Significance
The most recent common ancestor of Paratyphi A, one of the most common causes of enteric fever, existed approximately 450 y ago, centuries before that disease was clinically recognized. Subsequent changes in the genomic sequences included multiple mutations and acquisitions or losses of genes, including bacteriophages and genomic islands. Some of those evolutionary changes were reliably attributed to Darwinian selection, but that selection was only transient, and many genetic changes were subsequently lost because they rendered the bacteria less fit (purifying selection). We interpret the history of Paratyphi A as reflecting drift rather than progressive evolution and suggest that most recent increases in frequencies of bacterial diseases are due to environmental changes rather than the novel evolution of pathogenic bacteria.
Keywords: pathogen evolution, historical reconstruction, phylogeography
Abstract
Multiple epidemic diseases have been designated as emerging or reemerging because the numbers of clinical cases have increased. Emerging diseases are often suspected to be driven by increased virulence or fitness, possibly associated with the gain of novel genes or mutations. However, the time period over which humans have been afflicted by such diseases is only known for very few bacterial pathogens, and the evidence for recently increased virulence or fitness is scanty. Has Darwinian (diversifying) selection at the genomic level recently driven microevolution within bacterial pathogens of humans? Salmonella enterica serovar Paratyphi A is a major cause of enteric fever, with a microbiological history dating to 1898. We identified seven modern lineages among 149 genomes on the basis of 4,584 SNPs in the core genome and estimated that Paratyphi A originated 450 y ago. During that time period, the effective population size has undergone expansion, reduction, and recent expansion. Mutations, some of which inactivate genes, have occurred continuously over the history of Paratyphi A, as has the gain or loss of accessory genes. We also identified 273 mutations that were under Darwinian selection. However, most genetic changes are transient, continuously being removed by purifying selection, and the genome of Paratyphi A has not changed dramatically over centuries. We conclude that Darwinian selection is not responsible for increased frequency of enteric fever and suggest that environmental changes may be more important for the frequency of disease.
The most recent common ancestors (MRCA) of some bacterial pathogens such as Helicobacter pylori and Mycobacterium tuberculosis existed nearly 100,000 ya (1, 2), setting a lower limit for how long they have infected humans. Other MRCAs are much younger, ranging from ∼3,000 y for Yersinia pestis and Mycobacterium leprae (3, 4) to only decades for individual clones of Clostridium difficile, Staphylococcus aureus, and Shigella sonnei (5–7). However, the ages of most bacterial pathogens remain unknown. Here, we use genomic analyses of 149 isolates of Salmonella enterica serovar Paratyphi A to address the age and microevolutionary history of one of the major causes of enteric fever.
Enteric fever is a generic epidemiological designation for clinically similar syndromes of prolonged, systemic human salmonellosis that are called typhoid fever when caused by serovar Typhi, and paratyphoid fever when caused by serovars Paratyphi A, B, or C. Each of these serovars corresponds to a distinct, human-specific, genetically monomorphic bacterial population according to multilocus sequence typing (8). However, these four populations are not related to each other at the genetic level, nor do we know which convergent genetic features are responsible for causing similar disease syndromes.
The annual global burden of enteric fever has been estimated as 27 million cases of clinical disease and 200,000 deaths (9), almost all of which are caused by Typhi or Paratyphi A. It is not possible to reconstruct what the disease burden of enteric fever was in the past because of insufficiently discriminatory historical descriptions of clinical syndromes. Until the mid-19th century, enteric fever was not even reliably distinguished from typhus (10), which is caused by Rickettsia, and the distinction between serovars Typhi and Paratyphi A was first achieved in 1898 (11). At that time, Paratyphi A was relatively common in North America (12). Today, Paratyphi A accounts for a sizable fraction (14–64%) of all enteric fever in India, Pakistan, Nepal, Indonesia, and China (13–16), but has largely disappeared from Europe and North America, except for travelers returning from South and Southeast Asia (17, 18). Otherwise, little is known about its historical patterns of spread or its evolutionary history.
Phylogeographic reconstructions of genetically monomorphic pathogens can be achieved by comparative genomics (3, 4, 6, 19). However, only two complete Paratyphi A genomes (20, 21) and five draft genomes (16) have been described. Comparisons of the two complete genomes revealed multiple pseudogenes (20, 21), which were interpreted as reflecting adaptation to its human host, but it is not clear whether the formation of pseudogenes reflects continued adaptation, or is possibly only a transient phenomenon because it results in lessened fitness (22) associated with temporary adaptations to fluctuating environments (23). Indeed, the dynamics of changes in the contents of the pan-genome are unclear for almost all bacterial taxa, as are the selective forces that shape genomic content with time. Understanding such dynamics depends on algorithms that are suitable for large numbers of genomes, reliably attribute individual genetic polymorphisms to mutation (vertical descent) or homologous recombination, and can distinguish whether differences in genomic content reflect gain or loss. We used vertically acquired mutations identified by a novel algorithm to reconstruct the genealogy of Paratyphi A over the period of 450 y since its MRCA. In turn, this genealogy led to a reconstruction of the history of global transmissions since the mid-1800s. We have also mapped all genetic changes at the genomic level to that genealogy, thus showing that most are transient due to purifying selection, including multiple mutations that were attributed to Darwinian selection by a second new algorithm.
Results
Vertical Descent in Core Genomes.
We performed short-read genomic sequencing (Illumina) of 142 Paratyphi A strains. To strengthen dating estimates, we included 42 old strains (1917–1980) from the historical, global collection at the Institut Pasteur (Paris) (Dataset S1, tab 1). The remaining strains were isolated between 1997 and 2009 and represent the current global distribution of Paratyphi A, including isolates from India (42 genomes) and Pakistan (12), where Paratyphi A is now most common. For each of these strains, as well as for publicly available short reads from five additional Chinese isolates (16), we performed de novo assemblies and mapped the reads to those assemblies to avoid assembler errors that result in false SNP calls (24). The final set of 149 genomes, including the completed genomes of ATCC 9150 (20) and AKU 12601 (21), yielded a 4,073,403-bp core genome after removing repetitive DNA (henceforth core genome). Mapping of all polymorphic sites within the core genome to ATCC 9150 identified only 4,584 SNPs (Table 1), which is comparable to the diversity found in Y. pestis (3), serovar Typhi (25), and other genetically monomorphic bacterial pathogens (26). These SNPs yielded a single (unambiguous) maximum parsimony tree, except for several polytomies and a few homoplasies (repeated, independent, convergent mutations) (SI Appendix, Fig. S1). The same topology was obtained by maximum likelihood (SI Appendix, Fig. S2) and maximum clade credibility (Fig. 1B) analyses of 4,525 SNPs after excluding recombinant SNPs (see below).
Table 1.
Summary statistics for 149 genomes of S. enterica serovar Paratyphi A
| Number | |
| Genomes | 149 |
| Mean read coverage | 153 (62.5–631.6) |
| Mean genome length, bp | 4,573,587 (4,432,213–4,794,508) |
| Core genome, bp | 4,073,403 |
| Intact CDSs in core genome | 3,365 |
| Pseudogenes mutated before MRCA | 117 |
| Pseudogenes since MRCA | 300 |
| Nonrepetitive SNPs/ indels | 4,584/443 |
| Regions (length, bp) of recombination | 24 (419) |
| Recombinant SNPs/indels | 59/3 |
| Nonrepetitive, nonrecombinant SNPs/indels | 4,525/440 |
| Homoplastic SNPs/indels | 29/46 |
| Regions (length, bp) under selection | 76 (24,038) |
| SNPs/indels under selection | 165/108 |
| Accessory genome (genes) | 1,553 |
| Total length of accessory genes, bp | 1,178,142 |
| Accessory regions (acquisitions/Loss) | 82 (50/93) |
| Bacteriophages (acquisitions/Loss) | 23 (29/41) |
| Integrated plasmids (acquisitions/Loss) | 2 (2/0) |
| Plasmids (acquisitions/loss) | 11 (18/2) |
| Other genomic islands (acquisitions/loss) | 46 (1/50) |
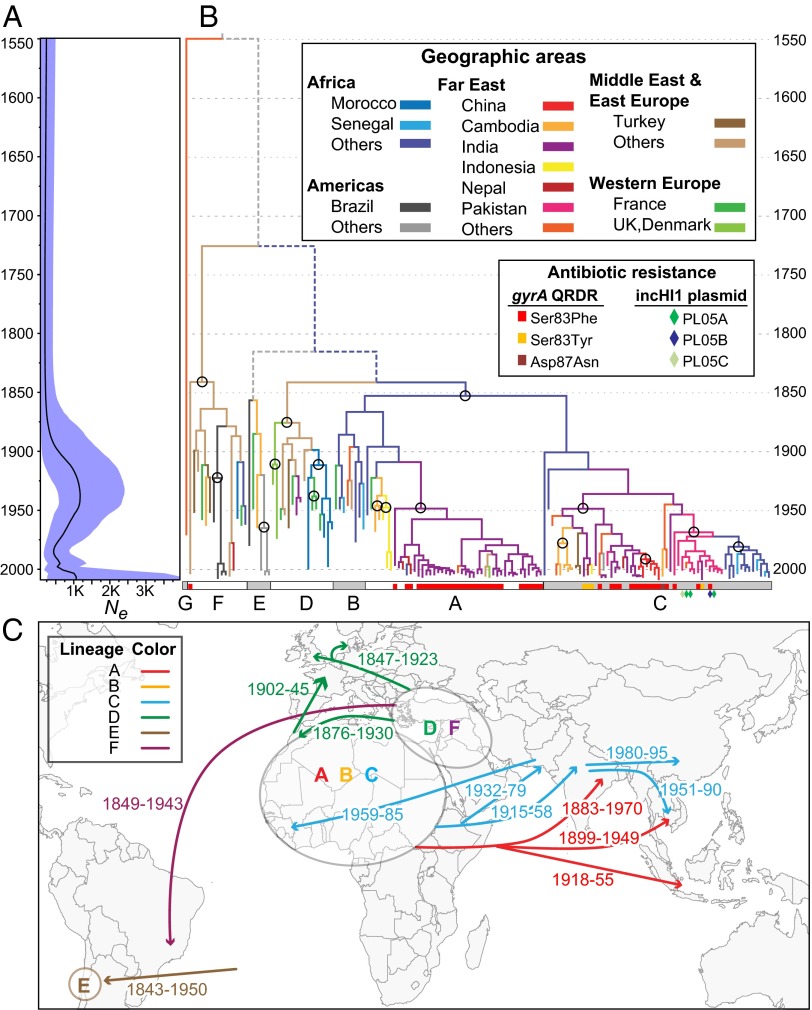
Fig. 1.
Phylogeny and demography of Paratyphi A. The 4,525 nonrepetitive, nonrecombinant SNPs in the core genome (Table 1) were analyzed with Beast 1.8 by using the model (exponential clock rate; Bayesian Skyline) of population dynamics among 14 combinations (Dataset S1, tab 2), which yielded the highest Bayes factors. (A) Bayesian skyline plot showing temporal changes since 1549 in effective population size (Ne) (black curve) with 95% confidence intervals (cyan). (B) Maximum clade credibility tree (asymmetric diffusion model, BSSVS, no transmission from Western Europe), colored by geographical sources of bacterial isolates (tips) and inferred historical sources (branches). Older transmissions that were supported by ≥80% of trees are indicated by circles, and depicted in C. Inner branches with lower levels of support are indicated by dashed, colored (≥50% support), or gray (<50%) lines. Lineages A..G are indicated at the base by alternating white and gray rectangles, which also present information on antibiotic resistance due to mutations in gyrA or the acquisition of an MDR IncHI1 plasmid (diamonds). (C) Sources (ovals) of lineages and associated geographic transmissions (arrows with CI95% of dates; Dataset S1, tab 3).
The low number of homoplasies indicates that homologous recombination has been very rare in Paratyphi A since its MRCA, or absent. This conclusion is seemingly contradictory to prior analyses (27) showing that extensive homologous recombination was previously common between Typhi and Paratyphi A, resulting in exceptionally similar stretches spanning one-quarter of their genomes. (Our independent analyses confirmed the low diversity between large portions of Typhi and Paratyphi A genomes.) We therefore attempted to quantify the number of polymorphic core SNPs within Paratyphi A that arose through homologous recombination with unrelated donors during recent microevolution. To this end, we developed a novel Hidden Markov Model (RecHMM), which detects the clusters of sequence diversity that mark recombination events within individual branches (SI Appendix, SI Materials and Methods). RecHMM yields comparable results to ClonalFrame (28), except that it is computationally more efficient and can be used for comparisons of hundreds of bacterial core genomes. RecHMM identified only 59 SNPs and 3 short insertions or deletions (indels) in a total of 419 bp scattered over 24 recombinant regions (Table 1 and Dataset S1, tab 4). Thus, whereas homologous recombination from an unrelated donor (Typhi) was extensive before the MRCA of Paratyphi A, it has essentially stopped during more recent history, and 99% (4,525/4,584) of SNPs in the core genome arose by mutation (vertical descent).
Historical Transmissions.
All three phylogenetic methods defined seven deep branches, designated lineages A..G (Fig. 1B), of which lineage G is quite distinct, and has only been isolated once (Hong Kong, 1971). According to the most probable Bayesian model for Beast 1.8 (29) (Dataset S1, tab 2), the MRCA of Paratyphi A existed in 1549 (CI95%: 1247–1703), and mutations have accumulated with a mean (exponential, relaxed) clock rate of 1.94 × 10−7 per nucleotide per year. At least 80% of the maximum credibility trees generated by Beast supported the presence in the mid-19th century of lineages A–C in Africa, lineages D and F in the Near East, and lineage E in South America (Fig. 1C and Dataset S1, tab 3). Subsequently, lineages A and C spread to South and Southeast Asia, and almost all modern isolates from those areas belong to those lineages. Lineage D spread to Morocco and Western Europe, whereas lineage F spread to South America, but both are now rare. Comparable results were obtained with Beast 2 (30), except that it calculated a 200 y older TMRCA (Dataset S1, tab 2). Thus, Paratyphi A is likely to have been a major cause of life-threatening disease in humans over the last 450–700 y, or longer, and has spread globally on multiple occasions.
The effective population size, Ne, of Paratyphi A stayed constant until the early 20th century, at which point Ne increased fivefold during the repeated spread of Paratyphi A between countries and continents (Fig. 1A). Ne dropped dramatically in the 1950s, possibly due to the introduction of antibiotics for the treatment of disease, but has risen again in the last decade. The frequency of transmissions between geographical areas has decreased since the mid-1950s, and most of those rare transmissions now involve lineages A and C (SI Appendix, Fig. S3).
Darwinian Selection Within the Accessory Genome.
We were intrigued by the homoplastic SNPs and short indels within the core genome (Table 1). Homoplasies are often a sign of recombination (31). However, only 3/29 homoplastic SNPs and 1/46 homoplastic short indels mapped within the 24 recombinant regions. An alternative source of homoplastic mutations is Darwinian selection for convergent mutations in the same gene, or the same nucleotide, on multiple, independent occasions, such as has been observed in laboratory evolution (32–34). However, genomic analyses of several genetically monomorphic pathogens have failed to reveal traces of Darwinian selection (3, 24, 35, 36), except that antibiotic resistance is associated with some lineages (5, 6). We did not identify any lineage-specific changes in antibiotic resistance in Paratyphi A. Reduced susceptibility to some antibiotics can be repeatedly acquired (convergent evolution) but then lost again due to the greater fitness of antibiotic-sensitive strains (37–40) (purifying selection). Indeed, one of the homoplastic sites in Paratyphi A is within the Quinolone Resistance Determining Region (QRDR) of the gyrA DNA gyrase gene, and, similar to serovar Typhi (37), mutations at that site (Ser83Phe, Ser83Tyr) that result in reduced sensitivity to fluoroquinolones were repeatedly acquired and lost within the core genome genealogy defined by the core SNPs (Fig. 1B). We also made similar observations for the presence or absence of IncHI1 plasmids associated with multiple drug resistance (41), which form part of the accessory genome (genes that were lacking in one or more genomes). These plasmids were present in individual isolates of lineage C from Pakistan, but other closely related isolates were antibiotic-sensitive and/or did not carry these plasmids, consistent with their repeated gain and loss (Fig. 1B).
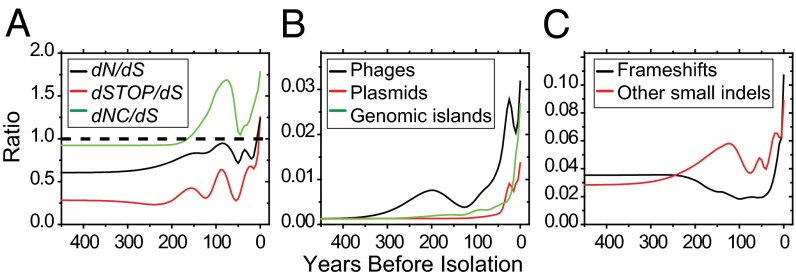
These observations stimulated an intensive search for additional Darwinian selection within the accessory and the core genome. The 1,560 genes (1.2 Mb; Dataset S1, tab 5) of the accessory genome were clustered in 82 large regions with strong homologies to bacteriophages (23 regions), plasmids in the cytoplasm (11) or integrated into the chromosome (2), and other genomic islands (46) (Table 1). Based on the core SNP genealogy, 143 individual events of gain or loss were observed for these 82 regions (SI Appendix, Fig. S1). Homoplastic gain might reflect Darwinian selection, but none of these acquisitions were significantly associated with any single genealogical branch (P > 0.05), and except for the plasmids, most mobile elements were lost even more frequently than they were acquired (Table 1). None of the bacteriophages carried any obvious cargo genes that might have changed the bacterial phenotype, and almost all acquisitions or losses were very recent, and restricted to terminal branches or the tips of the tree (Fig. 2B). These patterns are those expected for frequent transmissions of selfish DNA, followed by their loss within 100 y because of purifying selection.
Fig. 2.
Temporal mapping of ratios of genetic changes by mutation type (see SI Appendix, Fig. S4 for additional details). (A) Rates of nonsynonymous (dN), stop (dSTOP) and noncoding (dNC) mutations to rates of synonymous (dS) mutations. (B) Frequencies of acquisition/loss of regions in the accessory genome relative to SNPs. (C) Frequencies of indels relative to SNPs.
Because of frameshifts and stop codons, approximately one in 20 coding sequences is a pseudogene in Paratyphi A genomes ATCC 9150 (173 genes) (20) and AKU_12601 (204 genes) (21). This high frequency has been attributed to streamlining of functions that are unimportant for the infection of the human host (20, 21), a specialized form of Darwinian selection. Gene loss has also been shown to facilitate rewiring of existing enzymatic networks as an initial step in microevolution (23). Selection for gene loss may have occurred early in the evolution of Paratyphi A because 28% (117/417; Table 1) of the pseudogenes found among the 149 genomes were already present in the MRCA. In contrast, most of the other 72% occurred very recently (Fig. 2 A and C), and are restricted to only one or few isolates (SI Appendix, Fig. S5), once again suggesting a transient balance of opposed evolutionary forces due to streamlining/temporary adaptation and purifying selection. The frequent appearance of pseudogenes in the phylogeny demonstrates that their functions are not essential for the infection of humans, the only known host for Paratyphi A. However, their subsequent removal by purifying selection implies that the function of these genes is beneficial for infection and/or transmission between humans, even if they are not essential. Purifying selection also seems to remove many small indels (≤39 bp; Fig. 2C) and nonsynonymous mutations and mutations in noncoding regions (Fig. 2A), because these were also preferentially found in terminal nodes.
Transient Darwinian Selection in the Core Genome.
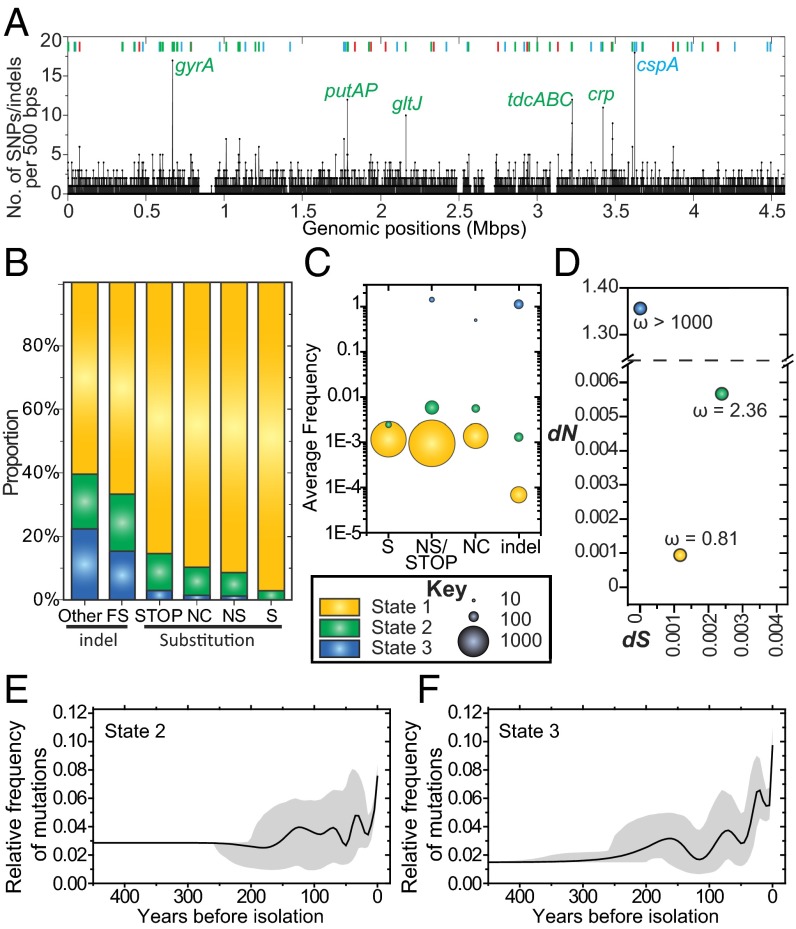
Our observations indicate that most gene acquisitions or losses and mutations are neutral because they are not uniformly present in entire lineages or sublineages, or are rapidly removed by purifying selection. However, they did not exclude the possible existence of rare mutations or short indels that do provide selective advantages. We therefore developed a second, novel Hidden Markov Model (DHMM) (SI Appendix, SI Materials and Methods) to identify regions of clustered SNPs/indels in the core genome, such as would be expected under Darwinian selection (42). Similar to other programs based on Hidden Markov Models, DHMM assigns clustered nucleotides to multiple “states,” but the emission parameters in DHMM were designed to ensure that some of these states correspond to regions that are under diversifying selection. Unlike other methods such as PAML (43) or a χ2 goodness-of-fit test (3), which are restricted to the diversity within genes and ignore intergenic regions, DHMM is suitable for the analysis of all genomic nucleotides. For the nonrecombinant, nonrepetitive core genome of Paratyphi A, DHMM assigned each individual nucleotide to one of three states, designated states 1, 2, and 3 (Fig. 3). State 1 contains 98% of all nucleotides (Dataset S1, tab 6) and likely corresponds to neutral mutations because ω (dN/dS) was 0.8 (Fig. 3D). State 1 included almost all sites with synonymous mutations, and decreasing proportions of sites with nonsynonymous, noncoding, and stop codon mutations, followed by frameshifts and other short indels (Fig. 3B). Mutations assigned to state 2 had a higher ω ratio (2.4; Fig. 3D), indicating moderate diversifying selection, and state 3 did not include any synonymous mutations at all (ω >1000). The vast majority of the 273 SNPs assigned to states 2 or 3 (Dataset S1, tab 7) were in state 2, whereas 69 homoplastic mutations were assigned to state 3 and three homoplastic synonymous mutations to state 1. We interpret these observations as reflecting two groups of polymorphisms with moderate (state 2) or extremely high (state 3) levels of Darwinian selection.
Fig. 3.
Properties of mutations in the nonrepetitive, nonrecombinant core genome assigned to states 1–3 by DHMM. (A) Chromosomal mapping (strain ATCC 9150) of all SNPs and short indels. Lines at the bottom represent densities of mutations in sequential 500-bp windows. Colored bars at the top represent 76 regions containing mutations in states 2 (red), 3 (blue), or both states (green). (B) Proportions of all categories of mutations (X axis) colored by state (Key). FS, frameshift; NC, noncoding; NS, nonsynonymous; S, synonymous; STOP, stop codons. (C) Average frequencies of SNPs and short indels by mutation type per nucleotide in each HMM state. The areas of circles represent the numbers of mutations. (D) Scatter plot of the rates of synonymous (dS) vs. nonsynonymous (dN) mutations and their ratio, ω. (E and F) Temporal mapping of the relative frequencies of mutations to all mutations in state 2 (E) or 3 (F) plus 95% confidence intervals.
The nucleotides in states 2 or 3 clustered in 76 regions spanning 24 kb scattered over the entire genome (Fig. 3A and Dataset S1, tab 8). Most of the 76 regions contained nucleotides from both states (34 regions) or only from state 3 (28). Genes in the 56 regions with attributable functions were associated with regulation; stress responses to osmotic, oxygen, temperature, or other stimuli; virulence factors; and carbon utilization (Table 2). Interestingly, only four of the 76 regions encode antibiotic resistance (ompC, acrA, acrD, and the QRDR region in gyrA). Thus, our data indicate that the multiple regions within the Paratyphi A core genome that are under Darwinian selection are primarily associated with metabolic functions. However, once again, almost all of these mutations have occurred recently (Fig. 3 E and F), indicating that they too are only transient and continuously being lost by purifying selection.
Table 2.
Functional assignments of regions containing sites in DHMM states 2 and 3
| State | No. of regions | Total length, bp | Function (number) |
| 2 | 14 | 7,300 | Unknown (4), Regulatory (4), Antibiotic resistance (2), Carbon utilization (2), Virulence factor (2), Osmotic stress (2), Other stress (1) |
| 2, 3 | 34 | 16,691 | Osmotic stress (11), Unknown (9), Regulatory (9), Carbon utilization (3), Oxygen stress (3), Virulence factor (4), Antibiotic resistance (2), RNA degradation (1), Temperature stress (1), Other stress (1) |
| 3 | 28 | 47 | Unknown (7), Regulatory (6), Carbon utilization (5), Virulence factor (5), Temperature stress (3), Osmotic stress (2), Oxygen stress (2), Other stress (2), RNA degradation (1) |
| Total | 76 | 24,038 | Unknown (20), Regulatory (19), Osmotic stress (15), Virulence factor (11), Carbon utilization (10), Oxygen stress (5), Temperature stress (4), Other stress (4), Antibiotic resistance (4), RNA degradation (2) |
Regions were counted once for each function they were assigned to, resulting in multiple counts for some regions. The individual genes and their putative functions within the 76 regions are listed in Dataset S1, tab 8.
Discussion
Until a few years ago, genetically monomorphic bacterial pathogens represented a technical challenge for genetic analyses because their genetic diversity is so low (44). With the advent of high-throughput, short-read sequencing, this technical challenge has disappeared, and comparative genomic analyses have yielded unambiguous genealogies for taxa with low frequencies of recombination (3, 6, 7, 24, 37, 45). For several taxa, their genealogies are indicative of phylogeographic histories of global dispersions, including Y. pestis (3, 45), S. sonnei (7), MRSA ST22 (6), and seventh pandemic Vibrio cholera (19). The results presented here for S. enterica serovar Paratyphi A allow similar conclusions. Only few SNPs were identified in a global sample, and most of those SNPs represent mutations that were acquired by vertical descent. The TMRCA of Paratyphi A was dated to ∼1549, and distinct lineages have spread globally since the mid-19th century. We note that our TMRCA dating has wide confidence intervals, partly because the dates of isolation of the bacterial strains that were available only span approximately 100 y. All TMRCA estimates are underestimates, partially because of lineage extinction, and both additional precision and a greater age estimate might be provided by ancient DNA analyses on Paratyphi A, which are yet to be performed.
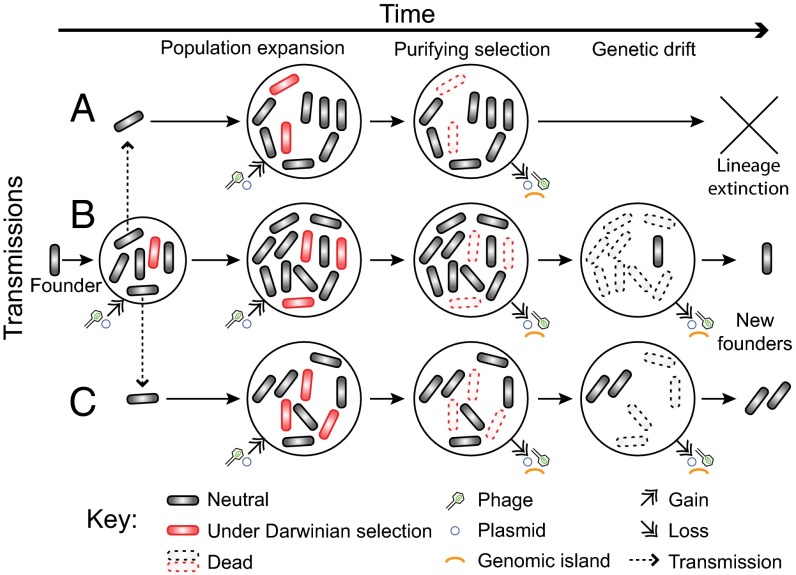
Many prior analyses were embedded within an intellectual framework of “emerging diseases” and Darwinian selection of particularly fit variants, and focus on the fixation of particular genetic changes in modern lineages, such as those that can lead to antibiotic resistance (6, 7). However, the null hypothesis is that successful genetic lineages with uniform genetic features are fixed by random events during nonadaptive processes, such as bottlenecks and genetic drift (46, 47), or reflect purifying selection (48) (Fig. 4). Some genetic changes that are commonly interpreted as representing Darwinian selection, such as increased antibiotic resistance, result in lessened fitness in the absence of secondary epistatic mutations (46, 49, 50). As a result, although antibiotic resistant variants of serovars Agona and Typhi have arisen on multiple occasions, these lineages have subsequently repeatedly thrown off antibiotic sensitive sublineages (24, 37). Antigenic variants of Neisseria meningitidis that allow immune escape are also usually only transient because host immunity to multiple antigens eliminates all but nonoverlapping antigen combinations (51). Geographic dispersions of bacterial pathogens can also result in the purification of genetic diversity because of the bottlenecks associated with small founding populations (45, 52, 53).
Fig. 4.
Cartoon of microevolutionary dynamics in Paratyphi A and other bacterial pathogens. Population expansions and transmissions between geographic areas (A, B, and C) are accompanied by genetic changes, some of which are neutral (gray) and others of which are under Darwinian selection (red). These genetic changes include various categories of mutations in the core and accessory genome, as well as the acquisition of bacteriophages and plasmids. Most deleterious genetic changes, including those under Darwinian selection, are removed by purifying selection (indicated as “Dead”). Others, including neutral changes, are lost because of genetic drift (also Dead), which is particularly effective during intermittent, random reductions in population size. The population size is particularly low during transmissions, which can even result in sequential founder effects due to bottlenecks.
Our observations, facilitated in part by the development of two novel HMM methods, extend beyond prior analyses, because they provide a temporal framework over centuries for all pangenomic changes. Mutations arise and genomic regions are acquired by horizontal gene transfer (Fig. 4), Darwinian selection results in the repeated rise of genotypes including mutations in genomic regions related to metabolism, virulence, and antibiotic resistance. Still other mutations result in genomic streamlining through the loss of genomic regions. And all these changes are occurring concurrently with the appearance of novel lineages and sublineages. However, instead of progressive evolution of greater fitness or virulence or fixation of antibiotic resistance (6, 54–56), genetic changes within Paratyphi A mimic Brownian motion or a drunkard’s walk. Almost all genetic changes seem to be either random, or are selected only transiently and are subsequently lost via purifying selection against less fit variants. Our data suggest that the crucial genomic contents of Paratyphi A that facilitate enteric fever in humans accumulated very early in its history and were already present in the MRCA; i.e., Paratyphi A has not become any more efficient at causing enteric fever over 500 y of microevolution. The same general conclusions seem to apply to multiple other bacterial pathogens with MRCAs dating several decades up to millennia because comparative genomics has also failed to identify signals of increased virulence or transmissibility during recent microevolution (3, 4, 24). These reflections imply that many epidemics and pandemics of bacterial disease in human history reflected chance environmental events, including geographical spread and/or transmissions to naïve hosts, rather than the recent evolution of particularly virulent organisms.
Materials and Methods
Whole-genome sequencing of 142 strains (Dataset S1, tab 9) was performed by using an Illumina HiSeq 2000 on 300-bp paired-end libraries in 96-fold multiplexes. Reads were assembled, validated, and aligned to the reference genome ATCC 9150, and SNPs in the core genome and genomic islands in the accessory genome were identified as described (24). Core genome SNPs were stripped of those attributed to recombination by RecHMM (https://sourceforge.net/projects/paratyphia/files/RecHMM) before reconstructing phylogenies and historical patterns of transmission. Mutations under Darwinian selection were identified with DHMM (https://sourceforge.net/projects/paratyphia/files/DHMM). Further methodological details are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Remco R. Bouckaert for assistance and advice with Beast 2; Yajun Song, Ronan Murphy, and Del Pickard for DNA preparation; Philippe Roumagnac for assistance with logistics; and Kathryn E. Holt and Camilla Mazzoni for very early analyses at the beginning of this project. M.A. and Z.Z. were initially supported by Science Foundation of Ireland Grant 05/FE1/B882. F.-X.W. is supported by the programme des Investissements d'Avenir no. ANR-10-LABX-62-IBEID.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence data have been deposited with the European Nucleotide Archive, www.ebi.ac.uk/ena (accession nos. ERR028897–ERR028999, ERR030042–ERR030144, ERR033909–ERR034063, ERR134160–ERR134255, and ERR237537–ERR237542; individual genome assemblies have been deposited under accession nos. PRJEB5545–PRJEB5690). The accession numbers for each strain are listed in Dataset S1, tab 9.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411012111/-/DCSupplemental.
References
- 1.Moodley Y, et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8(5):e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comas I, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45(10):1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui Y, et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci USA. 2013;110(2):577–582. doi: 10.1073/pnas.1205750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuenemann VJ, et al. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science. 2013;341(6142):179–183. doi: 10.1126/science.1238286. [DOI] [PubMed] [Google Scholar]
- 5.He M, et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45(1):109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holden MTG, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23(4):653–664. doi: 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt KE, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44(9):1056–1059. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Achtman M, et al. S. Enterica MLST Study Group Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012;8(6):e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82(5):346–353. [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DC. Gerhard’s distinction between typhoid and typhus and its reception in America, 1833-1860. Bull Hist Med. 1980;54(3):368–385. [PubMed] [Google Scholar]
- 11.Gwyn LB. On infection with a Para-Colon bacillus in a case with all the clinical features of typhoid fever. Johns Hopkins Hospital Bulletin. 1898;9(84):54–56. [Google Scholar]
- 12.Bainbridge FA. The Milroy lectures on paratyphoid fever and meat poisoning. Lancet. 1912;179(4620):705–709. [Google Scholar]
- 13.Ochiai RL, et al. Salmonella paratyphi A rates, Asia. Emerg Infect Dis. 2005;11(11):1764–1766. doi: 10.3201/eid1111.050168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karki S, Shakya P, Cheng AC, Dumre SP, Leder K. Trends of etiology and drug resistance in enteric fever in the last two decades in Nepal: A systematic review and meta-analysis. Clin Infect Dis. 2013;57(10):e167–e176. doi: 10.1093/cid/cit563. [DOI] [PubMed] [Google Scholar]
- 15.Punjabi NH, et al. Enteric fever burden in North Jakarta, Indonesia: A prospective, community-based study. J Infect Dev Ctries. 2013;7(11):781–787. doi: 10.3855/jidc.2629. [DOI] [PubMed] [Google Scholar]
- 16.Liang W, et al. Pan-genomic analysis provides insights into the genomic variation and evolution of Salmonella Paratyphi A. PLoS ONE. 2012;7(9):e45346. doi: 10.1371/journal.pone.0045346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta SK, et al. Laboratory-based surveillance of paratyphoid fever in the United States: Travel and antimicrobial resistance. Clin Infect Dis. 2008;46(11):1656–1663. doi: 10.1086/587894. [DOI] [PubMed] [Google Scholar]
- 18.Tourdjman M, et al. Unusual increase in reported cases of paratyphoid A fever among travellers returning from Cambodia, January to September 2013. Euro Surveill. 2013;18(39):18. doi: 10.2807/1560-7917.es2013.18.39.20594. [DOI] [PubMed] [Google Scholar]
- 19.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477(7365):462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClelland M, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36(12):1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 21.Holt KE, et al. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics. 2009;10:36. doi: 10.1186/1471-2164-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo CH, Ochman H. The extinction dynamics of bacterial pseudogenes. PLoS Genet. 2010;6(8):e1001050. doi: 10.1371/journal.pgen.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hottes AK, et al. Bacterial adaptation through loss of function. PLoS Genet. 2013;9(7):e1003617. doi: 10.1371/journal.pgen.1003617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Z, et al. Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet. 2013;9(4):e1003471. doi: 10.1371/journal.pgen.1003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt KE, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40(8):987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Achtman M. Insights from genomic comparisons of genetically monomorphic bacterial pathogens. Philos Trans R Soc Lond B Biol Sci. 2012;367(1590):860–867. doi: 10.1098/rstb.2011.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didelot X, Achtman M, Parkhill J, Thomson NR, Falush D. A bimodal pattern of relatedness between the Salmonella Paratyphi A and Typhi genomes: Convergence or divergence by homologous recombination? Genome Res. 2007;17(1):61–68. doi: 10.1101/gr.5512906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175(3):1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith JM. The detection and measurement of recombination from sequence data. Genetics. 1999;153(2):1021–1027. doi: 10.1093/genetics/153.2.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer JR, et al. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335(6067):428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 34.Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335(6067):457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 35.Holt KE, et al. High-throughput bacterial SNP typing identifies distinct clusters of Salmonella Typhi causing typhoid in Nepalese children. BMC Infect Dis. 2010;10:144. doi: 10.1186/1471-2334-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42(6):498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roumagnac P, et al. Evolutionary history of Salmonella typhi. Science. 2006;314(5803):1301–1304. doi: 10.1126/science.1134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kos VN, et al. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with Methicillin-resistant S. aureus hospital-acquired infection in the United States. MBio. 2012;3(3):e00112–e12. doi: 10.1128/mBio.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farhat MR, et al. Genomic analysis identifies targets of convergent positive selection in drug-resistant Mycobacterium tuberculosis. Nat Genet. 2013;45(10):1183–1189. doi: 10.1038/ng.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holt KE, et al. Multidrug-resistant Salmonella enterica serovar paratyphi A harbors IncHI1 plasmids similar to those found in serovar typhi. J Bacteriol. 2007;189(11):4257–4264. doi: 10.1128/JB.00232-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chattopadhyay S, Paul S, Kisiela DI, Linardopoulou EV, Sokurenko EV. Convergent molecular evolution of genomic cores in Salmonella enterica and Escherichia coli. J Bacteriol. 2012;194(18):5002–5011. doi: 10.1128/JB.00552-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 44.Achtman M. Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol. 2008;62:53–70. doi: 10.1146/annurev.micro.62.081307.162832. [DOI] [PubMed] [Google Scholar]
- 45.Morelli G, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42(12):1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong LI, Bloom JD. Epistatically interacting substitutions are enriched during adaptive protein evolution. PLoS Genet. 2014;10(5):e1004328. doi: 10.1371/journal.pgen.1004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nei M. Selectionism and neutralism in molecular evolution. Mol Biol Evol. 2005;22(12):2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gagneux S, et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312(5782):1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- 50.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312(5770):111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 51.Buckee CO, et al. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc Natl Acad Sci USA. 2008;105(39):15082–15087. doi: 10.1073/pnas.0712019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu P, et al. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc Natl Acad Sci USA. 2001;98(9):5234–5239. doi: 10.1073/pnas.061386098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holt KE, et al. Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc Natl Acad Sci USA. 2013;110(43):17522–17527. doi: 10.1073/pnas.1308632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sangal V, et al. Global phylogeny of Shigella sonnei strains from limited single nucleotide polymorphisms (SNPs) and development of a rapid and cost-effective SNP-typing scheme for strain identification by high-resolution melting analysis. J Clin Microbiol. 2013;51(1):303–305. doi: 10.1128/JCM.02238-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bos KI, et al. A draft genome of Yersinia pestis from victims of the Black Death. Nature. 2011;478(7370):506–510. doi: 10.1038/nature10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.