Abstract
Recent efforts in genomic research have enabled the characterization of molecular mechanisms underlying many types of cancers, ushering novel approaches for diagnosis and therapeutics. Melanoma is a molecularly heterogeneous disease, as many genetic alterations have been identified and the clinical features can vary. Although discoveries of frequent mutations including BRAF have already made clinically significant impact on patient care, there is a growing body of literature suggesting a role for additional mutations, driver and passenger types, in disease pathophysiology. Although some mutations have been strongly associated with clinical phenotypes of melanomas (such as physical distribution or morphologic subtype), the function or implications of many of the recently identified mutations remains less clear. The phenotypic and clinical impact of genomic mutations in melanoma remains a promising opportunity for progress in the care of melanoma patients.
Melanoma is a molecularly heterogeneous disease, with diverse genomic alterations and clinical phenotypes. Some mutations (e.g., BRAF) are well characterized, but the significance of others remains unclear.
There have been many recent advances in our understanding of melanoma, from a greater appreciation of epidemiologic trends and risk factors to an increased understanding of the molecular genomics and biology of melanoma. Studies suggest that a number of molecularly distinct changes have a role in the pathophysiology of melanoma, and melanomas encompass a heterogeneous group when considering many known genomic alterations and diverse clinical phenotypes. To date, there are clinical subtypes or “phenotypes” of melanoma that are associated with specific genomic changes, in addition to many identified genomic mutations that lack a clear clinical correlation.
Genomic studies are able to inform us about the disease phenotype, in addition to improving our understanding of disease pathogenesis. The various phenotypes of melanoma are characterized by clinical features, such as bodily distribution or risk factors. Cutaneous, uveal, acral, and mucosal melanomas (Fig. 1) have divergent clinical courses and are associated with distinct mutations, and risk factors such as skin phototype or UV exposure pattern are also associated with distinct alterations in genetic mutations. A melanoma phenotype can also be classified based on histopathologic morphology such as superficial spreading, nodular, desmoplastic, etc. In this review, we examine a number of genomic mutations that are associated with this molecularly heterogeneous disease while reviewing the known genotype–phenotype associations of malignant melanoma and use of genomic information for the treatment of malignant melanoma.
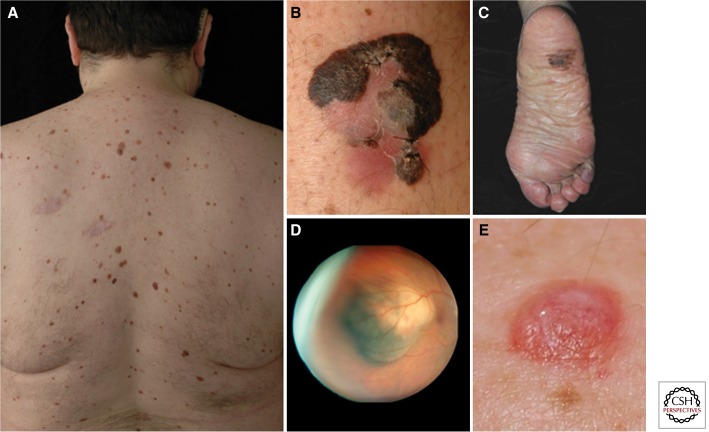
Figure 1.
Clinical melanoma phenotypes. (A) A patient with multiple clinically atypical moles. Because ∼80% of all acquired nevi harbor BRAFV600E mutations, hundreds of BRAF-activating events occur in such nevi-prone individuals without progression to melanoma. It is thought that melanocyte growth arrest results from oncogene-induced senescence. Common forms of cutaneous melanoma include (B) superficial spreading melanoma, which is known to harbor BRAF or NRAS mutations, and (C) acral lentiginous melanoma, which can be driven by KIT mutations. (D) Ocular melanomas are associated with activating GNAQ/GNA11 mutations and inactivating changes in the BAP1; patients with germline BAP1 mutations also show a higher risk of both cutaneous and ocular melanomas and develop unique nevoid-melanoma-like melanocytic proliferations (BAPoma) on the skin (E). (Ocular melanoma courtesy of Ivana Kim, Mass Eye and Ear Infirmary. Figure based on modified data from Tsao et al. 2012.)
MELANOMA EPIDEMIOLOGY
Melanoma is an aggressive malignancy borne of melanocytes, the pigment-generating cells of the skin. Malignant melanoma is responsible for ∼60% of deaths from skin cancers, and both incidence and prevalence of this disease have increased over the past few decades; meanwhile, overall mortality rates have remained somewhat stable. According to the American Cancer Society’s projections, in 2013, an estimated 76,690 new cases of malignant melanoma will be diagnosed (in addition to 61,300 cases of melanoma in situ), with a projection of 9480 deaths from malignant melanoma (Siegel et al. 2013). Melanoma is one of only a few cancer types profiled with a projected increase in incidence in 2013 among both men and women (Siegel et al. 2013).
With increased incidence of melanoma has come a greater interest and appreciation of risk factors for disease. Major risk factors for melanoma include exposure to UV radiation, family history, nevi (dysplastic, large number, or giant congenital nevi), increased age, fair skin phototype, occupation, and body mass index (Rigel 2010). Intrinsic risk factors, such as genetic and phenotypic predisposition, are inherent to the patient and cannot be modified; however, it is important to identify patients who have increased risk so they can adjust behaviors and exposures to extrinsic risk factors. Extrinsic risk factors include environmental, behavioral, or exposures that can be minimized, such as occupation and UV exposure, recreational tanning, medications, and chemical exposures.
MELANOMAGENESIS
The inciting events for melanoma pathophysiology are not well understood. Melanocytes over time can aggregate and form “nests” to create a nevus. Historically, it was thought that nevi undergo changes that lead to dysplastic features and potential evolution to melanoma. However, in most melanomas, there is no evidence of a preexisting nevus. On histopathologic diagnosis of melanoma, the frequency of the presence of a nevus component reported in the literature ranges from 10.8% to 57.6% (Crucioli and Stilwell 1982; Sagebiel 1993), although it is possible that the detection of preexisting nevus is limited by sampling or limitations in assessment, as suggested by the wide range. The molecular trigger for de novo or transformative initiation of a melanoma is not known.
The clinical course of a melanoma is dependent on growth control, which can be examined on a cellular level, and provides insights regarding the genes and pathways that are mutated or otherwise disrupted in melanoma. A number of genes and signaling pathways are involved in cellular proliferation and growth, and have been implicated in the pathophysiology of melanoma as described in detail below and depicted in Figure 2. At the surface of melanocytes are growth factor receptors that are stimulated by growth factors or other ligands to trigger proliferation and division of cells. Many growth factors and molecular signals act via members of the tyrosine kinase family, which trigger a variety of signaling cascades including NRAS, BRAF, and PI3K, to activate signals for survival, proliferation, etc. In contrast, there are a number of brakes for proliferation (such as p16) and damage checkpoints (p53) that can be mutated to allow proliferation and growth to proceed without regulation.
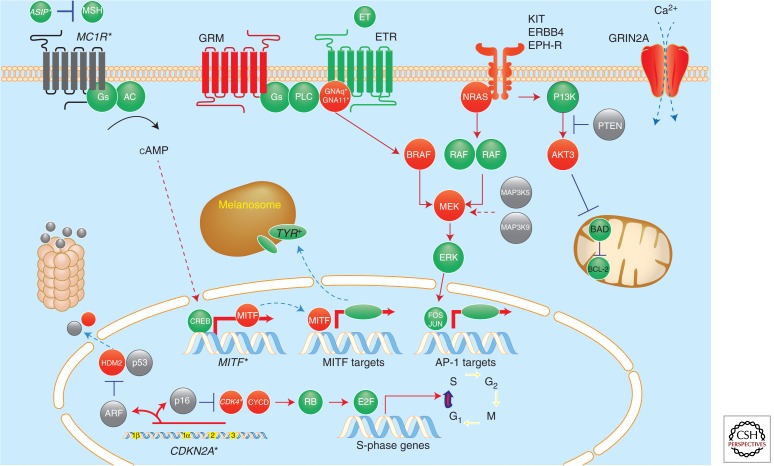
Figure 2.
A molecular map of melanoma. Heritable loci with risk alleles or single-nucleotide polymorphisms are shown in italics with asterisks (e.g., CDKN2A*). Red and gray colors indicate somatic alterations that result in gain of function (i.e., oncogenes such as BRAF) or loss of function (i.e., tumor-suppressor genes such as PTEN), respectively. (Figure based on modified data from Tsao et al. 2012.)
On a tissue level, the growth and clinical behavior of a melanoma is likely dependent on the local environment, including fibroblasts, vascular supply, and immune system. Melanoma, like all tumors, is dependent on angiogenesis for blood supply, the local matrix and stroma that may enable growth, invasion, or metastasis, and interplay with immune cells such as lymphocytes that circulate, recognize, target, and potentially eliminate tumors.
Each of these cellular, tissue, or environmental factors may contribute singly or in concert to the onset or progression of melanoma, as evidenced by genetic mutations that have been identified among melanomas. It is interesting to note the associations of specific mutations with clinical phenotypes, whether behavioral, hereditary, or associated with melanoma, at a specific distribution on the body. Some genomic mutations may actively promote oncogenic behavior (“driver mutations”), compared to others that are biologically silent (“passenger mutations”), which may suggest carcinogenic exposure while not actively contributing to a cell’s malignant behavior. The genomics of melanoma have also been important in the development of therapies to target driver mutations, a strategy that is becoming increasingly important in the era of personalized medicine.
MELANOMA PHENOTYPES WITH KNOWN GENETIC ASSOCIATIONS
Loci Associated with Hereditary Melanomas
Germline melanoma susceptibility and prognostic genes were the subject of a recent comprehensive review by Ward et al. (2012) that highlighted high-penetrance genes (CDKN2A and CDK4), moderate penetrance genes (MC1R, discussed below), and reviewed the literature for low-penetrance genes, which have higher prevalence in the population and less profound effects on melanoma risk. The low-penetrance candidate melanoma susceptibility and prognostic genes include 51 genes that are relevant to pigmentation/nevus count, the immune-mediated response to tumors, DNA-repair, metabolism, and vitamin D receptor polymorphisms, as summarized in Table 1. Many of these variants were derived from genome-wide association studies and have yet to be credentialed on a functional level. We will thus focus on some of the higher-penetrance genes that have been described.
Table 1.
High-, moderate-, and low-penetrance candidate melanoma susceptibility and prognostic genes
| High-penetrance genes |
CDKN2A CDK4 BAP1 TERT |
| Moderate-penetrance genes |
MC1R MITF(E318K) |
| Low-penetrance genes Pigmentation/nevus count genes |
ASIP, TYR, TYRP1, OCA2, SLC45A2 (MATP), MYO7A, NID1, KIT, KITLG, IRF, HERC2, PAX3, EDNRB, ADTB3A, CHS1, MLANA, ATRN, SOX10, HPS, MGRN1, MYO5A, SLC24A4, PLA2G6 |
| Immune genes | IL-10, IL-1β, TNF-α, LT-α, IL-6R, IFN-γ, HLA class II allele DQB1*0301, ICAM-1 |
| DNA-repair genes | XPD/ERCC2, ERCC1, XPF, XRCC3, MGMT, XRCC1, MDM2, APEX1, TERT1, TRF1, TERT-CLPTMIL |
| Metabolism genes | CYP2D6, GSTM1, GSTT1, GSTP1 |
| Vitamin D receptor polymorphisms | Fokl, Bsml, Taql, Apal, A-1012G |
Data adapted from Ward et al. 2012.
CDKN2A, cyclin-dependent kinase inhibitor 2A; CDK4, cyclin-dependent kinase 4; BAP1, BRCA1-associated protein 1 gene; TERT, telomerase reverse transcriptase; MC1R, melanocortin 1 receptor; MITF, microphthalmia-associated transcription factor.
Approximately 10% of newly diagnosed melanoma patients have an affected primary family member (de Snoo and Hayward 2005). Such cases may represent familial melanomas with germline mutations, and the best-characterized familial mutations involve the p16 pathway, which regulates the cell cycle. Among patients with the familial atypical multiple mole melanoma syndrome, there is early onset of disease, development of multiple primary melanomas, and increased numbers of clinically atypical nevi.
The cyclin-dependent kinase inhibitor 2A (CDKN2A) gene locus encodes tumor-suppressor proteins (p16 and p14), which function in cell-cycle arrest. Mutations in p16 have been identified in 41% of familial cutaneous melanoma cases (Goldstein et al. 2006). More than 30 variants of p16 pathway mutations in familial melanoma samples have been detected using a microsphere-based array assay (Lang et al. 2011).
Patients with CDKN2A mutations carry an increased risk of melanoma of 70% over their lifetimes (de Snoo and Hayward 2005), along with increased risk of pancreatic cancer, nervous system tumors, and uveal melanoma (Goldstein et al. 2006). The prevalence of CDKN2A mutations is higher among the Greek population (which has a relatively low baseline risk of melanoma) compared to other reported cohorts, suggesting the strength of genetic factors in melanomagenesis (Nikolaou et al. 2011).
The cyclin-dependent kinase 4 (CDK4) gene locus encodes an enzyme that is inhibited by binding of p16 and is also found to be mutated among clusters of melanoma families (Zuo et al. 1996), although CDK4 mutations are more rare than CDKN2A gene mutations. A study of 17 known families with CDK4 germline mutations showed that there are no phenotypically distinguishing features from CDKN2A families (Puntervoll et al. 2013).
Several other more rare hereditary conditions confer an increased risk of melanoma and one of the best studied is xeroderma pigmentosum, a rare autosomal dominant disease caused by mutation of one of eight nucleotide excision repair pathway genes. Nucleotide excision repair is critical for the body’s response to UV-induced damage, and impairment of this pathway confers a 1000-fold increase of skin cancers including melanoma (Budden and Bowden 2013). Germline mutations in the BRCA1-associated protein 1 gene (BAP1) predispose to melanocytic tumors (Wiesner et al. 2011) and have been identified in patients with melanomas, melanocytic BAP1-mutated atypical intradermal tumors, and malignant mesotheliomas (Carbone et al. 2012). A syndrome showing cutaneous/ocular melanoma, atypical melanocytic proliferations, and other internal neoplasms (COMMON syndrome) is associated with germline-inactivating mutations of BAP1 (Njauw et al. 2012). A cutaneous proliferation of this syndrome, a “BAPoma,” is depicted in Figure 1E.
Heritable Variants that Modulate Pigmentation and Melanoma Risk
The melanocortin 1 receptor (MC1R) gene encodes a G-protein-coupled receptor that activates adenylate cyclase to up-regulate microphthalmia-associated transcription factor (MITF) on binding of α-melanocyte stimulating hormone (α-MSH), thereby regulating the production of pigment (Garcia-Borron et al. 2005). MC1R is known to have variant alleles that are responsible for physical features including red hair, pale skin, inability to tan, and a tendency to freckle. These features are classified as Fitzpatrick skin phototype I, and MC1R variants are found in >80% of such individuals (Valverde et al. 1995).
The variant MC1R pigmentary phenotype confers an extrinsic environmental melanoma risk factor for these patients, as they are especially susceptible to UV exposure and sunburns. A lifetime of intermittent intense UV exposure is associated with BRAF mutation-containing melanomas, described in greater detail below. Among an Italian cohort, patients with one or two MC1R variants were found to have a five- to 15-fold increased risk of BRAF-mutant melanomas regardless of actinic damage, and there were no BRAF-negative melanomas among this population (Fargnoli et al. 2008). Landi et al. (2006) found BRAF mutations among >80% of patients with two variant MC1R alleles, compared to only ∼30% of patients with wild-type MC1R. MC1R variant alleles were found to be significant among Fitzpatrick skin phototype I patients, such that the presence of three active MC1R alleles (R151C, R160W, and D294H) confers a doubled risk of melanoma for each active allele a patient carries. This increased risk is also observed among medium- and dark-skinned individuals who carry one of these three variant MC1R alleles (Palmer et al. 2000).
MC1R variants likely play a role in melanoma pathogenesis by both pigmentary and nonpigmentary mechanisms (such as generation of reactive oxygen species). A meta-analysis of nine MC1R variants identified associations with melanoma without relation to pigment (Raimondi et al. 2008). MC1R mutations have been shown to confer increased melanoma risk independent of UV exposure (Mitra et al. 2012), which suggests a separate mechanism of carcinogenesis. Mitra et al. (2012) used a mouse model to show that the selective absence of pheomelanin synthesis was protective against the development of melanoma, and their data suggest that pheomelanin is carcinogenic because of increased oxidative damage. As a transcription factor, MITF has an important role in regulating the expression of downstream proteins that are relevant in melanoma. One such example is BCL2A1, a BCL2 family member that is amplified in 30% of melanomas, has a role in promoting melanomagenesis in vitro, and protects against apoptosis (Haq et al. 2013). Expression of BCL2A1 was shown to correlate with poorer clinical responses to BRAF inhibitors (Haq et al. 2013), which illustrates the complex relationship of MITF, BCL2 family member regulation of apoptosis, and clinical response to mitogen-activated protein kinase (MAPK)-pathway inhibition, which is described in further detail below.
Although treatments have not been developed to target the increased melanoma risk conferred by MC1R mutations, it has been suggested that novel MSH compounds may be used as radiolabeled peptides for therapeutic targeting of MC1R, which is overexpressed in melanoma and showed promising in vivo targeting features (Eberle et al. 2010).
The downstream MITF transcription factor is a lineage-specific master regulator of melanocyte development, which is amplified in 20%–40% of metastatic melanomas (Garraway et al. 2005; Gast et al. 2010). The MITF E318K variant was found to alter the sumoylation pattern of MITF and predispose carriers to familial and sporadic melanoma. Furthermore, this risk allele is associated with multiple primary melanomas, positive family history, increased numbers of nevi, and nonblue eye color (Yokoyama et al. 2011).
Somatic Changes Associated with Common Forms of Melanoma
Evidence for UV Mutagenesis
Although UV radiation is strongly associated with an increased risk of melanoma, and an intermittent UV radiation exposure pattern is associated with BRAF mutation (described further below), it is interesting that signature UV mutations (pyrimidine dimers) are identified less frequently within mutated oncogenes in melanomas compared to nonmelanoma skin cancers, and the most common BRAF mutations do not harbor characteristic pyrimidine dimer mutations despite a posited role for UV exposure in melanomagenesis. Whole genome sequencing of a human melanoma, which was compared to a matched lymphoblastoid cell line from the same patient, identified thousands of mutations, many of which were pyrimidine dimers that suggested UV exposure (Pleasance et al. 2010). However, relatively few mutations were identified within gene-coding regions, suggesting that the UV-signature mutations did not impact the melanoma’s driver mutations, but were more prevalent among passenger mutations. This could be explained by an active transcription-coupled nucleotide excision repair system, a statistical artifact based on a single patient or method of sequencing, or alternative sources of mutagenesis such as reactive oxygen species are more relevant than UV in melanomagenesis.
A 2012 comprehensive permutation-based analysis of driver mutations used a new framework to control for gene-specific basal mutation rates and explored the effect of misrepair of DNA damage attributed to UV-exposure, specifically identifying C > T (UVB-associated) and G > T (UVA-associated) mutations. The investigators found that among 262 driver mutations in 21 genes, 46% were caused by UV-associated mutations, and the percentage increased to 67% when driver mutations in BRAF or NRAS were excluded (Hodis et al. 2012), and this approach identified six novel melanoma genes (PPP6C, RAC1, SNX31, TACC1, STK19, and ARID2). These results suggest that functionally activating events (e.g., BRAF circa 1799 G > A) are structurally constrained and highly codon specific. In other words, random UV-induced mutations along BRAF are likely to inactivate, rather than activate, the BRAF proto-oncogene and are thus ineffective in stimulating cancer growth. On the other hand, tumor suppressors, such as CDKN2A and TP53, tend to harbor widespread deleterious changes caused by random UV mutagenesis (Hocker and Tsao 2007).
To more specifically examine sun-exposed melanomas, a sequencing study of exomes of 147 melanomas enabled the identification of markedly increased signature UV mutations (C > T somatic mutations) among sun-exposed melanomas in comparison to acral, mucosal, and uveal melanomas that are generally shielded from UV exposure (Krauthammer et al. 2012).
BRAF
Mutation of the BRAF gene is associated with melanomas occurring on skin with little histopathologic evidence of chronic sun damage (such as solar elastosis), and is more often found among younger patients with intermittent intense sun exposure (Junkins-Hopkins 2010). BRAF encodes a serine/threonine protein kinase that is an important regulator of the RAS/RAF/MAPK/extracellular signal-regulated kinase (ERK) signaling pathway (Fig. 2) that impacts cellular proliferation, differentiation, and survival among many diverse cellular functions. In 2002, mutations in BRAF were discovered to be harbored among a range of human cancers, including malignant melanoma (Davies et al. 2002).
More than 60% of cutaneous melanomas harbor a mutation in BRAF (Smalley 2010) and, among these, more than 80% feature the specific V600E amino acid substitution mutation (Davies et al. 2002). This mutation causes constitutively active kinase activity and activation of MEK and ERK signaling. BRAF mutations are most often found among patients without chronic actinic damage (Curtin et al. 2005), and with a younger age (6 vs. 63 years old) on diagnosis of distant metastasis (Long et al. 2011).
Although the prevalence among melanomas makes the BRAF mutation an appealing therapeutic target, interestingly, BRAF mutations are also found among banal nevi. In 2003, Pollock et al. (2003) identified BRAF mutations among 80% of primary melanomas, 68% of melanoma metastases, and 82% of nevi tested suggesting that the activating BRAF mutation may have a role in initiation of melanocytic proliferation, but is not sufficient for melanomagenesis. Consistently, BRAF mutations have been identified in 85% of 20 eruptive melanocytic nevi that underwent BRAF sequencing (John and Smalley 2011).
Despite the fact that BRAF mutations are found among banal melanocytic proliferations, the selective inhibition of mutated BRAF showed promise in initial studies treating patients with metastatic melanoma, with complete or partial tumor regression among the majority of patients treated (Flaherty et al. 2010). A phase III study of vemurafenib, a Food Drug Administration-approved inhibitor of V600E, compared to conventional chemotherapy with dacarbazine showed improvement in overall survival (6-mo survival of 84% vs. 64%) and prolonged progression-free survival (5.3 vs. 1.6 mo) (Chapman et al. 2011). Longer-term follow up studies revealed a mean overall survival time of 16 mo among patients undergoing treatment with vemurafenib (Sosman et al. 2012), although disease recurrence is noted as novel resistance pathways such as selection for (or acquisition of) additional mutations have limited the long-term effectiveness of vemurafenib (Fedorenko et al. 2011).
Tumor genomic profiling has been proposed as a technique to identify resistance pathways to BRAF inhibition in melanoma and was used to identify MEK1 downstream kinase mutation as a mechanism for therapeutic resistance (Wagle et al. 2011). MAP kinase kinase (MEK, MAP2K) is an important downstream signaling member that yields ERK phosphorylation and cell-survival signals. The most prevalent MEK mutations cause constitutive phosphorylation of ERK and were identified among 8% of melanomas (Nikolaev et al. 2012). Other mechanisms of secondary resistance include mutational activation of NRAS (Nazarian et al. 2010), emergence of a PLX-resistant alternative BRAF splice product (Poulikakos et al. 2011), and BRAF amplification (Shi et al. 2012). Additionally, activation of various receptor tyrosine kinases, such as platelet-derived growth factor receptor (PDGFR) (Nazarian et al. 2010), IGF-1R (Villanueva et al. 2010), and FGFR3 (Yadav et al. 2012) has also been reported.
Recently, combinations of therapies targeting both BRAF and MEK have been used to improve outcomes. A phase III trial of 322 patients with metastatic melanoma harboring activating V600 mutations in BRAF showed that trametinib (a selective MEK inhibitor) improved the rates of progression-free survival (4.8 vs. 1.5 mo) and overall survival at 6 mo (81% vs. 67%) compared to patients receiving chemotherapy with dacarbazine or paclitaxel (Flaherty et al. 2012b). The combination of BRAF and MEK inhibition among patients with metastatic melanoma and BRAF V600 mutations was studied in phase I and II trials of combinations of dabrafenib (a BRAF selective inhibitor) and trametinib, which were found to be safely combined with improvement of progression-free survival (9.4 vs. 5.8 mo) and improved rate of complete or partial response (76% vs. 54%) compared to monotherapy with dabrafenib (Flaherty et al. 2012a).
Although combinations of targeted therapies may improve overall response, it is important to note that many of the relevant oncogenic signals converge on common cellular pathways, such as MAPK and PI3K. Wilson et al. (2012) postulated that receptor tyrosine kinase ligands upstream of targeted pathways may provide a resistance mechanism and showed that hepatocyte growth factor confers resistance to vemurafenib in BRAF-mutant cells. In this manner, an increase of any receptor tyrosine kinase ligand upstream of a targeted pathway may confer resistance to inhibitors.
G-Regulatory Proteins
G-regulatory proteins of the RAS family are activated by guanosine-5′-triphosphate (GTP) binding and interact with many signaling pathways in the cell, including MAPK and PI3K pathways (Fig. 2), to cause diverse cellular outcomes including cell-cycle-progression and survival signals. In addition to the GNAQ and GNA11 G-regulatory protein encoding genes associated with ocular melanoma described in detail below, mutations in RAS proteins causing constitutive activation have been identified in melanoma. NRAS mutations were identified in 18% of melanomas (Lee et al. 2011), although NRAS mutations are also found in a large number of congenital nevi (Bauer et al. 2007). Although some associate NRAS mutations with chronic actinic damage and nodular melanomas, NRAS mutations were not found to be specific for a melanoma subtype and were noted among melanomas from non-sun-damaged skin (Curtin et al. 2005). The majority of RAS mutations in melanomas are NRAS mutations, although the Sanger Institute’s COSMIC database reports HRAS and KRAS mutations in melanomas at low frequency (cancer.sanger.ac.uk/cancergenome/projects/cosmic/).
NRAS mutations are generally found among melanomas without BRAF mutations (Curtin et al. 2005), and activating mutations in NRAS confer resistance to BRAF-targeted therapy, as reviewed elsewhere (Nikolaou et al. 2012). Although direct inhibition of mutant NRAS has not been effective, the targeting of downstream pathways such as MAPK pathway inhibitors may prove to be a useful treatment strategy in the future. Studies in melanoma cell lines suggest that combined targeting of MEK and PI3K/AKT/mammalian target of rapamycin (mTOR) pathways is needed to effectively inhibit NRAS-mutant melanoma, and may provide an alternative therapeutic approach (Posch et al. 2013).
An activating mutation in RAC1, which encodes a small G-protein member of the Rac subfamily of Rho GTPases that regulates cellular events including activation of protein kinases and cell growth, has also been described. The RAC1 activating mutation was identified among 9.2% of sun-exposed melanomas (Krauthammer et al. 2012). A zebrafish model showed that a constitutively active RAC1 mutant was unable to initiate melanocyte neoplasia, independently or when combined with the BRAF-V600E mutation; however, the RAC1-activating mutant was able to accelerate tumor nodule formation, in concert with the overexpression of Rac activator T-cell lymphoma invasion and metastasis 1 (Tiam1), which is overexpressed in melanomas of both zebrafish and humans (Dalton et al. 2013). In vitro studies suggested that mutated BRAF regulates Tiam/Rac activity to modulate cadherin levels and facilitate melanoma invasion (Monaghan-Benson and Burridge 2012).
CELL-SURFACE RECEPTORS
A number of tyrosine kinases have been implicated in melanoma pathophysiology, including c-Kit (as described above), ERBB family members, Ephrin type-A receptor 2 (EphA2), and PDGFR), along with nonreceptor tyrosine kinases such as Src family kinases and GRIN21/GRM3.
In 2009, mutational analysis of the protein tyrosine kinase gene family in melanoma allowed the identification of 30 somatic mutations of kinase domains of 19 protein tyrosine kinases, revealing ERBB4 mutations in 19%, and FLT1 and PTK2B mutations in 10% of patients with melanomas (Prickett et al. 2009). A recent screen of 109 melanomas, including cell lines, primary melanomas, and paired metastases, indicated that ERBB4 mutations were found in 21% of melanoma cell lines, 2.1% of primary melanomas, and 3.6% of melanoma metastases, corresponding to one patient, and suggesting that ERBB4 melanoma mutations may be rare (Manca et al. 2013).
Additional ERBB family members have also been implicated in melanoma. ERBB2 and ERBB3 are tyrosine kinase receptors that are activated by neuregulin-1 and have been found to be mutated in melanoma cell lines, promoting cell growth and survival in in vitro studies (Zhang et al. 2013).
The epidermal growth factor receptor and nonreceptor tyrosine kinase Src family kinase-STAT3 signaling pathway are up-regulated in cell lines that are resistant to BRAF inhibitors, and this pathway was found to stimulate invasion and metastasis and may potentially mediate resistance (Girotti et al. 2013).
The receptor tyrosine kinase PDGFR is not known to be mutated in melanoma; however, PDGFR-β may underlie a mechanism of resistance to BRAF-targeted therapies. Shi et al. (2011) used in vitro assays to show that melanoma cells resistant to BRAF targeted therapies via PDGFR-β had up-regulation of phosphorylated ERK and AKT, and showed cross talk with the PI3K/AKT/mTOR and MAPK pathways.
EphA2, a member of the Eph family of receptor tyrosine kinases, is highly expressed by melanoma, up-regulated by UV radiation, able to induce apoptosis, and thought to potentially be able to uncover an aggressive melanoma population (Udayakumar et al. 2011). Evidence supporting a role for EphA2 in tumorigenesis includes in vitro studies in which the down-regulation of EphA2 resulted in reduced invasion, proliferation, clonogenicity, and vasculogenic mimicry; analysis of a panel of human melanoma tumor cell lines showed a correlation between EphA2 expression and metastatic potential (Margaryan et al. 2009).
Glutamate receptors are nonkinase receptors that have been associated with tumor progression, and exon capture analysis has implicated mutations of these receptors in melanoma. GRIN2A, which encodes an ionotropic ligand-gated glutamate receptor, was mutated in 33% of melanoma samples (Wei et al. 2011). GRM3, which encodes a G-coupled glutamate receptor, was found to be mutated in 16.3% of melanoma samples, and to regulate MEK phosphorylation (Prickett et al. 2011). In vitro experiments using selumetinib (a selective inhibitor of MEK1/2) caused death of mutant GRM3 melanoma cells (Prickett et al. 2011), suggesting a role for MEK inhibition among melanomas with GRM3 mutations.
PI3K/AKT/mTOR PATHWAY MUTATIONS
Similar to the MAPK signaling pathway above, the PI3K/AKT/mTOR pathway is a crucial cellular signaling pathway that promotes growth and survival. Although no PI3K mutations have been identified in melanoma, downstream component Akt3 and PI3K negative-regulator PTEN have been implicated in melanoma. In addition, a somatic mutation-profiling study reported novel PI3K pathway mutations in melanomas, including MTOR, IRS4, PIK3R1, PIK3R4, PIK3R5, and NFKB1 (Shull et al. 2012).
AKT family member activation mutations are found in up to 43%–60% of melanomas (Stahl et al. 2004). Akt3 was found to be the predominantly active isoform that is increased among primary and metastatic melanomas compared to dysplastic nevi (Stahl et al. 2004), and comparative genomic hybridization showed amplifications of AKT among melanomas (Bastian et al. 1998). Dysregulation of tumor suppressor PTEN also impacts the PI3K/AKT/mTOR pathway in melanomas by up-regulating PI3K signaling. Loss of PTEN expression was noted in 38% of primary and 58% of metastatic melanomas (Birck et al. 2000). Mutations in PTEN are found concurrently with mutations in BRAF, but not NRAS, which is able to independently activate PI3K signaling as described above (Tsao et al. 2004). Regulators of PI3K pathway members also impact melanomagenesis. PREX2 is a PTEN-regulating protein that was found to accelerate tumor formation of in vivo immortalized human melanocytes (Berger et al. 2012). PREX2 was first identified on sequencing of genomes of 25 metastatic melanomas and matched germline DNA; PREX2 was subsequently identified in 14% of an independent extension cohort of 107 human melanomas (Berger et al. 2012).
PI3K pathway inhibitors may prove useful to treat melanomas with mutations in PTEN and AKT; specifically, mTOR inhibitors, which function downstream from Akt in the PI3K pathway, have been used in early studies. Studies of dual inhibition of BRAF and the PI3K/AKT/mTOR pathway in cells showed induction of melanoma cell apoptosis via a mechanism independent of MEK signaling (Sanchez-Hernandez et al. 2012). In vitro experiments combining BRAF, MEK, and PI3K/AKT/mTOR inhibitors were able to overcome acquired resistance to BRAF inhibitor dabrafenib (Greger et al. 2012). However, phase II studies performed with combinations of everolimus with temozolomide (an alkylating agent) and temsirolimus or tipifarnib with sorafenib (a multikinase inhibitor) failed to show promising clinical activity for metastatic melanoma (Margolin et al. 2012; Dronca et al. 2013).
REGULATORY PROTEINS
Tumor suppressors, including p16, p14, p16INK4a/retinoblastoma, and p53, are likely to play a role in senescence and proliferation pathways and may interact with other mutations in melanoma, as reviewed elsewhere (Swick and Maize 2012). Regulatory protein Hdm2 (also termed Mdm2) has important roles in the RB1 and TP53 pathways and is known to be amplified in a small proportion (3%) of melanomas (Muthusamy et al. 2006). In vitro studies showed that suppression of melanoma growth via MEK inhibition was potentiated by Hdm2 antagonism (and rescue of p53 function) (Ji et al. 2012). Another tumor suppressor, neurofibromin 1, was shown in a mouse model to cooperate with BRAF mutations in melanomagenesis by preventing oncogene-induced senescence (Maertens et al. 2013). Poly (ADP-ribose) polymerase 1 (PARP1) is an enzyme involved in single-strand DNA repair, and a nested case-control study of DNA repair pathway genes in malignant melanoma patients with matched controls identified a PARP1 gene variant to have increased melanoma risk (Zhang et al. 2011).
Regulation of Tumorigenesis: Non-Protein-Coding Regions of Genes
Although many studies have focused on protein-coding regions of genes to identify oncogenes, somatic mutations in regulatory regions of genes may also be important in tumorigenesis. Mutations in the core promoter of telomerase reverse transcriptase (TERT), which encodes for the telomerase catalytic subunit, were identified in 71% of 70 melanomas examined, and reporter assays found these mutations to increase transcriptional activity from the TERT promoter (Huang et al. 2013). Chromatin factors may also play an important role in the regulation of tumorigenesis; a zebrafish model of melanoma was used to identify SETDB1, a histone methyltransferase, to be amplified and accelerate melanoma onset (Ceol et al. 2011).
GENETIC MUTATIONS ASSOCIATED WITH SITE-SPECIFIC MELANOMAS
Uveal Melanoma
Uveal melanoma (also termed ocular melanoma) represents 3.1% of all melanomas (Singh et al. 2001) and has an aggressive clinical course, with up to 50% of lesions metastasizing, often to the liver (Singh et al. 2005). Uveal melanoma also has very high mortality; one series of 145 patients with metastases from uveal melanoma showed only 13% of patients surviving at 1 yr (Gragoudas et al. 1991).
Somatic mutations of GNAQ or GNA11 have been identified in 83% of uveal melanomas, which generally lack mutations in BRAF, NRAS, and KIT (Van Raamsdonk et al. 2010). GNAQ and GNA11 encode members of G-protein α-subunits, which are part of the G-protein-coupled receptor and regulate the MAPK signaling pathway. Mutations in these proteins inactivate a phosphatase, which would otherwise halt activation of the MAPK pathway. A large number of blue nevi contain similar mutations (Van Raamsdonk et al. 2010), in addition to lesions of Nevus of Ota (also called oculodermal melanocytosis). Nevus of Ota is a risk factor for uveal melanoma, conferring a 4.6% lifetime risk of ocular melanoma (Sahel and Albert 1993).
Although no treatment targets the G-protein α-subunits directly, targeted therapies directed at protein kinase C (PKC) in uveal melanoma cells harboring a GNAQ mutation inhibited the growth of uveal melanoma via PKC/Erk-1/2 and PCK/NK-κB pathways (Wu et al. 2012). Another approach that has been studied in vitro is inhibition of MEK and AKT pathways; MEK inhibitor selumetinib and AKT inhibitor MK2206 decreased cell viability in a synergistic manner and may hold therapeutic promise (Ambrosini et al. 2013).
Somatic-inactivating mutations of BAP1 have also been identified among patients with high-risk ocular melanomas among patients with COMMON syndrome as noted above (Njauw et al. 2012). A germline BAP1 mutation associated with uveal melanoma has also been identified (Hoiom et al. 2013).
ACRAL AND MUCOSAL MELANOMAS
Acral and mucosal melanomas are known to have increased genomic instability and chromosomal aberrations such as DNA losses or gains and changes in amplicons and total copy-number transitions (Curtin et al. 2005). Frequently, melanomas of these sites harbor a somatic activation of KIT, which encodes a transmembrane receptor tyrosine kinase. Kit is activated by binding of stem-cell factor ligand, which induces dimerization, autophosphorylation, and activation of a variety of downstream signaling pathways. Array comparative genomic hybridization data revealed copy number increases of KIT in 36% of acral and 39% of mucosal melanomas (Curtin et al. 2006). The overexpression of Kit was identified in 80% of primary vaginal melanomas (Vaysse et al. 2013).
The prevalence of KIT mutations at acral and mucosal sites, in addition to areas with chronic sun-damaged skin, raises questions regarding the role of KIT in melanomagenesis at these sites. Minor et al. (2012) found KIT mutations to be associated with a significantly shortened survival time among stage III or IV acral or mucosal melanoma patients, or those with cumulative sun-damaged skin. On the other hand, some melanomas do not respond to KIT inhibition despite the presence of KIT mutations (Carvajal et al. 2011), suggesting that alterations in KIT are not always therapeutically relevant.
Although KIT mutations are most commonly associated with melanomas of these sites, Puig-Butille et al. (2013) explored the role of RAS-pathway members in acral lentiginous melanoma and found alterations in some pathway members to be present in 87.5% of cases. Using 17 frozen primary acral melanomas, the investigators identified alterations in NRAS (17%), AURKA (37.5%), CCND1 or TERT (31.2%), and NRAS (25%) (Puig-Butille et al. 2013). Additional mutations in oncogenes and tumor suppressors were identified in acral melanoma cell lines, including BRAF, NRAS, KIT, PTEN, and TP53, in addition to genes that are typical of other types of cancer, including BRCA1 and BRCA2 (Furney et al. 2012).
Treatment of melanomas with KIT mutations has been attempted using kinase inhibitors such as imatinib, dasatinib, and others. Imatinib mesylate, a small molecule selective inhibitor of receptor tyrosine kinases, was used in a 2009 case report describing a patient with anal mucosal melanoma with a KIT-activating mutation that responded to imatinib; the report also reviewed 12 reported cases with no, partial, major, or complete responses to therapeutic inhibitors with activity targeting c-Kit including imatinib, sorafenib, sunitinib, and dasatinib (Satzger et al. 2010). A 2010 case report described a patient with two metastases of acral lentiginous melanoma, one of which contained the KIT V559A mutation and responded to imatinib, whereas wild-type gene metastatic disease did not respond (Terheyden et al. 2010). In 2011, a phase 2 study of imatinib in 25 melanoma patients with KIT mutations showed two complete responses of 94–95 wk, two durable partial responses of 53 and 89 wk, and two transient partial responses of 12 and 18 wk, suggesting a significant response potential for a subset of patients (Carvajal et al. 2011).
The multikinase inhibitors sorafenib and dasatinib target c-Kit in addition to other kinases, and case reports and small series show temporary clinical improvement (Quintas-Cardama et al. 2008; Handolias et al. 2010). Sorafenib has been studied in several phase I, II, and III clinical trials (as reviewed by Mangana et al. 2012) and subpopulations of patients experienced benefit from treatment, although biomarkers to identify these subpopulations are lacking.
Genomic Expression Patterns Associated with Aggressive Clinical Course
At the present time, there is no molecular marker for an aggressive clinical course of melanoma, although studies have sought to identify such a signature. Whole-genome expression profiling has been used to identify genes that are differently expressed among localized versus aggressive melanomas and may be used to predict melanoma clinical course. In 2006, Winnepenninckx et al. (2006) identified 254 genes whose expression was associated with melanoma metastases. Recently, a molecular grading scheme for melanoma was constructed using a copy DNA-mediated annealing, selection, extension, and ligation assay for assay-based RNA profiling of primary tumor specimens to classify high- and low-grade melanomas on a molecular basis, and found that high-grade melanomas had increased expression of proliferation and BRCA1/DNA damage signaling genes, whereas low-grade melanomas showed increased expression of immune genes (Harbst et al. 2012).
In 2013, a quantitative reverse transcriptase–polymerase chain reaction–based gene signature was identified, based on nine genes, to predict overall survival and distant metastasis-free patient survival independent of the American Joint Committee on Cancer staging at diagnosis. The queried genes are KRT9, KBTBD10, DCD, ECRG2, PIP, SCGB1D2, SCGB2A2, COL6A6, and HES6, and the genetic signatures were validated among a cohort of 44 additional melanomas (Brunner et al. 2013). Several of the genes associated with metastasis encode stromal proteins, suggesting the importance of the tumor’s microenvironment in the clinical course of melanoma. For example, α-catulin is a protein that down-regulates E-cadherin and promotes melanoma progression and invasion in vitro (Kreiseder et al. 2013).
It is likely that mutations associated with dedifferentiation or dysregulated growth are nonspecifically associated with an aggressive clinical course for any tumor type. This is likely the case for CCND1, the gene encoding Cyclin D1, which is a transcription regulator of cyclin-dependent kinases (CDK4 or CDK6) that alter the cell cycle. For melanomas, cyclin D1 protein expression is associated with Breslow thickness, metastasis, and shorter survival time (Vizkeleti et al. 2012). A reported aggressive case showed gains in CCND1 copy number by fluorescence in situ hybridization and corresponded to the presence of markedly pleomorphic cellular histopathology and an overall aggressive clinical course (Hawryluk et al. 2013). The association of cyclin D1 amplification with poor prognosis is not unique to melanoma, as this trend has been noted among neuroblastomas and tumors of the colorectal tract, breast, pancreatic duct, and types of lymphoma (Hawryluk et al. 2013). Another example is Nectin-like-5 (NECL-5), which regulates cell–cell junctions and has been implicated in malignant transformation in a variety of cell types and was found to be positively correlated with malignancy in melanoma (Bevelacqua et al. 2012).
CONCLUDING REMARKS
Clinical features may be suggestive of melanoma mutations, which ultimately can be assessed with mutational analysis, often providing insights into therapeutic options for patient management. Chemosensitivity assays and signaling pathway studies may also provide value in the assessment of therapeutic potential of targeted therapies. One reported case describes a patient with metastatic melanoma that lacked mutations in BRAF, NRAS, and KIT, although the MAPK pathway was found to be activated by assay of genomic DNA; a combination treatment with sorafenib and dacarbazine provided a 9-mo partial response for this patient (Passeron et al. 2011).
A number of clinical phenotypes remain without known genomic associations at this time. For example, lentigo maligna melanoma, aggressive melanomas in the elderly, melanomas of unknown primary site, desmoplastic melanomas, melanomas of pregnancy, and amelanotic melanomas do not have a known association with a specific genetic mutation. Far more striking is the number of genetic mutations that have been associated with melanoma, many of which are associated with unregulated growth and proliferation in a manner that is not specific to melanoma or known to be associated with a particular melanoma clinical phenotype. Many of these mutations have suggested potential targets to explore combinatorial therapy and prevent melanoma treatment resistance.
Downstream mediators of other cellular pathways including apoptosis, antiangiogenic, and immunological targets, although not associated with melanoma-specific gene mutations, will likely also play an important role in disease pathogenesis and management (Ko and Fisher 2011). Ultimately, a durable response to melanoma treatment may require multiple targeted therapies and engagement of the immune system.
ACKNOWLEDGMENTS
The authors gratefully acknowledge Dr. Matthew Hawryluk for his critical review and comments on this work. Funding for this effort is supported in part by K24 CA149202, the generous donors to the Massachusetts General Hospital Millennium Melanoma fund (both to H.T.), and the Dermatology Foundation Fellowship in Pediatric Dermatology (E.B.H.).
Footnotes
Editors: Anthony E. Oro and Fiona M. Watt
Additional Perspectives on The Skin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Ambrosini G, Musi E, Ho AL, de Stanchina E, Schwartz GK 2013. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol Cancer Ther 12: 768–776 [DOI] [PubMed] [Google Scholar]
- Bastian BC, LeBoit PE, Hamm H, Brocker EB, Pinkel D 1998. Chromosomal gains and losses in primary cutaneous melanomas detected by comparative genomic hybridization. Cancer Res 58: 2170–2175 [PubMed] [Google Scholar]
- Bauer J, Curtin JA, Pinkel D, Bastian BC 2007. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol 127: 179–182 [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevelacqua V, Bevelacqua Y, Candido S, Skarmoutsou E, Amoroso A, Guarneri C, Strazzanti A, Gangemi P, Mazzarino MC, D’Amico F, et al. 2012. Nectin like-5 overexpression correlates with the malignant phenotype in cutaneous melanoma. Oncotarget 3: 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birck A, Ahrenkiel V, Zeuthen J, Hou-Jensen K, Guldberg P 2000. Mutation and allelic loss of the PTEN/MMAC1 gene in primary and metastatic melanoma biopsies. J Invest Dermatol 114: 277–280 [DOI] [PubMed] [Google Scholar]
- Brunner G, Reitz M, Heinecke A, Lippold A, Berking C, Suter L, Atzpodien J 2013. A nine-gene signature predicting clinical outcome in cutaneous melanoma. J Cancer Res Clin Oncol 139: 249–258 [DOI] [PubMed] [Google Scholar]
- Budden T, Bowden NA 2013. The role of altered nucleotide excision repair and UVB-induced DNA damage in melanomagenesis. Int J Mol Sci 14: 1132–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M, Ferris LK, Baumann F, Napolitano A, Lum CA, Flores EG, Gaudino G, Powers A, Bryant-Greenwood P, Krausz T, et al. 2012. BAP1 cancer syndrome: Malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J Transl Med 10: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal RD, Antonescu CR, Wolchok JD, Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ, Chmielowski B, Lutzky J, et al. 2011. KIT as a therapeutic target in metastatic melanoma. JAMA 305: 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. 2011. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471: 513–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 364: 2507–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crucioli V, Stilwell J 1982. The histogenesis of malignant melanoma in relation to pre-existing pigmented lesions. J Cutan Pathol 9: 396–404 [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Brocker EB, LeBoit PE, et al. 2005. Distinct sets of genetic alterations in melanoma. N Engl J Med 353: 2135–2147 [DOI] [PubMed] [Google Scholar]
- Curtin JA, Busam K, Pinkel D, Bastian BC 2006. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 24: 4340–4346 [DOI] [PubMed] [Google Scholar]
- Dalton LE, Kamarashev J, Barinaga-Rementeria Ramirez I, White G, Malliri A, Hurlstone A 2013. Constitutive Rac activation is not sufficient to initiate melanocyte neoplasia but accelerates malignant progression. J Invest Dermatol 133: 1572–1581 [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. 2002. Mutations of the BRAF gene in human cancer. Nature 417: 949–954 [DOI] [PubMed] [Google Scholar]
- de Snoo FA, Hayward NK 2005. Cutaneous melanoma susceptibility and progression genes. Cancer Lett 230: 153–186 [DOI] [PubMed] [Google Scholar]
- Dronca RS, Allred JB, Perez DG, Nevala WK, Lieser EA, Thompson M, Maples WJ, Creagan ET, Pockaj BA, Kaur JS, et al. 2013. Phase II study of Temozolomide (TMZ) and Everolimus (RAD001) therapy for metastatic melanoma: A North Central Cancer Treatment Group study, N0675. Am J Clin Oncol 10.1097/COC.0b013e31827b45d4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AN, Bapst JP, Calame M, Tanner H, Froidevaux S 2010. MSH radiopeptides for targeting melanoma metastases. Adv Exp Med Biol 681: 133–142 [DOI] [PubMed] [Google Scholar]
- Fargnoli MC, Pike K, Pfeiffer RM, Tsang S, Rozenblum E, Munroe DJ, Golubeva Y, Calista D, Seidenari S, Massi D, et al. 2008. MC1R variants increase risk of melanomas harboring BRAF mutations. J Invest Dermatol 128: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko IV, Paraiso KH, Smalley KS 2011. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem Pharmacol 82: 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O’Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. 2010. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, Hamid O, Schuchter L, Cebon J, Ibrahim N, et al. 2012a. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 367: 1694–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, et al. 2012b. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367: 107–114 [DOI] [PubMed] [Google Scholar]
- Furney SJ, Turajlic S, Fenwick K, Lambros MB, MacKay A, Ricken G, Mitsopoulos C, Kozarewa I, Hakas J, Zvelebil M, et al. 2012. Genomic characterisation of acral melanoma cell lines. Pigment Cell Melanoma Res 25: 488–492 [DOI] [PubMed] [Google Scholar]
- Garcia-Borron JC, Sanchez-Laorden BL, Jimenez-Cervantes C 2005. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res 18: 393–410 [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. 2005. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436: 117–122 [DOI] [PubMed] [Google Scholar]
- Gast A, Scherer D, Chen B, Bloethner S, Melchert S, Sucker A, Hemminki K, Schadendorf D, Kumar R 2010. Somatic alterations in the melanoma genome: A high-resolution array-based comparative genomic hybridization study. Genes Chromosomes Cancer 49: 733–745 [DOI] [PubMed] [Google Scholar]
- Girotti MR, Pedersen M, Sanchez-Laorden B, Viros A, Turajlic S, Niculescu-Duvaz D, Zambon A, Sinclair J, Hayes A, Gore M, et al. 2013. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov 3: 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, Azizi E, Bianchi-Scarra G, Bishop DT, Bressac-de Paillerets B, et al. 2006. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 66: 9818–9828 [DOI] [PubMed] [Google Scholar]
- Gragoudas ES, Egan KM, Seddon JM, Glynn RJ, Walsh SM, Finn SM, Munzenrider JE, Spar MD 1991. Survival of patients with metastases from uveal melanoma. Ophthalmology 98: 383–389; discussion 390 [DOI] [PubMed] [Google Scholar]
- Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, Dickerson SH, Laquerre SG, Liu L, Gilmer TM 2012. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol Cancer Ther 11: 909–920 [DOI] [PubMed] [Google Scholar]
- Handolias D, Hamilton AL, Salemi R, Tan A, Moodie K, Kerr L, Dobrovic A, McArthur GA 2010. Clinical responses observed with imatinib or sorafenib in melanoma patients expressing mutations in KIT. Br J Cancer 102: 1219–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq R, Yokoyama S, Hawryluk EB, Jonsson GB, Frederick DT, McHenry K, Porter D, Tran TN, Love KT, Langer R, et al. 2013. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci 110: 4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbst K, Staaf J, Lauss M, Karlsson A, Masback A, Johansson I, Bendahl PO, Vallon-Christersson J, Torngren T, Ekedahl H, et al. 2012. Molecular profiling reveals low- and high-grade forms of primary melanoma. Clin Cancer Res 18: 4026–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk EB, Baran JL, Gerami P, Sepehr A 2013. “Monster cell” melanoma with pulmonary metastasis and cyclin D1 amplification. J Cutan Pathol 40: 61–65 [DOI] [PubMed] [Google Scholar]
- Hocker T, Tsao H 2007. Ultraviolet radiation and melanoma: A systematic review and analysis of reported sequence variants. Hum Mutat 28: 578–588 [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. 2012. A landscape of driver mutations in melanoma. Cell 150: 251–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiom V, Edsgard D, Helgadottir H, Eriksson H, All-Ericsson C, Tuominen R, Ivanova I, Lundeberg J, Emanuelsson O, Hansson J 2013. Hereditary uveal melanoma: A report of a germline mutation in BAP1. Genes Chromosomes Cancer 52: 378–384 [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA 2013. Highly recurrent TERT promoter mutations in human melanoma. Science 339: 957–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Njauw CN, Taylor M, Neel V, Flaherty KT, Tsao H 2012. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J Invest Dermatol 132: 356–364 [DOI] [PubMed] [Google Scholar]
- John JK, Smalley KS 2011. Identification of BRAF mutations in eruptive melanocytic nevi: New insights into melanomagenesis? Expert Rev Anticancer Ther 11: 711–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junkins-Hopkins JM 2010. Malignant melanoma: Molecular cytogenetics and their implications in clinical medicine. J Am Acad Dermatol 63: 329–332 [DOI] [PubMed] [Google Scholar]
- Ko JM, Fisher DE 2011. A new era: Melanoma genetics and therapeutics. J. Pathol 223: 241–250 [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, et al. 2012. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 44: 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiseder B, Orel L, Bujnow C, Buschek S, Pflueger M, Schuett W, Hundsberger H, de Martin R, Wiesner C 2013. α-Catulin downregulates E-cadherin and promotes melanoma progression and invasion. Int J Cancer 132: 521–530 [DOI] [PubMed] [Google Scholar]
- Landi MT, Bauer J, Pfeiffer RM, Elder DE, Hulley B, Minghetti P, Calista D, Kanetsky PA, Pinkel D, Bastian BC 2006. MC1R germline variants confer risk for BRAF-mutant melanoma. Science 313: 521–522 [DOI] [PubMed] [Google Scholar]
- Lang JM, Shennan M, Njauw JC, Luo S, Bishop JN, Harland M, Hayward NK, Tucker MA, Goldstein AM, Landi MT, et al. 2011. A flexible multiplex bead-based assay for detecting germline CDKN2A and CDK4 variants in melanoma-prone kindreds. J Invest Dermatol 131: 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Choi JW, Kim YS 2011. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: A meta-analysis. Br J Dermatol 164: 776–784 [DOI] [PubMed] [Google Scholar]
- Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, Hughes TM, Thompson JF, Scolyer RA, Kefford RF 2011. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol 29: 1239–1246 [DOI] [PubMed] [Google Scholar]
- Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, Bronson RT, McMahon M, Granter S, Flaherty K, et al. 2013. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov 3: 338–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca A, Lissia A, Cossu A, Rubino C, Ascierto PA, Stanganelli I, Palmieri G 2013. Mutations in ERBB4 may have a minor role in melanoma pathogenesis. J Invest Dermatol 133: 1685–1687 [DOI] [PubMed] [Google Scholar]
- Mangana J, Levesque MP, Karpova MB, Dummer R 2012. Sorafenib in melanoma. Expert Opin Investig Drugs 21: 557–568 [DOI] [PubMed] [Google Scholar]
- Margaryan NV, Strizzi L, Abbott DE, Seftor EA, Rao MS, Hendrix MJ, Hess AR 2009. EphA2 as a promoter of melanoma tumorigenicity. Cancer Biol Ther 8: 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin KA, Moon J, Flaherty LE, Lao CD, Akerley WL 3rd, Othus M, Sosman JA, Kirkwood JM, Sondak VK 2012. Randomized phase II trial of sorafenib with temsirolimus or tipifarnib in untreated metastatic melanoma (S0438). Clin Cancer Res 18: 1129–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor DR, Kashani-Sabet M, Garrido M, O’Day SJ, Hamid O, Bastian BC 2012. Sunitinib therapy for melanoma patients with KIT mutations. Clin Cancer Res 18: 1457–1463 [DOI] [PubMed] [Google Scholar]
- Mitra D, Luo X, Morgan A, Wang J, Hoang MP, Lo J, Guerrero CR, Lennerz JK, Mihm MC, Wargo JA, et al. 2012. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature 491: 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan-Benson E, Burridge K 2012. Mutant B-RAF regulates a Rac-dependent cadherin switch in melanoma. Oncogene 32: 4836–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy V, Hobbs C, Nogueira C, Cordon-Cardo C, McKee PH, Chin L, Bosenberg MW 2006. Amplification of CDK4 and MDM2 in malignant melanoma. Genes Chromosomes Cancer 45: 447–454 [DOI] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. 2010. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, Harshman K, Guipponi M, Bukach O, Zoete V, et al. 2012. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 44: 133–139 [DOI] [PubMed] [Google Scholar]
- Nikolaou V, Kang X, Stratigos A, Gogas H, Latorre MC, Gabree M, Plaka M, Njauw CN, Kypreou K, Mirmigi I, et al. 2011. Comprehensive mutational analysis of CDKN2A and CDK4 in Greek patients with cutaneous melanoma. Br J Dermatol 165: 1219–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou VA, Stratigos AJ, Flaherty KT, Tsao H 2012. Melanoma: New insights and new therapies. J Invest Dermatol 132: 854–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njauw CN, Kim I, Piris A, Gabree M, Taylor M, Lane AM, DeAngelis MM, Gragoudas E, Duncan LM, Tsao H 2012. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PloS ONE 7: e35295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JS, Duffy DL, Box NF, Aitken JF, O’Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA 2000. Melanocortin-1 receptor polymorphisms and risk of melanoma: As the association explained solely by pigmentation phenotype? Am J Hum Genet 66: 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passeron T, Lacour JP, Allegra M, Segalen C, Deville A, Thyss A, Giacchero D, Ortonne JP, Bertolotto C, Ballotti R, et al. 2011. Signalling and chemosensitivity assays in melanoma: Is mutated status a prerequisite for targeted therapy? Exp Dermatol 20: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. 2010. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, et al. 2003. High frequency of BRAF mutations in nevi. Nat Genet 33: 19–20 [DOI] [PubMed] [Google Scholar]
- Posch C, Moslehi H, Feeney L, Green GA, Ebaee A, Feichtenschlager V, Chong K, Peng L, Dimon MT, Phillips T, et al. 2013. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci 110: 4015–4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480: 387–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y 2009. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet 41: 1127–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Wei X, Cardenas-Navia I, Teer JK, Lin JC, Walia V, Gartner J, Jiang J, Cherukuri PF, Molinolo A, et al. 2011. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet 43: 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig-Butille JA, Badenas C, Ogbah Z, Carrera C, Aguilera P, Malvehy J, Puig S 2013. Genetic alterations in RAS-regulated pathway in acral lentiginous melanoma. Exp Dermatol 22: 148–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll HE, Yang XR, Vetti HH, Bachmann IM, Avril MF, Benfodda M, Catricala C, Dalle S, Duval-Modeste AB, Ghiorzo P, et al. 2013. Melanoma prone families with CDK4 germline mutation: Phenotypic profile and associations with MC1R variants. J Med Genet 50: 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintas-Cardama A, Lazar AJ, Woodman SE, Kim K, Ross M, Hwu P 2008. Complete response of stage IV anal mucosal melanoma expressing KIT Val560Asp to the multikinase inhibitor sorafenib. Nat Clin Pract Oncol 5: 737–740 [DOI] [PubMed] [Google Scholar]
- Raimondi S, Sera F, Gandini S, Iodice S, Caini S, Maisonneuve P, Fargnoli MC 2008. MC1R variants, melanoma and red hair color phenotype: A meta-analysis. Int J Cancer 122: 2753–2760 [DOI] [PubMed] [Google Scholar]
- Rigel DS 2010. Epidemiology of melanoma. Semin Cutan Med Surg 29: 204–209 [DOI] [PubMed] [Google Scholar]
- Sagebiel RW 1993. Melanocytic nevi in histologic association with primary cutaneous melanoma of superficial spreading and nodular types: Effect of tumor thickness. J Invest Dermatol 100: 322S–325S [DOI] [PubMed] [Google Scholar]
- Sahel JA, Albert DM 1993. Intraocular melanomas. Cancer Treat Res 65: 161–199 [DOI] [PubMed] [Google Scholar]
- Sanchez-Hernandez I, Baquero P, Calleros L, Chiloeches A 2012. Dual inhibition of V600EBRAF and the PI3K/AKT/mTOR pathway cooperates to induce apoptosis in melanoma cells through a MEK-independent mechanism. Cancer Lett 314: 244–255 [DOI] [PubMed] [Google Scholar]
- Satzger I, Kuttler U, Volker B, Schenck F, Kapp A, Gutzmer R 2010. Anal mucosal melanoma with KIT-activating mutation and response to imatinib therapy—Case report and review of the literature. Dermatology 220: 77–81 [DOI] [PubMed] [Google Scholar]
- Shi H, Kong X, Ribas A, Lo RS 2011. Combinatorial treatments that overcome PDGFRβ-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res 71: 5067–5074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, Ng C, Chodon T, Scolyer RA, Dahlman KB, et al. 2012. Melanoma whole-exome sequencing identifies V600EB-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun 3: 724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull AY, Latham-Schwark A, Ramasamy P, Leskoske K, Oroian D, Birtwistle MR, Buckhaults PJ 2012. Novel somatic mutations to PI3K pathway genes in metastatic melanoma. PloS ONE 7: e43369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A 2013. Cancer statistics, 2013. CA Cancer J Clin 63: 11–30 [DOI] [PubMed] [Google Scholar]
- Singh AD, Shields CL, Shields JA 2001. Prognostic factors in uveal melanoma. Melanoma Res 11: 255–263 [DOI] [PubMed] [Google Scholar]
- Singh AD, Bergman L, Seregard S 2005. Uveal melanoma: Epidemiologic aspects. Ophthalmol Clin North Am 18: 75–84 [DOI] [PubMed] [Google Scholar]
- Smalley KS 2010. Understanding melanoma signaling networks as the basis for molecular targeted therapy. J Invest Dermatol 130: 28–37 [DOI] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. 2012. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 366: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP 2004. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res 64: 7002–7010 [DOI] [PubMed] [Google Scholar]
- Swick JM, Maize JC Sr 2012. Molecular biology of melanoma. J Am Acad Dermatol 67: 1049–1054 [DOI] [PubMed] [Google Scholar]
- Terheyden P, Houben R, Pajouh P, Thorns C, Zillikens D, Becker JC 2010. Response to imatinib mesylate depends on the presence of the V559A-mutated KIT oncogene. J Invest Dermatol 130: 314–316 [DOI] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, Yang G, Haluska FG 2004. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol 122: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Chin L, Garraway LA, Fisher DE 2012. Melanoma: From mutations to medicine. Genes Dev 26: 1131–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udayakumar D, Zhang G, Ji Z, Njauw CN, Mroz P, Tsao H 2011. EphA2 is a critical oncogene in melanoma. Oncogene 30: 4921–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde P, Healy E, Jackson I, Rees JL, Thody AJ 1995. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet 11: 328–330 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, et al. 2010. Mutations in GNA11 in uveal melanoma. N Engl J Med 363: 2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaysse C, Pautier P, Filleron T, Maisongrosse V, Rodier JF, Lavoue V, Reyal F, Thomas L, de la Fouchardiere A, Delannes M 2013. A large retrospective multicenter study of vaginal melanomas: Implications for new management. Melanoma Res 23: 138–146 [DOI] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, et al. 2010. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18: 683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizkeleti L, Ecsedi S, Rakosy Z, Orosz A, Lazar V, Emri G, Koroknai V, Kiss T, Adany R, Balazs M 2012. The role of CCND1 alterations during the progression of cutaneous malignant melanoma. Tumour Biol 33: 2189–2199 [DOI] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. 2011. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol 29: 3085–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KA, Lazovich D, Hordinsky MK 2012. Germline melanoma susceptibility and prognostic genes: A review of the literature. J Am Acad Dermatol 67: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Program NCS, Stemke-Hale K, Davies MA, et al. 2011. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet 43: 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, Obenauf AC, Murali R, Fried I, Griewank KG, Ulz P, Windpassinger C, Wackernagel W, Loy S, Wolf I, et al. 2011. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet 43: 1018–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. 2012. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487: 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, et al. 2006. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst 98: 472–482 [DOI] [PubMed] [Google Scholar]
- Wu X, Li J, Zhu M, Fletcher JA, Hodi FS 2012. Protein kinase C inhibitor AEB071 targets ocular melanoma harboring GNAQ mutations via effects on the PKC/Erk1/2 and PKC/NF-κB pathways. Mol Cancer Ther 11: 1905–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Zhang X, Liu J, Estrem S, Li S, Gong XQ, Buchanan S, Henry JR, Starling JJ, Peng SB 2012. Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J Biol Chem 287: 28087–28098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Woods SL, Boyle GM, Aoude LG, MacGregor S, Zismann V, Gartside M, Cust AE, Haq R, Harland M, et al. 2011. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature 480: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Qureshi AA, Guo Q, Han J 2011. Genetic variation in DNA repair pathway genes and melanoma risk. DNA Repair 10: 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wong P, Jiaqi D, Jacobs B, Borden EC, Bedogni B 2013. An ERBB3/ERBB2 oncogenic unit plays a key role in NRG1 signaling and melanoma cell growth and survival. Pigment Cell Melanoma Res 26: 408–414 [DOI] [PubMed] [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC 1996. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 12: 97–99 [DOI] [PubMed] [Google Scholar]