Significance
When cells sense changes in their environment, molecular pathways relay information to the transcriptional machinery, which induces gene expression changes that allow for adaptation to the new conditions. In this paper, the C-terminal domain (CTD) of the RNA polymerase II largest subunit is shown to be involved in adaptation of yeast cells to conditions of low phosphate levels and where galactose is the primary fuel source. Specifically, threonine residues within the repeated CTD structure are required for induction of genes involved in phosphate and galactose metabolism, by stimulating the eviction of the histone variant Htz1 from the gene promoters. This finding reveals a role for the CTD in chromatin remodeling that facilitates expression of specific genes.
Keywords: H2A.Z, SWRc
Abstract
The C-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNAP II) consists of repeated YSPTSPS heptapeptides and connects transcription with cotranscriptional events. Threonine-4 (Thr4) of the CTD repeats has been shown to function in histone mRNA 3′-end processing in chicken cells and in transcriptional elongation in human cells. Here, we demonstrate that, in budding yeast, Thr4, although dispensable for growth in rich media, is essential in phosphate-depleted or galactose-containing media. Thr4 is required to maintain repression of phosphate-regulated (PHO) genes under normal growth conditions and for full induction of PHO5 and the galactose-induced GAL1 and GAL7 genes. We identify genetic links between Thr4 and the histone variant Htz1 and show that Thr4, as well as the Ino80 chromatin remodeler, is required for activation-associated eviction of Htz1 specifically from promoters of the Thr4-dependent genes. Our study uncovers a connection between transcription and chromatin remodeling linked by Thr4 of the CTD.
In eukaryotic cells, RNA polymerase II (RNAP II) transcribes all protein coding genes as well as a number of noncoding RNA genes, including those for most snRNAs and microRNAs. Expression of RNAP II-transcribed genes is highly regulated, ensuring that the timing of production, quantity, and structure of the RNAs generated is appropriate. Much of this regulation occurs on the genes themselves, whose accessibility is limited by nucleosomes that can form densely packed chromatin (reviewed in refs. 1 and 2). A significant amount of regulation, however, targets RNAP II itself. Specifically, the C-terminal domain (CTD) of Rpb1, the largest subunit of RNAP II, is subject to a multitude of phosphorylation and dephosphorylation events that mark different steps of the RNAP II transcription cycle (recently reviewed in refs. 3–5). Although many of these modifications occur generally, some appear to play critical roles in expression of specific genes (6–8). The CTD consists of a tandemly repeated hepta-peptide, whose consensus sequence, Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, is conserved in yeast, which harbors 26 repeats, and vertebrates, with 52 repeats. The emerging picture places this unusual structure in a highly important role in coordinating transcription by RNAP II with other events in the generation and maturation of RNAs.
Since its discovery, numerous studies have focused on the role of the entire CTD and the function of individual amino acids within its repeats. At promoters, the unphosphorylated CTD interacts directly with subunits of the Mediator complex (9), which is required for Mediator’s role in stimulating transcription (10). Additionally, early studies determined that the CTD is required for full activation by some promoter-bound activators (11–13) whereas it was later shown that the CTD can mediate the influence of activators on pre-mRNA processing (14–16).
Several genome-wide analyses uncovered a pattern of CTD phosphorylation that occurs once RNAP II clears promoters (e.g., refs. 17 and 18). Phosphorylation of Ser5 occurs near the 5′ end of genes where it enables recruitment of the capping machinery to the nascent pre-mRNA (19, 20), as well as recruitment of Set1, a COMPASS-associated methyltransferase that modifies histone H3-Lys4 around the 5′ end of transcriptionally active genes (21, 22). Toward the 3′ end of genes, Ser5 phosphorylation decreases, Ser2 phosphorylation increases, and CTD repeats that are dually phosphorylated on Ser5 and Ser2 can bind the Set2 methyltransferase, thereby targeting histone H3 for trimethylation on Lys36 (23). Furthermore, Ser2 phosphorylation functions in the recruitment of splicing, 3′ processing, and termination factors (reviewed in refs. 4 and 5). Like Ser5, Ser7 is phosphorylated near the beginning of the transcription cycle, and, although it is detected on both protein-coding and noncoding genes, mutation of Ser7 to Ala in human Rpb1 caused defects in expression specifically of snRNA genes (7). These studies demonstrate that the CTD not only participates in the production and processing of RNA but also leaves behind chromatin modifications that demarcate regions of transcriptionally active genes.
We previously characterized the role of threonine-4 (Thr4) of the CTD repeats by generating chicken DT40 cells that express a mutant form of Rpb1 in which each Thr4 was replaced with Val (8). Cells expressing the Thr-Val (T4V) form of the CTD were not able to survive past 24 h, likely due to defects that were detected in 3′-end processing specifically of histone pre-mRNAs. In metazoans, histone mRNA 3′ processing involves a mechanism that is distinct from the typical polyadenylation of other pre-mRNAs, whose processing and expression were not affected by the Thr4 mutation. In another study, ChIP-seq analysis determined that Thr4 phosphorylation increases toward the 3′ end of RNAP II-transcribed genes and that Thr-to-Ala mutation caused defects in transcription elongation in a human cell line (24). In budding yeast, histone pre-mRNAs are not processed by a distinct mechanism, leading to the question of whether Thr4 plays gene-specific and/or general roles in transcription in that organism. Thr4 mutation in fission yeast was shown to be without effect on growth (25, 26).
Here, we address the role of Thr4 in Saccharomyces cerevisiae by characterizing a strain that expresses a T4V mutant form of Rpb1. The mutant strain is viable and shows no general transcriptional defect in normal growth conditions but is unable to grow in low phosphate or galactose-containing medium. Linking Thr4 with phosphate and galactose metabolism, we found that T4V cells are unable to fully repress phosphate-regulated (PHO) genes under normal growth conditions and to fully activate PHO5 and the galactose-inducible GAL1 and GAL7 genes under their respective inducing conditions. We identify genetic links between T4V and genes encoding the SWR1 and INO80 chromatin remodeling complex subunits, as well as the gene encoding the target of these complexes, the histone variant Htz1. Connecting our observations, we demonstrate that eviction of Htz1 from promoter-proximal nucleosomes, a process associated with gene activation, is defective in T4V cells, specifically for the genes requiring Thr4 for activation, and we implicate the Ino80 complex in Thr4-mediated loss of Htz1 from these genes. Our study, therefore, links Thr4 of the yeast CTD with promoter-associated chromatin remodeling necessary for control of activated transcription.
Results
Thr4 Is Phosphorylated on RNAP II Transcribed Genes but Is Not Essential for Viability in Rich Medium.
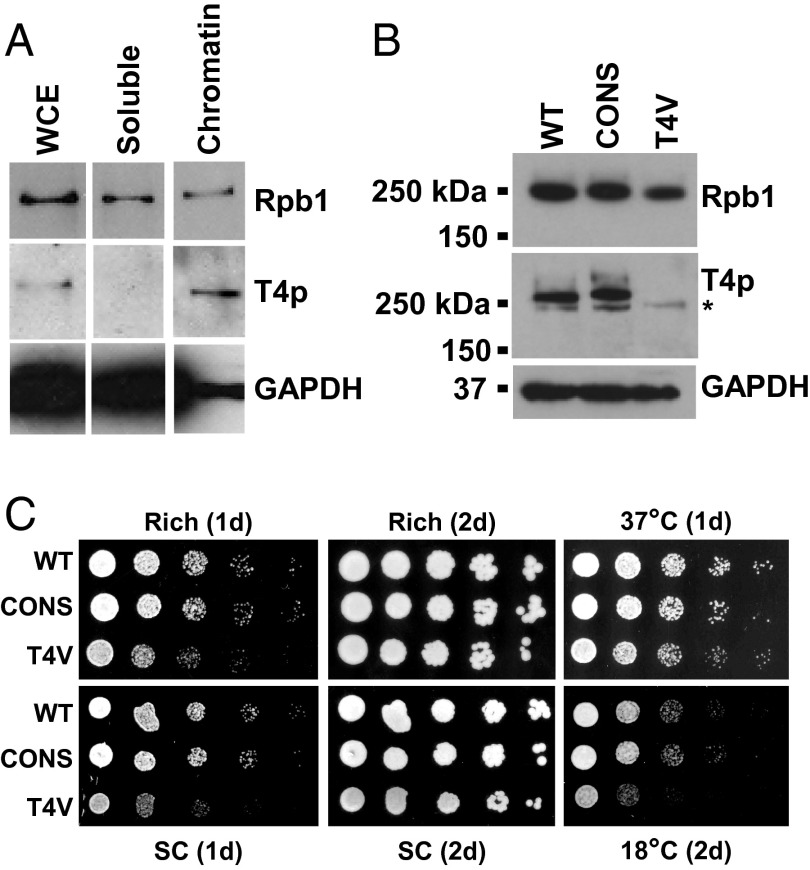
In our previous study, we determined that phosphorylated Thr4 (P-Thr4) is present on Rpb1 in chicken and human cells, and in budding yeast (8). To characterize Thr4 phosphorylation in yeast, we first conducted chromatin fractionation using yeast extracts and found by Western blot (WB) with a P-Thr4 specific antibody that Thr4 is phosphorylated specifically on chromatin-associated RNAP II (Fig. 1A), suggesting that Thr4 functions during transcription. To explore this possibility, we constructed a yeast strain in which Thr4 on each repeat of the CTD was replaced by Val, analogous to what was done in DT40 cells (8). This strain, named T4V, expresses only Rpb1 containing 24 repeats of the sequence YSPVSPS. In addition to replacing Thr with Val, this mutant also eliminated the naturally occurring variation found in some repeats of the CTD (27). To account for any effects that might result from this loss of nonconsensus residues instead of the Thr-Val switch, we constructed a control strain that expressed an Rbp1 with 26 consensus CTD repeats, named CONS. Fig. 1B confirms by WB that Thr4 was phosphorylated in the WT and CONS strains, but not in the T4V strain, and that Rpb1 levels in T4V, CONS, and WT strains were equivalent (SI Appendix, Fig. S5A). Comparison of growth of the three strains showed that T4V displayed a transient and modest growth defect, compared with WT and CONS, on both rich and synthetic complete (SC) medium and that this defect was exacerbated at low temperatures (18 °C) (Fig. 1C). Chromatin immunoprecipitation (ChIP) analysis showed that P-Thr4 was detected on constitutively active and activated inducible genes and that the T4V mutation did not significantly affect RNAP II levels on constitutive genes (SI Appendix, Fig. S1 A–D).
Fig. 1.
Thr4 of the yeast CTD is phosphorylated on chromatin-associated RNAP II and is not essential for viability. (A) Chromatin fractionation of yeast extract followed by Western blot analysis with an RNAP II antibody (Rpb1), an antibody that recognizes phosphorylated Thr4 on CTD repeats (T4p) (8), and a GAPDH antibody. (B) Western blot analysis of yeast extracts derived from WT, CONS, or T4V strains analyzed with the same antibodies as in A. The asterisk (*) indicates a nonspecific cross-reacting band detected at longer exposures. (C) Yeast spot assay comparing growth of WT, CONS, and T4V strains, with serial fivefold dilutions, on rich or synthetic complete (SC) medium at 30 °C, or on SC medium at indicated temperature, for indicated number of days.
Thr4 Is Required for Repression of Phosphate-Regulated Genes in Normal Medium.
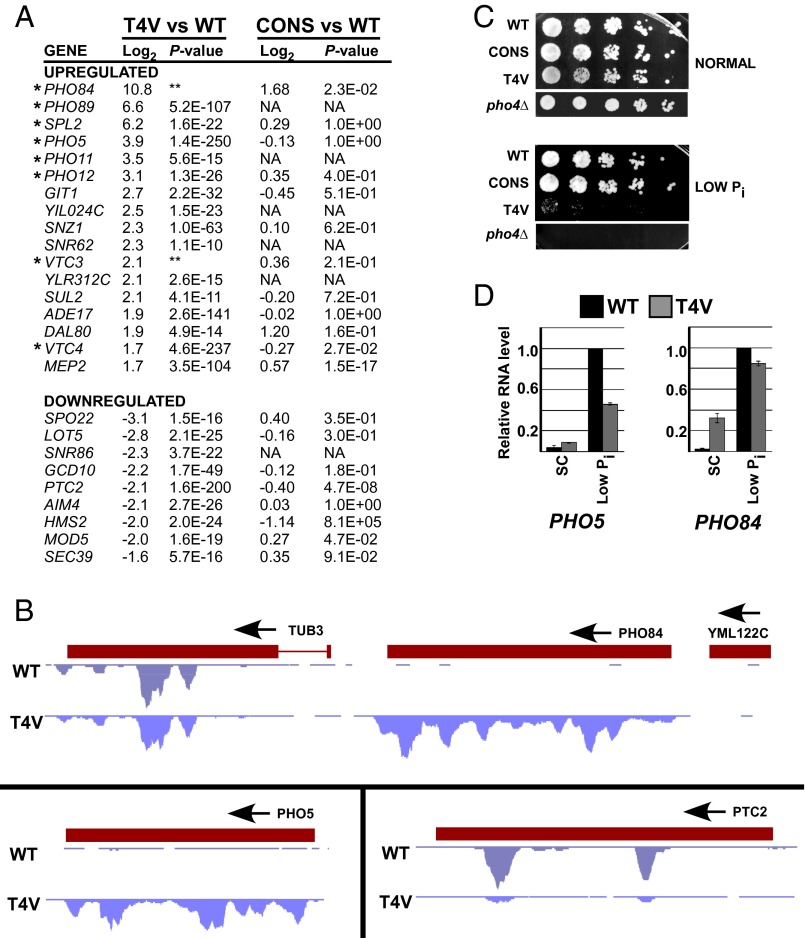
Our observation that Thr4 is not essential for growth indicates that it is not generally required for RNAP II transcription. However, to determine whether Thr4 is involved in transcription of specific genes, we first performed global mRNA sequencing (RNA-seq) in WT and T4V strains grown in SC medium. A summary of the most significantly up- and down-regulated genes is shown in Fig. 2A, and the full results are in Dataset S1. Seventeen genes displayed an average increased expression of at least threefold (log2 > ∼1.6) whereas nine genes showed an average drop in expression of at least threefold (log2 < ∼1.6). A diagram of WT and T4V RNA-seq read distributions over the chromosomal loci for up-regulated genes PHO84 (with unaffected flanking genes) and PHO5, and down-regulated gene PTC2, is shown in Fig. 2B. Interestingly, approximately half of the up-regulated genes are involved in phosphate metabolism (Fig. 2A, indicated by asterisks). Specifically, these genes, hereafter referred to as PHO genes, are also up-regulated under growth conditions of low inorganic phosphate (Pi) levels (28). RT-PCR analysis on a subset of the regulated genes validated the RNA-seq results and confirmed that the T4V mutation resulted in up-regulation of PHO genes (SI Appendix, Fig. S2).
Fig. 2.
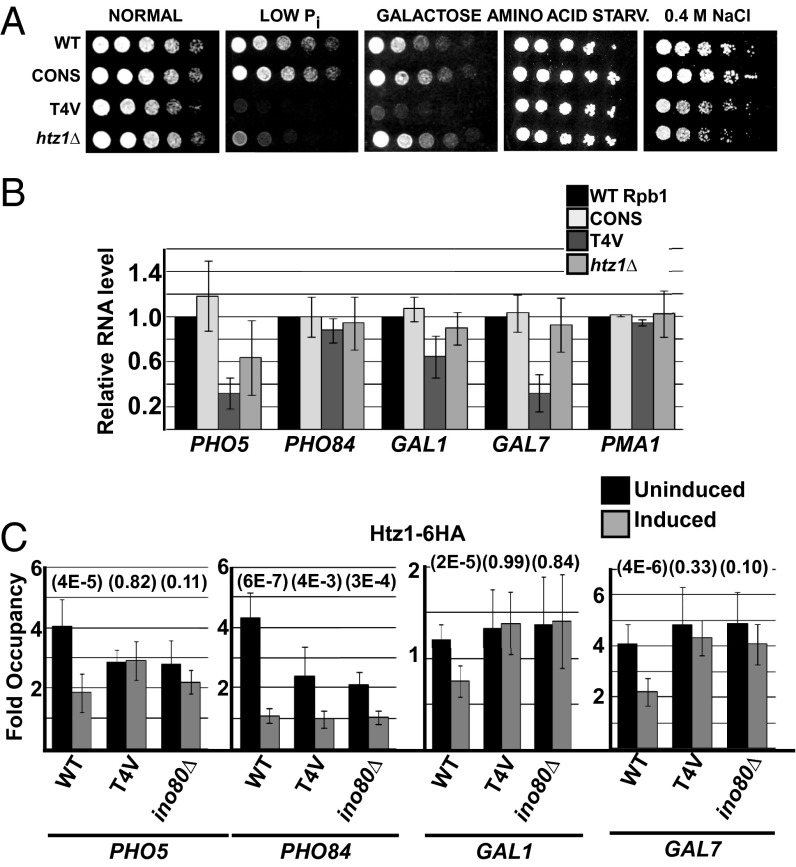
Thr4 is required to maintain repression of PHO genes under normal growth conditions. (A) Summary of most significantly up- and down-regulated genes from RNA-seq analysis performed in WT and T4V strains grown in SC medium. Average change in expression of genes (log2) in T4V over WT strains and P value (Fisher’s exact test) were determined from two sets of data and are indicated here (refer to Dataset S1 for a complete list and details). Corresponding log2 and P values from the WT versus CONS RNA-seq analysis (Dataset S2) are also shown. Genes previously demonstrated to be up-regulated during conditions of low Pi (28) are indicated (*). P values that are beyond the limit of our analysis are indicated with double asterisk (**). NA, genes whose expression was not detectable in the sample. (B) Read distributions from the WT versus CONS RNA-seq analysis for selected genes including PHO84 (with flanking genes shown), PHO5, and PTC2. (C) Spot assay comparing growth of indicated strains and serial fivefold dilutions on normal or low Pi medium. (D) RT-PCR analysis of PHO5 and PHO84 RNA levels in SC or low Pi media in WT or htz1∆ strains.
To determine whether these changes in gene expression were specific to the T4V mutation and not due to the loss of nonconsensus CTD repeats, we performed an additional RNA-seq analysis comparing mRNAs from CONS and WT strains. This analysis showed no similarity with the T4V vs. WT RNA-seq analysis (SI Appendix, Fig. S3). Although PHO84 mRNA levels were approximately threefold higher in CONS than in WT (log2 of 1.68), this increase was dramatically less than in T4V (>1,000-fold, log2 of 10.8), and no derepression of other PHO genes was observed in the CONS strain (Fig. 2A and Dataset S2). Some PHO-regulated genes, however, also appeared up-regulated in the CONS strain in the RT-PCR validation (SI Appendix, Fig. S2). An explanation for this apparent discrepancy stems from the fact that the RT-PCR analysis was performed with random-primed RNA, which detected polyA− as well polyA+ transcripts whereas the RNA-seq analysis detected only polyA+ RNAs. Thus, a parsimonious interpretation of these results is that loss of nonconsensus CTD residues resulted in production of polyA− transcripts, which could be sense and/or antisense, from some PHO genes, whereas mutation specifically of Thr4 led to up-regulation of polyA+ sense transcripts from all PHO genes analyzed. Furthermore, RNA-seq of a strain expressing Rpb1 with a mutant CTD composed entirely of otherwise consensus repeats in which all Tyr1 residues were changed to Phe (Y1F) revealed no up-regulation of the PHO genes. Our results, therefore, demonstrate that the CTD is involved in repressing PHO gene transcription and that Thr4 functions specifically in preventing the production of polyadenylated PHO mRNAs during normal growth.
To extend the above results, we examined growth of the T4V strain in low Pi medium. Strikingly, whereas WT and CONS strains grew normally on this medium, T4V was inviable (Fig. 2C). T4V sensitivity to low Pi medium was observed in both S288C and W303 background strains. Although both the RNA-seq and RT-PCR analyses showed significant derepression of PHO-regulated genes in the T4V strain, derepressed transcription levels for PHO5 and PHO84 were still significantly lower than observed during normal induction of these genes in low Pi conditions (Fig. 2D). Our observations, therefore, indicate that, although Thr4 is required for normal expression of only a small number of genes under normal growth conditions, it is necessary for repression of PHO-regulated genes and plays an essential role in the cellular response to low phosphate levels.
Thr4 Is Genetically Linked to HTZ1 and the SWR1 and INO80 Chromatin Remodeling Complexes.
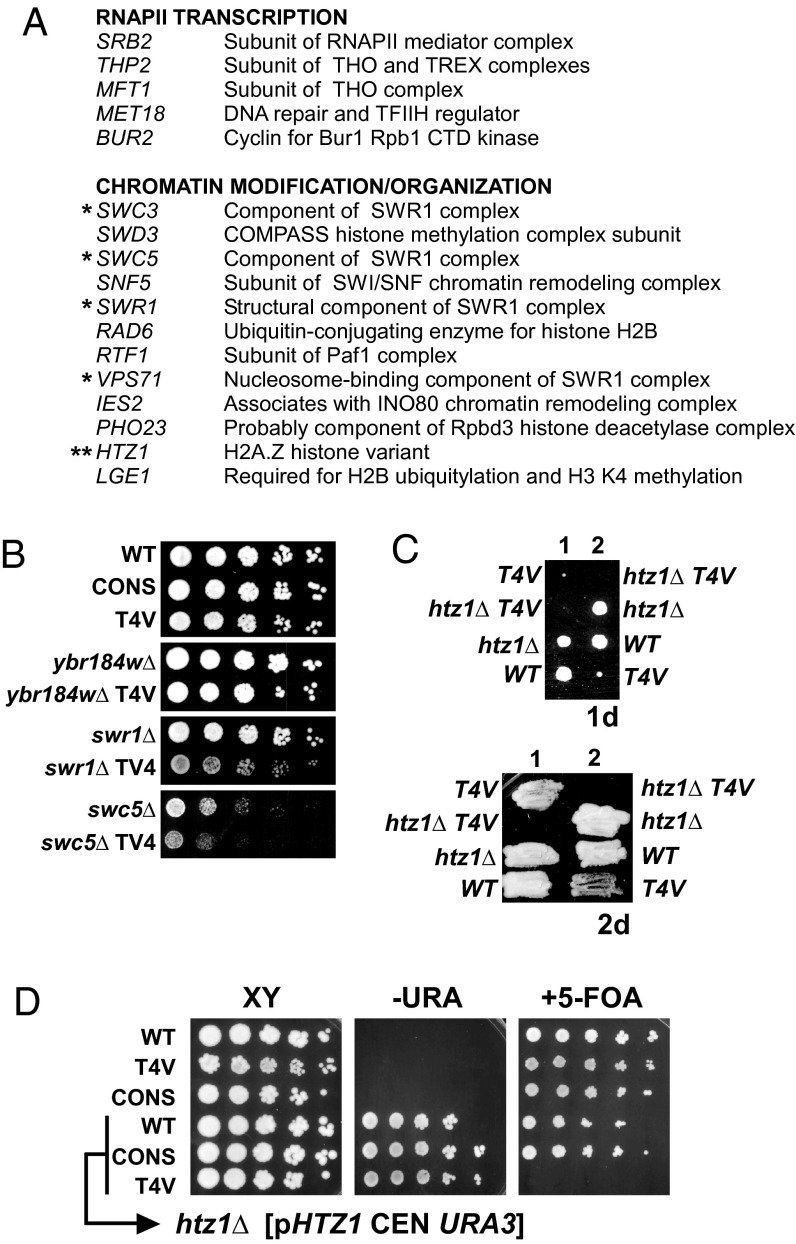
To identify pathways that mediate the effects of Thr4 on growth and transcription, we carried out genome-scale yeast genetic interaction assays [synthetic genetic array (SGA)] (29) with the T4V and CONS strains. Genes whose deletion in combination with the T4V mutation (but not with the CONS mutation) resulted in the most dramatic growth defect in both of two independent SGA analyses are listed in SI Appendix, Fig. S4, and the subset of these genes that is involved in transcription and chromatin regulation is indicated in Fig. 3A. Notably, several components of the SWR1 chromatin remodeling complex were among the strongest genetic interactions with the T4V mutation, which we confirmed by comparing the growth of independently derived double-mutant strains with single-mutant strains (Fig. 3B). The SWR1 complex replaces conventional histone H2A with the variant Htz1 (also known as H2A.Z) in nucleosomes (30–32), specifically those associated with promoters of transcriptionally inactive genes (33–35). Additionally, the gene encoding Ies2, a subunit of another chromatin remodeling complex, the INO80 complex (INO80c), showed a strong genetic interaction with T4V (Fig. 3A) (most other genes encoding INO80c subunits are essential and were therefore not represented in the screen). INO80c can exchange Htz1/H2B dimers for H2A/H2B in in vitro assays and is required for normal localization of Htz1 across the genome, suggesting that it functions in cells to remove Htz1 from activated promoters (36).
Fig. 3.
Thr4 is linked genetically to Htz1 and its regulators. (A) List of genes showing synthetic genetic interaction with T4V mutation in two SGA screens. Genes displaying strongest interactions and involved in RNAP II transcription or chromatin regulation are shown (refer to SI Appendix, Fig. S2 for a complete list). (B) Confirmation of genetic interaction between T4V and SWR1 and SWC5 components of SWR1 complex. Strains of indicated genotypes were generated by recombinant transformation and growth compared by spot assay on SC medium with serial fivefold dilutions. YBR184W showed no interaction with T4V in SGA screen and serves as a control. (C) Tetrad dissection analyses demonstrating synthetic lethal phenotype between T4V and HTZ1. Growth after 24 h is shown. After 48 h, colonies were streaked onto rich-medium plates, and growth is shown after additional 48 h. Genotype of inviable double mutant was inferred based on expected segregation pattern. (D) WT, T4V, and CONS strains were transformed with plasmid (pHTZ1 CEN URA3), and then endogenous HTZ1 was deleted by homologous recombination. Original and product strains were spotted on media shown, including 5-fluoroorotic acid (5-FOA), which allows growth only in the absence of URA3 expression (i.e., if the plasmid is lost).
The HTZ1 gene was also a strong hit in the SGA screen. Consistent with this genetic interaction, HTZ1 and the T4V mutation showed a synthetic lethal interaction in multiple independent tetrad dissection analyses (Fig. 3C). T4V spores derived from HTZ1/htz1Δ WT-RPB1/T4V double heterozygotes displayed a more severe growth defect than normal after dissection (compare Fig. 3C, Upper with Fig. 1C; and see also Fig. 5B) although, like other T4V strains, the defect was transient and T4V approached WT growth after ∼48 h (Fig. 3C, Lower). Additionally, attempts to delete HTZ1 in a haploid T4V strain by homologous recombination produced no viable transformants. In another approach, we deleted HTZ1 in WT, CONS, or T4V strains harboring an HTZ1 expression plasmid (with a URA3 marker) and then eliminated the plasmid by growth on 5-FOA. Only the T4V htz1Δ background showed no growth on 5-FOA (Fig. 3D), supporting the synthetic genetic interaction between HTZ1 and T4V. These results together strongly implicate Thr4 in Htz1-related pathways.
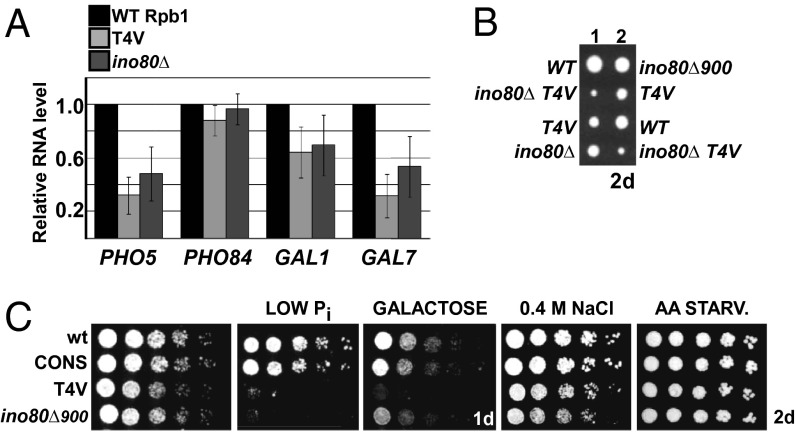
Fig. 5.
Evidence that Thr4 dependence is linked to the INO80 complex. (A) RT-PCR analysis of mRNA levels of indicated genes under respective induced conditions in indicated strains. Data are represented as mean ± SD of three independent experiments. (B) Tetrad dissection analyses demonstrating negative synthetic genetic interaction between T4V and INO80. Growth after 48 h is shown. (C) Growth comparison of indicated stains and serial fivefold dilutions in indicated media.
Thr4 Is Required for Expression of PHO5, GAL1, and GAL7 and for Eviction of Htz1 from Their Promoters During Induction.
Htz1 is typically found within the two nucleosomes that flank the nucleosome-free region at RNAP II promoters (34, 37), and, although it is not required for maintaining a repressed state, its removal from these nucleosomes stimulates gene activation (33, 35). Correspondingly, genomic analyses localized Htz1 primarily at repressed gene promoters, and its occupancy is inversely proportional to transcription levels (33, 35, 37, 38). Relating Htz1 function with our observed role for Thr4 in PHO gene regulation, a previous study (39) demonstrated that HTZ1 acts synthetically with genes encoding the SWI/SNF chromatin-remodeling complex to activate expression of PHO5, which encodes the cell’s major acid phosphatase (40). We compared growth of htz1∆ and T4V cells on low Pi medium and found that the two strains displayed strong growth defects, indicating that both Thr4 and Htz1 indeed play critical roles in the PHO pathway (Fig. 4A). We then determined whether Thr4 and Htz1 are needed for expression of genes induced during low Pi conditions and found that both T4V and htz1∆ strains showed reduced expression of PHO5 whereas induction of another PHO gene, PHO84, was not affected (Fig. 4B).
Fig. 4.
Thr4 is required for full expression and Htz1 eviction from promoters of specific genes. (A) Spot assay comparing growth of indicated strains and serial fivefold dilutions on indicated medium. Amino acid starvation medium was previously described (63). (B) RT-PCR analysis of mRNA levels of indicated genes under the respective inducing conditions, with PMA1 levels analyzed as control, grown in SC medium. Data are represented as mean ± SD of three independent experiments. (C) ChIP analysis of Htz1-6HA occupancy at promoter regions of indicated genes, uninduced or induced as indicated, in WT, T4V, and ino80∆ strains. Data are represented as mean ± SD of three independent experiments, and P values for each pair of values (uninduced and induced) are indicated in parentheses above bars.
We also tested for growth defects on other media in which gene induction is necessary for growth. We found that T4V was also highly sensitive to medium containing galactose as the sole carbon source but grew normally on amino acid-depleted or high osmolarity media (Fig. 4A). Low Pi and galactose sensitivity was specific to the T4V mutation because the CONS and strain did not show specific growth defects on these media (Fig. 4A). Galactose sensitivity correlated with reduced expression of the galactose-induced GAL1 and GAL7 genes in T4V (Fig. 4B) whereas expression of the amino acid starvation-induced ARG1 and high osmolarity-induced STL1 genes was not affected. Notably, a previously described strain expressing Rpb1 with a CTD truncated to ∼13 repeats showed normal activation of the GAL1 promoter (11), further demonstrating that the observed defects are specific for the Thr4 mutation. In all cases, reduced transcription of genes in T4V correlated with less RNAP II recruited to the genes’ promoters, as determined by ChIP analysis (SI Appendix, Fig. S1D). The htz1∆ strain also grew normally on amino acid-depleted and high osmolarity media, but, although Htz1 is known to play a role in activation of GAL1 (39, 41, 42), our htz1∆ strain (and several additional clones derived from the W303 parental strain) failed to display a significant growth defect on galactose (Fig. 4A) (Discussion). Taken together, our results indicate that Thr4 is required for full induction of a subset of inducible genes and suggest that the role of Thr4 in induction is related to Htz1 function.
Eviction of Htz1 from promoter-associated nucleosomes is thought to stimulate gene activation by exposing promoter DNA (35). To address whether impaired activation of PHO5, GAL1, and GAL7 in T4V cells reflected defective eviction of Htz1 from their respective promoters, we used strains expressing Htz1 as a 6x-HA epitope-tagged derivative (SI Appendix, Fig. S5A) and performed ChIP experiments with an HA antibody. Fig. 4C shows that, in WT cells, induction of PHO5, PHO84, GAL1, and GAL7 resulted in a significant loss of Htz1 from their respective promoter regions, confirming that Htz1 eviction corresponds with activation of these genes. Consistent with our gene-expression analysis, the T4V mutation effectively abolished Htz1 loss from the PHO5, GAL1, and GAL7 promoters during their induction, but not from PHO84 (Fig. 4C). Defective Htz1 eviction on these genes is specific to the CTD Thr4 mutation because altering the CTD to all consensus repeats (CONS) did not affect Htz1 loss on the GAL1 promoter (SI Appendix, Fig. S5B). At the uninduced PHO5 and PHO84 promoters, less Htz1 was detected in T4V cells than in WT (Fig. 4C), probably caused by nucleosome loss coinciding with derepression of these genes due to the Thr4 mutation (Discussion). Despite the lower levels of Htz1 in the T4V mutant, significant Htz1 eviction was observed for PHO84, but not for PHO5. These observations strongly link Htz1 promoter dynamics to Thr4 and indicate that Thr4 is required for Htz1 eviction from some inducible genes, thereby enabling their full induction.
INO80c Functions in Thr4-Mediated Stimulation of Htz1 Eviction.
We next examined whether Thr4 stimulates Htz1 loss from specific activated promoters through an INO80c-dependent mechanism. Indeed, Htz1 ChIP analysis showed that Htz1 loss from promoters of PHO5, GAL1, and GAL7 during induction was defective in cells expressing a nonfunctional, truncated version of the Ino80 ATPase subunit of the complex (ino80Δ900) (Fig. 4C). Consistent with previous observations that Ino80 plays a general role in regulating Htz1 occupancy on the genome (36), ino80Δ900 cells showed reduced Htz1 occupancy on both PHO5 and PHO84 promoters in normal growth conditions (Fig. 4C). During induction of these genes, however, loss of Ino80 resulted in impaired Htz1 eviction for PHO5 but not PHO84, paralleling the results seen with the T4V mutation (Fig. 4C). In all cases, impaired Htz1 loss from promoters resulted in reduced induction of the respective genes (Fig. 5A). These observations, therefore, implicate both Thr4 and Ino80 in clearing Htz1 from a specific set of induced promoters and suggest that Ino80 mediates the effects of Thr4 on induction of these genes (i.e., genes showing a requirement for Thr4 for induction are genes that show Ino80-dependence for Htz1 eviction). In support of this notion, in addition to the genetic interaction we detected between T4V and IES2 in the SGA screen (Fig. 3A), we detected a negative synthetic genetic interaction between INO80 and the T4V mutation (Fig. 5B). Furthermore, like T4V, cells lacking Ino80 were unable to grow on low Pi medium, were highly sensitive to galactose, but were essentially unaffected by amino acid depletion or high osmolarity (Fig. 5C). Therefore, our data implicate Ino80 in Thr4-stimulated loss of Htz1 from PHO5, GAL1, and GAL7 promoters, pointing to an unexpected role for the CTD in chromatin remodeling.
Discussion
Here, we have provided evidence that Thr4 of the CTD plays a critical but selective role in transcription in budding yeast. Like our previous analysis of Thr4 function in chicken cells (8), we found that this highly conserved CTD residue does not play a general role in transcription, but functions in expression of specific genes. Thr4 is required for both repression of PHO genes and for full activation of a subset of inducible genes, including PHO5, GAL1, and GAL7. Although it is likely that Thr4 maintains PHO gene repression by regulating chromatin structure, we found that its role in activation involves stimulating the eviction of Htz1 specifically from promoters of the Thr4-sensitive genes. To carry out this function, we provided evidence that Thr4 recruits or stimulates INO80c, whose chromatin remodeling activity acts to remove Htz1 from those promoters. Below, we discuss these previously unidentified functions of the CTD.
Our data implicate Thr4 in both repression and activation of specific genes, and chromatin remodeling appears to be at play in both cases. Repression of PHO-regulated genes is accomplished by blocking access of the basic helix–loop–helix transcription factor Pho4 to its recognition elements upstream of the PHO genes, by keeping it out of the nucleus and through interference with positioned nucleosomes on its binding sites (43). Consequently, disruption of chromatin structure at PHO genes leads to derepression. For example, in the absence of Set1, loss of H3K4 methylation causes reduced nucleosome occupancy around the PHO5 promoter, and, as a result, PHO5 becomes derepressed (44). Under repressive conditions, Cbf1, another basic helix–loop–helix protein, binds chromatin at many of the same sites that become bound by Pho4 under conditions of low Pi (45). Cbf1 is required for normal nucleosomal positioning (46), and, in its absence, PHO genes become derepressed, even under normal Pi conditions, when most Pho4 is cytoplasmic (43). Interestingly, there is a high degree of overlap between genes that we found are up-regulated in T4V with genes up-regulated in CBF1-deleted cells (43), suggesting that a similar defect in nucleosomal arrangement occurs in T4V cells. Indeed, the reduced levels of Htz1 at the PHO5 and PHO84 promoters that we detected in T4V cells in noninducing conditions likely reflect reduced nucleosome occupancy at these derepressed genes. It is likely, then, that Thr4 contributes to maintaining the nucleosomal structure necessary for repression of PHO genes.
Induction of PHO5 and GAL1, two of the Thr4-sensitive genes we identified, has been studied in some detail, and Htz1 is known to play a role in this process. Although Htz1 is evicted from the GAL1 promoter upon induction (39, 41) and is quickly redeposited upon returning to a repressed state (47), its effects are on GAL1 activation, not repression. Cells lacking HTZ1 show reduced recruitment of TBP, RNAP II, Mediator, SAGA, and Swi/Snf, with reduced GAL1 expression as a consequence (39, 41, 42, 47). The effect is transient (47), however, and, in some cases, RNAP II is efficiently recruited to the induced GAL1 promoter, and GAL1 expression occurs in the absence of Htz1 (our results and refs. 39 and 48), indicating that other pathways can sometimes compensate for the lack of Htz1. Htz1 eviction also occurs at the PHO5 promoter upon its induction (39, 49). Nucleosome disassembly at the PHO5 promoter was shown to be rate-limiting for its expression (50), and efficient PHO5 induction is dependent on Htz1 (our results and ref. 39). We have now shown that Thr4 is required for eviction of Htz1 from GAL1 and PHO5, as well as GAL7 promoters. The consequences of the T4V mutation on expression of these genes and growth on low Pi and galactose-containing media are more severe than in htz1∆ cells, indicating that, beyond promoting Htz1 eviction, Thr4 likely stimulates expression through additional pathways. Nonetheless, our data, supported by genetic interactions between T4V and HTZ1 and its regulators, connect transcription to chromatin remodeling by stimulating Thr4-dependent Htz1 eviction.
Our data also implicate INO80c in Thr4 function. In addition to roles in double-strand break repair and stalled replication fork recovery (51), INO80c is required for full induction of a number of genes, including reporter genes containing upstream activation sequences of the PHO5 or GAL1 promoters (52, 53). INO80c associates with many RNAP II promoters and is recruited to genes when they are activated (54, 55), including PHO5, where it was shown to be required for nucleosome remodeling and gene induction (52). In the absence of Ino80, Htz1 localization across the genome is perturbed, with increased occupancy at some loci and reduced occupancy at others, thereby linking INO80c with Htz1 dynamics (36). More specifically, Ino80 is required for Htz1 eviction from the KAR4 promoter during induction, demonstrating that it can function in clearing promoter-proximal nucleosomes of Htz1 coincident with gene activation (36). Indeed, we have now shown that additional genes, specifically those that are sensitive to the T4V mutation (i.e., PHO5, GAL1, and GAL7), also require Ino80 for efficient Htz1 loss during their activation. Based on our findings, we expect that other genes found to be dependent on Ino80 for Htz1 eviction will also show Thr4 sensitivity.
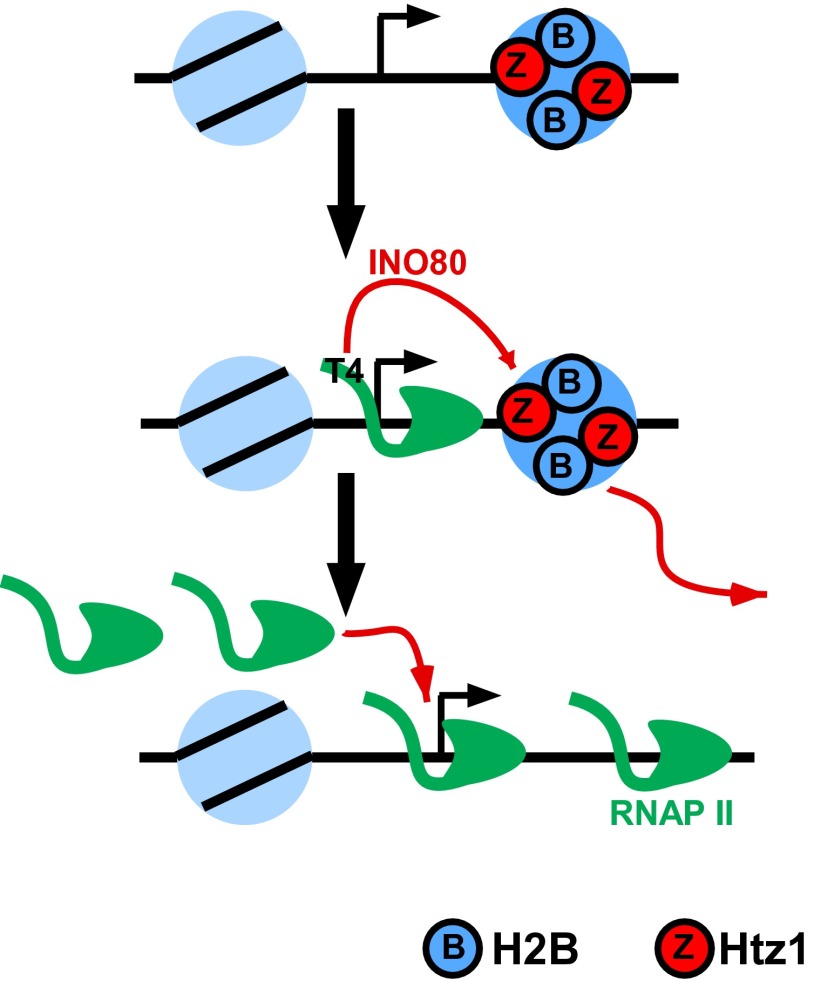
We have presented several lines of evidence linking Thr4 with Ino80 function. Based on these connections, we propose a model (Fig. 6) in which, upon activation, a recruited molecule of RNAP II triggers eviction of Htz1 from promoter-proximal nucleosomes by stimulating the INO80 complex, or facilitating its recruitment, to newly activated promoters, in a manner dependent on Thr4 or its phosphorylation. We were unable to detect an Ino80–Rpb1 interaction by coimmunoprecipitation, suggesting that recruitment or stimulation of INO80c by Thr4 is transient and/or mediated by other factors. However, failure to detect such an interaction is not surprising as many known CTD interactions are not detectable in this type of assay. For example, capping enzymes, polyadenylation factors, and the CTD kinases Ctk1 and Kin28 were not detected in a tandem-affinity purification of Rpb1 even though they are all well-characterized CTD binding proteins (56). Importantly, transcription and RNAP II have been shown to be important for promoter-proximal nucleosome eviction (57). We now implicate Thr4 and INO80c in this process, which we propose facilitates further recruitment of additional RNAP II molecules. In vitro studies showed that INO80c evicts Htz1/H2B dimers from nucleosomal octamers and that, in some cases, eviction was part of an INO80c-dependent Htz1/H2B exchange for H2A/H2B (36) whereas, in another in vitro study, INO80c was not able to replace Htz1/H2B dimers with H2A/H2B dimers in nucleosomal arrays (58). In our model, however, we suggest that Thr4-mediated recruitment or stimulation of INO80c at specific promoters causes a loss of Htz1 that destabilizes promoter-proximal nucleosomes, thereby increasing accessibility to promoter DNA and facilitating further rounds of RNAP II recruitment and increased transcription (33, 35). Consequently, H2A-containing nucleosomes (which presumably form at promoters in the absence of Htz1) are not efficiently destabilized by INO80c, resulting in reduced gene activation.
Fig. 6.
Model indicating role for Thr4 of yeast CTD repeats in promoting Htz1 eviction. On repressed genes, promoter-proximal nucleosomes contain Htz1/H2B dimers (depicted as encircled Z and B). During activation, an initially recruited RNAP II molecule stimulates INO80 activity or recruitment through Thr4 or Thr4 phosphorylation, thereby enabling INO80-mediated eviction of Htz1/H2B from nucleosomes. Htz1 eviction destabilizes nucleosomes, facilitating their loss and increasing DNA access for further rounds of transcription. When transcription ends, in the absence of Thr4, INO80 no longer acts, and Htz1/H2B is redeposited at promoter-proximal nucleosomes by the SWR1 complex.
Defects in repression or activation that result from the T4V mutant CTD might arise from an inability to phosphorylate the mutated Thr residues. Genomic ChIP analysis determined that the average Thr4 phosphorylation pattern in human cells increases toward the 3′ end of genes (24). In yeast, a genomic ChIP (59), as well as our ChIP analysis of individual genes, showed that most Thr4 phosphorylation is detected on Rpb1 associated with the middle portion of genes, with only low levels detected at promoters. This distribution might indicate that the Thr residue itself is important for promoter-specific functions, including those we report in this study.
A recent study examined the transcriptomes of fission yeast strains harboring a variety of CTD substitutions (25). Consistent with our findings, mutation of Thr4 (to Ala; T4A) in that study affected expression of only a small number of genes, including orthologs of PHO5 and PHO84. However, whereas our T4V mutation resulted in derepression of these genes, in fission yeast expressing the T4A mutant, expression of the orthologous genes was significantly decreased. The different effect of Thr4 mutation might reflect differences between the two species. For example, Ser2 has been shown to be required for viability in S. cerevisiae but not in Schizosaccharomyces pombe during normal growth (26, 60). Differences between the CTD constructs could also contribute (e.g., T4A and 14 consensus heptads in ref. 29, compared with T4V and 24 heptads in our study). Nonetheless, the fact that PHO5 and PHO84 (and orthologs) are among only a small number of genes whose expression relies on Thr4 in both budding and fission yeast strains strongly points to conserved roles for this CTD residue in regulating transcription of PHO genes.
An interesting question is whether our findings on Thr4 function extend to metazoan organisms. As mentioned above, two studies have characterized the role of Thr4 and its phosphorylation in vertebrate species, in which it plays functions necessary for viability (8, 24). Furthermore, INO80c functions in promoting eviction of transcription start site-associated nucleosomes containing the mammalian Htz1 ortholog H2A.Z during gene activation in mouse embryonic stem cells during differentiation, indicating that the same basic INO80c-mediated mechanism is at play throughout eukaryotes (61). It remains to be seen whether the role we uncovered for Thr4 in mediating this mechanism in yeast is conserved in higher species. In any event, our results show that Thr4 of the yeast CTD plays an important role in coupling transcription with promoter-associated nucleosomal remodeling, providing yet another function for the multifunctional CTD.
Materials and Methods
A detailed description of yeast strain construction and growth conditions, ChIP, RNA and protein analyses, and RNA sequencing can be found in SI Appendix, SI Materials and Methods. Briefly, T4V and CONS strains were constructed by homologous recombination in yeast strains using CTD constructs prepared for our previous study (8). SGA analysis, comparing CONS and T4V strain genetic interactions, was as previously described (62). RT-PCR analyses were performed with random-primed total RNA for cDNA synthesis whereas RNA-seq experiments used polyA+ RNA. Primer sequences used for cloning, ChIP, and RT-PCR are available upon request.
Supplementary Material
Acknowledgments
We thank Elizabeth Miller and Silvere Pagant (Columbia University) for assistance with the SGA screen. This work was supported by National Institutes of Health Grants R01GM097174 (to J.L.M.) and R01GM084089 (to B.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1412802111/-/DCSupplemental.
References
- 1.Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics. 2012;190(2):351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. Biochim Biophys Acta. 2013;1829(1):84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corden JL. RNA polymerase II C-terminal domain: Tethering transcription to transcript and template. Chem Rev. 2013;113(11):8423–8455. doi: 10.1021/cr400158h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829(1):55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 5.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26(19):2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drogat J, Hermand D. Gene-specific requirement of RNA polymerase II CTD phosphorylation. Mol Microbiol. 2012;84(6):995–1004. doi: 10.1111/j.1365-2958.2012.08071.x. [DOI] [PubMed] [Google Scholar]
- 7.Egloff S, et al. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318(5857):1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334(6056):683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai KL, et al. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol. 2013;20(5):611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myers LC, et al. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12(1):45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allison LA, Ingles CJ. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci USA. 1989;86(8):2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber HP, et al. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374(6523):660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 13.Scafe C, et al. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347(6292):491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 14.Nagaike T, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. RNA Biol. 2011;8(6):964–967. doi: 10.4161/rna.8.6.17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem. 2002;277(45):43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- 16.Rosonina E, Bakowski MA, McCracken S, Blencowe BJ. Transcriptional activators control splicing and 3′-end cleavage levels. J Biol Chem. 2003;278(44):43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- 17.Bataille AR, et al. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45(2):158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 18.Kim H, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat Struct Mol Biol. 2010;17(10):1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCracken S, et al. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11(24):3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11(24):3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: Linking transcriptional elongation to histone methylation. Mol Cell. 2003;11(3):721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 22.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11(3):709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 23.Kizer KO, et al. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol. 2005;25(8):3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hintermair C, et al. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31(12):2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwer B, Bitton DA, Sanchez AM, Bahler J, Shuman S. Individual letters of the RNA polymerase II CTD code govern distinct gene expression programs in fission yeast. Proc Natl Acad Sci USA. 2014;111(11):4185–4190. doi: 10.1073/pnas.1321842111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwer B, Shuman S. Deciphering the RNA polymerase II CTD code in fission yeast. Mol Cell. 2011;43(2):311–318. doi: 10.1016/j.molcel.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 28.Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell. 2000;11(12):4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 30.Kobor MS, et al. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2(5):E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogan NJ, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12(6):1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 32.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303(5656):343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 33.Li B, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci USA. 2005;102(51):18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19(3):360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123(2):219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144(2):200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raisner RM, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123(2):233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guillemette B, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3(12):e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103(3):411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 40.Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72(6):323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- 41.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol Cell Biol. 2001;21(18):6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halley JE, Kaplan T, Wang AY, Kobor MS, Rine J. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 2010;8(6):e1000401. doi: 10.1371/journal.pbio.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, O’Shea EK. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Mol Cell. 2011;42(6):826–836. doi: 10.1016/j.molcel.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang SS, Zhou BO, Zhou JQ. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol Cell Biol. 2011;31(15):3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431(7004):99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent NA, Eibert SM, Mellor J. Cbf1p is required for chromatin remodeling at promoter-proximal CACGTG motifs in yeast. J Biol Chem. 2004;279(26):27116–27123. doi: 10.1074/jbc.M403818200. [DOI] [PubMed] [Google Scholar]
- 47.Gligoris T, Thireos G, Tzamarias D. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol Cell Biol. 2007;27(11):4198–4205. doi: 10.1128/MCB.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santisteban MS, Hang M, Smith MM. Histone variant H2A.Z and RNA polymerase II transcription elongation. Mol Cell Biol. 2011;31(9):1848–1860. doi: 10.1128/MCB.01346-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14(5):667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 50.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133(4):716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao Y, Shen X. INO80 subfamily of chromatin remodeling complexes. Mutat Res. 2007;618(1-2):18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbaric S, et al. Redundancy of chromatin remodeling pathways for the induction of the yeast PHO5 promoter in vivo. J Biol Chem. 2007;282(38):27610–27621. doi: 10.1074/jbc.M700623200. [DOI] [PubMed] [Google Scholar]
- 53.Ebbert R, Birkmann A, Schüller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol Microbiol. 1999;32(4):741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- 54.Jónsson ZO, Jha S, Wohlschlegel JA, Dutta A. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol Cell. 2004;16(3):465–477. doi: 10.1016/j.molcel.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 55.Klopf E, et al. Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2009;29(18):4994–5007. doi: 10.1128/MCB.01858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 57.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20(1):90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luk E, et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143(5):725–736. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayer A, et al. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336(6089):1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 60.West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140(4):1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, et al. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151(7):1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- 63.Rosonina E, Duncan SM, Manley JL. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010;24(12):1242–1252. doi: 10.1101/gad.1917910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.