Significance
Various intrinsically disordered proteins form structure on binding to partner proteins. The mechanism of this process is still poorly understood, and kinetic measurements will be important in furthering understanding. Here, we have performed the first comparison of the kinetics for various different intrinsically disordered partners with a protein [CREB binding protein–kinase-inducible domain interacting (KIX)] that is key in deciding which proteins are expressed within a cell. We compare the rate constants in the presence and absence of a ligand at a second binding site on KIX, thereby providing insight into the mechanism of allostery between the two sites.
Keywords: intrinsic disorder, dynamic allostery, coupled folding and binding, induced fit, intrinsically disordered protein
Abstract
The kinase-inducible domain interacting (KIX) domain of CREB binding protein binds to multiple intrinsically disordered transcription factors in vivo at two distinct sites on its surface. Several reports have been made of allosteric communication between these two sites in this well-characterized model system. In this work, we have performed fluorescence stopped-flow measurements to investigate the kinetics of binding of five KIX binding proteins. We find that they all have similar association and dissociation rate constants for complex formation, despite their wide range of intrinsic helical propensities. Furthermore, by careful arrangement of pseudofirst-order conditions, we have been able to show that both association and dissociation rate constants are decreased when a partner is bound at the alternative site. These decreases suggest that positive allosteric effects are not mediated by structural changes in binding sites but rather, through a more general mechanism, largely mediated through dissociation, which we propose is largely related to changes in the flexibility of the KIX domain itself.
Intrinsically disordered proteins (IDPs) are proteins without a defined structure that nonetheless play important biological roles. Although originally it was thought that such proteins were rare exceptions to the structure–function paradigm, bioinformatic analyses have shown that they are relatively common, particularly in eukaryotes, where proteins with intrinsically disordered regions constitute around 30% of the proteome (1). IDPs are heavily represented in processes such as transcription and signaling, especially where multiple partners are involved, at hubs in protein interaction networks (1, 2). One key and much studied hub protein involved in transcriptional regulation within the cell is CREB binding protein (CBP) and its paralogue p300. CBP is a transcriptional activator that acts as a scaffold (or bridge) for several constituents of the transcriptional machinery (3). The small 87-aa folded kinase-inducible domain interacting (KIX) domain of CBP binds to many different transcription factors in vivo, including cMyb, MLL, CREB, cJun, p53, FOXO3a, BRCA1, and SREBP (3), most of which are intrinsically disordered. The expression levels and interactions between these components play an important role in determining the expression profile of the cell. Mutations in these transcription factors and CBP are known to be associated with disease, particularly cancer (3). In addition, viral transcription factors, such as PBX-1 E2A-PBX1, HTLV-1 Tax, HIV-1 Tat, and HTLV-1 basic leucine zipper (HBZ), are known to bind to KIX, thereby using it for transcription of viral proteins or blocking its function (3). NMR structures of CBP KIX in complex with several of its partners (4–9) (Fig. 1) and when complexed with a small molecule (10) are available, but there is no structure for the KIX domain alone. All of the structures show the peptides to contain short α-helical elements of structure bound to one or both of two hydrophobic binding surfaces on KIX. Structures are available for two ternary complexes that show that the two KIX ligands do not interact directly (5, 8) (Fig. 1 C and D and Fig. S1). Nevertheless, there have been several reports of positive cooperativity in binding of ligands by KIX (8, 11, 12). For example, the complex between cMyb and KIX.MLL is more stable than the complex of cMyb with KIX alone (5). This phenomenon has been investigated by both NMR and simulations (8, 13–17), but here, we use kinetics studies that have the potential to provide insights into the mechanistic basis of the allosteric effect.
Fig. 1.
NMR structures for KIX complexed with various ligands. (A) cMyb [pink; Protein Data Bank (PDB) ID code 1SB0] and pKID (pale green; PDB ID code 1KDX) bound to the cMyb site of KIX (blue and light blue). (B) MLL (pale yellow; PDB ID code 2LXS) and E2A (cyan; PDB ID code 2KWF) bound to the MLL site of KIX (blue and light blue). (C and D) Ternary complexes (displayed in darker shades) of (C) cMyb, KIX, and MLL (PDB ID code 2AGH) and (D) pKID, KIX, and MLL (PDB ID code 2LXT) are aligned with the two relevant binary complexes from A and B. The 20 lowest energy structures are displayed, and the structures are aligned according to the KIX protein alone.
In this work, we have investigated the binding kinetics of peptides from five separate transcription factors to KIX: two that bind at the cMyb site (pKID and cMyb) and three that bind at the MLL site (MLL, E2A-PBX1, and HBZ). Four of the binding partners investigated (4, 7–9) have corresponding complex structures published (Fig. 1). The final protein, HBZ, is known from competition studies with cMyb and MLL to bind at the MLL site (11). Furthermore, we report the kinetic effects of a partner bound at one site on IDP binding at the other site. Using this approach, we reveal that the major source of the allostery between the sites in each case is actually through a reduction in the dissociation rate constant. Given the apparently general nature of the result combined with previous NMR results and simulations that show reduced structural fluctuations within KIX on binding pKID, MLL, and cMyb, we hypothesize that changes in flexibility of the KIX protein on partner binding make a key common contribution to the observed allosteric effects.
Results
Properties of KIX Binding Partners.
N-terminally FITC-labeled versions of cMyb25, pKID, E2A, and MLL corresponding to the peptides used in generating the NMR structures were made using solid-phase peptide synthesis. In addition, peptides of the transactivation domains (TADs) of cMyb and HBZ were recombinantly expressed and labeled with Alexa594 through maleimide chemistry at a natural cysteine within each sequence. The binding partners display a range of sizes (19–121 aa) and sequences (Fig. S2) as well as net charges (−8.0 to −3.0) and charge distributions. All peptides contain a transcription factor binding motif ΦXXΦΦ, and the final hydrophobic residue (Φ) in each case is a leucine that inserts into a small and somewhat deeper hydrophobic pocket (Fig. S2).
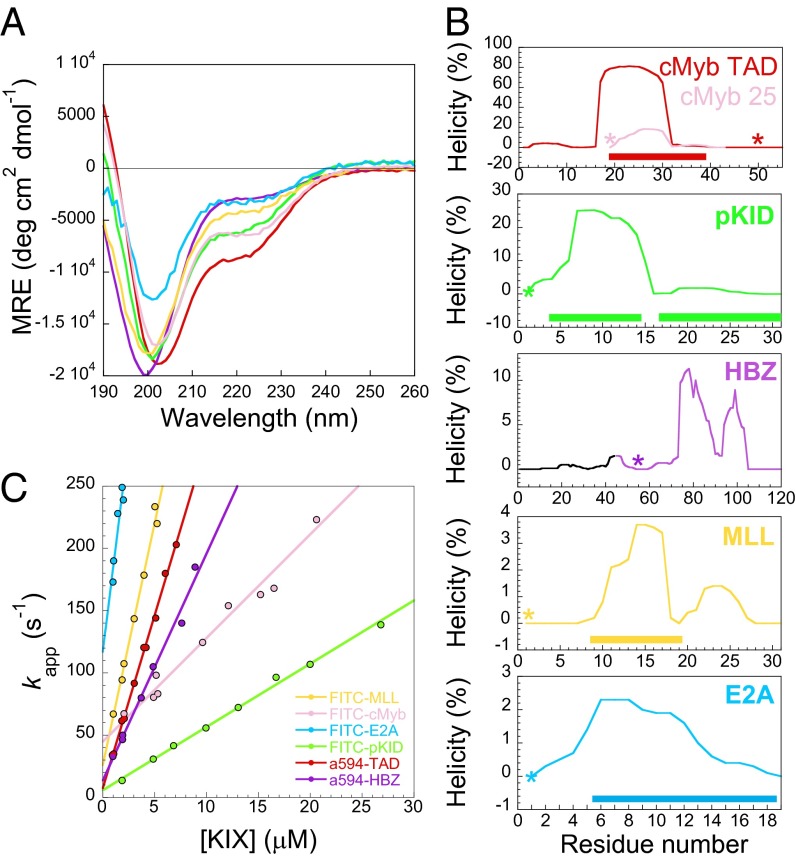
CD spectra (Fig. 2A) showed that all of the binding partners investigated in this study are predominantly disordered, including the viral protein HBZ, which has not previously been reported as an IDP (although the HBZ TAD is fused with a long, presumably disordered tail). However, it is not clear whether this disorder is spread over the whole peptide or confined to particular areas, which makes it difficult to compare the degree of disorder for the binding residues of each of the peptides. In lieu of residue-specific information, we can examine predicted structural information from the helical prediction software AGADIR (18). AGADIR suggests that the peptides will display a wide range of helicities in the binding regions from 1–2% for E2A to 80% for cMybTAD (Fig. 2B) under our experimental conditions. The predicted overall helicity by AGADIR roughly agreed with that estimated from CD using various methods (19–21) but was generally lower, especially for the more disordered proteins (Fig. S3).
Fig. 2.
Secondary structural properties and association kinetics for various KIX binding partners. (A) CD spectra indicate a range of helical contents. Data are plotted as Mean Residual Ellipticity (MRE), and are the average of three independent experiments. (B) AGADIR predictions of helicity. Solid rectangles indicate residues in helix within the complex. Note the different scales on the y axes. *Position of fluorophores in labeled peptides. (C) Association rates with KIX under pseudofirst-order conditions (with KIX in excess). Data for FITC-cMyb obtained from Shammas et al. (22).
On mixing with KIX, there is an increase in helicity for all of the peptides (Fig. S4), even for cMybTAD, which is predicted to have a much more helical binding region in its free form. However, there is a smaller change in helicity on binding of cMybTAD than reported previously for cMyb25 (by around a factor of two) (22), which likely indicates that the binding region is more helical in its unbound form, consistent with the AGADIR prediction. The difference spectrum for HBZ is much larger than that for the others (Fig. S4), which may indicate more extensive helical formation in this case for either HBZ itself or KIX. Thus, all of these interactions may be accurately described as coupled folding and binding reactions.
Association Kinetics for KIX Binding Partners.
Labeled peptide was mixed rapidly with an excess of KIX in a stopped-flow apparatus, and the fluorescence intensity (and/or anisotropy) change on binding was monitored. Kinetic traces were fit to single exponentials to extract apparent rate constants (in seconds−1) for all peptides, with the exception of FITC-pKID, which fit poorly to a single exponential and was, instead, fit to a single exponential with a linear drift term (this peptide has previously been reported to associate through a three-state induced fit mechanism with a populated intermediate) (SI Materials and Methods). These experiments were performed with various concentrations of KIX (Fig. 2B) to extract the apparent association rate constants (k+; in molar−1seconds−1), which are given by the gradients of the straight-line fits in Fig. 2B and shown in Fig. 3.
Fig. 3.
Rate constants for association between KIX and partner peptides (closed circles) correlate with peptide charge under our standard experimental conditions (R = −0.77, P = 0.04, n = 6). Squares represent rates extrapolated to infinite ionic strength, where long-range electrostatic effects are excluded. Squares with crosses represent association with KIX alone, and squares with diamonds represent association with KIX already bound to another partner. Data for FITC-cMyb obtained from ref. 22.
The relation between the peptide charge and k+ (Fig. 3, circles) suggests that the rate constants might be remarkably similar in the absence of attractive long-range electrostatic effects, despite the very different sequences, helical propensities, and sizes of the binding partners. To investigate electrostatic effects further, we performed association experiments at a range of ionic strengths (Fig. S5) to allow us to estimate the basal rate constants under such conditions for cMyb25, cMybTAD, and MLL. In all three cases, association rate constants were lowered with increasing salt concentrations, and the basal rate constants were within approximately a factor of two of each other (Fig. 3, squares). Interestingly the basal rate constants for FITC-cMyb25 and A594-cMybTAD are the same within error, despite the much larger predicted helical propensity within the binding region of the latter and its observed higher overall helicity. Indeed, there seems to be little relation between overall initial helicity and association rate constant (R = −0.22, P = 0.68, n = 6). In fact, the highest rate constant is observed for E2A, the peptide that is predicted to have the least helical binding region in the unbound state.
Dissociation Kinetics for Partners from KIX.
To estimate the dissociation rate constants, we performed out-competition assays with a range of concentrations of competing ligand. A mixture of labeled peptide and KIX was mixed rapidly with an unlabeled protein that bound to the same site. All kinetic traces fit well to single exponential functions. The apparent rate constants (kapp) are shown as a function of the concentration of unlabeled ligand in Fig. 4, Right, blue circles and Fig. S6. The asymptote in such plots represents the dissociation rate constant (k−). Because the complexes in these studies have relatively high Kd values, the apparent rate constants do not become independent of protein concentration, even when very large excesses of protein are used for out-competition, and therefore, we fit the data to Eq. 2 to obtain a better estimate of the true k−. This observed dissociation rate constant may be a combination of unfolding, folding, and unbinding rate constants. The values of k− for each peptide vary more considerably than k+ values, ranging from 1.04 ± 0.02 s−1 for FITC-pKID to 133 ± 6 s−1 for E2A (i.e., over two orders of magnitude).
Fig. 4.
Observed rates from pseudofirst-order association experiments and competition dissociation studies for the interaction of labeled peptides with KIX and KIX bound to other ligands. Apparent association and dissociation rate constants are given by the gradients of (Left) straight-line fits and (Right) asymptotes, respectively.
Having estimates for apparent k+ and k− allowed us to calculate an equilibrium constant (Kd = k−/k+). In general, we found these estimates to agree favorably with those obtained directly from equilibrium measurements (Table S1).
Allosteric Effects on Kinetics.
We then proceeded to investigate the effect of having a partner bound at the other site on the kinetics. Labeled peptide was rapidly mixed with solutions of KIX bound to another (unlabeled) ligand, again maintaining pseudofirst-order conditions. KIX protein was over 98% bound to unlabeled ligand (according to estimated Kd values). Numerical simulations performed (assuming each reaction to be a simple two-state process) suggested resultant relative errors of less than 2% in k+. We observed in all cases that having a partner bound at the other site actually slows association (Fig. 4 and Table S2). This slowing is not necessarily surprising, because binding the first partner can mean that the KIX complex actually becomes negatively charged overall, swapping the expected long-range electrostatic forces from attractive to repulsive, which could potentially mask an allosteric effect on association that would be present in vivo where the proteins have different global charges. However, a decrease in k+ is also observed for FITC-MLL and A594-HBZ binding to KIX.pKID compared with KIX, although pKID is globally uncharged at this pH (Q = −0.3). Furthermore, investigation of the ionic strength dependence of k+ (Fig. S5) with and without the partner bound indicates that it is not the explanation. Even at infinite ionic strengths, where long-range electrostatics effects are eliminated, the association is similar or still somewhat slowed (Fig. 3, diamonds in squares and Table S1). Interestingly, the long-range electrostatic forces seem to remain attractive, even with a partner already bound, which suggests that it is the properties of the binding surface rather than the global charges that are important for rate enhancement.
Next, we compared k− for ternary complexes with those for binary complexes. The dissociation experiments were performed as before but with an excess of (unlabeled) second ligand present in the solution containing KIX complex. A range of dissociation rate constants was observed, which were 2- to 40-fold lower than those determined previously without ligand bound at the second site (Fig. 4, Right) (Table S2). Combining the reduction in k+ with that in k− allows us to estimate an apparent cooperativity factor (α) for ligand binding. In each case, the decrease in k− was larger than that in k+, resulting in values of α that are all >1, which indicates a positive allosteric effect. The values that we obtained were similar to those previously reported in the literature (Table S2).
Discussion
Mechanism of Coupled Folding and Binding with KIX.
The binding of proteins to the KIX domain seems to be characterized by high-association rate constants (Fig. 3) considering the relatively high ionic strength of our solutions (I ∼ 0.26 M). Fast association could reflect the role of these interactions in vivo, allowing transcriptional apparatus to be quick to adjust to changing cellular conditions. Interestingly, it seems that the k+ for each of the peptides might actually be remarkably similar when corrected for long-range electrostatic interactions, despite their divergent sequences, and even binding sites. The basal k+ values for cMyb25 (22), cMybTAD, and MLL obtained here in the absence of electrostatic rate enhancement (7.7 ± 0.5, 6.8 ± 2.2, and 3.4 ± 0.6 μM−1s−1, respectively) and very recently, MLL with a mutated version of KIX (where the binding site is made more hydrophobic) (23) are higher than any other reported values for ordered or disordered protein interactions (22), and they are of the order of magnitude (105–106 M−1s−1) typically described as representing diffusion-limited processes, where all correctly oriented collisions result in complex formation. Given the remarkably high k+, it seems unlikely that substantial conformational selection (of the peptides or lowly populated forms of KIX) is taking place (24).
We found that there was a poor correlation between k+ and residual helical content, even for cMyb25 and cMybTAD, which have identical sequences in the binding region. This finding is in contrast to recent work with the activation domain of activator for thyroid hormone and retinoid receptors (ACTR) and nuclear coactivator binding domain (NCBD) of CBP, where a strong positive correlation was observed (25). These differences presumably relate to differences in the mechanism of coupled folding and binding and most specifically, the degree of helical structure formation in the transition state. The poor correlation between helicity and k+ that we observed would not be expected for a conformational selection mechanism and therefore, is consistent with previous reports for both cMyb and pKID undergoing folding after binding (9, 22, 26). In the case of cMyb25, we have previously shown that there is still an additional activation energy barrier for this reaction, because k+ shows a dependence on temperature above that expected from diffusion considerations alone (22). This additional temperature dependence could represent an energetic barrier for folding. FITC-pKID has previously been reported to undergo an induced fit mechanism with a populated intermediate in NMR studies and simulations (9, 27), and we did observe two separate kinetic phases in association experiments. The fastest of these two phases (under our experimental conditions and concentration range) is dependent on the protein concentration and corresponds to a binding rate. It is slightly lower (5.0 ± 0.1 μM−1s−1) than the kon value reported previously for pKID from NMR relaxation dispersion measurements performed at various pKID/KIX ratios (6.3 μM−1s−1) (9); however, a lower value is expected, because our measurements are performed at a lower temperature. The second rate, which is around 5–20 s−1, does not seem to be concentration-dependent and is likely to represent the subsequent folding/structural rearrangement step. Again, this rate is lower than that reported by Sugase et al. (9) at a higher temperature, which ranges from 30 to 3,000 s−1 depending on the residue examined.
Although the k+ values only differ by an order of magnitude and are likely to be highly similar after long-range electrostatic attraction is accounted for, the differences in k− are more substantial, ranging over more than two orders of magnitude. If the peptide is required to unfold before it can dissociate from the complex, then one might reason that the peptides with the highest helical propensity will be the ones with the lowest dissociation rates. We checked for such a correlation; however, it was not statistically significant (P = 0.07, n = 6). It is possible that the effect is simply being masked by more specific interactions considering the relatively small sample size.
Effect of Charge on Association Rate Constants.
Peptides with higher (negative) global charges associate faster with KIX, the reaction being enhanced by favorable long-range electrostatics that can be removed by screening with salt. However, changing the global charge of KIX to be negative by binding another protein at the other site does not seem to alter the long-range electrostatics to be repulsive. In fact, on fitting the data collected over a range of ionic strengths (Fig. S5) to a Debye–Huckel-like model, the parameters previously identified as describing the magnitudes of and distances between the charges remained the same within error for FITC-MLL association. This indifference shows the importance of the charges within the binding interface of the protein, which remain very similar in the presence of the second ligand (Figs. S7 and S8) and is highly reminiscent of the result reported for the interaction between the two folded proteins barnase and barstar, where the same Debye–Huckel-like fit gave parameters that best matched the charges and distances between the interfaces of the two proteins rather than the global values (28). Note that, since we submitted this work, the association of a shorter version of MLL to a mutated version of KIX (Y631W) has been reported (23). This peptide has three fewer negatively charged residues (just outside of the helical region in the complex) than our longer version and association rates are actually enhanced by salt (23). Thus, we emphasize the importance of considering charges when comparing protein association kinetics.
Cooperativity of Binding.
Previous equilibrium studies of allostery in the KIX system have provided estimates of the cooperativity of binding between several different pairs of proteins (Table S1). In general, we showed good agreement with these when we considered the changes in k+ and k− observed when a second partner was bound. It should be noted that, in the typical case, the cooperativity of binding should be the same for a pair of proteins when considered in either direction. For cMyb and HBZ as well as pKID and MLL, the cooperativity does not appear symmetrical. Unlike in the previous equilibrium studies, our proteins cannot form a closed thermodynamic cycle, because in some experiments, a partner will have a fluorophore attached, whereas in others, it will not, providing a likely, simple explanation of our observations.
MLL binding to KIX has been found to alter the structure of KIX somewhat to resemble more closely a lowly populated (7%) subspecies of KIX posited to have a preorganized cMyb/pKID binding site and be the binding competent form (13) [i.e., Brüschweiler et al. (13) suggest allostery to be mediated through enhanced conformational selection of KIX (the folded protein)]. In a subsequent study, Brüschweiler et al. (8) present structural data detailing a rearrangement of the hydrophobic core of KIX.MLL on binding pKID, which chemical shift data suggest may also be present in the binding-competent excited state of KIX.MLL. The excited state has been investigated recently using molecular dynamics simulations with enhanced sampling methods (15), showing its structural similarity with the ternary KIX.MLL.pKID complex. MLL binding to KIX was observed to lead to an opening of the hydrophobic core of the KIX, allowing transition to the excited (binding-favored) state. These studies could be interpreted to suggest that association of cMyb/pKID with KIX will be accelerated when MLL is bound at the other site. We have tested this prediction here but seen no evidence in support of it. Given the extremely fast k+, a mechanism involving enhancing association seems unlikely, and indeed, we have observed no increase as a result of any partner being bound. In fact, we observed a decrease in all cases. In a recent investigation of cMyb binding in the presence of MLL, a similar effect was seen (23). Some of this decrease is likely to be because of less favorable long-range electrostatic effects on binding another protein, but we also see a decrease in k+ for MLL and HBZ binding to KIX when pKID (net charge ∼ −0.3) is already bound and a decrease in k+ in the two cases where we have corrected for changes in long-range electrostatic effects by performing experiments at multiple ionic strengths. The (relatively small) decrease in k+ could correspond to less optimal binding surfaces on KIX as a result of conformational changes on binding the first ligand or even an advantage of flexibility in KIX when binding. Regardless, our data seem inconsistent with an allosteric mechanism mediated through enhanced conformational selection of an activated species of KIX.
Proposed Allosteric Mechanism.
Given that the allosteric effect is manifested in all eight cases through k−, a general mechanistic explanation involving the KIX protein itself seems likely. A convenient and highly plausible explanation, thus, is in the dynamic allosteric phenomenon (29–31), which has been observed previously in the CAP and PDZ protein systems (32, 33). In this case, the entropic cost would be paid to stiffen the KIX protein on binding the first ligand and not on binding the second, thereby making binding of the second ligand more favorable. There is already excellent experimental support for this hypothesis in backbone amide parameters, which show KIX to be a generally dynamic protein on the picosecond-to-nanosecond timescale. Binding of ligands has been observed to stiffen the backbone of the entire KIX protein. Stiffening of the backbone in helices 1–3 was observed on binding pKID or MLL, with no additional stiffening taking place on binding a second ligand (8). Furthermore, we can examine whether the observed free energy changes are consistent with such an explanation. The original paper describing such allostery estimates that a complex may be stabilized by a few kilojoules per mole through the effect (34). We can calculate the change in free energy between the bound state (B) and the transition state (TS) for dissociation as a result of having a second ligand bound (ΔΔGB − TS) using the relative change in observed k− according to Eq. 1:
| [1] |
where k−ternary and k−binary represent the dissociation rate constants for a ligand in the presence and absence of another ligand, respectively.
Excluding the interaction between cMybTAD and HBZ (discussed below), the values range from 1.6 to 4.9 kJ/mol, which could be consistent with this explanation. Previously, this mechanism has been suggested to account for the enhanced stability of the MLL complex with KIX when pKID is already bound, because changes in order parameters for residues close to the MLL binding site were observed on pKID binding to KIX (8); also, no activated species were observed in relaxation dispersion studies (13). Although there were no changes in order parameters near the cMyb binding site when MLL bound to KIX (8), we note that it does not exclude a dynamic allosteric mechanism also operating in this direction, because significant changes were observed in the backbone dynamics in all of the helices. Dynamic allostery in its original sense is, by its very nature, a purely thermodynamic effect. It simply requires a reduced entropic cost for ligand binding (when another is already bound), and therefore, an increased free energy difference between the free and bound states (31, 34). The location of the stiffening observed on binding the first ligand may, therefore, be unimportant as long as there is an overall entropic loss as a result. This generalized thermodynamic effect could act to increase k+ and/or decrease k− depending on the proportion of stiffening that has occurred in the transition state. In the absence of additional complicating factors, because our data show effects mostly on k−, they would suggest that KIX is less constrained in the transition state for binding than it is in the complex (i.e., the KIX is similar in flexibility in its free form and the transition state, and more constrained in the complex). This suggested mechanism compares with the cracking mechanism described previously by Miyashita et al. (35) for ligand binding to adenylate kinase, where a relatively inflexible protein undergoes a large conformational change to another inflexible state, but as suggested here, this state is reached through a flexible/disordered transition state (where particularly strained residues are unfolded) (35, 36).
In an accompanying work, Law et al. (16) have performed Gō-like molecular dynamics simulations of free KIX and KIX.cMyb, KIX.MLL, and KIX.cMyb.MLL complexes. Law et al. (16) observe a positive allosteric effect in both directions arising largely from a decrease of k− when the second ligand is bound and associated with an entropic change in both cases (TΔS = 2.5 kJ/mol). The stiffening observed in KIX on binding is predominantly located in the L12 loop and the C terminus of helix-α3, and in the case of MLL binding, it is associated with a compression of the hydrophobic core (16). The relative decreases in k− (4.4 ± 1.0 and 3.3 ± 0.8 for cMyb and MLL, respectively) agree well with those that we obtained experimentally (3.9 ± 0.1 and 4.1 ± 0.1, respectively). Their results, therefore, seem very consistent with our proposed explanation of our experimental results. Unlike us, however, Law et al. (16) observe a small increase in k+ as a result of having a partner bound at the other site. This apparent discrepency is most likely because of limitations in the resolution of the simulation methodology.
Previously, the largest reported allosteric effect has been between cMyb and HBZ, and this pair is that for which we observed the largest allosteric effect on k−. Because there is no NMR structure where HBZ is bound, it is not possible to exclude a specific and direct interaction between the two binding partners. The smallest decrease in k− is observed for pKID when MLL is bound—inspection of the binary and ternary complexes (Fig. 1 and Fig. S1) shows that the position of pKID is shifted significantly on binding MLL, removing some of its interactions with the KIX protein. Unlike for cMyb and MLL, which have only small changes in interaction area on formation of ternary complexes (Table S1), that for pKID decreases from 760 to 599 Å2 on KIX.pKID binding MLL. This decrease might explain the smaller allosteric effect observed in this case.
Conclusion
By performing these experiments for four different pairs of KIX binding partners, we have assembled eight estimates for how association and dissociation rate constants are changed in the presence of a second ligand at the alternate site. In all of the cases that we investigated, the allostery between sites arises from a reduction in the dissociation rate constant rather than an increase in the association rate constant, and importantly, again in all cases, association rates were actually lowered. Given that there have been several reports of positive allostery between KIX binding ligands and none of negative allostery (37), a generic mechanistic explanation, such as dynamic changes in KIX on ligand binding, seems desirable. Of course, for each pair of partners, we would also expect more specific effects to be superimposed on this generic thermodynamic effect (enthalpic and entropic). These specific effects might be best understood by comparison of the interactions between the ligand and KIX in its binary and ternary complexes.
Materials and Methods
Expression, Purification, Labeling, Concentration Determination, and CD Analysis of Proteins.
A detailed description, including estimation of parameters such as charge, is in SI Materials and Methods.
Calculating Association and Dissociation Rate Constants.
Kinetics experiments were performed using an SX20 Applied Photophysics fluorescence stopped-flow spectrophotometer. Experiments with FITC- and Alexa594-labeled peptides were performed using 515- and 610-nm long-pass cutoff filters and excitation wavelengths of 493 and 593 nm, respectively. Either fluorescence intensity or fluorescence anisotropy (using an FP1 accessory) was used as a probe of the reaction progress. Under each condition, 10–60 traces were collected and averaged. Data collected before the first 1 ms were removed before fitting. All kinetics experiments were performed at 10 °C. For association experiments, 0.25 μM labeled peptide was rapidly mixed in a 1:1 ratio with 2–50 μM KIX or 2–10 μM KIX with 40–200 μM another (unlabeled) ligand. For dissociation experiments, a solution of 0.25 μM labeled peptide with 1 μM KIX (with or without 30–100 μM of another unlabeled ligand) was mixed rapidly with 0–200 μM unlabeled peptide.
Kinetic traces were fit to a single exponential decay function to extract an apparent rate constant, with the exception of FITC-pKID association traces (Discussion and Figs. S9 and S10). Apparent association rate constants (k+) were obtained as the gradient as straight-line fits in Figs. 2C and 4, Left. Apparent dissociation rate constants (k−) were obtained by fitting Eq. 2 to the plots displayed in Fig. 4, Right and Fig. S6:
| [2] |
where [Unlab] represents the concentration of unlabeled ligand used to displace the labeled ligand and Kd,unlab represents the equilibrium dissociation constant between the unlabeled ligand and KIX protein.
Complex stabilities were determined using the equilibrium approach as described in SI Materials and Methods.
Ionic Strength Studies.
In ionic strength studies, 0.25 μM labeled peptide in Mops buffer was mixed 1:1 with 8 μM KIX (with or without 100 μM unlabeled second ligand) in Mops buffer. Mops buffers were 20 mM Mops adjusted to each ionic strength with sodium chloride. The pH of each buffer was 7.30 ± 0.02 at 10.0 °C. Because the experiment was performed under pseudofirst-order conditions, an estimate for k+ was extracted from
| [3] |
where k− was the same as that calculated earlier in phosphate buffer. This method assumes that k− does not vary considerably with ionic strength, which has been shown previously for FITC-cMyb (22). We tested applicability for FITC-MLL by comparison with k+ values obtained using a standard pseudofirst-order approach (Fig. S5C, open circles). The dependence of k+ on ionic strength (I) was modeled as has been done previously using a Debye–Huckel-like approach (22, 28). Data in Fig. S5 were fit to Eq. 4, which is a rearranged version of the equation presented by Vijayakumar et al. (28) to extract the basal association rate at infinite ionic strength (kI=∞):
| [4] |
where and are used as free-fitting parameters. In these equations, QA and QB represent the net charge of the proteins or binding surfaces, R represents the distance between them, k is the Boltzmann constant, T is the temperature, ε is the dielectric constant of the solvent, NA is Avagadro’s constant, and e is the unit charge.
Supplementary Material
Acknowledgments
The authors thank Joseph Rogers for useful discussions and Isabelle Lemasson for providing the HTLV-1 basic leucine zipper plasmid. This work was supported by Wellcome Trust Grant WT095195MA. J.C. is a Wellcome Trust Senior Research Fellow.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1405815111/-/DCSupplemental.
References
- 1.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 3.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14(13):1553–1577. [PubMed] [Google Scholar]
- 4.Denis CM, et al. Structural basis of CBP/p300 recruitment in leukemia induction by E2A-PBX1. Blood. 2012;120(19):3968–3977. doi: 10.1182/blood-2012-02-411397. [DOI] [PubMed] [Google Scholar]
- 5.Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J Biol Chem. 2002;277(45):43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, et al. Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment. Proc Natl Acad Sci USA. 2012;109(16):6078–6083. doi: 10.1073/pnas.1119073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J Mol Biol. 2004;337(3):521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 8.Brüschweiler S, Konrat R, Tollinger M. Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core. ACS Chem Biol. 2013;8(7):1600–1610. doi: 10.1021/cb4002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447(7147):1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 10.Wang N, et al. Ordering a dynamic protein via a small-molecule stabilizer. J Am Chem Soc. 2013;135(9):3363–3366. doi: 10.1021/ja3122334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook PR, Polakowski N, Lemasson I. HTLV-1 HBZ protein deregulates interactions between cellular factors and the KIX domain of p300/CBP. J Mol Biol. 2011;409(3):384–398. doi: 10.1016/j.jmb.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramírez JA, Nyborg JK. Molecular characterization of HTLV-1 Tax interaction with the KIX domain of CBP/p300. J Mol Biol. 2007;372(4):958–969. doi: 10.1016/j.jmb.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brüschweiler S, et al. Direct observation of the dynamic process underlying allosteric signal transmission. J Am Chem Soc. 2009;131(8):3063–3068. doi: 10.1021/ja809947w. [DOI] [PubMed] [Google Scholar]
- 14.Korkmaz EN, Nussinov R, Haliloğlu T. Conformational control of the binding of the transactivation domain of the MLL protein and c-Myb to the KIX domain of CREB. PLOS Comput Biol. 2012;8(3):e1002420. doi: 10.1371/journal.pcbi.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palazzesi F, Barducci A, Tollinger M, Parrinello M. The allosteric communication pathways in KIX domain of CBP. Proc Natl Acad Sci USA. 2013;110(35):14237–14242. doi: 10.1073/pnas.1313548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law SM, Gagnon JK, Mapp AK, Brooks CL., III Prepaying the entropic cost for allosteric regulation in KIX. Proc Natl Acad Sci USA. 2014;111:12067–12072. doi: 10.1073/pnas.1405831111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang N, Lodge JM, Fierke CA, Mapp AK. Dissecting allosteric effects of activator–coactivator complexes using a covalent small molecule ligand. Proc Natl Acad Sci USA. 2014;111:12061–12066. doi: 10.1073/pnas.1406033111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz V, Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. Nat Struct Biol. 1994;1(6):399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 19.Chen YH, Yang JT, Martinez HM. Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion. Biochemistry. 1972;11(22):4120–4131. doi: 10.1021/bi00772a015. [DOI] [PubMed] [Google Scholar]
- 20.Morriset JD, David JSK, Pownall HJ, Gotto AM. Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry. 1973;12(7):1290–1299. doi: 10.1021/bi00731a008. [DOI] [PubMed] [Google Scholar]
- 21.Muñoz V, Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. III. Temperature and pH dependence. J Mol Biol. 1995;245(3):297–308. doi: 10.1006/jmbi.1994.0024. [DOI] [PubMed] [Google Scholar]
- 22.Shammas SL, Travis AJ, Clarke J. Remarkably fast coupled folding and binding of the intrinsically disordered transactivation domain of cMyb to CBP KIX. J Phys Chem B. 2013;117(42):13346–13356. doi: 10.1021/jp404267e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toto A, Giri R, Brunori M, Gianni S. The mechanism of binding of the KIX domain to the mixed lineage leukemia protein and its allosteric role in the recognition of c-Myb. Protein Sci. 2014;23(7):962–969. doi: 10.1002/pro.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kiefhaber T, Bachmann A, Jensen KS. Dynamics and mechanisms of coupled protein folding and binding reactions. Curr Opin Struct Biol. 2012;22(1):21–29. doi: 10.1016/j.sbi.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Iešmantavičius V, Dogan J, Jemth P, Teilum K, Kjaergaard M. Helical propensity in an intrinsically disordered protein accelerates ligand binding. Angew Chem Int Ed Engl. 2014;53(6):1548–1551. doi: 10.1002/anie.201307712. [DOI] [PubMed] [Google Scholar]
- 26.Gianni S, Morrone A, Giri R, Brunori M. A folding-after-binding mechanism describes the recognition between the transactivation domain of c-Myb and the KIX domain of the CREB-binding protein. Biochem Biophys Res Commun. 2012;428(2):205–209. doi: 10.1016/j.bbrc.2012.09.112. [DOI] [PubMed] [Google Scholar]
- 27.Turjanski AG, Gutkind JS, Best RB, Hummer G. Binding-induced folding of a natively unstructured transcription factor. PLOS Comput Biol. 2008;4(4):e1000060. doi: 10.1371/journal.pcbi.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vijayakumar M, et al. Electrostatic enhancement of diffusion-controlled protein-protein association: Comparison of theory and experiment on barnase and barstar. J Mol Biol. 1998;278(5):1015–1024. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- 29.Tsai CJ, del Sol A, Nussinov R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378(1):1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kern D, Zuiderweg ERP. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. 2003;13(6):748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Hilser VJ, Wrabl JO, Motlagh HN. Structural and energetic basis of allostery. Annu Rev Biophys. 2012;41:585–609. doi: 10.1146/annurev-biophys-050511-102319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petit CM, Zhang J, Sapienza PJ, Fuentes EJ, Lee AL. Hidden dynamic allostery in a PDZ domain. Proc Natl Acad Sci USA. 2009;106(43):18249–18254. doi: 10.1073/pnas.0904492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13(9):831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper A, Dryden DTF. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11(2):103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 35.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci USA. 2003;100(22):12570–12575. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitford PC, Miyashita O, Levy Y, Onuchic JN. Conformational transitions of adenylate kinase: Switching by cracking. J Mol Biol. 2007;366(5):1661–1671. doi: 10.1016/j.jmb.2006.11.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thakur JK, Yadav A, Yadav G. Molecular recognition by the KIX domain and its role in gene regulation. Nucleic Acids Res. 2014;42(4):2112–2125. doi: 10.1093/nar/gkt1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.