Abstract
Objectives
HIV-infected controllers have provided novel insights into mechanisms of viral control. We investigated the degree to which HIV DNA and RNA are present in gut-associated lymphoid tissue (GALT) of controllers.
Design
Cross-sectional cohort study.
Methods
Colorectal biopsy pieces were obtained from 5 untreated non-controllers, 5 ART-suppressed subjects, and 9 untreated controllers.
Results
Rectal HIV DNA was lower in controllers (median 496 copies/106 CD4+ T cells) than in untreated non-controllers (117483 copies/106 CD4+ T cells, p=0.001) and ART-suppressed subjects (6116 copies/106 CD4+ T cells, p=0.004). Similarly, rectal HIV RNA was lower in controllers (19 copies/106 CD4+ T cells) than in non-controllers (15210 copies/106 CD4+ T cells, p=0.001) and ART-suppressed subjects (1625 copies/106 CD4+ T cells, p=0.0599). Rectal HIV RNA/DNA ratios were not statistically different between the 3 groups.
Conclusions
Despite being able to maintain very low plasma HIV RNA levels in the absence of antiretroviral therapy, HIV-infected controllers have readily measurable levels of HIV DNA and RNA in GALT. As expected, controllers had lower rectal HIV DNA and RNA compared to untreated non-controllers and ART-suppressed individuals. Compared to the mechanisms of “natural” viral control of controllers, long-term antiretroviral therapy does not reduce the total HIV reservoir to the level of controllers.
Keywords: HIV, controllers, gut-associated lymphoid tissue, GALT, viral reservoir
Introduction
Antiretroviral therapy (ART) decreases HIV-associated morbidity and mortality but does not completely restore health [1, 2]. A small proportion of HIV-infected individuals (“controllers”) are able to maintain low plasma viremia in the absence of ART [3-5]. They present a unique opportunity to better understand HIV persistence and viral control. Multiple studies have examined the potential virologic and host factors associated with viral control [6-9]. We and others have previously shown that: (1) most controllers have detectable plasma viremia and cell-associated RNA and DNA in peripheral blood mononuclear cells (PBMCs) if ultrasensitive assays are used [10-12]; and (2) a significant proportion of controllers are infected with replication-competent virus [13, 14] and not with virus that contains significant genetic defects [15]. However, these studies have thus far been limited to measurements in blood. Given the increasing recognition that the interactions between the host and virus during low-level viremic states are more apparent in tissues than in blood [16-19], we measured HIV DNA and RNA in gut-associated lymphoid tissue (GALT) of controllers, and compared these measurements to those in untreated and ART-suppressed non-controllers.
Methods
Subjects were identified from the University of California San Francisco (UCSF) SCOPE cohort. Colorectal biopsies were obtained from 5 untreated non-controllers (plasma RNA >10,000 copies/mL), 5 ART-suppressed non-controllers (plasma RNA <40 copies/mL for ≥12 months), and 9 untreated controllers (plasma RNA ≤1000 copies/mL for ≥12 months). All subjects provided written informed consent. This study was approved by the UCSF Committee on Human Research.
For each subject, 30 colorectal biopsy specimens were obtained 10-20cm from the anal verge using 3mm jumbo forceps. Eighteen to 24 biopsy pieces were placed directly into 10mL RPMI-1640 media containing fetal calf serum (15%), penicillin (100U/mL), streptomycin (100ug/mL), and L-glutamine (2mM). Biopsy pieces were dissociated to a cell suspension by collagenase digestion and mechanical disruption [20]. One aliquot of cells was set aside for flow cytometry and stained with CD45-FITC, CD3-APC and CD4-PE (BD biosciences) for 15min at 25C. Propidium iodide was added to stain non-viable cells and samples were run on an Accuri C6; the total number of viable mononuclear cells, and the proportion and absolute number of viable CD45+ leukocytes and CD4+ T cells was determined. Another aliquot of cells was frozen at -80C for subsequent nucleic acid extraction.
Total DNA was extracted from rectal cells using Trireagent (BD Bioscience) and further purified using the QIAgen Pure Gene kit. DNA concentrations and purity were assessed using a ND-1000 Spectrophotometer (NanoDrop). Three replicates of up to 500ng of DNA were assayed for HIV DNA using a modification of a published TaqMan PCR assay that uses primers (HXB2 positions 522-543, 626-643) and probe (559-584) from the LTR region [21]. Reaction volume was 50uL with 10pmol of each primer, 10pmol of probe, and 25uL of 2x TaqMan Gene Expression Master Mix (Applied Biosystems). Cycling conditions were: 50C for 2min, 95C for 10min, then 60 cycles of 95C for 15sec and 59C for 1min. External standards were prepared from DNA extracted from known numbers of 8E5 cells (NIH AIDS Reagent Program). HIV DNA copy numbers were normalized to cellular input into the PCR, as determined by DNA mass (assuming 1ug total DNA corresponds to 160,000 cells). Results were further normalized by the percent of all cells that were CD3+CD4+ by flow cytometry and expressed as copies/106 CD4+T cells.
Total RNA was extracted from rectal cells using Trireagent (BD Bioscience), treated with DNase (2.5uL RNase-free DNase [QIAgen] and 10uL buffer RDD in a total of 100uL for 15min at 25C), and purified with the QIAgen RNeasy protocol with minor modifications (precipitation with 700uL of 100% EtOH and washing with RPE). RNA concentrations and purity were assessed using a ND-1000 Spectrophotometer. Three replicates of up to 500ng of RNA were assayed for total processive HIV RNA transcripts using primers and probe from the LTR region (as above). Reaction volume was 50uL with 10pmol of each primer, 10pmol of probe, 25uL of TaqMan RNA-to-Ct 1-Step mix (Applied Biosystems), and 1.25uL of 40x RT. Cycling conditions were: 48C for 20min, 95C for 5min, then 60 cycles of 95C for 15sec and 59C for 1min. Genomic HIV RNA standards were prepared from lab stocks of NL4-3 virions by extracting RNA and quantifying HIV RNA via replicate measurements using the Abbot Real Time assay. HIV RNA copy numbers were normalized to cellular input into the PCR, as determined by RNA mass (assuming that 1ng RNA correspond to 1000 cells [22]), which has been shown to correlate with levels of GAPDH RNA [17]. Results were further normalized by the percent of all cells that were CD3+CD4+ by flow cytometry and expressed as copies/106 CD4+T cells.
All statistical analyses were conducted with GraphPad Prism version 5.04. Virologic parameters were compared between unmatched groups using the Wilcoxon rank sum test.
Results
The median plasma RNA was 3.3×104 copies/mL for untreated non-controllers, <40 copies/mL for ART-suppressed non-controllers (median duration of viral suppression 8.6 years), and 58 copies/mL for controllers (Table 1).
Table 1. Baseline characteristics.
| Untreated Non-controllers | ART-suppressed Non-controllers | Controllers | |
|---|---|---|---|
|
| |||
| (n=5) | (n=5) | (n=9) | |
| ART | No | Yes | No |
| Plasma HIV RNA (copies/mL) | 32,997 (11,660 - 235,492) | <40 (<40 - <40) | 58 (40 - 95) |
| CD4+ T cell count (cells/mm3) | 424 (414 - 487) | 660 (565 - 785) | 708 (528 - 872) |
| Nadir CD4+ T cell count (cells/mm3) | 382 (372 - 400) | 270 (50 - 416) | 486 (379 - 506) |
| Age (years) | 42 (40 - 44) | 58 (52 - 62) | 51 (48 - 55) |
| Gender (% male) | 100% | 100% | 89% |
Data represent medians and interquartile ranges (IQR). ART=antiretroviral therapy.
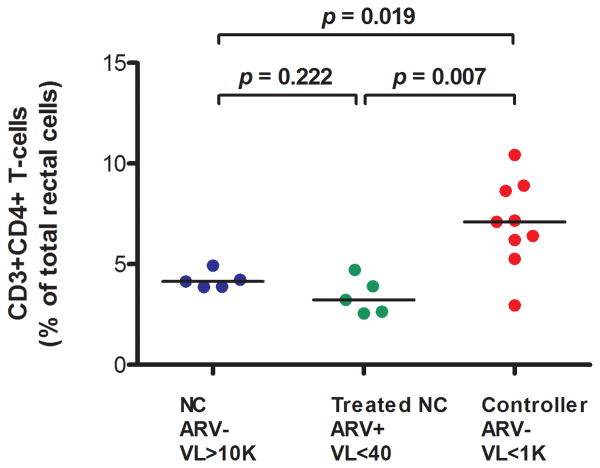
Rectal CD4+ T cell content (percent of total rectal cells) was higher in controllers (median 7.1%) than in untreated non-controllers (4.1%, p=0.019) or ART-suppressed subjects (4.3%, p=0.007) (Figure 1A).
Figure 1A. Rectal CD4+ T cell content (percent of total rectal cells).
ARV=antiretroviral therapy. VL=plasma HIV RNA (copies/mL). NC=Non-controller.
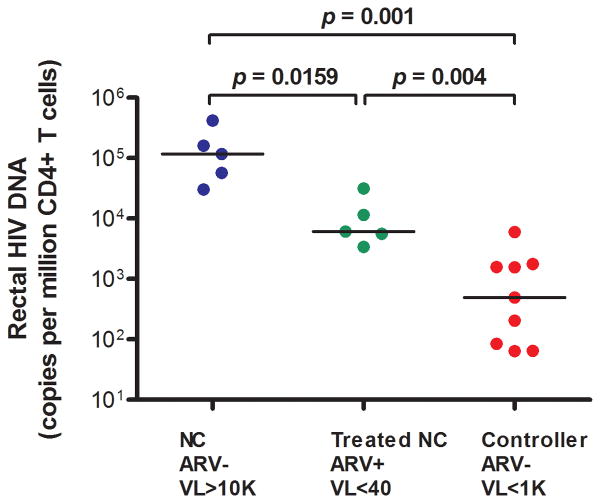
Rectal HIV DNA/106 rectal cells (“tissue burden”) was lower in controllers (median 44 copies/106 rectal cells) than in untreated non-controllers (4,558 copies/106 rectal cells; p=0.0033) or ART-suppressed subjects (254 copies/106 rectal cells, p=0.0112). Similarly, when normalized to 106 CD4+ T cells (“HIV per CD4+ T cell”), rectal HIV DNA was lower in controllers (median 496 copies/106 CD4+ T cells) than in untreated non-controllers (117,483 copies/106 CD4+ T cells, p=0.001) or ART-suppressed subjects (6,116 copies/106 CD4+ T cells, p=0.004) (Figure 1B).
Figure 1B. Rectal HIV DNA (copies/million CD4+ T cells).
ARV=antiretroviral therapy. VL=plasma HIV RNA (copies/mL). NC=Non-controller.
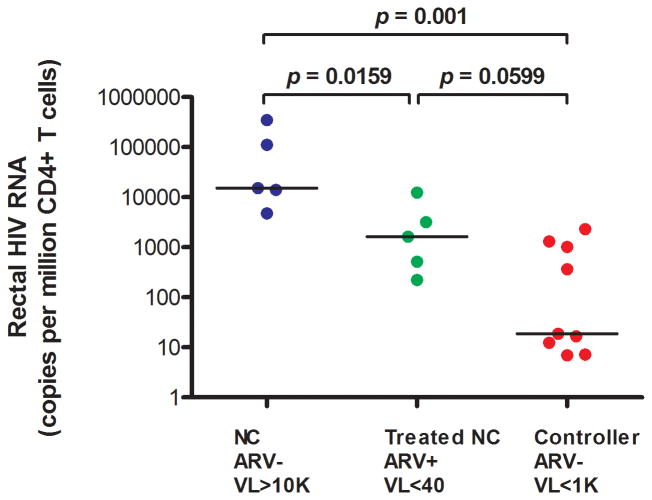
Rectal HIV RNA/106 rectal cells was lower in controllers (median 2 copies/106 rectal cells) than in untreated non-controllers (694 copies/106 rectal cells, p=0.001), but the difference between controllers and ART-suppressed subjects (70 copies/106 rectal cells) did not reach statistical significance (p=0.1119). However, when normalized to 106 CD4+ T cells, rectal HIV RNA was lower in controllers (median 19 copies/106 CD4+ T cells) than in non-controllers (15,210 copies/106 CD4+ T cells, p=0.001) or ART-suppressed subjects (1,625 copies/106 CD4+ T cells, p=0.0599) (Figure 1C).
Figure 1C. Rectal HIV RNA (copies/million CD4+ T cells).
ARV=antiretroviral therapy. VL=plasma HIV RNA (copies/mL). NC=Non-controller.
Rectal HIV RNA/DNA ratios (a measure of average transcription per infected cell) were not statistically different between the 3 groups (median 0.19 controllers, 0.25 untreated non-controllers, and 0.29 ART-suppressed subjects).
Discussion
Given the potential limitations of ART [1, 2, 23], there is a growing interest in developing curative approaches in which viral control is maintained in the absence of any therapy. HIV-infected controllers may prove to be an informative model for developing such strategies. We performed extensive virologic measurements in a cohort of controllers, focusing for the first time on GALT, where much of the viral reservoir is presumed to reside.
First, we observed that untreated non-controllers have an extremely high burden of HIV DNA in the rectum, corresponding to an average of one copy for every 10 CD4+ T cells. This calculation, which assumes that most or all of the HIV DNA is in CD4+ T cells and is evenly distributed, should be verified in sorted and terminally diluted CD4+ T cells. However, if this approximation is true, it suggests that in most untreated non-controllers, a large proportion of CD4+ T cells in the rectum may be infected with HIV.
Second, we observed that HIV-infected controllers have readily measurable levels of HIV DNA and RNA in the rectum, despite being able to maintain very low levels of plasma RNA in the absence of ART. As expected, controllers had higher rectal CD4+ T cell numbers and lower rectal HIV DNA and RNA levels compared to untreated non-controllers. However, we did not detect a difference in HIV RNA/DNA ratios. These data suggest that in controllers, the mechanisms of viral control result in a lower total frequency of HIV-infected cells but may not reduce the average HIV transcription rate per infected cell.
Controllers also had lower rectal HIV DNA and RNA levels compared to ART-suppressed non-controllers, but HIV RNA/DNA ratios were similar between the two groups. These data suggest that compared to the mechanisms responsible for “natural” host-mediated control of viral replication, long-term ART does not reduce the total HIV reservoir to the level of controllers. Strong, polyfunctional mucosal responses are at least partially responsible for the ability of controllers to limit the total HIV reservoir size [24, 25].
Finally, it was notable that the degree of heterogeneity in rectal HIV DNA and RNA measurements appeared to be greater in controllers, compared to untreated non-controllers or ART-suppressed subjects. This is consistent with previous immunologic/virologic studies of controllers. Although grouped into the same phenotypic group based upon suppression of plasma viremia to low levels, multiple studies have shown that controllers are in fact a rather heterogeneous group. For example, although controllers are enriched for several HLA class I alleles, not all controllers have protective HLA alleles [7, 8]. Similarly, replication-competent virus has been recovered from a significant proportion [13, 14], but not all, HIV-infected controllers [26]. The observed heterogeneity in rectal HIV DNA and RNA measurements undoubtedly reflects that multiple factors are contributing to “natural” viral control.
Our study has several limitations, including a relatively small sample size and variability in duration of HIV infection, immunodeficiency, and ART regimen for ART-suppressed subjects. Mucosal sampling was limited to the colorectum, and HIV measurements were limited to unsorted cells. Future studies should characterize the viral reservoir in controllers in other important locations within the gut (including ileum) [17] and other lymphoid tissues (lymph nodes), and should describe the relationship between reservoir size and immunologic correlates. Finally, our HIV assays measured total HIV DNA and RNA and did not allow us to distinguish between unintegrated and integrated forms of DNA, or between genomic and messenger RNA. Recent reports have shown that controllers have large excesses of unintegrated HIV DNA and 2-LTR circles in PBMCs, suggesting that they may have an intrinsic ability to block HIV integration [27, 28].
Despite the apparent ability of controllers to limit the spread of infection (as measured by rectal HIV DNA), the paradoxically high relative level of transcription (as demonstrated by HIV RNA/DNA ratios that were similar to untreated non-controllers) suggests that controllers may have productive infection and/or ongoing viral replication that could theoretically be reduced by ART. We have previously shown that controllers have higher levels of immune activation compared to ART-suppressed subjects [29], some controllers with high levels of immune activation progress immunologically to AIDS despite maintenance of virologic control [29], and controllers have higher levels of atherosclerosis compared to HIV-negative subjects [30]. Thus, the ability to control HIV to low levels may be incomplete and/or dissociated from control of immune activation or end-organ damage. Prospective treatment studies are currently underway by our group to define the role of viral replication and the virologic and immunologic effects of ART in these individuals.
Acknowledgments
HH conceived and designed the study, recruited and enrolled study subjects, conducted statistical analyses, and wrote the manuscript. MS and PWH obtained gut biopsy samples and edited the manuscript. ES processed gut biopsy samples and edited the manuscript. KH, LG, MC, and RH recruited study subjects. JNM and SGD provided conceptual advice and edited the manuscript. JKW and SAY performed HIV RNA and DNA analyses on gut biopsy samples and edited the manuscript.
Source of Funding: HH and SGD have received research support from Merck, Inc., and Gilead, Inc. This work was supported by grants from the National Institute of Allergy and Infectious Diseases (R01 AI087145, K23 AI075985, K24 AI069994, R56 AI091573, R01 NS051132), the National Cancer Institute (K23 CA157929), the Delaney AIDS Research Enterprise (DARE; U19 AI0961090), California HIV/AIDS Research Program (grant number ID08-SF-004), American Foundation for AIDS Research (106710-40-RGRL), the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763), the UCSF Clinical and Translational Research Institute Clinical Research Center (UL1 RR024131), the Center for AIDS Prevention Studies (P30 MH62246), the CFAR Network of Integrated Systems (R24 AI067039), and the U.S. Department of Veterans Affairs (1 IK2 CX000520-01, 5101BX001048). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
MS, ES, KH, LG, MC, RH, PWH, JNM, JKW, SAY: No conflicts.
This study was presented in part at the 19th Conference on Retroviruses and Opportunistic Infections, March 2012, Seattle, Washington, USA (abstract #362).
References
- 1.Phillips AN, Neaton J, Lundgren JD. The role of HIV in serious diseases other than AIDS. Aids. 2008;22:2409–2418. doi: 10.1097/QAD.0b013e3283174636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;5:e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubert JB, Burgard M, Dussaix E, Tamalet C, Deveau C, Le Chenadec J, et al. Natural history of serum HIV-1 RNA levels in 330 patients with a known date of infection. The SEROCO Study Group Aids. 2000;14:123–131. doi: 10.1097/00002030-200001280-00007. [DOI] [PubMed] [Google Scholar]
- 4.Lambotte O, Boufassa F, Madec Y, Nguyen A, Goujard C, Meyer L, et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005;41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 5.Lefrere JJ, Mariotti M, Morand-Joubert L, Thauvin M, Roudot-Thoraval F. Plasma human immunodeficiency virus RNA below 40 Copies/mL is rare in untreated persons even in the first years of infection. J Infect Dis. 1999;180:526–529. doi: 10.1086/314906. [DOI] [PubMed] [Google Scholar]
- 6.Emu B, Sinclair E, Favre D, Moretto WJ, Hsue P, Hoh R, et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005;79:14169–14178. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emu B, Sinclair E, Hatano H, Ferre A, Shacklett B, Martin JN, et al. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J Virol. 2008;82:5398–5407. doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. Journal of Infectious Diseases. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 9.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatano H, Delwart EL, Norris PJ, Lee TH, Dunn-Williams J, Hunt PW, et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol. 2009;83:329–335. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pereyra F, Palmer S, Miura T, Block BL, Wiegand A, Rothchild AC, et al. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J Infect Dis. 2009;200:984–990. doi: 10.1086/605446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Connell KA, Brennan TP, Bailey JR, Ray SC, Siliciano RF, Blankson JN. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol. 2010;84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J, et al. Isolation and Characterization of Replication-Competent HIV-1 from a Subset of Elite Suppressors. J Virol. 2007;81:2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamine A, Caumont-Sarcos A, Chaix ML, Saez-Cirion A, Rouzioux C, Delfraissy JF, et al. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study) Aids. 2007;21:1043–1045. doi: 10.1097/QAD.0b013e3280d5a7ac. [DOI] [PubMed] [Google Scholar]
- 15.Miura T, Brockman MA, Brumme CJ, Brumme ZL, Carlson JM, Pereyra F, et al. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J Virol. 2008;82:8422–8430. doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J Infect Dis. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 17.Yukl SA, Gianella S, Sinclair E, Epling L, Li Q, Duan L, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yukl SA, Shergill AK, McQuaid K, Gianella S, Lampiris H, Hare CB, et al. Effect of raltegravir-containing intensification on HIV burden and T-cell activation in multiple gut sites of HIV-positive adults on suppressive antiretroviral therapy. Aids. 2010;24:2451–2460. doi: 10.1097/QAD.0b013e32833ef7bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatano H, Jain V, Hunt PW, Lee TH, Sinclair E, Do TD, et al. Cell-based measures of viral persistence are associated with immune activation and Programmed Cell Death Protein 1 (PD-1)-expressing CD4+ T cells. J Infect Dis. 2012 doi: 10.1093/infdis/jis630. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shacklett BL, Yang O, Hausner MA, Elliott J, Hultin L, Price C, et al. Optimization of methods to assess human mucosal T-cell responses to HIV infection. J Immunol Methods. 2003;279:17–31. doi: 10.1016/s0022-1759(03)00255-2. [DOI] [PubMed] [Google Scholar]
- 21.Kumar AM, Borodowsky I, Fernandez B, Gonzalez L, Kumar M. Human immunodeficiency virus type 1 RNA Levels in different regions of human brain: quantification using real-time reverse transcriptase-polymerase chain reaction. J Neurovirol. 2007;13:210–224. doi: 10.1080/13550280701327038. [DOI] [PubMed] [Google Scholar]
- 22.Fischer M, Huber W, Kallivroussis A, Ott P, Opravil M, Luthy R, et al. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissues. J Clin Microbiol. 1999;37:1260–1264. doi: 10.1128/jcm.37.5.1260-1264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lodwick RK, Sabin CA, Porter K, Ledergerber B, van Sighem A, Cozzi-Lepri A, et al. Death rates in HIV-positive antiretroviral-naive patients with CD4 count greater than 350 cells per microL in Europe and North America: a pooled cohort observational study. Lancet. 2010;376:340–345. doi: 10.1016/S0140-6736(10)60932-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferre AL, Hunt PW, Critchfield JW, Young DH, Morris MM, Garcia JC, et al. Mucosal immune responses to HIV-1 in elite controllers: a potential correlate of immune control. Blood. 2009;113:3978–3989. doi: 10.1182/blood-2008-10-182709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferre AL, Lemongello D, Hunt PW, Morris MM, Garcia JC, Pollard RB, et al. Immunodominant HIV-specific CD8+ T-cell responses are common to blood and gastrointestinal mucosa, and Gag-specific responses dominate in rectal mucosa of HIV controllers. J Virol. 2010;84:10354–10365. doi: 10.1128/JVI.00803-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kloosterboer N, Groeneveld PH, Jansen CA, van der Vorst TJ, Koning F, Winkel CN, et al. Natural controlled HIV infection: preserved HIV-specific immunity despite undetectable replication competent virus. Virology. 2005;339:70–80. doi: 10.1016/j.virol.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Graf EH, Mexas AM, Yu JJ, Shaheen F, Liszewski MK, Di Mascio M, et al. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog. 2011;7:e1001300. doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buzon MJ, Seiss K, Weiss R, Brass AL, Rosenberg ES, Pereyra F, et al. Inhibition of HIV-1 integration in ex vivo-infected CD4 T cells from elite controllers. J Virol. 2011;85:9646–9650. doi: 10.1128/JVI.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, et al. Relationship between T Cell Activation and CD4(+) T Cell Count in HIV-Seropositive Individuals with Undetectable Plasma HIV RNA Levels in the Absence of Therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsue P, Hunt PW, Martin JN, Schnell A, Kalapus C, Deeks SG. Role of ART, viral replication, and HIV infection in atherosclerosis. 15th Conference on Retroviruses and Opportunistic Infections; Boston, MA. 2008. [Google Scholar]