Significance
The catalytic cycle of many enzymes often requires that several, spatially remote enzyme structural elements undergo functionally important conformational changes. To maximize and regulate the efficiency of catalysis, therefore, considerable evidence suggests that small protein enzymes can coordinate the conformational changes of multiple structural elements. Whether and how larger, multicomponent biomolecular machines such as the ribosome also exhibit such coordination, however, remains unknown. Here, we demonstrate that the ribosome coordinates multiple, functionally important conformational changes to maximize and regulate the efficiency with which it translocates along its messenger RNA template. Our findings suggest that such coordination is likely to be a general and important mechanism through which all biomolecular machines maximize and regulate their functional efficiencies.
Abstract
One of the most challenging unanswered questions regarding the structural biology of biomolecular machines such as the two-subunit ribosome is whether and how these machines coordinate seemingly independent and random conformational fluctuations to maximize and regulate their functional efficiencies. To address this question, we have used ribosome mutagenesis or a ribosome-targeting antibiotic to predictably perturb the dynamics of intersubunit rotation, a structural rearrangement of the ribosome that is essential for the translocation and ejection of ribosome-bound tRNAs during translation. Concomitantly, we have used single-molecule fluorescence resonance energy transfer (smFRET) to characterize the effects of these perturbations on the dynamics of ribosomal L1 stalk movements and ribosome-bound tRNA reconfigurations, conformational changes that are likewise essential for the translocation and ejection of tRNAs during translation. Together with the results of complementary biochemical studies, our smFRET studies demonstrate that the ribosome uses cooperative conformational changes to maximize and regulate the efficiency with which it translocates and ejects tRNAs during translation. We propose that the ribosome employs cooperative conformational changes to efficiently populate global conformational states that are productive for translation, that translation factors exploit this cooperativity as part of their mechanisms of action, and that antibiotics exploit it to maximize the potency with which they inhibit translation. It is likely that similar cooperative conformational changes underlie the function and regulation of other biomolecular machines.
During the catalytic cycle of many enzymes, multiple, spatially distant enzyme structural elements must often undergo functionally important conformational changes (1). Consistent with the view that such structural rearrangements must be rapidly organized and executed to maintain catalytic efficiency, many recent studies strongly suggest that small, monomeric protein enzymes have evolved complex networks of cooperative conformational changes that coordinate the inherently stochastic conformational fluctuations of multiple structural elements in a manner that is optimal for catalysis (2). Within the context of energy landscape theory (3), such enzymes can be thought of as having evolved dynamic energy landscapes that bias enzyme conformational sampling in such a manner to maximize the efficiency of catalysis (2). Related proposals suggest that binding of allosteric effectors to enzymes remodels such energy landscapes, altering enzyme conformational sampling as part of the mechanisms through which these effectors regulate enzymatic activity (4).
Unfortunately, the conformational dynamics of large, macromolecular complexes remain much more challenging to characterize than those of small, monomeric proteins (5), making it very difficult to elucidate the role that cooperative conformational changes play in the function and regulation of biomolecular machines such as the ribosome (6). Following each round of aminoacyl–tRNA incorporation and peptide bond formation by the translating ribosome, the resulting ribosomal pretranslocation (PRE) complex must rapidly translocate the newly deacylated tRNA from the ribosomal peptidyl–tRNA binding (P) site to the ribosomal deacylated (or exit) tRNA binding (E) site and the newly formed peptidyl–tRNA from the ribosomal aminoacyl–tRNA binding (A) site to the P site. Concurrent with translocation of the tRNAs, the PRE complex advances along the mRNA by precisely one codon and ultimately ejects the E-site tRNA, producing a ribosomal posttranslocation (POST) complex that is ready for the next round of the elongation cycle.
During the first step of translocation, the A- and P-site tRNAs are reconfigured from their classical P/P (denoting the small ribosomal subunit P site/large ribosomal subunit P site) and A/A configurations into their hybrid P/E and A/P configurations. Numerous studies suggest that formation of the hybrid tRNA configuration is accompanied by large-scale conformational changes of the ribosome itself (6–9). Indeed, relative to X-ray crystallographic and cryogenic electron microscopy (cryo-EM) structures of ribosomal complexes carrying classically configured tRNAs, structures of ribosomal complexes carrying hybrid-configured tRNAs exhibit a ratchet-like relative rotation of the ribosomal subunits as well as a closure of the L1 stalk element of the large subunit [defined as 23S ribosomal RNA (rRNA) helices 76, 77, and 78 (H76–78) and ribosomal (r)-protein L1] that allows the L1 stalk to establish a physical interaction with the P/E-configured tRNA (10, 11). Notably, single-molecule fluorescence resonance energy transfer (smFRET) studies have shown that in the absence of elongation factor G (EF-G), the guanosine triphosphatase translation factor that promotes further steps along the translocation pathway, PRE complexes and PRE complex analogs lacking an A-site tRNA (PRE–A complexes) undergo thermally activated, stochastic, and reversible fluctuations between classical and hybrid tRNA configurations (classical⇆hybrid) (12), nonrotated and rotated subunit orientations (NR⇆R) (13), and open and closed L1 stalk conformations (L1o⇆L1c) (14, 15). Unfortunately, however, whether and how these stochastic tRNA, intersubunit, and L1 stalk dynamics are coordinated within PRE/PRE–A complexes to facilitate translocation and/or allosteric regulation of translocation remains unknown, severely limiting our understanding of the fundamental physical processes that drive and control the rapid and precise translocation of the tRNAs through the ribosome during protein synthesis.
To determine whether tRNA, intersubunit, and L1 stalk movements are coordinated within PRE/PRE–A complexes and to characterize the structural basis for such cooperative conformational changes, here we have used smFRET to characterize the L1 stalk and tRNA dynamics of Escherichia coli PRE–A complexes assembled using ribosomes that have been strategically mutagenized or treated with a ribosome-binding inhibitor to predictably perturb intersubunit rotation. Together with the results of complementary translocation studies, our findings directly demonstrate that the ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translocation and E-site tRNA ejection during translation. Using structural and phylogenetic analyses of ribosomes to rationalize our results leads us to propose a structure-based model for the observed cooperativity. Collectively, the work presented here strongly suggests that coordination of spatially remote conformational changes is a fundamental aspect of the mechanism and regulation of translocation, other steps of the translation elongation cycle, other stages of protein synthesis, and other biomolecular machines.
Results and Discussion
Intersubunit Rotation Can Be Predictably Perturbed by Disrupting Electrostatic Interactions at the Subunit Interface.
Intersubunit rotation requires extensive remodeling of a collection of highly conserved, noncovalent, intersubunit RNA•RNA, RNA•r-protein, and r-protein•r-protein interactions known as intersubunit bridges (11). Intersubunit rotation and the attendant remodeling of intersubunit bridges have been shown to play critical roles in enabling and regulating translocation (16, 17). Indeed, deletion of the functionally critical bridge B1b, which undergoes the largest rearrangement during intersubunit rotation (18), has been shown to increase the propensity of PRE complexes to undergo EF-G–independent translocation (19) and disruption of single interactions across several intersubunit bridges, including bridge B1b, has been shown to alter the rate of PRE complex translocation and/or the propensity of PRE complexes to slip by one nucleotide and shift their reading frame on the mRNA during translocation (17, 19, 20). Similarly, the tuberactinomycin family of antibiotic inhibitors of protein synthesis (e.g., viomycin, capreomycin, etc.) acts by binding to bridge B2a and strongly inhibiting translocation, presumably by blocking the remodeling of bridge B2a that accompanies intersubunit rotation (8).
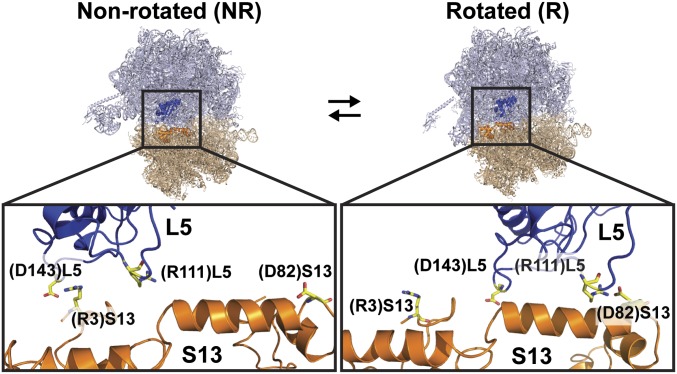
Given its particularly critical and well-studied role in enabling and regulating translocation, we began by developing a series of mutant ribosomes in which bridge B1b was systematically mutagenized to predictably perturb intersubunit rotation. In E. coli, bridge B1b is composed of interactions between r-protein S13 on the small, or 30S, subunit and r-protein L5 on the large, or 50S, subunit. By comparing the structures of ribosomal complexes in the NR- and R-subunit orientations (11), we identified two highly conserved (Table S1), charged amino acids in S13, arginine 3 [(R3)S13] and aspartic acid 82 [(D82)S13], whose electrostatic interactions with oppositely charged residues in L5 are dramatically remodeled upon intersubunit rotation (Fig. 1). When ribosomal complexes are in their NR-subunit orientation, (R3)S13 is involved in an electrostatic interaction with the highly conserved (Table S1) aspartic acid 143 of L5 [(D143)L5], whereas (D82)S13 is positioned too far away to interact with any residues within the 50S subunit. Upon intersubunit rotation, however, this electrostatic interaction is disrupted such that when ribosomal complexes are in their R-subunit orientations, (R3)S13 is now too far away to interact with any residues within the 50S subunit and (D82)S13 is involved in an electrostatic interaction with the highly conserved (Table S1) arginine 111 of L5 [(R111)L5]. Because (R3)S13–(D143)L5 and (D82)S13–(R111)L5 interactions are observed in all available high-resolution structures of ribosomal complexes in the NR- or R-subunit orientations, respectively, disruption of (R3)S13–(D143)L5 or (D82)S13–(R111)L5 interactions should primarily alter the stabilities of the NR- or R-subunit orientations and the corresponding rates of intersubunit rotation, respectively, in PRE/PRE–A complexes. Guided by these analyses, we set out to perturb intersubunit rotation by constructing and purifying wild-type 30S subunits (WT), 30S subunits lacking S13 [(–)S13], and 30S subunits reconstituted with recombinantly expressed and purified wild-type S13 [(WT)S13] or each of five S13 mutants [(R3A)S13, (R3D)S13, (D82A)S13, (D82K)S13, and (R3D/D82K)S13] (SI Materials and Methods).
Fig. 1.
Intersubunit bridge B1b. The structures of bridge B1b observed in ribosomal complexes in the NR-subunit [Protein Data Bank (PDB) IDs: 3R8S and 3R8T] (Left) and R-subunit (PDB IDs: 3R8N and 3R8O) (Right) orientations are shown. The zoomed-in views highlight the (R3)S13–(D143)L5 and (D82)S13–(R111)L5 interactions that are formed across bridge B1b in ribosomal complexes in the NR and R orientations, respectively. S13 (orange) and L5 (dark blue) are labeled and shown in cartoon representation and (R3)S13, (D82)S13, (R111)L5, and (D143)L5 are labeled, depicted in stick representations, and colored according to atom type.
All PRE–A Complexes Exhibited Dynamic L1o⇆L1c and L1◦tRNA⇆L1•tRNA Equilibria.
L1 stalk and tRNA dynamics of PRE/PRE–A complexes can be monitored using two previously reported smFRET signals. The first signal, denoted smFRETL1–L9, derives from Cy3 FRET donor-labeled r-protein L9 [L9(Cy3)] and Cy5 FRET acceptor-labeled r-protein L1 [L1(Cy5)] and reports on movements of the L1 stalk between its L1o and L1c conformations (14). The second signal, denoted smFRETL1-tRNA, derives from Cy3-labeled P-site tRNA [tRNA(Cy3)] and L1(Cy5) and reports on rearrangements of the PRE/PRE–A complex between a conformation in which the L1 stalk is in its L1o conformation and is too far away to physically interact with the P/P-configured tRNA (L1◦tRNA) and a conformation in which the L1 stalk is in its L1c conformation and is able to physically interact with the P/E-configured tRNA (L1•tRNA) (21). Using each of the 30S subunits described in the previous section, we therefore assembled eight PRE–A complexes harboring L1(Cy5)- and L9(Cy3)-labeled 50S subunits for smFRETL1–L9 experiments and eight PRE–A complexes harboring L1(Cy5)-labeled 50S subunits and tRNAPhe(Cy3) for smFRETL1-tRNA experiments (SI Materials and Methods and Fig. 2).
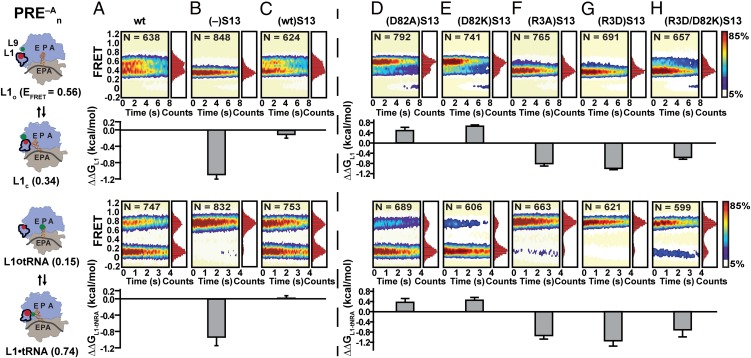
Fig. 2.
(A–H) Steady-state smFRET measurements and changes in the free-energy differences of PRE−A complexes formed using (A) WT, (B) (–)S13, (C) (WT)S13, (D) (D82A)S13, (E) (D82K)S13, (F) (R3A)S13, (G) (R3D)S13, and (H) (R3D/D82K)S13 30S subunits. Cartoon representations of PRE–A complexes labeled using the smFRETL1–L9 (Upper Row) and smFRETL1-tRNA (Lower Row) labeling schemes are displayed at Left. The 30S subunit (tan); 50S subunit (light blue); L1 stalk; mRNA (gray curve); A, P, and E sites; and P-site tRNA (orange) are depicted and labeled. Surface contour plots of the time evolution of population FRET were generated by superimposing individual smFRETL1–L9 vs. time trajectories (Upper Row) and smFRETL1-tRNA vs. time trajectories (Lower Row) for each PRE–A complex. Contours are colored from white to red, denoting the lowest to highest population levels (population color bar at Far Right) and N is the number of smFRET trajectories used to construct each contour plot. The one-dimensional EFRET histogram corresponding to each surface contour plot is plotted along the right-hand y axis of the surface contour plots. Bar graphs depicting the changes in the free-energy difference between L1o and L1c (ΔΔGL1, Upper Row) and L1◦tRNA and L1•tRNA (ΔΔGL1-tRNA, Lower Row) for each PRE–A complex relative to (columns A–C) and for each PRE–A complex carrying an S13-reconstitued 30S subunit relative to (columns D–H) were generated using the ΔΔGL1 and ΔΔGL1-tRNA values in Tables S2 and S3.
smFRET imaging of PRE−A complexes was accomplished using total internal reflection fluorescence microscopy as previously described (14, 21) (SI Materials and Methods). Consistent with previous studies, the smFRETL1–L9 signals from all PRE–A complexes exhibited fluctuations between a FRET state with a FRET efficiency (EFRET) = 0.54 ± 0.02, assigned to the L1o conformation of the L1 stalk, and a FRET state with an EFRET = 0.35 ± 0.01, assigned to the L1c of the L1 stalk, thus reporting on the L1o⇆L1c equilibrium (Fig. 2 and Fig. S1A) (14). Likewise consistent with previous studies, the smFRETL1-tRNA signals from all PRE–A complexes exhibited fluctuations between a FRET state with an EFRET = 0.15 ± 0.02, assigned to the L1◦tRNA conformation of the PRE–A complex, and a FRET state with an EFRET = 0.74 ± 0.03, assigned to the the L1•tRNA conformation of the PRE–A complex, thus reporting on the L1◦tRNA⇆L1•tRNA equilibrium (Fig. 2 and Fig. S1B) (21).
L1-Stalk and P-Site tRNAPhe Dynamics Are Coupled to Intersubunit Rotation.
Relative to (where the subscript denotes the identity of the 30S subunit of the PRE–A complex), exhibits dramatic shifts of the L1o⇆L1c and L1◦tRNA⇆L1•tRNA equilibria toward L1c and L1•tRNA, respectively, increasing the equilibrium constants (KL1 and KL1-tRNA, respectively) by sixfold each (Tables S2 and S3). These sixfold increases in KL1 and KL1-tRNA correspond to free-energy difference changes between L1o and L1c (ΔΔGL1) and L1◦tRNA and L1•tRNA (ΔΔGL1-tRNA) in vs. of ∼ –1.0 kcal⋅mol–1 (where the negative values indicate shifts of the equilibria toward L1c and L1•tRNA, respectively) (Fig. 2 A and B and Tables S2 and S3). Survival probability analyses (SI Materials and Methods) reveal that the observed equilibrium shifts are predominantly driven by fivefold increases in the rates of L1o→L1c (kL1o→L1c) and L1◦tRNA→L1•tRNA (kL1◦tRNA→L1•tRNA) that are further augmented by 30% decreases in the rates of L1c→L1o (kL1c→L1o) and L1•tRNA→L1◦tRNA (kL1•tRNA→L1◦tRNA) (Tables S2 and S3). A similar comparison demonstrates that in vitro reconstitution of recombinant wild-type S13 into (–)S13 30S subunits yields a PRE–A complex [i.e., ] with thermodynamic and kinetic properties that are virtually indistinguishable from those of PRE–A complexes formed using completely wild-type 30S subunits (i.e., ) (Fig. 2C, Fig. S1C, and Tables S2 and S3). Because we have purified 30S subunits that incorporated S13 during our reconstitutions from those that did not (SI Materials and Methods and Fig. S2) (19), we can directly attribute the differences in the thermodynamic and kinetic properties of PRE–A(–)S13 vs. and to the absence of S13. Interestingly, PRE complexes formed using (–)S13 30S subunits exhibit increased rates of EF-G–independent translocation (19), demonstrating that within the context of PRE complexes containing wild-type 30S subunits, S13 acts to suppress premature, EF-G–independent translocation. Our data reveal that S13 accomplishes this by shifting the conformational equilibria of the PRE complex toward L1o and L1◦tRNA such that the PRE complex is in a conformation that is incompatible with translocation.
Because S13 deletion disrupts both S13–L5 and S13–P-site tRNA interactions (22), we next sought to investigate how single S13 substitution mutations designed to exclusively disrupt S13–L5 interactions modulate the thermodynamic and kinetic properties of PRE–A complexes. From the structural analyses described above (Fig. 1), we expected that and , which contain mutations designed to disrupt the favorable (D82)S13–(R111)L5 interaction observed in the R-subunit orientation while having no effect on the NR-subunit orientation, would primarily destabilize the R-subunit orientation relative to [note that the thermodynamic and kinetic properties of all PRE−A complexes harboring S13 mutants are reported relative to ]. As shown in Fig. 2 D and E and Tables S2 and S3, and both exhibited twofold decreases of KL1 and KL1-tRNA, corresponding to ΔΔGL1 and ΔΔGL1-tRNA values of ∼ +0.4 kcal⋅mol–1 that favor the L1o and L1◦tRNA conformations. In both cases, the equilibrium shifts were almost exclusively driven by twofold increases in kL1c→L1o and kL1•tRNA→L1◦tRNA with no detectable changes in kL1o→L1c and kL1◦tRNA→L1•tRNA (Tables S2 and S3). It is remarkable that disruption of a single, noncovalent, electrostatic interaction across the subunit interface of the ribosome can alter the thermodynamic and kinetic properties of the entire PRE–A complex; it is clear from this observation that the collection of noncovalent interactions that comprise the intersubunit bridges has evolved to ensure the metastability of the PRE complex.
The effects of disrupting the (D82)S13–(R111)L5 interactions, which are discussed in greater detail in SI Results and Discussion, demonstrate that disruption of an S13–L5 interaction that is predicted to destabilize the R-subunit orientation and consequently result in an increased rate of R→NR transitions (kR→NR), also destabilizes the L1c conformation of the L1 stalk and the P/E conformation of the P-site tRNAPhe, resulting in increases in kL1c→L1o and kL1•tRNA→L1◦tRNA. Correspondingly, disruption of an S13–L5 interaction that is predicted to have no effect on the NR-subunit orientation or the rate of NR→R transitions (kNR→R) has virtually no effect on the stability of the L1o conformation of the L1 stalk and the P/P conformation of the P-site tRNA or the corresponding kL1o→L1c and kL1◦tRNA→L1•tRNA. Taken together, these observations are consistent with a model in which L1 stalk and P-site tRNAPhe dynamics are coupled to intersubunit rotation in PRE/PRE–A complexes (21). It is important to note, however, that the identity of the P-site tRNA can influence the conformational equilibria of PRE/PRE–A complexes (21, 23) and that the PRE–A complexes investigated here all contained exclusively tRNAPhe at the P site. Thus, future experiments using tRNAs other than tRNAPhe will be needed to determine whether and how the coupling of P-site tRNA dynamics to intersubunit rotation observed here depends on the identity of the P-site tRNA.
Based on the structural analysis described above and in Fig. 1, we likewise expected that and , which were designed to disrupt the favorable (R3)S13–(D143)L5 interaction observed in the NR-subunit orientation while having no effect on the R-subunit orientation, would primarily result in a destabilization of the NR-subunit orientation. As shown in Fig. 2 F and G and Tables S2 and S3, and both exhibited four- to sixfold increases in KL1 and KL1-tRNA, corresponding to ΔΔGL1 and ΔΔGL1-tRNA values of ∼ –1.0 kcal⋅mol–1 that favor the L1c and L1•tRNA conformations. In both cases, the equilibrium shifts were primarily driven by two- to fivefold increases in kL1o→L1c and kL1◦tRNA→L1•tRNA that were augmented by 30–40% decreases in kL1c→L1o and kL1•tRNA→L1◦tRNA (Tables S2 and S3).
The effects of disrupting the (R3)S13–(D143)L5 interactions, which are discussed in greater detail in SI Results and Discussion, demonstrate that disruption of an S13–L5 interaction that is predicted to destabilize the NR-subunit orientation and consequently result in an increase in kNR→R, also destabilizes the L1o conformation of the L1 stalk and the P/P configuration of the P-site tRNAPhe, resulting in increases in kL1o→L1c and kL1◦tRNA→L1•tRNA. Once again, these observations are consistent with a model in which L1-stalk and P-site tRNAPhe dynamics are coupled to intersubunit rotation in PRE/PRE–A complexes (21). In contrast to the data obtained using and , however, the data obtained using and exhibited small, but detectable and reproducible, decreases in kL1c→L1o and kL1•tRNA→L1◦tRNA (Tables S3 and S4). These small effects likely result from an (R3)S13 mutation-mediated increase in the stability of the R-subunit orientation or a change in the stability of one or more of the transition-state subunit orientations that are sampled during R→NR transitions (SI Results and Discussion and Fig. S3). Interestingly, , which harbors an (R3D/D82K)S13 double mutation, exhibits thermodynamic and kinetic properties that are essentially the sum of those observed for and , strongly suggesting that, within wild-type ribosomes, (R3)S13–(D143)L5 and (D82)S13–(R111)L5 interactions largely act independently to regulate the intersubunit, L1 stalk, and tRNA dynamics of PRE complexes (SI Results and Discussion; Fig. 2 E, G, and H; and Tables S2 and S3).
The Architecture of the Ribosome Itself Couples L1-Stalk Dynamics to Intersubunit Rotation.
The results presented in Fig. 2 demonstrate that destabilization of the NR-subunit orientation also destabilizes the L1o conformation of the L1 stalk and increases kL1o→L1c, whereas destabilization of the R-subunit orientation also destabilizes the L1c conformation of the L1 stalk and increases kL1c→L1o. This cooperativity between intersubunit rotation and L1-stalk dynamics is remarkable given that the (R3)S13 mutations are ∼20 Å away from the nearest residue within the L1 stalk (D102 in L1) in the R-subunit orientation and ∼90 Å away from the nearest residue within the L1 stalk (G2112 in H76) in the NR-subunit orientation (24, 25). Because structures of P-site tRNA-bound ribosomal complexes reveal that P/P-configured tRNAs directly contact residues in S13 and L5 whereas P/E-configured tRNAs directly contact residues in S13 and in the L1 stalk (10, 11), it is possible that P/P⇆P/E transitions serve to physically couple intersubunit rotation with L1-stalk movements. To test whether a P-site tRNA is strictly required for the cooperativity that we observe between intersubunit rotation and L1-stalk movements, we prepared vacant 70S ribosomes using wild-type 30S subunits and L1–L9-labeled 50S subunits, and lacking any bound tRNAs (70SWT, where the subscript identifies the 30S subunit in the vacant 70S ribosome) (SI Materials and Methods).
Compared with , 70SWT exhibited an eightfold decrease in KL1, dramatically shifting the L1o⇆L1c equilibrium toward L1o (Fig. 2A, Fig. S4A, and Tables S3 and S4). Taken together with previous smFRET studies demonstrating that, relative to a PRE–A complex similar to , the NR⇆R equilibrium in vacant 70S ribosomes is shifted by a similar factor of 8 toward the NR-subunit orientation (13), this result already strongly suggests that the architecture of the ribosome itself serves to physically couple L1-stalk movements to intersubunit rotation. To test this possibility further, we prepared a series of seven vacant 70S ribosomes, using the same 30S subunits that were used to prepare the PRE–A complexes described above. Notably, all of the vacant 70S ribosomes that we prepared exhibited differences in KL1, ΔΔGL1, kL1o→L1c, and kL1c→L1o compared with 70S(WT)S13 that mirrored those observed when the corresponding PRE–A complexes were compared with (Tables S3 and S4).
To confirm that the changes in L1-stalk dynamics that we observe upon disruption of S13–L5 interactions across bridge B1b are indeed due to changes in intersubunit rotation, we conducted a series of smFRETL1–L9 experiments, using and 70SWT in which we altered the dynamics of intersubunit rotation by perturbing a different intersubunit bridge, using a ribosome-targeting antibiotic. The tuberactinomycin antibiotic viomycin is a potent translocation inhibitor that binds within bridge B2a and stabilizes the ribosome in its R-subunit orientation (26), thereby perturbing intersubunit rotation in a manner that is distinct and independent of disrupting S13–L5 interactions across bridge B1b. Consistent with the results obtained by disrupting S13–L5 interactions, analysis of the smFRETL1–L9 experiments conducted as a function of increasing viomycin concentration demonstrates that the viomycin-mediated stabilization of the R-subunit orientation of and 70SWT also stabilizes the L1c conformation of the L1 stalk (Fig. S4B and Table S5). Thus, viomycin inhibits translocation both by directly stabilizing the R-subunit orientation and by allosterically stabilizing the L1c conformation of the L1 stalk and, presumably, the P/E configuration of the P-site tRNA; it is these allosteric effects of viomycin that are likely responsible for the potency with which this antibiotic inhibits translocation.
Collectively, the results of smFRETL1–L9 experiments conducted on vacant 70S ribosomes in which intersubunit rotation was perturbed by either the disruption of S13–L5 interactions across bridge B1b or the binding of viomycin at bridge B2a demonstrate that the architecture of the ribosome has evolved to physically couple L1-stalk dynamics to intersubunit rotation. Comparative structural analysis (11) suggests that the cooperativity we observe arises from steric clashes between the L1-stalk and 30S-subunit components that exclude the L1 stalk from adopting the L1c conformation within the NR-subunit orientation and from favorable interactions between the L1-stalk and 30S-subunit components that stabilize the L1c conformation of the L1 stalk within the R-subunit orientation (SI Results and Discussion and Fig. S5). Comparison of the ΔΔGL1 values measured in the presence vs. the absence of the P-site tRNAPhe (i.e., the PRE–A complexes vs. the vacant 70S ribosomes) using the disruption of S13–L5 interactions, however, demonstrates that, although the architecture of the ribosome itself physically couples L1-stalk dynamics to intersubunit rotation, the presence of a P-site tRNAPhe within a PRE/PRE–A complex serves to increase the strength of this coupling (Fig. 2, Fig. S4A, and Tables S3 and S4). As noted above, however, the identity of the P-site tRNA can influence the conformational equilibria of PRE/PRE–A complexes (21, 23) and future experiments using tRNAs other than tRNAPhe will therefore be needed to determine whether and how tRNA-mediated modulation of the cooperativity between L1-stalk dynamics and intersubunit rotation depends on the identity of the P-site tRNA.
Changes in the Relative Stabilities of Two Global Conformations of the PRE Complex Regulate the Rate and Extent of EF-G–independent Translocation.
As described above, the smFRET data obtained using and reveal that S13 suppresses EF-G–independent translocation in wild-type PRE complexes (19) by shifting the conformational equilibria of the PRE complex toward L1o, L1◦tRNA, and, presumably, NR such that the PRE complex is in a conformation that is incompatible with translocation. In addition, we have characterized single substitution mutations within S13 [e.g., (D82K)S13] that likewise shift the conformational equilibrium of the PRE–A complex toward L1o and L1◦tRNA by stabilizing an NR/L1o/(P/P) global conformation of the PRE–A complex in which the ribosome, L1 stalk, and P-site tRNAPhe occupy the NR-subunit orientation, the L1o conformation, and the P/P configuration, respectively. Conversely, we have characterized single substitution mutations within S13 [e.g., (R3D)S13] that shift the conformational equilibrium of the PRE–A complex toward L1c and L1•tRNA by stabilizing an R/L1c/(P/E) global conformation of the PRE–A complex in which the ribosome, L1 stalk, and P-site tRNAPhe occupy the R-subunit orientation, the L1c conformation, and the P/E configuration, respectively.
To test whether the stabilities of these two global conformations of the PRE–A complex regulate the propensity of PRE complexes to undergo EF-G–independent translocation, we prepared PRE complexes analogous to , , , , and [i.e., PRE(WT)S13, PRE(–)S13, PRE(R3D)S13, PRE(D82K)S13, and PRE(R3D/D82K)S13] and assessed the propensity of each of these PRE complexes to undergo EF-G–independent translocation. To do this, we used two independent biochemical assays: a puromycin reactivity assay and a tripeptide synthesis assay (SI Materials and Methods and Fig. S6). The results of these experiments demonstrate that, relative to PRE(WT)S13, PRE complexes exhibiting a shift toward R/L1c/(P/E) [i.e., PRE(–)S13, PRE(R3D)S13, and PRE(R3D/D82K)S13] display a corresponding increase in the rate and extent of EF-G–independent translocation, whereas the PRE complex exhibiting a shift toward NR/L1o/(P/P) [i.e., PRE(D82K)S13] displays a corresponding decrease in the rate and extent of EF-G–independent translocation. Collectively, these results demonstrate that S13 indeed suppresses EF-G–independent translocation, at least in part, by stabilizing the NR/L1o/(P/P) global conformation of PRE complexes via bridge B1b. Similar results were obtained when we varied the experimental conditions (i.e., the Mg2+ concentration) to alter the relative stabilities of the NR/L1o/(P/P) and R/L1c/(P/E) global conformations of the PRE complex within a representative PRE complex, PRE(R3D)S13, and assessed the propensity of this PRE complex to undergo EF-G–independent translocation (Fig. S6). Thus, any factor that can modulate the relative stabilities of the NR/L1o/(P/P) and R/L1c/(P/E) global conformations of PRE complexes is also likely to be able to control the propensity of PRE complexes to undergo EF-G–independent translocation. Indeed, EF-G itself functions, at least in part, by dramatically stabilizing the R/L1c/(P/E) global conformation of the PRE complex (13, 21).
Cooperative Conformational Changes Enable the Ribosome to Maximize and Regulate the Efficiency of Translation.
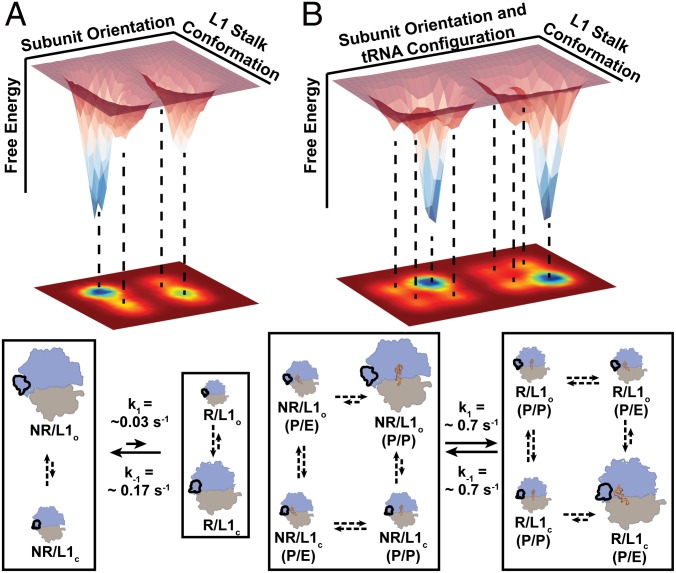
Numerous experimental and computational studies strongly suggest that the architectures and corresponding conformational free-energy landscapes of small monomeric protein enzymes have evolved such that complex networks of cooperative conformational changes facilitate catalysis and allosteric regulation (1, 2). Our findings here demonstrate that the architecture and corresponding conformational free-energy landscape of the ribosome have evolved such that cooperative intersubunit rotation and L1-stalk dynamics ensure that vacant 70S ribosomes can efficiently sample NR/L1o and R/L1c global conformations (Fig. 3A). The presence of a P-site tRNAPhe within a PRE–A complex remodels this landscape, further increasing the cooperativity between intersubunit rotation and L1-stalk dynamics and enabling the PRE–A complex to efficiently sample NR/L1o/(P/P) and R/L1c/(P/E) global conformations (Fig. 3B). Within the context of a PRE complex, such cooperativity maximizes the efficiency with which the PRE complex samples the R/L1c/(P/E) global conformation that is productive for translocation, thereby ensuring that the translating ribosome can undergo rapid translocation during the elongation cycle. Likewise, because POST complexes predominantly sample the NR-subunit orientation (13), such cooperativity maximizes the efficiency with which the POST complex samples the NR/L1o/(P/P) global conformation in which the L1 stalk occupies the L1o conformation that enables ejection of the deacylated tRNA that was just translocated into the E site from the ribosome (27), thereby ensuring that the translating ribosome can rapidly progress into the next round of the elongation cycle.
Fig. 3.
Cooperativity of intersubunit rotation and L1-stalk dynamics in vacant ribosomes and intersubunit rotation, L1-stalk, and P-site tRNA dynamics in PRE–A complexes. (A) A simplified, 3D schematic representation (Top) and corresponding 2D projection (Middle) of the conformational free energy landscape governing the intersubunit and L1-stalk dynamics of vacant 70S ribosomes. Valleys in the free-energy landscape correspond to relatively low-energy conformations of the ribosome (structural cartoons in the kinetic scheme, Bottom). Conformational changes occurring along the subunit orientation and the L1-stalk conformation axes of the plot are indicated by the solid and dashed arrows in the kinetic schemes (Bottom), respectively. Although numerous intersubunit orientations and L1-stalk conformations can be sampled along each axis, for simplicity we depict only sampling of the relatively low-energy NR- and R-intersubunit orientations and L1o and L1c conformations of the L1 stalk. This defines the four relatively low-energy global conformations of the vacant 70S ribosome denoted in the kinetic scheme. (B) Same as A, except using a PRE–A complex instead of a vacant 70S ribosome and, for simplicity, depicting only the relatively low-energy P/P and P/E configurations of the P-site tRNA, thereby defining the eight relatively low-energy conformations of the PRE–A complex denoted in the kinetic scheme. Note that, for clarity, subunit orientation and tRNA configuration are plotted on the same axis.
In addition to maximizing the efficiency of mechanical processes such as the translocation and ejection of tRNAs, cooperative conformational changes can also be exploited for the allosteric regulation of protein synthesis by translation factors and antibiotic inhibitors. Previously, we (14) and others (15) have shown that in addition to stabilizing the R-subunit orientation (13), binding of EF-G to PRE–A complexes stabilizes the L1c conformation of the L1 stalk. This occurs despite the fact that EF-G does not directly contact either the L1 stalk or the intervening P-site tRNA within the PRE–A complex (14). The present study now allows us to rationalize this observation, strongly suggesting that EF-G uses the cooperative conformational changes that we have characterized here to allosterically regulate the dynamics of the L1 stalk by directly modulating the dynamics of intersubunit rotation. Similarly, the results of the viomycin experiments shown in Fig. S4B demonstrate that antibiotic inhibitors of translocation can exploit the cooperative conformational changes that we observe here as part of the mechanism through which they so strongly inhibit translocation. More generally, cooperative conformational changes such as the ones that we have characterized here are likely to play important roles in regulating the efficiency of many of the other steps of protein synthesis as well as the efficiency of the numerous biological processes that are carried out by other biomolecular machines.
Methods
smFRET Experiments.
All smFRET experiments were performed in Tris-polymix buffer [50 mM Tris-OAc (pH25 °C = 7.0), 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 10 mM β-mercaptoethanol, 5 mM putrescine-HCl, and 1 mM spermidine, free base] containing 15 mM Mg(OAc)2 and supplemented with an oxygen-scavenging system (protocatechuic acid/protocatechuate-3,4-dioxygenase) (28) and a triplet-state quencher mixture [1 mM 1,3,5,7-cyclooctatetraene (Sigma-Aldrich) and 1 mM 3-nitrobenzyl alcohol (Fluka)] (29). Further details regarding data acquisition, processing, and analysis are in SI Materials and Methods.
EF-G–Independent Translocation Experiments.
Puromycin reactivity and tripeptide synthesis assays were performed in translocation buffer [50 mM Tris⋅HCl (pH25 °C = 7.5), 70 mM NH4Cl, 30 mM KCl, 1 mM DTT] containing 7 mM MgCl2. Further details regarding data acquisition, processing, and analysis are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank R. Green for providing the E. coli strains MG1655 and S13MG1, J. Frank for providing the cryo-EM reconstruction of the E. coli 70S ribosome, G. Böel and J. Luff for help with the phylogenetic and sequence analysis, and J.-W. van de Meent for help with the kinetic analysis of the smFRET data. We also thank D. MacDougall, S. Mitra, C. Kinz-Thompson, and N. Bailey for helpful comments on the manuscript. This work was supported by grants to R.L.G. from the Burroughs Wellcome Fund (Career Award in the Biomedical Sciences 1004856), the US National Science Foundation (MCB 0644262), and the US National Institute of General Medical Sciences (GM 084288).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401864111/-/DCSupplemental.
References
- 1.Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301(5637):1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 2.Hammes GG, Benkovic SJ, Hammes-Schiffer S. Flexibility, diversity, and cooperativity: Pillars of enzyme catalysis. Biochemistry. 2011;50(48):10422–10430. doi: 10.1021/bi201486f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 4.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4(8):474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 5.Frank J. Molecular Machines in Biology: Workshop of the Cell. Cambridge, UK: Cambridge Univ Press; 2011. [Google Scholar]
- 6.Frank J, Gonzalez RL., Jr Structure and dynamics of a processive Brownian motor: The translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci USA. 2007;104(50):19671–19678. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: One step closer to the molecular mechanism. ACS Chem Biol. 2009;4(2):93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tinoco I, Jr, Gonzalez RL., Jr Biological mechanisms, one molecule at a time. Genes Dev. 2011;25(12):1205–1231. doi: 10.1101/gad.2050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32(2):190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101(35):12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30(5):578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci USA. 2009;106(37):15702–15707. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA. 2009;106(8):2571–2576. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104(12):4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Fredrick K. Contribution of intersubunit bridges to the energy barrier of ribosomal translocation. Nucleic Acids Res. 2013;41(1):565–574. doi: 10.1093/nar/gks1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114(1):123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 19.Cukras AR, Green R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J Mol Biol. 2005;349(1):47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Vila-Sanjurjo A, O’Connor M. Mutations in the intersubunit bridge regions of 16S rRNA affect decoding and subunit-subunit interactions on the 70S ribosome. Nucleic Acids Res. 2011;39(8):3321–3330. doi: 10.1093/nar/gkq1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30(3):348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 23.Fei J, Richard AC, Bronson JE, Gonzalez RL., Jr Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol. 2011;18(9):1043–1051. doi: 10.1038/nsmb.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin H, Kelley AC, Ramakrishnan V. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci USA. 2011;108(38):15798–15803. doi: 10.1073/pnas.1112185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 26.Ermolenko DN, et al. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat Struct Mol Biol. 2007;14(6):493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 27.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285(5436):2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 28.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94(5):1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez RL, Jr, Chu S, Puglisi JD. Thiostrepton inhibition of tRNA delivery to the ribosome. RNA. 2007;13(12):2091–2097. doi: 10.1261/rna.499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.