SUMMARY
To understand the relationships between the non-GCIMP glioblastoma (GBM) subgroups, we performed mathematical modeling to predict the temporal sequence of driver events during tumorigenesis. The most common order of evolutionary events is 1) chromosome (chr) 7 gain and chr10 loss, followed by 2) CDKN2A loss and/or TP53 mutation, and 3) alterations canonical for specific subtypes. We then developed a computational methodology to identify drivers of broad copy number changes, identifying PDGFA (chr7) and PTEN (chr10) as driving initial non-disjunction events. These predictions were validated using mouse modeling, showing that PDGFA is sufficient to induce proneural-like gliomas, and additional NF1 loss converts proneural to the mesenchymal subtype. Our findings suggest most non-GCIMP-mesenchymal GBMs arise as, and evolve from, a proneural-like precursor.
Keywords: Copy number alteration, Glioblastoma, GBM subgroups, Neurofibromatosis type I disease, Mathematical modeling, Bioinformatic analyses, PDGFA, RCAS/tv-a system
INTRODUCTION
Glioblastomas (GBMs) are the most common and malignant central nervous system (CNS) neoplasms and arise initially as grade 4 tumors (primary GBM) or evolve from lower grade gliomas (secondary GBM). Deep molecular analyses of primary GBMs have divided them into four classes denoted proneural (PN), mesenchymal (MES), classical (CL), and neural (NL) (Brennan et al., 2009; Phillips et al., 2006; Verhaak et al., 2010). However, in some cases the division is blurred in that a sample might show patterns of more than one subtype (Brennan et al., 2009; Phillips et al., 2006; Sottoriva et al., 2013; Verhaak et al., 2010). These subclasses can be associated with canonical mutations such as PDGFRA amplification in PN-GBM, loss of NF1 in MES-GBM, and amplification of EGFR in CL-GBM. In the case of NL-GBMs, particular molecular abnormalities remain unidentified.
One of the best-studied GBM subclasses is PN-GBM, some of which carrying mutant IDH1 or IDH2 and have the methylator phenotype denoted by GCIMP (Noushmehr et al., 2010). Although the transcriptomal patterns for GCIMP tumors resemble those of the other PN-GBMs (non-GCIMP), their biology is significantly different in that the GCIMP GBMs display global hypermethylation of CpG islands, characteristic copy number alterations, and prolonged patient survival, suggesting that these tumors represent high-grade versions of grade 2 and 3 diffuse gliomas, which are commonly GCIMP, and thus represent secondary GBMs (Noushmehr et al., 2010; Ohgaki and Kleihues, 2012).
The existence of different GBM subtypes raises questions regarding their natural history and the temporal sequence in which individual alterations arise (Bhat et al., 2013; Phillips et al., 2006; Sottoriva et al., 2013). Furthermore, it is unclear whether these subgroups are fundamentally different tumors from their inception, or whether they evolve from a common glioma precursor.
Given the evidence of spontaneous conversion of human PN to MES tumors and observed subtype mosaicism within the same tumor (Phillips et al., 2006; Sottoriva et al., 2013), the GBM subtypes might be directionally variable from one subtype to another. Finally, it remains unclear whether the existence of subgroups has therapeutic implications (Bhat et al., 2013). For example, the subtype-defining mutations may be promising therapeutic targets if they represent initiating genetic events in glioma evolution but their inhibition would unlikely provide lasting therapeutic benefit if they represent late events.
Chromosome instability is common in human cancers, it is characterized by widespread chromosome (chr) copy number changes, such as gain or loss of whole chr or fractions thereof (Lengauer et al., 1998). Gain of chr7 as well as loss of chr10 and parts of chr9 are very frequent in gliomas. Alteration of chr7 frequently includes focal amplification of the EGFR locus (The Cancer Genome Atlas Research Network, 2008). Nevertheless, forced expression of the wild-type EGFR or the constitutively active EGFRvIII mutant alone does not form gliomas in mice (Holland et al., 1998; Zhu et al., 2009), suggesting that EGFR might not be an initiator of this tumor type.
Genome-wide high throughput sequencing efforts have led to the identification of many genomic alterations in cancer genomes, but it remains a challenge to identify genuine driver gene(s) from the many genes located within a large chromosomal event. Here we utilized a combined computational and experimental approach to investigate the natural history of human non-GCIMP GBMs.
RESULTS
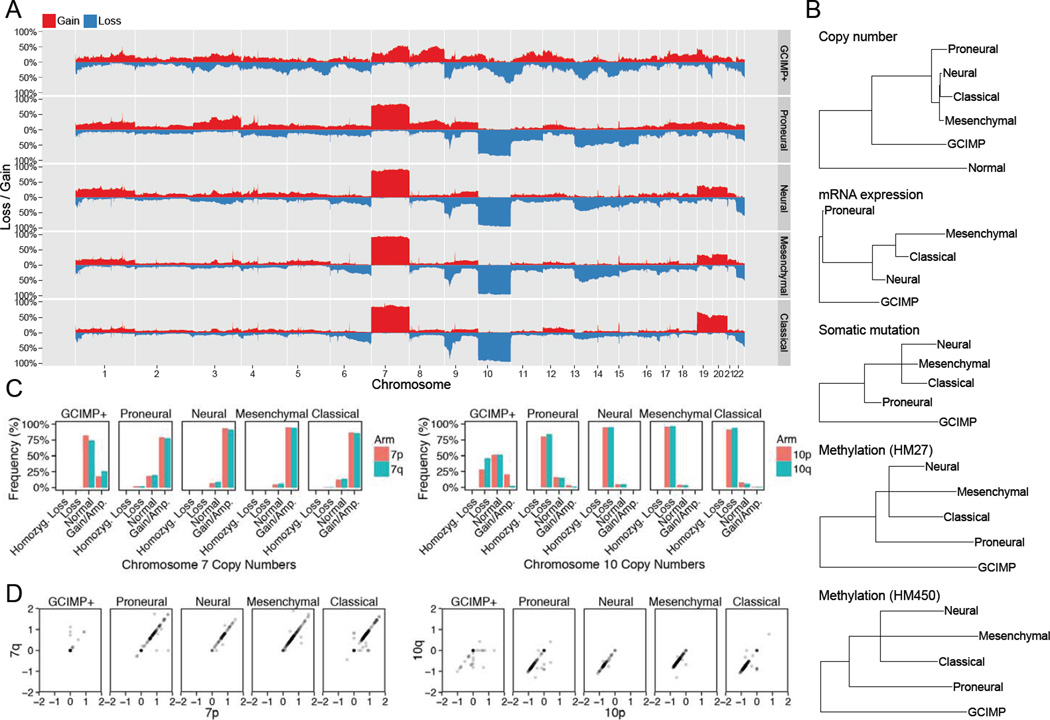
We first utilized genomic data from The Cancer Genome Atlas (TCGA) to investigate the frequencies and types of genomic alterations in GBM (The Cancer Genome Atlas Research Network, 2008). The overall pattern of broad gains and losses in the GCIMP tumors showed some similarities to that in the non-GCIMP PN-GBM (Noushmehr et al., 2010) but were strikingly similar between the non-GCIMP PN-GBM and other non-GCIMP subtypes (Figure 1A and Table S1). These patterns suggest that all non-GCIMP GBM are very similar with respect to genomic alterations acquired during their evolution, and distinct from the GCIMP tumors. We then performed phylogenetic analysis of copy number, mRNA expression, somatic mutations and promoter methylation data of the non-GCIMP subgroups and found that PN-GBMs located separately on the phylogenetic trees generated by each of these data types, suggesting that they are distinct from other non-GCIMP subtypes (Figure 1B).
Figure 1. Somatic copy number alterations and their frequencies in GBM.
(A) Genome plot visualizes frequencies of copy number gains (red) and losses (blue) along the genome in GCIMP tumors and the 4 non-GCIMP subtypes (proneural, neural, mesenchymal and classical GBM). (B) Phylogenetic analysis in all GBM samples determining the putative evolutional order in GBM subtypes using all data types (copy number, mRNA expression, somatic mutation and methylation) and the Neighbor Joining algorithm. Artificial samples with exactly 2 copies in the whole genome serve as normal copy number control. For DNA methylation analysis, both HM27 and HM450 platform is shown. (C) Histograms showing the frequencies of whole arm gains and losses of both chr7 and chr10 in each subtype separately. (D) Copy numbers of the p and q arms of chr7 and chr10 for all TCGA samples stratified by GBM subtype. See also Figure S1 and Tables S1–S3
Gain of several copies of chr7 and loss of one copy of chr10 were the most frequent events in non-GCIMP GBMs, with frequencies of 86% and 90%, respectively (Figures 1A and 1C). GCIMP tumors almost never lose the entire chr10 but show similar frequencies of loss (26%) and amplification (24%) of 10p (Figures 1A and 1C). Note that gain of chr7 refers to the gain of a few copies of the whole chr7, which is different from amplification of any specific gene on chr7, and loss of chr10 refers to the loss of a whole copy of chr10 but leaving the other copy intact, which is not synonymous with focal deletion of any gene on chr10. These events are distinct from the commonly reported high-level amplification and deletion of specific genes in GBMs such as PDGFRA, EGFR or MET (The Cancer Genome Atlas Research Network, 2008).
When investigating 7p and 7q separately for all GBM subtypes, we found that 7p gain ranged from 81% for PN-GBMs to 95% for MES-GBMs while 7q gain ranged from 79% for PN-GBMs to 93% for MES-GBMs (Figure 1C). In general, 7p and 7q gain were balanced (Figure 1D), with the exception of GCIMP tumors, in which 7q gain occurred in 24% of tumors and 7p gain in 15%. These data imply that for non-GCIMP GBMs, chr7 is gained as the entire chromosome. Balanced chr7 gain is present in 82% of all non-GCIMP GBM samples and represents the most common chromosomal abnormality in these tumors. Non-GCIMP cases with normal copy number of both chr7 and chr10 were rare (2%) (Figure S1A, Table S2 and S3) and had a similar age and survival distribution as GCIMP tumors (Figure S1B–C), suggesting that some of these non-GCIMP cases are potentially misclassified GCIMP tumors (Figure S1D).
Mathematical modeling suggests gain of chr7 and loss of chr10 as first events in all non-GCIMP GBM subtypes
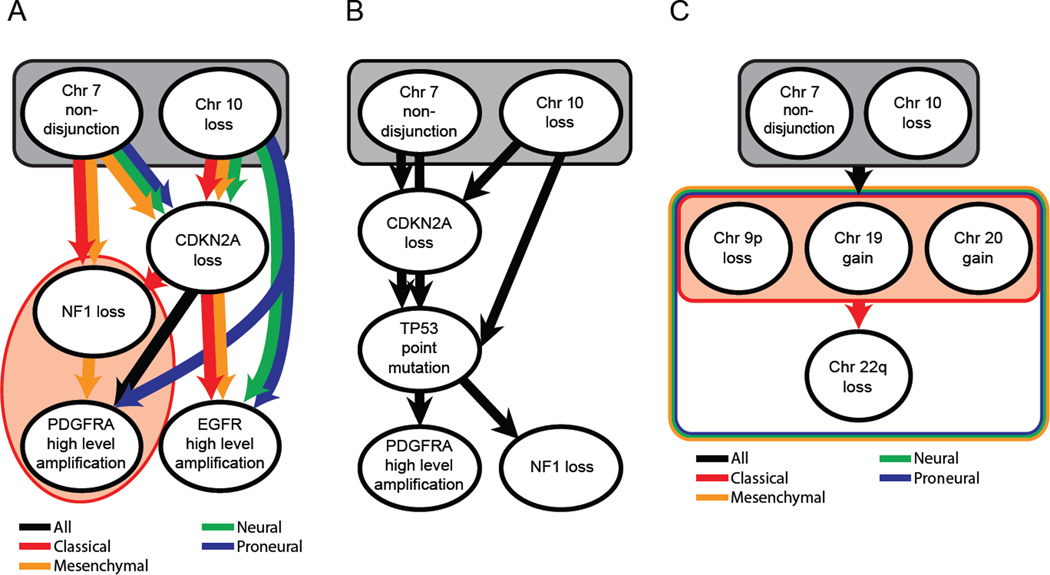
The high prevalence and even distribution of chr7 gain and chr10 loss in all non-GCIMP GBM subtypes raises the question of when these events occur during GBM development. Thus, we utilized a computational methodology, Retracing Evolutionary Steps In Cancer (RESIC), to determine the temporal sequence of genetic alterations during tumorigenesis from cross-sectional genomic data of a population of tumors at their fully transformed stage (See also Supplemental Experimental Procedures) (Attolini et al., 2010; Cheng et al., 2012). Applying RESIC to TCGA data (The Cancer Genome Atlas Research Network, 2008) to examine the temporal order of the known subtype-specific alterations EGFR (CL-GBM), NF1 (MES-GBM) and PDGFRA (PN-GBM) (Verhaak et al., 2010), together with gain of chr7 and loss of chr10, we identified chr7 gain together with chr10 loss as the first events in all subtypes of GBM (Figures 2A, S2A and S2B). Loss of CDKN2A was another early event in all subtypes. Further analyses based on samples with mutational information only (n=85 non-GCIMP samples) revealed that TP53 mutations arose after CDKN2A loss (Figures 2B and S2C). Finally, when considering all frequent chromosome arm-level events, we found that chr7 gain and chr10 loss arise before any other broad gains or losses (Figures 2C and S2D). An almost identical ordering of these events was inferred using a different method (Beerenwinkel and Sullivant, 2009) (Figure S2E).
Figure 2. Temporal sequence of events in glioma development.
(A) Order of events for subtype-specific copy number alterations per subtype. (B) Order of events for all GBM samples for which TP53 point mutation data was available (n=85). (C) Order of chromosome level copy number alterations. Black ovals represent distinct mutational events. Arrows represent an ordering of events detected by RESIC. Rectangles containing events represent sets of events where RESIC cannot distinguish an order of events. Black denotes orderings that are shared across subtypes. Red, Yellow, Green and Blue denotes classical, mesenchymal, neural and proneural subtype specific orderings, respectively. Orderings shared by multiple subtypes but not all subtypes are denoted by multicolored arrows and rectangles. See also Figure S2.
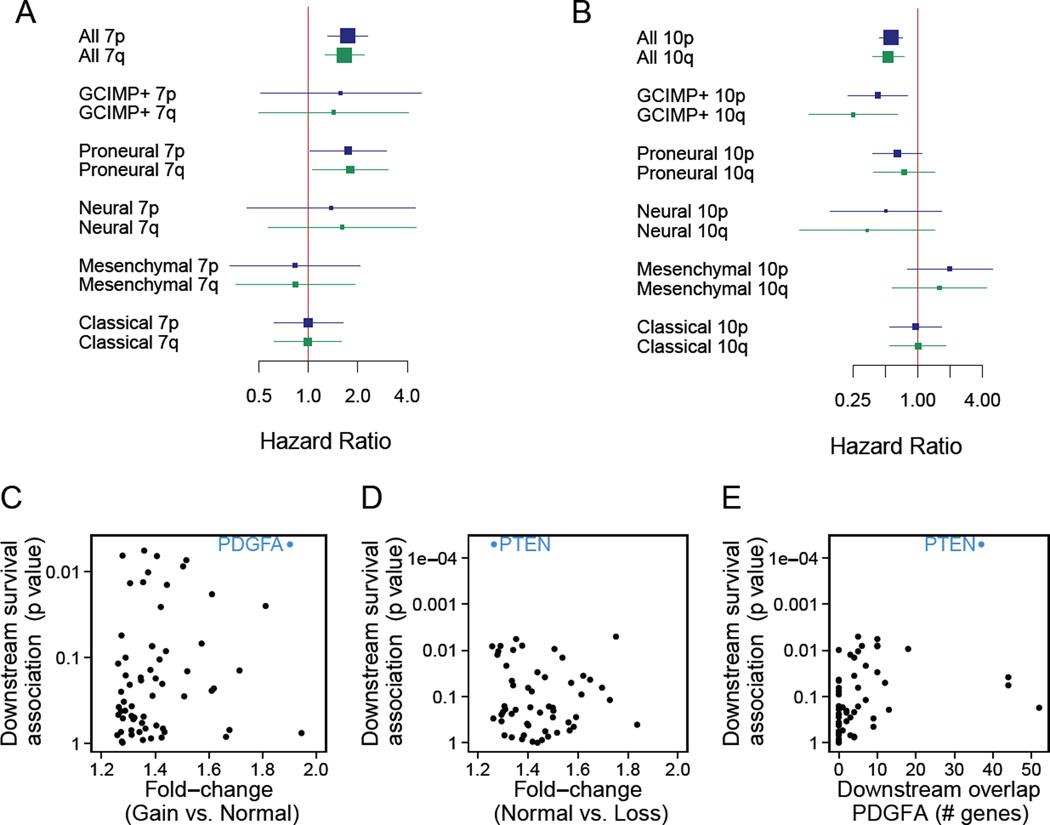
To delineate the clinical implications of these arm-level events, we investigated the association between chr7 gain or chr10 loss and GBM patient survival. Copy numbers of chr7 and chr10 correlate negatively and positively, respectively, with survival (Figures 3A and B). This finding arises primarily because these events do not occur as frequently in GCIMP tumors, and patients with these tumors survive longer on average, than non-GCIMP GBMs (Noushmehr et al., 2010). However, even in the non-GCIMP GBMs, chr7 gain correlates with poor survival of patients, but only for PN-GBM cases (Figure 3A). These data suggest that a gene or several genes on chr7 drive the gain of copy number early in GBM evolution and promote the aggressive character in PN-GBM. By contrast, the lack of association of chr10 loss with overall survival might be due to haplosufficiency of genes on chr10 (Figure 3B).
Figure 3. Drivers of chr7 and chr10 alterations in the proneural subtype.
(A-B) Forest plots showing the Hazard Ratios (HR, squares) of chr7 gain (A) and chr10 loss (B) and their confidence intervals. HR significantly larger than 1 (A) or lower than 1 (B) signify a copy number change associated with poor prognosis. Square size is proportional to the sample size. (C, D) Genes on chr7 (C) and chr10 (D) ranked by association of downstream genes with overall survival, with the corresponding p values shown on the y-axis. Genes were considered with a fold-change (x-axis) in expression of larger than 1.25 (averaged over both Affymetrix and Agilent platforms) when comparing patients with normal and altered chr7 and chr10 copy numbers. The top-ranking hits are in blue. (E) The overlap in downstream genes of PDGFA in the KEGG pathway database with the top-ranking genes on chr10 (x-axis). See also Figure S3 and Table S4–6.
Computational analysis suggests PDGFA and PTEN as major drivers of non-disjunction events
We thus aimed to identify the gene(s) on chr7 and chr10 that drive the acquisition of these arm-level events, lead to the poor survival of patients, and are potential targets for therapies. We designed a computational algorithm consisting of two steps. The first step ranks genes based on the extent of correlation between their copy number and expression levels. Genes are first sorted by the fold-change of expression between tumors that do and do not harbor the arm-level event. Genes with anti-correlation of copy number and mRNA levels are excluded. In the second step, the resulting gene list is further filtered by the degree of correlation between a gene’s expression level and patient survival.
When applying the first step to genes on chr7 for all non-GCIMP GBM samples, we identified a set of genes with the highest expression response to increased copy number predicted to occur with gain of chr7 (Table S4). Note that EGFR was not selected in this first filtering because low-level gain of EGFR, as found with whole chr7 gain, is not correlated with its increased expression (p = 0.19, Student t-test EGFR normal vs. gain, Figure S3A). High-level amplification of EGFR is predicted by RESIC to be a late event (Figure 2A) and such focal amplification of EGFR correlates significantly with its expression level (p < 0.001, Student t-test EGFR gain vs. focal amplification, Figure S3A). Almost all identified point mutations in EGFR as well as EGFRvIII were found in samples with high-level focal gene amplification (Figure S3A and Table S1). These data further support that EGFR may not be a driver of initial oncogenic events in this tumor type, but rather contributes to tumor progression or maintenance through high expression driven by focal amplification arising at later stages of tumor formation.
The second step then showed direct correlation between expression levels and survival was uninformative because many genes on chr7 displayed a correlation between their copy number and expression; thus their expression also correlates with survival in the PN-GBM (Table S4). Therefore, we investigated the correlation between survival and the expression levels of pathways downstream of genes on chr7. When applying this filter to chr7, we found that, of the 190 genes whose expression fold-change between cases of normal and gained chr7 copy number was at least 1.25, 13 genes displayed a correlation with survival in PN-GBMs of p < 0.05 (Table S5). Figure 3C summarizes the two filters applied to chr7. This combined ranking approach identified PDGFA as the highest-ranking gene on chr7 (Figures S3B, Table S5 and S6), i.e., expression of PDGFA downstream genes was associated with overall survival and chr7 gain increased PDGFA mRNA levels significantly. Thus, PDGFA most likely is the gene providing the strongest selective advantage to gaining extra copies of chr7.
We then sought to identify potential drivers of chr10 loss. Application of the two steps outlined above is only appropriate for chromosomal gains, since the correlation between expression levels and patient outcomes necessitates a degree of variability in copy number across patients. Chr7 gain meets this requirement since patients may harbor 3, 4, or more copies of chr7. By contrast, chr10 loss results in a single remaining copy of chr10 in almost all cases (Figures 1C and D), thus necessitating an alternative strategy. As loss of chr10 and gain of chr7 almost always co-occur (p = 0.001, Fisher’s exact, Figures 1A and C), we hypothesized that the phenotypic effect of a main driver of chr10 loss might amplify the effects of the driver of chr7 gain and vice versa, therefore likely activating the same pathways and resultant changes in gene expression. We thus also ranked the genes on chr10 by the association of the expression levels of their downstream genes with survival (Table S4 and S5). The tumor suppressor PTEN displayed a highly significant survival association between the expression levels of its downstream genes and survival (false discovery rate [FDR] < 0.02, Figures 3D and S3C, Table S5) and also a large overlap of downstream genes of PDGFA (Figure 3E), suggesting it may be a significant driver of chr10 loss in these tumors. In non-GCIMP GBM subtypes other than PN-GBMs, we did not identify any such strong pathway enrichments of chr7 and chr10, consistent with the lack of correlation of chr7 gain with survival in these GBM subtypes (Figure 3A). Our computational approach thus suggests PDGFA and PTEN as primary drivers of the chr7 and chr10 non-disjunction events, respectively. Note that this analysis does not suggest that these genes alone are sufficient for driving these events.
Mouse modeling suggests that the predicted first events in glioma evolution are sufficient to induce gliomagenesis
Our computational analyses predict that elevated expression of PDGFA represents the strongest initial driving event in glioma evolution. This is noteworthy because of the well-described role of PDGF signaling in GBM, especially in PN-GBM (Brennan et al., 2009; The Cancer Genome Atlas Research Network, 2008; Verhaak et al., 2010). Note that the elevated expression of PDGFA as a likely driver is not the same as the frequently reported high-level focal amplification of PDGFRA found primarily in PN-GBM. The PDGF signaling network is transmitted by four ligands (A, B, C and D isoforms) and two protein tyrosine kinase receptors (α and β). Overexpression of PDGFB generates BB dimers that activate both PDGFRα and β while overexpression of PDGFA only activates PDGFRα (Andrae et al., 2008; Heldin and Westermark, 1999). In animal modeling systems, signaling driven by PDGFB alone is sufficient to form GBMs, and additional loss of Tp53, Cdkn2a (Ink4a-Arf) or Pten accelerates tumor formation (Dai et al., 2001; Fomchenko et al., 2011; Hu et al., 2005; Shih et al., 2004; Squatrito et al., 2010). Although the PDGFB protein level is elevated in many PN-GBMs, it does not correlate with the PDGFB mRNA level in these tumors (Brennan et al., 2009). Moreover, the alteration of PDGFB is rare in GBMs (Verhaak et al., 2010), suggesting that other PDGFs such as PDGFA could potentially drive oncogenesis in this GBM subtype. However, it remains to be determined whether elevated PDGFA is sufficient to initiate gliomas, either alone or in combination with other early events such as CDKN2A, TP53 or PTEN loss (Figures 2A, B, S3C and D) (Jackson et al., 2006; Nazarenko et al., 2011).
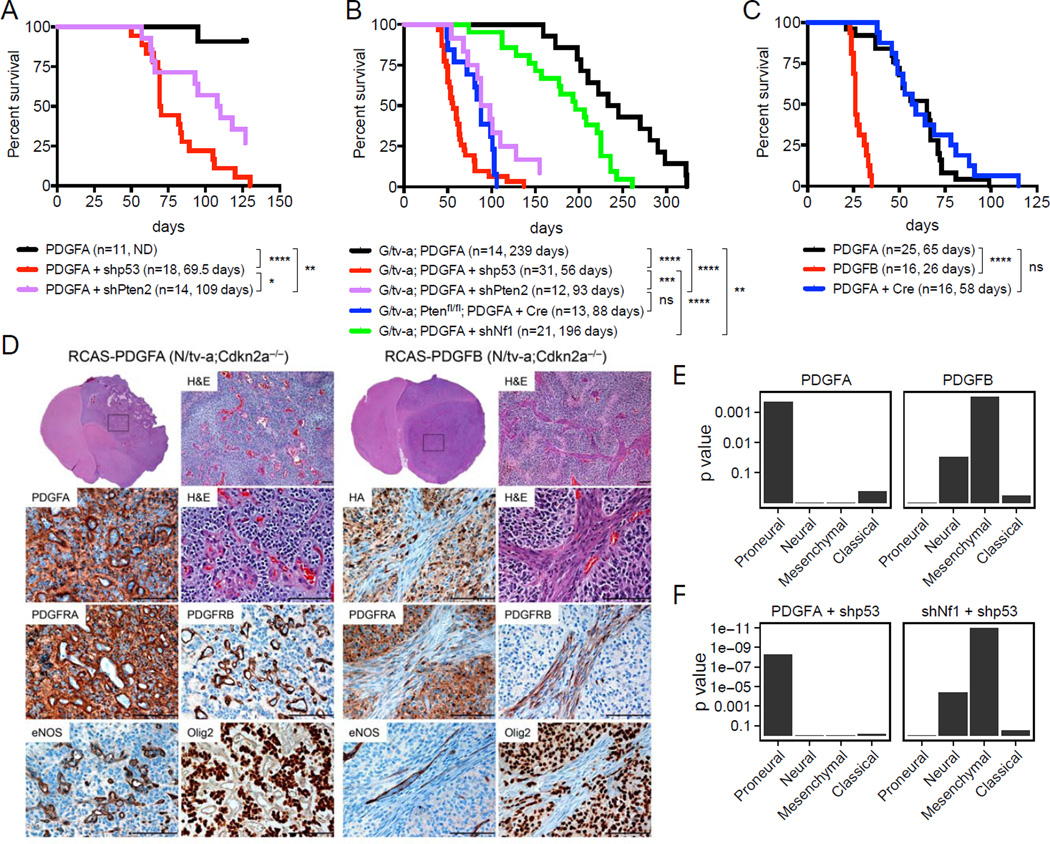
We thus tested the ability of genes predicted to drive human GBM evolution to induce gliomas in vivo using the RCAS/tv-a system that allows post-natal cell type specific gene transfer (Holland et al., 1998; Holland and Varmus, 1998). To determine whether PDGFA alone is sufficient for glioma formation, we infected neonatal N/tv-a mice (Nestin-expressing cell of origin, n=11) and G/tv-a mice (GFAP-expressing cell of origin, n=14) with an RCAS retroviral vector expressing human PDGFA (RCAS-PDGFA) (Figures S4A and B). All N/tv-a mice were sacrificed at 4 months, when the first mice developed symptoms of intracranial pathology. Eight of 11 mice harbored brain tumors of various sizes demonstrating histological features of grade 2 human oligodendrogliomas including round nuclei, perinuclear halos, strong Olig2 and lower GFAP expression (Figures 4A and data not shown). All G/tv-a mice formed tumors ranging from 159 to 323 days post injection; many developed high-grade gliomas with histologic features of human oligodendroglioma (Figures 4B and S4C). These data indicate that elevated PDGFA expression is sufficient to initiate gliomagenesis in vivo.
Figure 4. PDGFA induces gliomas with similar expression pattern to human proneural GBMs in mice. (A-C).
Kaplan-Meier survival curves showing symptom-free survival of PDGFA- or PDGFB-induced gliomas in Nestin (N)/tv-a (A), GFAP (G)/tv-a or G/tv-a;Ptenfl/fl (B), N/tv-a;Cdkn2a−/−;Ptenfl/fl mice (C). Tumors were generated by the injection of the indicated RCAS virus into neonatal brains. (*) p < 0.05, (**) p < 0.005, (***) p < 0.001, (****) p < 0.0001, ns: not significant. (D) Representative H&E and immunohistochemical analysis of the PDGFA- and PDGFB-induced gliomas in N/tv-a;Cdkn2a−/−;Ptenfl/fl mice. Boxes denote the enlarged-region. Scale bars, 100 µm. (E) GSEA of the RCAS-PDGFA-and RCAS-PDGFB-induced gliomas in N/tv-a;Cdkn2a−/−;Ptenfl/fl mice. Gene expression profiles were compared between the PDGFA- (n=7) and PDGFB- (n=5) induced gliomas based on the TCGA subtype signatures (Verhaak et al., 2010). Bar plots visualize enrichment p values (y-axis) of the four subtype signatures in the ranking of genes by differential expression (PDGFA versus PDGFB). A low p value indicates consistent expression with the subtype, i.e., many genes known to be up-regulated in the subtype are up and down-regulated genes are down. (F) GSEA of the RCAS-PDGFA/shp53 and RCAS-shNf1/shp53-induced gliomas in N/tv-a and G/tv-a mice. See also Figure S4 and Table S7
Pten loss shortens survival of mice with PDGFA-induced gliomas, but is not sufficient to induce gliomas alone
RESIC analysis predicted that combined loss of chr10 and gain of chr7 are the earliest events in non-GCIMP GBM subtypes. Previous results using the RCAS/tv-a system have shown loss of Pten alone insufficient for glioma formation in mice (Hu et al., 2005), suggesting loss of Pten might play a supportive role in PDGFA responsive cells, rather than serving as the tumor initiator. To confirm the supportive role of Pten loss in this context, we created an RCAS vector expressing mRFP and a short hairpin designed to knock down Pten expression (RCAS-shPten) (Figures S4A, D and E). Infection of both wild-type N/tv-a (n=3) and G/tv-a (n= 11) mice with this vector alone did not result in brain tumors by approximately 4 months. However, consistent with the RESIC prediction, simultaneous Pten loss significantly shortened the survival of mice with PDGFA-induced gliomas and elevated the grade of the tumors (Figures 4A, B and S4F, G and H). These features were also observed in a cremediated Pten deletion model (Figures 4B and S4F).
PDGFA-induced tumors resemble PN-GBMs, whereas PDGFB-induced tumors display a more MES-GBM character
RESIC analysis also predicted that loss of CDKN2A occurs after gain of chr7 (elevation of PDGFA expression) in these tumors. Therefore, we injected neonatal N/tv-a;Cdkn2a(Ink4a–Arf)−/− mice with RCAS-PDGFA (n=25) (Figure 4C). Tumors occurred in these mice with 100% penetrance and median survival of 65 days. These tumors demonstrated histological features of human oligodendroglioma and displayed a high-grade histology with uniformly abundant pseudopalisading necrosis, hypervascularity and brisk microvascular formation throughout the tumor (Figures 4C, D and S4I). Interestingly, additional loss of Pten in a Cdkn2a−/− background did not further shorten tumor latency (Figure 4C).
We then compared gliomas induced by PDGFA and PDGFB in newborn and adult N/tv-a;Cdkn2a−/−mice. We found that PDGFA generated tumors with a longer latency than PDGFB, and that gliomas induced by either ligand arose in newborn mice faster than in adults (Figures 4C and S4J) (Squatrito et al., 2010). The histology of tumors induced by PDGFA and PDGFB was generally similar; however, the vasculature and stroma were notably different (Figures 4D and S4K). PDGFB-induced gliomas showed spindled vasculature and atypical stroma (PDGFB activates both α receptors in the tumor cells as well as β receptors in the perivascular stroma), while tumors generated by PDGFA (that activates only the α receptor) showed glomeruloid microvascular proliferation more similar to that seen in human GBM (Figures 4D and S4K) (Helmy et al., 2012; Hermansson et al., 1988).
Subsequently, we profiled these tumors by gene expression analysis to determine which human GBM subtype they most closely resembled and found that PDGFA-induced gliomas in the mouse more closely resembled PN-GBMs in humans while the PDGFB-induced tumors shifted towards MES-GBMs (Figure 4E). The Olig2-expressing tumor cells from PDGFB-induced gliomas, which constitute the bulk of the tumor (Figure 4D), show a strongly proneural expression profile (Dougherty et al., 2012). Therefore, the MES character of the PDGFB-induced gliomas possibly reflects the abundant atypical perivascular stroma and other non-tumor stromal cell types found in these tumors (Figures 4D, E and S4K).
Loss of TP53 as well as PTEN or CDKN2A in PDGFA-induced gliomas results in a highly lethal disease in mice
CDKN2A loss frequently occurs in human PN-GBMs, but this GBM subgroup also commonly harbors mutations in TP53 (Verhaak et al., 2010). Therefore, we determined if loss of Tp53 would cooperate with PDGFA-induced gliomas in a similar way as Cdkn2a loss. We generated an RCAS vector expressing both mRFP and a short hairpin designed to knock down Tp53 expression (RCAS-shp53) (Figures S4A, L-P). Infecting mice with this vector alone did not generate gliomas by approximately 1 year; however, when combined with RCAS-PDGFA, high-grade gliomas formed in 100% of mice with a median survival of 69.5 days in wild-type N/tv-a mice (n=18) and 56 days in wild-type G/tv-a mice (n=31), similar to PDGFA in a Cdkn2a−/− background (Figures 4A–C). The tumors demonstrated histological features of human oligodendroglioma and displayed high-grade histology similar to PDGFA-induced tumors with Cdkn2a loss (Figure S4Q). Further the gene expression profiles of the gliomas induced by PDGFA with loss of Tp53 showed a significant enrichment of PN-GBM genes (Figure 4F) as well as downstream orthologs of human PDGFA identified in above computational analysis (Figures 3C, Figure S4R and Table S6). These observations suggest that loss of the tumor suppressor genes PTEN, CDKN2A or TP53 are sufficient to enhance PDGFA-induced gliomas resulting in highly lethal gliomas.
Simultaneous loss of Nf1 and Tp53 induces MES-GBM in the RCAS/tv-a model
RESIC analysis predicts that the same early events in PN-GBM also occur in MES-GBM but NF1 loss is late in the evolution of these tumors. In MES-GBMs (as well as all non-GCIMP GBMs), NF1 loss is associated with TP53 mutations, and less commonly with CDKN2A loss (Figure S5A) (Verhaak et al., 2010). Further, published mouse GBM models resulting from germline homozygous deletions of Nf1 and Tp53 show median survival of approximately 150–200 days (Reilly et al., 2000; Zhu et al., 2005) but they do not recapitulate human MES-GBM expression pattern (Liu et al., 2011). Other groups recently generated an H-RasV12-shp53 glioma model mimicking Nf1;Tp53 loss and reported H-RasV12-shp53-driven GBMs to show MES gene expression (Friedmann-Morvinski et al., 2012). However, there is no mouse model with Nf1;Tp53 loss reported to create a GBM with MES gene expression. More importantly, all of these GBM models were generated using technologies different from what we used, rendering it difficult to compare them. Therefore, to allow direct comparison between our PDGFA models and mouse models of MES-GBM, we engineered the RCAS/tv-a system to generate MES-GBM from normal cells. We then set out to initially reproduce the published effect of combined loss of Nf1 and Tp53.
We created an RCAS vector expressing GFP and a short hairpin designed to knock down Nf1 expression (RCAS-shNf1) (Figures S4A, L, N, O and S5B–C). Infection of both wild-type N/tv-a (n=20) and G/tv-a (n= 12) mice with this vector alone did not generate tumors by approximately 1 year; however, infection with the combination of RCAS-shNf1 and RCAS-shp53 led to the formation of gliomas in 100% of mice with a median survival of 146 days for G/tv-a (n=52) and 221 days for N/tv-a (n=32) mice (Figures 5A, B, S4O and S5D–G). These latencies are similar to those published for gliomas formed by germline models of combined Nf1 and p53 loss (Reilly et al., 2000; Zhu et al., 2005). Histologic analysis of the G/tv-a and N/tv-a tumors showed stronger expression of the MES markers vimentin and CD44, and the MES master regulators pSTAT3 and C/EBPβ in the G/tv-a tumors than the N/tv-a tumors (Figure 5B and data not shown) (Carro et al., 2010). Gene expression profiling of these tumors (G/tv-a: n=7, N/tv-a: n=11) demonstrated that they were more MES than those driven by PDGFA (Figure 4F), and that gliomas arising in G/tv-a mice had a more MES-like expression pattern than those arising in N/tv-a mice (Figure 5C). Collectively, these data suggest it is possible to generate MES gliomas from normal cells in post-natal mice with a similar latency to that seen with germline strategies that delete both genes (Reilly et al., 2000; Zhu et al., 2005). While Nf1 loss in N/tv-a;Cdkn2a−/−;Ptenfl/fl mice was considerably less efficient than loss of Tp53 for the formation of gliomas, as supported by the fact that NF1 loss is associated with TP53 mutations, and less commonly CDKN2A loss (Figures S5A, E and Table S7) (Verhaak et al., 2010).
Figure 5. Simultaneous loss of Nf1 and Tp53 induces MES-gliomas in the RCAS/tv-a model.
(A) Kaplan-Meier survival curves showing symptom-free survival and relative tumor grade of the RCAS-shNf1/shp53-induced gliomas in N/tv-a and G/tv-a mice. The percentage of tumors exhibiting WHO grades II (G2), III (G3), and IV (GBM) histological features are shown for each genotype. (B) Representative H&E and immunohistochemical analysis for the indicated protein of the RCAS-shNf1/shp53 induced-glioma in G/tv-a mice. The box denotes the enlarged-region. Scale bars, 100 µm. (C) GSEA of the RCAS-shNf1/shp53 induced-gliomas in G/tv-a and N/tv-a mice. See also Figure S5.
Additional loss of NF1 converts the PDGFA-induced PN phenotype to a MES gene expression pattern
The tumor latency for PDGFA-induced gliomas versus those induced by Nf1 loss is striking. In the mouse model, the loss of both Nf1 and Tp53 occurs simultaneously while in humans there would be multiple separate mutational events in a normal cell that would lead to loss of function of both alleles of NF1 and mutation of TP53. Such an evolutionary pathway to MES-GBMs formation, although possible, might be much slower than for the PN-GBMs, which requires elevated expression from a single gene such as PDGFA. Moreover, the importance of the chronological order of mutations in MES glioma formation is underscored by the fact that in mice, loss of Nf1 is less efficient at enhancing gliomas than loss of Tp53 in the context of forced PDGFA expression (Figure 4B). Finally, global copy number and RESIC analysis predicts that MES-GBMs initially arise from tumors with the same alterations as PN-GBMs and lose NF1 later (Figure 2).
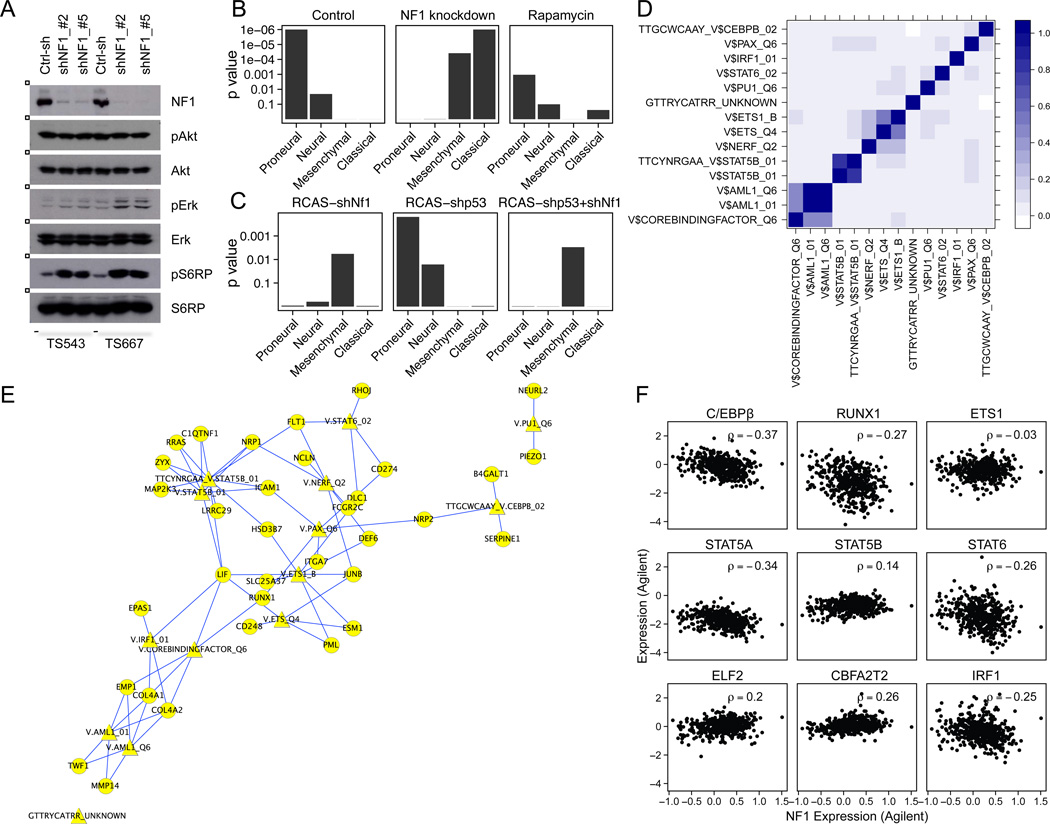
Therefore, we sought to determine if NF1 loss in PN-GBM could induce a PN- to MES-GBM transition. We obtained two PN-GBM cell lines that contain amplification of PDGFRA and by expression profiling fit the criteria for the PN subtype (Figures 6A, B and S6A, B) (Ozawa et al., 2010). Expression profiling of the lines before and after NF1 knockdown showed that the original PN expression pattern was shifted toward a MES or CL pattern in both cases (Figures 6B and S6B–C). Moreover, knockdown of Tp53 or Nf1 alone was sufficient to induce PN and MES gene enrichment in murine neurosphere culture, respectively (Figures 6C and S6D). These results suggest that NF1 function regulates transcriptional pathways necessary for mesenchymal conversion. To examine whether NF1 directly regulates the expression of mesenchymal transcriptional factors (TFs), we performed a TF enrichment analysis in murine neurosphere (Figure S6E), murine glioma (Figure S6F) and TCGA (Figure S6G) data sets. We identified 14 TF gene sets (MSigDB 4.0), corresponding to 9 different TFs, that were significantly (FDR < 0.05) associated with NF1 loss in the three datasets (Figures 6D and S6E–G). Some of these 14 TF gene sets associated with NF1 loss presented high similarity to each other (Figure 6D) and displayed significant overlap with recently reported mesenchymal TF target genes (Carro et al., 2010) (Figure 6E). Notably, NF1 loss was associated significantly with two main mesenchymal TFs, C/EBPβ and RUNX1 (Figure 6F). By contrast, perturbations of STAT3 and C/EBPβ expression did not affect NF1 expression in human GBM cells and mouse stem cells (Figures S6H and I), implying that these MES-associated TFs are downstream of NF1 loss. ShNf1 induced-tumors showed a statistically significant association between loss of NF1 and gene expression changes related to the mTOR pathway (Figure S6J). Furthermore, loss of NF1 did not appreciably elevate Erk or Akt phosphorylation (p) but did elevate pS6RP, suggesting that NF1 loss may affect mTOR activity in an Akt independent manner (Figure 6A). Thus, we treated the two NF1 knockdown cell lines with the mTOR inhibitor rapamycin (Figure S6K) and observed a marked but variable shift from MES/CL toward PN expression, suggesting that blockade of mTOR rescues the effect of NF1 loss in PN-GBM lines (Figure 6B).
Figure 6. AdditionalNF1 loss induces proneural to mesenchymal conversion in vitro and in vivo.
(A) Western blot analysis of two human GBM cell lines (TS543 and TS667) expressing controlor NF1-shRNAs (target sequence #2 and #5) with the indicated antibodies. (B) GSEA of human GBM cell lines. Gene expression profiles were compared between untreated cells, 0.1% DMSO or 1nM Rapamycin-treated cells for 5 hours in two different cell lines (TS543 and TS667) expressing controlor NF1-shRNAs. Each sample was analyzed in triplicate. Control-shRNA samples of the TS543 and TS667 cells also have technical duplicate samples. (C) GSEA of murine neurosphere lines. The gene expression profiles were analyzed in between N/tv-a; neurosphere lines expressing RCAS-shGL2, -shNf1, -shp53 or -shNf1/shp53. The subtype gene signature enrichment was determined by comparing control-shRNA sample (shGL2) with each shRNA samples, respectively. Each sample was analyzed in triplicate. (D) The overlap index of the 14 significant gene sets, corresponding to 9 different TFs; high overlaps are shown in dark blue. (E) Cytoscape visualization of the overlap of the 14 significant TF gene sets and the GBM mesenchymal gene signature (Carro et al., 2010). Diamond shaped nodes represent the 14 TF gene sets. Circular nodes represent TF target genes in both the mesenchymal gene signature and the 14 TF gene sets. TFs and target genes are connected by blue edges. The p value for the overlap of the mesenchymal gene signature and the 14 TFs was p = 0.005 (hypergeometric distribution). (F) Correlation of MES-TF expression with NF1 expression in the TCGA Agilent data. p values for C/EBPβ, RUNX1, STAT5A, STAT6 and IRF1 were p < 0.001 anti-correlation with NF1 expression. See also Figure S6.
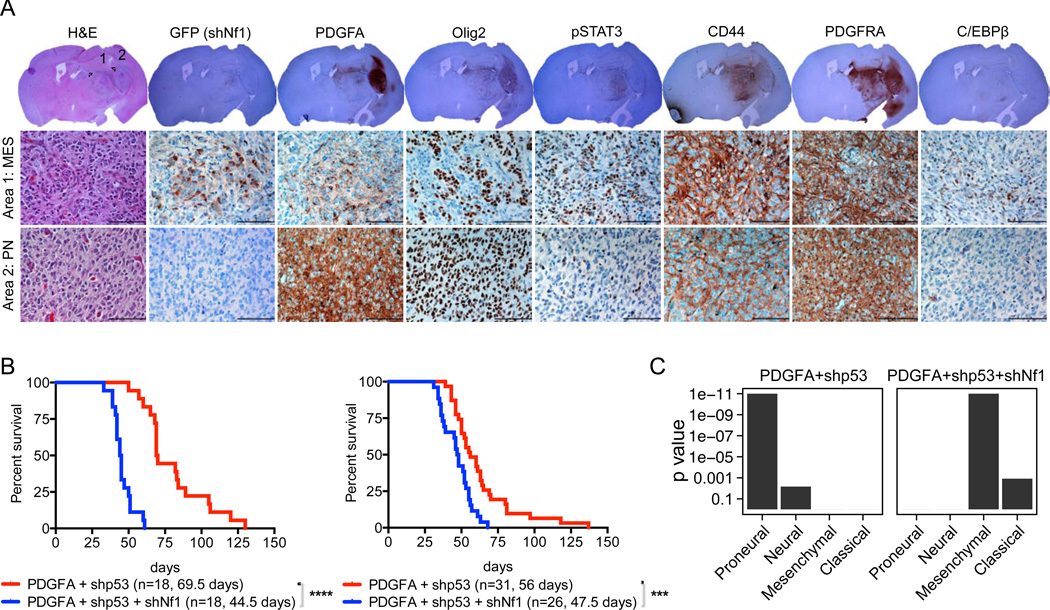
Finally, we determined if NF1 loss could convert PN-GBMs to MES-GBMs in mice. To test sequential loss, we initially generated PDGFA/shp53 tumors by concurrent stereotactic injection of the two vectors and then secondary infection with the GFP-shNf1 vector into the tumors using the same stereotactic coordinates 2 to 4 weeks later. The RCAS-GFP-shNf1 virus was regionally incorporated in 15 out of 20 mouse gliomas (Table S7). Many of the GFP-positive regions were detected in low PDGFA expression areas and/or the tumor periphery. Higher expression for MES-related markers such as CD44, pSTAT3 and C/EBPβ was preferably associated with GFP-positive regions (i.e. NF1 knockdown) and/or adjacent areas (Figure 7A). For simultaneous loss of NF1, we compared the GBMs generated by the double combination of RCAS-PDGFA and RCAS-shp53 (G/tv-a: n=31, N/tv-a: n=18, see above) to the triple combination of RCAS-PDGFA, RCAS-shp53 and RCAS-shNf1 (G/tv-a: n=26, N/tv-a: n=18). The additional loss of Nf1 in the triply-infected mice resulted in gliomas occurring significantly faster than the doubly-infected mice, both from the G/tv-a (Figure 7B right panel) and the N/tv-a (Figure 7B left panel) cell of origin. The expression patterns for the two populations showed that the doubly-infected mice generated gliomas with a PN gene expression pattern (Figures 4F and 7C) while the gliomas from the triply infected mice shifted toward MES (Figure 7C).
Figure 7. Additional Nf1 loss induces proneural to mesenchymal conversion in vivo.
(A) Representative H&E and immunohistochemical analysis for the indicated protein of the RCAS-PDGFA/shp53 induced-glioma secondary incorporating the RCAS-GFP-shNf1 virus in G/tv-a mice. Circles 1 and 2 represent mesenchymal and proneural tumor lesions, respectively. Scale bars, 100 µm. (B) Kaplan-Meier survival curves showing symptom-free survival of PDGFA/shp53 or PDGFA/shp53/shNf1 induced-gliomas in N/tv-a (Left panel) and G/tv-a (Right panel) mice. Tumors were generated by simultaneous injection of the relevant RCAS virus into neonatal pups brain. Survival curve of the RCAS-PDGFA/shp53 induced-tumors from Figures 4A and 4B were also shown in the figure for the comparison. (***) p = 0.0004, (****) p < 0.0001. (C) GSEA of the RCAS-PDGFA/shp53 and the RCAS-PDGFA/shp53/shNf1 induced-gliomas in N/tv-a and G/tv-a mice.
DISCUSSION
There has been much discussion about the implications of the molecular subgroups of GBM. The data presented here connects the GBM subtypes in an evolutionary framework where the primordial tumors are PN in character with the other subgroups evolving from them. Amplification of PDGFRA occurs primarily in tumors that remain PN. Mathematical modeling suggests that chr7 gain and chr10 loss are very common early genetic events in all subtypes of non-GCIMP GBM. Further, we found that the selective advantage for additional copies of chr7 is primarily, but potentially not exclusively, driven by elevated expression of PDGFA, and a primary driver for loss of chr10 is reduced expression of PTEN. These predictions were supported by mouse modeling data indicating that overexpression of PDGFA was sufficient to induce gliomas that were enhanced by PTEN loss to give rise to GBM with PN character. Note that chr7 encodes many other genes with elevated copy number by the gain of chr7 that may contribute to oncogenesis and therapeutic resistance. Similarly, there could be genes on chr10 additional to PTEN whose loss enhances oncogenesis or alters therapeutic response early in GBM evolution.
Loss of chr10 that occurs in nearly all non-GCIMP GBMs is represented by one copy loss. Therefore it is intriguing to know if loss of function of the remaining allele is essential in non-GCIMP GBM formation. Non-silent somatic point mutation in the remaining PTEN allele is found in 37.9% of all cases with single copy of chr10 and homozygous deletion was identified in 10% of non-GCIMP GBMs. Given that PTEN expression is also regulated by promoter methylation and miRNA, the remaining PTEN allele might be also inactivated by alternative mechanisms in non-GCIMP GBMs (Baeza et al., 2003; Huse et al., 2009; Wiencke et al., 2007). Further, the level of PTEN mRNA expression decreased from diploid to hemi- and homozygous loss in non-GCIMP GBMs. In mice, heterozygous loss of Pten was sufficient to induce glioma in combination with Tp53 loss, with or without Nf1 loss and the homozygous loss increased the aggressiveness of tumors (Kwon et al., 2008; Zheng et al., 2008). Thus heterozygous loss of PTEN appears enough for initiating an abrogation of gene function in GBM patients.
Mouse models can demonstrate that specific cell types, gene combinations and sequences of events are sufficient to form tumors, but they alone do not prove that human tumors form that way. The mathematical analysis of human GBM data presented here indicates that a specific set and order of gene abnormalities represents the major evolutionary pathway towards non-GCIMP GBM. The mouse modeling shows that this specific predicted series of events is sufficient to form PN-like GBM in mice, and that this character can be shifted to a MES one with subsequent loss of Nf1, predicted to be a late event in the evolution of MES-GBM in humans. Further, given the highly similar copy number patterns for most non-GCIMP GBM subtypes, it is likely that most non-GCIMP GBM subtypes arise similarly.
Our studies suggest that cell(s) harboring both chr7 gain and chr10 loss are the most likely cell of origin for gliomas. However, given the existence of a few cases of GBM in the TCGA dataset without copy number alterations of both chromosomes, there clearly are exceptions. Indeed, as shown by our Nf1/Tp53 loss model, it is possible that MES-GBMs could arise de novo via a different cell of origin and evolutionary pathway. Furthermore, the relatively rare patients without chr10 loss have significantly more whole chromosome 12 gains (FDR < 0.01) and patients without chr7 gain have significantly higher odds of PDGFRA somatic mutation (p = 0.05, Fisher’s exact). This might further indicate that pathways affected by chr7 gain and chr10 loss can be interrupted with similar consequence by alterations on other chromosomes. None-the-less, the majority of non-GCIMP GBMs is abnormal for chr7 and chr10 and most likely arises from a PN-like precursor lesion.
This series of molecular events early in the development of GBM is different from that seen in other cancer types induced by point mutations or translocations with the generation of a single oncogene in an otherwise normal background, such as CML (Hehlmann et al., 2007). The acquisition of multiple pro-oncogenic alterations by copy number gain and loss at the outset of tumor formation would reduce the likelihood of success for targeted therapy against any specific signaling targets, especially those acquired late in the tumor’s evolution. The data argue that PDGFA is the gene on chr7 that most likely provides the strongest selection for initial gain of chr7 at the beginning of non-GCIMP GBM formation. However, other genes on chr7 such as CUL1, CBLL1, ASNS, ARPC1A, and GLI3 might have similar but less robust effects and could collectively trump PDGFA expression later in GBM evolution. It remains to be elucidated how our findings relate to other tumor types that also show gains of large portions of chr7 and loss of chr10.
The chromosomal abnormalities early during GBM evolution are interesting in light of the observation that aneuploidy for at least one chromosome occurs in as many as 10% of all cells in the normal adult brain (Iourov et al., 2006; Iourov et al., 2009) and is even higher during development, reaching 30–35% (Bushman and Chun, 2013; Yurov et al., 2007). This higher prevalence in the brain than in other organs has been suggested to contribute to neuronal diversity (Bushman and Chun, 2013). However, from this diverse set of errors in chromosomal number throughout the normal brain, only those cells with loss of one copy of chr10 and gain of variable numbers of chr7 give rise to non-GCIMP GBMs. Given the prevalence of aneuploid cells in the normal brain, it is likely that many cells with both gain of chr7 and loss of chr10 exist in each individual. Therefore, other mechanisms may contribute to the fact that GBMs are not more prevalent. If aneuploidy in the CNS functions to generate neuronal diversity, we might speculate that the risk of developing GBM may be one price paid for complex CNS function.
EXPERIMENTAL PROCEDURES
Datasets of human GBM samples
We obtained segmented copy-number data, raw and normalized mRNA expression data, methylation data and somatic mutation data from the TCGA (The Cancer Genome Atlas Research Network, 2008). All analyses were done in R/Bioconductor (Gentleman et al., 2004). Detail of all bioinformatic analyses is described in Supplemental Experimental Procedures.
Phylogenetic analysis
To examine the relationship between GBM subgroups, we applied the Neighbor-Joining (NJ) algorithm (Saitou and Nei, 1987) independently to the TCGA GBM Agilent expression, SNP copy number, promoter methylation and somatic mutation data as previously described (Riester et al., 2010).
RESIC analysis
To determine the temporal sequence of genetic alterations, two whole chromosome events, chr7 non-disjunction and chr10 loss, as well as subtype-defining events (focal EGFR gain and amplification, PDGFRA gain and amplification, and NF1 loss), we applied the RESIC algorithm to the GBM dataset as previously described (Attolini et al., 2010; Cheng et al., 2012).
Pathway analysis
To prioritize genes in large copy number alterations, we developed a computational method. The algorithm identifies genes from (i) disease-related pathways for which (ii) copy number gain has profound impact on mRNA levels.
Transcription factor analysis
To examine whether NF1 loss had an impact on expression of certain TFs, Gene Set Enrichment Analysis (GSEA) was performed with MSigDB 4.0, which provides 615 putative TF/target gene sets (c3 category) (Subramanian et al., 2005).
Gene expression analysis for murine brain tumors, neurosphere and human GBM cell lines
Total RNAs were labeled by the Illumina protocol followed by hybridization to the BeadChip (Illumina, Inc. San Diego, CA). Raw data were processed at Genomics Core Lab of MSKCC and then normalized with the lumi R package (Du et al., 2008). For the GSEA, the Verhaak et al subtype signatures were used (Verhaak et al., 2010).
Vector constructs
The human PDGFA cDNA was purchased from OriGene Technologies, Inc. and was subcloned into RCAS-Y vector (Dunn et al., 2000). For the generation of RCAS-shRNA vector, shRNAs were initially assembled in pSUPER.retro.puro vector (OligoEngine) or pENTR-mRFP-H1 vector. The shRNAs containing H1 promoter were PCR-amplified and then inserted into the RCAS-Y vector. For the pENTR-mRFP-H1 vector, LR recombination was used to transfer the shRNA containing H1 and mRFP into RCAS-Destination vector (DV) (Loftus et al., 2001). See also Supplemental Experimental Procedures.
Generation of murine brain tumors
All animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committees of MSKCC and FHCRC and followed NIH guidelines for animal welfare. The RCAS/tv-a system used in this work has been described previously (Holland et al., 2000; Holland et al., 1998; Holland and Varmus, 1998; Hu et al., 2005; Uhrbom et al., 2004). GFAP (G)/tv-a, G/tv-a;Ptenfl/fl, G/tv-a+/−;Tg-Ef luc, Nestin (N)/tv-a (agouti) or N/tv-a;Cdkn2a(Ink4a-Arf)−/−;Ptenfl/fl mice were used for the RCAS-mediated gliomagenesis in this study. After injection of the relevant RCAS virus, mice were observed until they developed brain tumor related-symptoms unless there was a particular note. See also Supplemental Experimental Procedures.
H&E staining and immunohistochemistry
Mouse brains were prepared for paraffin and sectioning and stained with hematoxylin and eosin as described previously (Hambardzumyan et al., 2009; Holland et al., 2000). Briefly, immunohistochemical staining was performed with an automated staining processor (Hambardzumyan et al., 2008). The antibodies used in this study are described in Supplemental Experimental Procedures. The histological diagnosis and tumor grade was established based on the World Health Organization (WHO) criteria (Huse et al., 2009; Louis et al., 2007).
Western blot analysis
Cells were cultured, lysed with NF1 lysis buffer (Yunoue et al., 2003) and processed for Western blotting by standard methods. The antibodies used in this study were described in Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Most non-GCIMP GBMs evolve from a PN-like precursor glioma.
The initiating events for GBM are chromosomal non-disjunction events.
The primary drivers of these arm-level genomic events are PDGFA and PTEN.
SIGNIFICANCE.
Deep molecular analyses have divided non-GCIMP GBMs into four classes but the relationship between these subtypes is unknown. Here we combined computational and mathematical analyses of human GBM data with mouse modeling and found that these tumors appear to be initiated by broad genomic alterations driven partly by increased PDGFA expression and PTEN loss, and evolve from a common glioma precursor that is lethal and proneural in character. The acquisition of secondary molecular subtype characteristics by EGFR and PDGFRA amplification or NF1 loss is achieved late in GBM evolution. Our findings provide insight into the natural history of GBMs and therapeutic strategies targeting these late events.
ACKNOWLEDGEMENTS
We thank Robert ‘Jim’ Finney and Qunchao Zhang for technical assistance, Amanda Conlon, Christine Ferreira, Jenny Zhang, Raquel Sanchez and Desert Horse-Grant for secretarial assistance, Jeffrey Zhao, Maryam Hassimi and Agnes Viale for microarray analysis, members of Holland and Michor labs, Dr. Armida Fabius, Dr. Shigehisa Kitano, Dr. Atsushi Kawaguchi, Dr. Tatsuyuki Kakuma and Dr. Mitsutoshi Nakada for helpful discussions. The results in this article are analyzed using data generated by the TCGA project (http://cancergenome.nih.gov). We thank Dr. Hideyuki Saya and Dr. Shinji Kuninaka, Keio University for pSUPER-GL2, Nf1 and p53 shRNA constructs, to Dr. Cameron Brennan for human GBM cell lines. This work was supported by 2009 and 2010 Brain Tumor Center Grant, MSKCC to T.O., NIH grants U54CA163167-01, U01CA141502-01, and RO1CA100688to E.C.H., 2009 and 2010 and U54CA143798 to F.M. and E.C.H.
Abbreviations
- Chr
Chromosome
- CL
Classical
- CNS
Central nervous system
- GBM
Glioblastoma
- GCIMP
glioma-CpG island methylator phenotype
- GSEA
gene set enrichment analysis
- MES
mesenchymal
- NL
Neural
- NF1
Neurofibromatosis 1
- PN
proneural
- RESIC
Retracing Evolutionary Steps In Cancer
- shRNA
short hairpin RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All microarray data was deposited in Gene Expression Omnibus (GSE45874).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and seven tables and can be found with this article online at XXXX.
REFERENCES
- Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attolini CS, Cheng YK, Beroukhim R, Getz G, Abdel-Wahab O, Levine RL, Mellinghoff IK, Michor F. A mathematical framework to determine the temporal sequence of somatic genetic events in cancer. Proc Natl Acad Sci U S A. 2010;107:17604–17609. doi: 10.1073/pnas.1009117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta neuropathologica. 2003;106:479–485. doi: 10.1007/s00401-003-0748-4. [DOI] [PubMed] [Google Scholar]
- Beerenwinkel N, Sullivant S. Markov models for accumulating mutations. Biometrika. 2009;96:645–861. [Google Scholar]
- Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al. Mesenchymal Differentiation Mediated by NF-kappaB Promotes Radiation Resistance in Glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C, Momota H, Hambardzumyan D, Ozawa T, Tandon A, Pedraza A, Holland E. Glioblastoma subclasses can be defined by activity among signal transduction pathways and associated genomic alterations. PLoS One. 2009;4:e7752. doi: 10.1371/journal.pone.0007752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman DM, Chun J. The genomically mosaic brain: Aneuploidy and more in neural diversity and disease. Seminars in cell & developmental biology. 2013 doi: 10.1016/j.semcdb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YK, Beroukhim R, Levine RL, Mellinghoff IK, Holland EC, Michor F. A Mathematical Methodology for Determining the Temporal Order of Pathway Alterations Arising during Gliomagenesis. PLoS Comput Biol. 2012;8:e1002337. doi: 10.1371/journal.pcbi.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Celestino JC, Okada Y, Louis DN, Fuller GN, Holland EC. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes Dev. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty JD, Fomchenko EI, Akuffo AA, Schmidt E, Helmy KY, Bazzoli E, Brennan CW, Holland EC, Milosevic A. Candidate pathways for promoting differentiation or quiescence of oligodendrocyte progenitor-like cells in glioma. Cancer Res. 2012;72:4856–4868. doi: 10.1158/0008-5472.CAN-11-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dunn KJ, Williams BO, Li Y, Pavan WJ. Neural crest-directed gene transfer demonstrates Wnt1 role in melanocyte expansion and differentiation during mouse development. Proc Natl Acad Sci U S A. 2000;97:10050–10055. doi: 10.1073/pnas.97.18.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fomchenko EI, Dougherty JD, Helmy KY, Katz AM, Pietras A, Brennan C, Huse JT, Milosevic A, Holland EC. Recruited cells can become transformed and overtake PDGF-induced murine gliomas in vivo during tumor progression. PLoS One. 2011;6:e20605. doi: 10.1371/journal.pone.0020605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Amankulor NM, Helmy KY, Becher OJ, Holland EC. Modeling Adult Gliomas Using RCAS/t-va Technology. Transl Oncol. 2009;2:89–95. doi: 10.1593/tlo.09100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22:436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehlmann R, Hochhaus A, Baccarani M. Chronic myeloid leukaemia. Lancet. 2007;370:342–350. doi: 10.1016/S0140-6736(07)61165-9. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- Helmy K, Halliday J, Fomchenko E, Setty M, Pitter K, Hafemeister C, Holland EC. Identification of Global Alteration of Translational Regulation in Glioma In Vivo. PLoS One. 2012;7:e46965. doi: 10.1371/journal.pone.0046965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M, Nister M, Betsholtz C, Heldin CH, Westermark B, Funa K. Endothelial cell hyperplasia in human glioblastoma: coexpression of mRNA for platelet-derived growth factor (PDGF) B chain and PDGF receptor suggests autocrine growth stimulation. Proc Natl Acad Sci U S A. 1988;85:7748–7752. doi: 10.1073/pnas.85.20.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25:55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev. 1998;12:3675–3685. doi: 10.1101/gad.12.23.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Varmus HE. Basic fibroblast growth factor induces cell migration and proliferation after glia-specific gene transfer in mice. Proc Natl Acad Sci U S A. 1998;95:1218–1223. doi: 10.1073/pnas.95.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Pandolfi PP, Li Y, Koutcher JA, Rosenblum M, Holland EC. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma. Neoplasia. 2005;7:356–368. doi: 10.1593/neo.04595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iourov IY, Liehr T, Vorsanova SG, Kolotii AD, Yurov YB. Visualization of interphase chromosomes in postmitotic cells of the human brain by multicolour banding (MCB) Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2006;14:223–229. doi: 10.1007/s10577-006-1037-6. [DOI] [PubMed] [Google Scholar]
- Iourov IY, Vorsanova SG, Liehr T, Yurov YB. Aneuploidy in the normal, Alzheimer’s disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiology of disease. 2009;34:212–220. doi: 10.1016/j.nbd.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhao D, Chen J, Alcantara S, Li Y, Burns DK, Mason RP, Lee EY, Wu H, Parada LF. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Larson DM, Watkins-Chow D, Church DM, Pavan WJ. Generation of RCAS vectors useful for functional genomic analyses. DNA research : an international journal for rapid publication of reports on genes and genomes. 2001;8:221–226. doi: 10.1093/dnares/8.5.221. [DOI] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta neuropathologica. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I, Hedren A, Sjodin H, Orrego A, Andrae J, Afink GB, Nister M, Lindstrom MS. Brain abnormalities and glioma-like lesions in mice overexpressing the long isoform of PDGF-A in astrocytic cells. PLoS One. 2011;6:e18303. doi: 10.1371/journal.pone.0018303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. The Definition of Primary and Secondary Glioblastoma. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- Ozawa T, Brennan CW, Wang L, Squatrito M, Sasayama T, Nakada M, Huse JT, Pedraza A, Utsuki S, Yasui Y, et al. PDGFRA gene rearrangements are frequent genetic events in PDGFRA-amplified glioblastomas. Genes Dev. 2010;24:2205–2218. doi: 10.1101/gad.1972310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- Riester M, Stephan-Otto Attolini C, Downey RJ, Singer S, Michor F. A differentiation-based phylogeny of cancer subtypes. PLoS Comput Biol. 2010;6:e1000777. doi: 10.1371/journal.pcbi.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shih AH, Dai C, Hu X, Rosenblum MK, Koutcher JA, Holland EC. Dose-dependent effects of platelet-derived growth factor-B on glial tumorigenesis. Cancer Res. 2004;64:4783–4789. doi: 10.1158/0008-5472.CAN-03-3831. [DOI] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Brennan CW, Helmy K, Huse JT, Petrini JH, Holland EC. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18:619–629. doi: 10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrbom L, Nerio E, Holland EC. Dissecting tumor maintenance requirements using bioluminescence imaging of cell proliferation in a mouse glioma model. Nat Med. 2004;10:1257–1260. doi: 10.1038/nm1120. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiencke JK, Zheng S, Jelluma N, Tihan T, Vandenberg S, Tamguney T, Baumber R, Parsons R, Lamborn KR, Berger MS, et al. Methylation of the PTEN promoter defines low-grade gliomas and secondary glioblastoma. Neuro Oncol. 2007;9:271–279. doi: 10.1215/15228517-2007-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunoue S, Tokuo H, Fukunaga K, Feng L, Ozawa T, Nishi T, Kikuchi A, Hattori S, Kuratsu J, Saya H, Araki N. Neurofibromatosis type I tumor suppressor neurofibromin regulates neuronal differentiation via its GTPase-activating protein function toward Ras. J Biol Chem. 2003;278:26958–26969. doi: 10.1074/jbc.M209413200. [DOI] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;2:e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, Charest A. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci U S A. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.