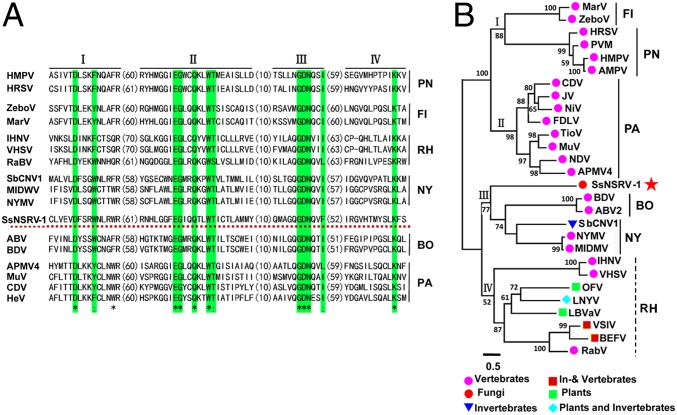
The largest protein encoded by ORF V contains a conserved mononegavirus domain—Mononeg_RNA_pol (pfam00946, 1.75e-86) (Fig. 2A)—and has significant sequence similarity with RNA polymerases from viruses in Bornaviridae, Filoviridae, Rhabdoviridae, and Paramyxoviridae, and the newly classified Nyamiviridae, which are all members of the order Mononegavirales. The four conserved motifs, I–IV, found in mononegaviruses were identified in this putative protein, reflecting the important roles in viral replication and maintenance (Fig. 3A). This protein has also been rarely detected when analyzing the nucleocapsid protein using the peptide mass fingerprinting (PMF) technique (Table S2).
Fig. 3.
(A) Amino acid sequence alignment of core RdRp motifs of SsNSRV-1 and selected viruses from Mononegavirales. The asterisks indicate identical amino acid residues and the colons represent chemically similar amino acid residues. Identical and chemically highly similar amino acid residues are shaded in green, and red, dotted horizontal line shows SsNSRV-1. (B) Phylogenetic analysis of SsNSRV-1. A phylogram of the core RdRp motifs of SsNSRV-1 and the selected mononegaviruses from Filoviridae, Pneumoviridae, Paramyxoviridae, Bornaviridae, Nyamiviridae, and Rhabdoviridae families are shown. The position of SsNSRV-1 is indicated with a red star. For abbreviations of virus names and viral protein accession numbers used in phylogenetic analysis, see Table S4. BO, Bornaviridae; FI, Filoviridae; NY, Nyamiviridae; PA, Paramyxoviridae; PN, Pneumoviridae; RH, Rhabdoviridae.
ORF II encodes the nucleoproteins (NPs) of SsNSRV-1. Two proteins (p43 and p41) isolated from purified nucleocapsids of SsNSRV-1 (Fig. 1G) were subjected to PMF analysis. The results showed that p43 and p41 generated a total of 33 and 23 peptide fragments, respectively (Table S2). Of these peptide fragments, 32 from p43 matched the peptide sequence encoded by ORF II (Table S2), accounting for 67.2% of the entire coverage (375 aa). Furthermore, 22 peptide fragments from p41 also matched ORF II-encoding peptide sequence (Table S2). Therefore, the SsNSRV-1 nucleocapsid may have two forms of nucleoproteins (NP), or protein p41 is a degradation product occurring during virion purification.
The protein encoded by ORF I was detected in the protein sample extracted from crude virion preparations (Table S2), and this protein is a possible membrane protein based on analysis with RaptorX web server (23). It is still not clear whether other viral genes code for viral structural proteins due to the extreme fragility of the virions. To determine their possible roles, all of the ORFs were analyzed to determine the isoelectric points (pI values), potential phosphorylation sites, and potential glycosylation sites in their products. All six deduced proteins of SsNSRV-1 had multiple phosphorylation and glycosylation sites. Proteins encoded by ORFs I–IV are acidic, and the protein encoded by ORF IV has the lowest pI (3.67), whereas proteins encoded by ORFs V (pI 8.83) and VI (pI 9.3) are alkaline.
SsNSRV-1 Is Phylogenetically Closely Related to Bornaviruses and Nyaviruses.
We performed maximum-likelihood phylogenetic analysis based on the amino acid core sequences of the L protein of SsNSRV-1 and other selected viruses, including representative strains of species in each genus in the Mononegavirales (the five families: Bornaviridae, Filoviridae, Paramyxoviridae, Rhabdoviridae, and Nyamiviridae), along with the segmented (−)ssRNA viruses, lettuce big-vein associated virus (LBVaV), and orchid fleck virus (OFV) (24–26). The results showed that SsNSRV-1 is most closely related to bornaviruses and nyaviruses (Fig. 3B); however, compared with Borna disease, Nyamanini, and midway viruses, it differs largely in genome size and gene organization.
Our phylogram showed that the current mononegaviruses were divided into two major clusters, which can be further split into four evolutionary lineages, I–IV (Fig. 3B). Lineages I and II were grouped into the first major cluster, and all of the viruses in this cluster possess large genomes and can cause dreadful diseases in vertebrates including humans. The remaining viruses have smaller genome sizes and formed the second major cluster, with varied hosts, including vertebrates, plants, and nematodes. SsNSRV-1 was also included in this cluster. In the second major cluster, although SsNSRV-1, bornaviruses, and nyaviruses made up the third lineage (lineage III), and the rhabdoviruses and the two segmented (−)RNA viruses (OFV and LBVaV) comprised the fourth lineage (lineage IV), SsNSRV-1 showed only a distant phylogenetic relationship with all other viruses in lineage IV.
Hypovirulence and Associated Traits of S. sclerotiorum Infected by SsNSRV-1.
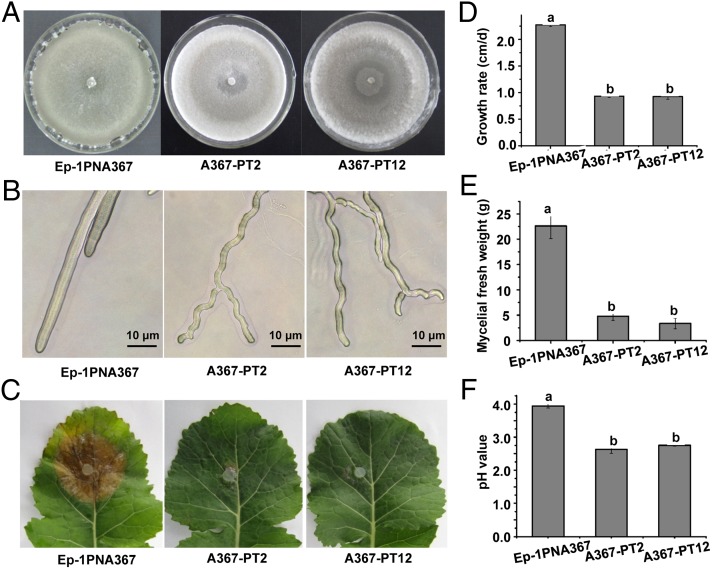
Using PEG-mediated methods, the virulent strain Ep-1PNA367 was successfully transfected with SsNSRV-1 virions, and this was confirmed by both viral RNA extraction and RT-PCR amplification as shown in Fig. S4. The newly SsNSRV-1–infected isolates displayed debilitation symptoms, including slow growth on potato dextrose agar (PDA), loss of the ability to produce sclerotia, and loss of pathogenicity on rapeseed (Fig. 4 A, C, and D). Moreover, we found that almost all of the hyphal tips of these newly transfected strains extended in a zigzag style, indicating the possible loss of growth polarity (Fig. 4B). Although the biomass of these newly infected strains has significantly decreased (Fig. 4E), the strains accumulated more acidic substances because the pH value of the culture filtrate was significantly lower than that of virus-free strain Ep-1PNA367 (Fig. 4F). The biological traits of strain AH98, which was naturally coinfected by SsNSRV-1 and SsHV-1, were slightly different from that of the SsNSRV-1–infected Ep-1PNA367. It appears that strain AH98 did not significantly lose growth polarity (Fig. S1).
Fig. 4.
Biological characterization of the SsNSRV-1–infected strain Ep-1PNA367 of S. sclerotiorum. (A) Colonial morphology of virus-infected strains growing on PDA at 20 °C for 10 d. (B) Hyphal tips of the SsNSRV-1–infected strain. (C) SsNSRV-1–infected strains lost virulence on detached leaves of rapeseed. Photos and data were taken at 3 d postinoculation. (D) Growth rate of SsNSRV-1–infected strains on PDA at 20 °C. (E) Biomass of SsNSRV-1–infected strains. (F) The pH values of filtrates of the SsNSRV-1–infected strain grown in 100 mL PDB in a flask for 6 d. Error bars indicate the SD from five sample means. Means followed by the different letters on the top of each column are significantly different at the P < 0.05 level of confidence according to Duncan's multiple range test.
Wide Distribution of SsNSRV-1 in Nature.
SsNSRV-1 is widely distributed in different climatological and geographical environments in China. Besides the Anhui Province from where the SsNSRV-1–infected strain AH98 was isolated, SsNSRV-1 was successfully detected in six strains of S. sclerotiorum, which were sampled from Heilongjiang, Hubei, Jiangxi, and Shaanxi Provinces. The Heilongjiang Province is in the northeast, the Jiangxi Province is in the southeast, and Shaanxi Province is in the northwest. The distance between Heilongjiang and Jiangxi Provinces is about 3,000 km, and these two provinces have very distinct climates. Notably, the SsNSRV-1–infected strain HLJ-048 was isolated from diseased soybean, whereas the other strains were isolated from diseased rapeseed.
Discussion
Kondo et al. (10) have presented strong evidence that (−)ssRNA viruses may occur in nature and potentially infect fungi. However, important questions about genome structure, virion morphology, transcription strategy, and infectivity remained to be answered. Here, we purified virions of a (−)ssRNA virus from the hypovirulent strain AH98 of S. sclerotiorum and characterized its genomic structure, transcription strategy, phylogenetic relationship, and biological impacts on its host. We have thus demonstrated that (−)ssRNA viruses do indeed infect fungi in nature.
All virions of members in the order Mononegavirales are enveloped with a phospholipid membrane originating from the host, but they are pleomorphic. Even among viruses belonging to the same family, they are always distinct in virion shape, e.g., paramyxoviruses (27). The virions of SsNSRV-1 seem to have a filamentous morphology that resembles Ebola virus virions. In view of the fact that nucleocapsids were not directly observed, it is presumed that a substructure, most likely a membrane, envelops the helical nucleocapsids (Fig. 1A). Moreover, putative enveloped virions-like structures were also observed in ultrathin hyphal sections (Fig. S2). The helical nucleocapsids would be released when this membrane-like substructure is damaged (Fig. 1A). However, the internal nucleocapsids in SsNSRV-1 virions are arranged differently from those of Ebola virus. The nucleocapsids of SsNSRV-1 seem to be organized in a helical style within the virions (Fig. 1A), whereas in the Ebola virus, the nucleocapsids are arranged linearly (15).
Despite the differences in virion shape and nucleocapsid organization, the structure and characteristics of SsNSRV-1 nucleocapsids are strikingly similar to those of almost all of the mononegaviruses. For example, the viral nucleocapsids have helical structures with widely variable conformation and can serve directly as the templates for replication and transcription. Therefore, among mononegaviruses, there is an extensive, conserved, and regular protein scaffold, which confers a unique feature distinct from all other virus families and plays critical roles in RNA transcription, replication, and virus assembly for adapting to the changing cellular microenvironment.
The genomic arrangement of SsNSRV-1 does not follow the basic five-gene pattern (N-P-M-G-L) of mononegaviruses (22). First, the genome of SsNSRV-1 has six ORFs, which were confirmed by Northern blot analysis, RT-PCR amplification, and 5′ and 3′ RACE analyses. Second, the NP may have two forms and both were encoded by ORF II instead of ORF I (Fig. 1G and Table S2). Two forms of NP have occurred in the Marburg and bornaviruses. In the Marburg virus, the 94-kDa form of the NP is phosphorylated, whereas the 92-kDa form is nonphosphorylated (28). In bornavirus, the p40 and p38 are generated by alternative initiation of translation and differ by 13 aa at the N terminus, where the nuclear localization sequence is located (29). Third, the ORF IV of SsNSRV-1, whose position corresponds to the glycoprotein G of mononegaviruses, is unlikely to encode G protein, not only because it is the smallest gene of SsNSRV-1 (186 bp), but also because of the lack of a putative signal peptide, a transmembrane segment, an external ectodomain, and an internal endodomain, which are universal structures of viral G protein (30). In rhabdoviruses, the G protein is thought to play a critical part in membrane fusion during cell entry by changing its conformation and causing rearrangement of the cellular membrane (31). However, considering the special environment and features of the fungal hosts, SsNSRV-1 may lack the G protein gene. Some mycoviruses, although phylogenetically related to animal or plant viruses, often lack nonessential genes, including genes for coat protein and movement protein (32–34). And fourth, there is an additional ORF VI following the L gene (ORF V), which is absent in all extant mononegaviruses. We compared SsNSRV-1 with the sequences assembled by Kondo et al. (10) in genome organization and putative ORFs. Interestingly, downstream of the L-like ORFs, all the sequences generated by Kondo et al. (10) have an ORF corresponding to the position of ORF VI in the SsNSRV-1 genome, although they do not have any significant similarity with each other. Moreover, there is a significant similarity between the NP of SsNSRV-1 and the ORF I of ShTSA3-L or the ORF II of ShTSA3-L. The SsNSRV-1 L protein also shows a significant similarity to all three L-like ORFs, whereas the remaining ORFs of SsNSRV-1 had no sequence similarity to those virus-like sequences. Therefore, fungal negative-stranded viruses may have special gene patterns that are distinct from the basic gene patterns (N-P-M-G-L) of most monopartite (−)ssRNA viruses. This finding may be indicative of another coevolutionary history with the fungal hosts.
Currently, the order Mononegavirales contains five families, and all are collectively divided into four evolutionary lineages based on phylogenetic analysis (Fig. 3B). Although the fungal negative-stranded virus SsNSRV-1 is clustered with bornaviruses and nyaviruses, it forms a separate subclade that is different from these two groups of mononegaviruses. This finding suggests that this fungal negative-stranded virus may represent another lineage in the order Mononegavirales, and a previously unidentified virus family, tentatively designated Mycomononegaviridae, may need to be created to encompass SsNSRV-1 and other fungal mononegaviruses that are yet to be discovered.
SsNSRV-1 can significantly reduce the growth rate and sclerotial production and can confer hypovirulence to S. sclerotiorum (Fig. 4 A–E). Although hypovirulent strains Ep-1PNA367-PT2 and Ep-1PNA367-PT12 have much less biomass than Ep-1PNA367 (Fig. 4E), the pH values of filtrates of the two strains infected by SsNSRV-1 were significantly lower than that of strain Ep-1PNA367 (Fig. 4F). This result suggests that the two strains can accumulate comparable amounts of oxalic acid, which is an essential pathogenicity determinant for S. sclerotiorum (35). Therefore, SsNSRV-1 is likely to affect other pathogenesis pathways rather than oxalic acid in S. sclerotiorum.
Briefly, we have isolated and characterized a fungal (−)ssRNA virus (SsNSRV-1) in S. sclerotiorum, which is phylogenetically related to viruses in the families Bornaviridae and Nyamiviridae, and we found this virus can successfully transfect the host’s protoplasts under the mediation of PEG. The discovery of this (−)ssRNA mycovirus may provide insights into the global virus ecology and evolution as well as the interactions between S. sclerotiorum and SsNSRV-1, and may also provide a system for examining fungal factors involved in (−)ssRNA viral replication and maintenance. Furthermore, because many (−)ssRNA viruses are serious human pathogens, this system is likely to provide a less hazardous path to study the replication of a (−)ssRNA virus and to establish a system for screening antiviral compounds against (−)ssRNA viruses.
Materials and Methods
Fungal Strains and Maintenance.
S. sclerotiorum strain AH98 was isolated from a sclerotium obtained from a diseased rapeseed (Brassica napus) plant from the Anhui Province. The virus-free strain Ep-1PNA367 was a single-ascospore-isolation progeny of a mycovirus-infected hypovirulent strain of Ep-1PN (33). The two strains and other strains used to analyze the natural distribution of SsNSRV-1 were cultured on PDA at 20–22 °C and stored on PDA slants at 4 °C.
Purification and Observation of Virus Particles.
The viral particles were isolated from strains AH98 and Ep-1PNA367-PT2 in accordance with the method described by Yu et al. (4) with minor modification. These strains were grown at 20 °C for 7–10 d on sterilized cellophane films placed on PDA or in potato dextrose broth (PDB). Approximately 30 g mycelia were ground in liquid nitrogen and mixed with three volumes of 0.1 M phosphate buffer (pH 7.0) containing 0.5% mercaptoethanol (vol/vol), and then the mixture was gently shaken on ice for 30 min. The mixture was separated with high-speed centrifugation (12,096 × g for 30 min), and supernatant collected to extract virions with ultracentrifugation. The virus particles were stained with 2% (wt/vol) phosphotungstic acid (PTA) solution (pH 7.4) and observed using a transmission electron microscope (Model Tecnai G2 20; FEI Company). To detect the morphology of virions in the hyphal cells, ultrathin fungal sections of the strain AH98 and Ep-EPNA367-PT2 were prepared to detect the morphology of SsNSRV-1 virions under TEM. These ultrathin fungal sections were prepared and observed in accordance with the method described by Wu et al. (36).
PMF Analysis of Viral Protein.
The purified virus particles after sucrose gradient centrifugation were boiled for 10 min and then loaded on 12% (vol/vol) polyacrylamide gel amended with 1% SDS. After electrophoresis, the gel was stained with Coomassie brilliant blue R250. The resulting protein bands were individually cut from the gel and subjected to PMF analysis at BGI Co., Ltd. To avoid missing any proteins during virion preparation, protein samples directly extracted from crude virion preparations (after ultracentrifugation) and 20% and 25% (wt/vol) sucrose gradient components (∼300 μg) were subjected to PMF analysis.
Nucleic Acid Extraction, cDNA Cloning, and Sequencing.
Viral genome RNA samples from virus nucleocapsids were extracted using phenol/chloroform extraction. Fungal total RNA samples were extracted using RNAiso, according to the RNA extraction reference (TaKaRa). All RNA samples were stored at −70 °C before use. RNA samples, either from virus particles or from mycelia mass, were used for cDNA cloning and terminal determination in accordance with the procedures described by Xie et al. (14). The terminal sequences of the viral genome were confirmed in accordance with methods described by Potgieter et al. (37) and Briese et al. (38). Primers and other oligonucleotides used for in this study are listed in Table S3.
Sequence and Phylogenetic Analysis.
Sequence analysis and assembly were performed using DNAMAN 6.0 software (LynnonBiosoft). The deduced amino acid sequences of SsNSRV-1 and the selected mononegaviruses were aligned using the online Constraint-based Multiple Protein Alignment Tool (www.ncbi.nlm.nih.gov/tools/cobalt/). The best-fit model of protein evolution (LG+I+G+F) was determined by maximum likelihood analyses using ProtTest 2.4 (39) (http://darwin.uvigo.es/software/prottest_server.html), based on the Akaike Information Criterion. Our phylogenetic tree was generated with PhyML 3.0, using the appropriate substitution mode (40). The tree was visualized using MEGA Version 5.0 programs (41).
RT-PCR, Northern Blot Analysis, and 5′ and 3′ RACE Analyses.
To confirm the characteristics of the genomic nucleic acid from the viral particles, 1.5 μg nucleic acid sample were treated with DNase I, S I nuclease, and RNase A and followed by RT-PCR using gene-specific primers. To detect the ORFs of SsNSRV-1, 1.5 μg total RNA from strain AH98 were reverse transcribed using oligo(dT) primer and the product amplified using gene-specific primers. The actin gene of S. sclerotiorum was used as the control. Northern blot analysis was done following the methods of Streit et al. (42) with minor modifications. DNA fragments were amplified by primer pairs based on the SsNSRV-1 genome sequence and were labeled with digoxigenin (DIG) as described in the user manual (GE Healthcare). The chemiluminescent signals of the probe-RNA hybrids were detected using a CDP-Star kit (GE Healthcare).
The 5′ and 3′ RACE techniques were used to determinate the transcription strategy of SsNSRV-1. The total RNA samples were extracted from mycelia growing on PDA for 4–6 d, and then the poly(A) RNA samples were isolated with a Dynabeads mRNA DIRECT Kit (Life Technologies). These poly(A) RNA samples were subjected to 5′ and 3′ RACE experiments using the methods described by Elizabeth et al. (43) and Bekal et al. (12), respectively. The primers and other oligonucleotides used for RT-PCR and Northern blot and RACE analyses are listed in Table S3.
Protoplast Preparation and Transfection Assays.
The Ep-1PNA367 strain was used as the recipient. The preparation of fungal protoplasts and the introduction of purified viruses into the protoplast preparations followed Yu et al. (4). The newly transfected isolates were confirmed by RT-PCR amplification using gene-specific primer pairs. The primers used here are listed in Table S3.
Biological Properties and Microscopic Observation of Fungal Strains.
Mycelial growth, colony morphology, and virulence tests on the detached leaves of rapeseed were assessed according to Yu et al. (4). To observe the morphology of hyphal tips, all tested strains were grown on PDA for 1–2 d and typical hyphal tips of each strain were carefully examined and photographed using a digital camera (Eclipse 55i; Nikon). To determine the pH values of the filtrates, four mycelial agar plugs (5 mm in diameter) were inoculated in a 250 mL-flask containing 100 mL PDB and incubated in a shaker at 100 rpm for 6 d. After centrifugation at 12,096 × g for 5 min, the resulting precipitate was used for analysis of mycelial fresh weight, and the supernatant was used for pH determination. All of the data collected were subjected to statistical analysis with a statistical software Statistical Analysis System (SAS), version 8.0 (ftp://legacy.gsfc.nasa.gov/xmm/software/sas/).
Detection of SsNSRV-1 in S. sclerotiorum Isolates from Different Regions of China.
Fifty hypovirulent isolates of S. sclerotiorum collected from different regions of China—the provinces of Heilongjiang, Shaanxi, Sichuan, Hunan, Hubei, Anhui and Jiangxi—were used for the detection of SsNSRV-1. The total RNA of these strains was extracted and subjected to cDNA synthesis using random hexanucleotides. Two primer pairs were designed to detect SsNSRV-1, and the actin gene was used as the control. The primers are listed in Table S3.
Supplementary Material
Acknowledgments
We thank Said A. Ghabrial (University of Kentucky) and Tom Hsiang (University of Guelph) for editing assistance, and the anonymous reviewers for constructive and helpful comments. This research was supported by China National Funds for Distinguished Young Scientists (Grant 31125023), the Special Fund for Agro-Scientific Research in the Public Interest of China (Grant 201103016), and China Agriculture Research System (Grant CARS-13).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.I.H. is a guest editor invited by the Editorial Board.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. KJ186782).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401786111/-/DCSupplemental.
References
- 1.Ghabrial SA, Suzuki N. Viruses of plant pathogenic fungi. Annu Rev Phytopathol. 2009;47:353–384. doi: 10.1146/annurev-phyto-080508-081932. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostakis SL. Biological control of chestnut blight. Science. 1982;215(4532):466–471. doi: 10.1126/science.215.4532.466. [DOI] [PubMed] [Google Scholar]
- 3.Chiba S, et al. A novel bipartite double-stranded RNA Mycovirus from the white root rot Fungus Rosellinia necatrix: Molecular and biological characterization, taxonomic considerations, and potential for biological control. J Virol. 2009;83(24):12801–12812. doi: 10.1128/JVI.01830-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu X, et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci USA. 2010;107(18):8387–8392. doi: 10.1073/pnas.0913535107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X, et al. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci USA. 2013;110(4):1452–1457. doi: 10.1073/pnas.1213755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawe AL, Nuss DL. Hypovirus molecular biology: From Koch’s postulates to host self-recognition genes that restrict virus transmission. Adv Virus Res. 2013;86:109–147. doi: 10.1016/B978-0-12-394315-6.00005-2. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, et al. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol Biol. 2012;12:91. doi: 10.1186/1471-2148-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin YH, et al. A novel quadripartite dsRNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix. Virology. 2012;426(1):42–50. doi: 10.1016/j.virol.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Wu M, et al. Characterization of a novel bipartite double-stranded RNA mycovirus conferring hypovirulence in the phytopathogenic fungus Botrytis porri. J Virol. 2012;86(12):6605–6619. doi: 10.1128/JVI.00292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo H, Chiba S, Toyoda K, Suzuki N. Evidence for negative-strand RNA virus infection in fungi. Virology. 2013;435(2):201–209. doi: 10.1016/j.virol.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. Biology of plant rhabdoviruses. Annu Rev Phytopathol. 2005;43:623–660. doi: 10.1146/annurev.phyto.43.011205.141136. [DOI] [PubMed] [Google Scholar]
- 12.Bekal S, Domier LL, Niblack TL, Lambert KN. Discovery and initial analysis of novel viral genomes in the soybean cyst nematode. J Gen Virol. 2011;92(Pt 8):1870–1879. doi: 10.1099/vir.0.030585-0. [DOI] [PubMed] [Google Scholar]
- 13.Bolton MD, Thomma BPHJ, Nelson BD. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol Plant Pathol. 2006;7(1):1–16. doi: 10.1111/j.1364-3703.2005.00316.x. [DOI] [PubMed] [Google Scholar]
- 14.Xie J, et al. A novel mycovirus closely related to hypoviruses that infects the plant pathogenic fungus Sclerotinia sclerotiorum. Virology. 2011;418(1):49–56. doi: 10.1016/j.virol.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Ellis DS, et al. Ultrastructure of Ebola virus particles in human liver. J Clin Pathol. 1978;31(3):201–208. doi: 10.1136/jcp.31.3.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mavrakis M, Kolesnikova L, Schoehn G, Becker S, Ruigrok RWH. Morphology of Marburg virus NP-RNA. Virology. 2002;296(2):300–307. doi: 10.1006/viro.2002.1433. [DOI] [PubMed] [Google Scholar]
- 17.Iseni F, Barge A, Baudin F, Blondel D, Ruigrok RWH. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J Gen Virol. 1998;79(Pt 12):2909–2919. doi: 10.1099/0022-1317-79-12-2909. [DOI] [PubMed] [Google Scholar]
- 18.Mountcastle WE, Compans RW, Caliguiri LA, Choppin PW. Nucleocapsid protein subunits of simian virus 5, Newcastle disease virus, and Sendai virus. J Virol. 1970;6(5):677–684. doi: 10.1128/jvi.6.5.677-684.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda T, Hagiwara K, Sagara H, Kawaoka Y. Characterization of the Ebola virus nucleoprotein-RNA complex. J Gen Virol. 2010;91(Pt 6):1478–1483. doi: 10.1099/vir.0.019794-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heggeness MH, Scheid A, Choppin PW. Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: reversible coiling and uncoiling induced by changes in salt concentration. Proc Natl Acad Sci USA. 1980;77(5):2631–2635. doi: 10.1073/pnas.77.5.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruigrok RWH, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14(4):504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Pringle CR, Easton AJ. Monopartite negative strand RNA genomes. Semin Virol. 1997;8(1):49–57. [Google Scholar]
- 23.Källberg M, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7(8):1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mihindukulasuriya KA, et al. Nyamanini and midway viruses define a novel taxon of RNA viruses in the order Mononegavirales. J Virol. 2009;83(10):5109–5116. doi: 10.1128/JVI.02667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasaya T, Kusaba S, Ishikawa K, Koganezawa H. Nucleotide sequence of RNA2 of Lettuce big-vein virus and evidence for a possible transcription termination/initiation strategy similar to that of rhabdoviruses. J Gen Virol. 2004;85(Pt 9):2709–2717. doi: 10.1099/vir.0.80061-0. [DOI] [PubMed] [Google Scholar]
- 26.Kondo H, Maeda T, Shirako Y, Tamada T. Orchid fleck virus is a rhabdovirus with an unusual bipartite genome. J Gen Virol. 2006;87(Pt 8):2413–2421. doi: 10.1099/vir.0.81811-0. [DOI] [PubMed] [Google Scholar]
- 27.Lamb RA, Parks GD. In: Paramyxoviridae: The viruses and their replication. Fields Virology. 5th Ed. Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 1449–1496. [Google Scholar]
- 28.Becker S, Huppertz S, Klenk HD, Feldmann H. The nucleoprotein of Marburg virus is phosphorylated. J Gen Virol. 1994;75(Pt 4):809–818. doi: 10.1099/0022-1317-75-4-809. [DOI] [PubMed] [Google Scholar]
- 29.Kobayashi T, et al. Nuclear targeting activity associated with the amino terminal region of the Borna disease virus nucleoprotein. Virology. 1998;243(1):188–197. doi: 10.1006/viro.1998.9049. [DOI] [PubMed] [Google Scholar]
- 30.Steven AC, Spear PG. Biochemistry. Viral glycoproteins and an evolutionary conundrum. Science. 2006;313(5784):177–178. doi: 10.1126/science.1129761. [DOI] [PubMed] [Google Scholar]
- 31.Albertini AAV, Baquero E, Ferlin A, Gaudin Y. Molecular and cellular aspects of rhabdovirus entry. Viruses. 2012;4(1):117–139. doi: 10.3390/v4010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, et al. A novel mycovirus that is related to the human pathogen hepatitis E virus and rubi-like viruses. J Virol. 2009;83(4):1981–1991. doi: 10.1128/JVI.01897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie J, et al. Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J Gen Virol. 2006;87(Pt 1):241–249. doi: 10.1099/vir.0.81522-0. [DOI] [PubMed] [Google Scholar]
- 34.Preisig O, Moleleki N, Smit WA, Wingfield BD, Wingfield MJ. A novel RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua. J Gen Virol. 2000;81(Pt 12):3107–3114. doi: 10.1099/0022-1317-81-12-3107. [DOI] [PubMed] [Google Scholar]
- 35.Williams B, Kabbage M, Kim HJ, Britt R, Dickman MB. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 2011;7(6):e1002107. doi: 10.1371/journal.ppat.1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu M, Zhang L, Li G, Jiang D, Ghabrial SA. Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology. 2010;406(1):117–126. doi: 10.1016/j.virol.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Potgieter AC, et al. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol. 2009;90(Pt 6):1423–1432. doi: 10.1099/vir.0.009381-0. [DOI] [PubMed] [Google Scholar]
- 38.Briese T, et al. Genomic organization of Borna disease virus. Proc Natl Acad Sci USA. 1994;91(10):4362–4366. doi: 10.1073/pnas.91.10.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 40.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 41.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Streit S, Michalski CW, Erkan M, Kleeff J, Friess H. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat Protoc. 2009;4(1):37–43. doi: 10.1038/nprot.2008.216. [DOI] [PubMed] [Google Scholar]
- 43.Scotto-Lavino E, Du G, Frohman MA. 5′ end cDNA amplification using classic RACE. Nat Protoc. 2006;1(6):2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.