Abstract
One limitation of current biodegradable polymeric nanoparticles is their inability to effectively encapsulate and sustainably release proteins while maintaining protein bioactivity. Here we report the engineering of a PLGA-polycation nanoparticle platform with core-shell structure as a robust vector for the encapsulation and delivery of proteins and peptides. We demonstrate that the optimized nanoparticles can load high amounts of proteins (>20% of nanoparticles by weight) in aqueous solution by simple mixing via electrostatic interactions without organic solvents, forming nanospheres in seconds with diameter <200 nm. We also investigate the relationship between nanosphere size, surface charge, PLGA-polycation composition, and protein loading. The stable nanosphere complexes contain multiple PLGA-polycation nanoparticles, surrounded by large amounts of protein. This study highlights a novel nanoparticle platform and nanotechnology strategy for the delivery of proteins and other relevant molecules.
Keywords: nanoparticle, PLGA, polycation, protein delivery, structure-function
Since the 1970s, protein therapy has become a promising strategy for the effective treatment of cancers, diabetes, cardiovascular diseases, and many other disorders.1-6 However, the safe and effective delivery of proteins to the desired disease locations or intracellular sites remains a major challenge due to the intrinsic properties of most proteins, such as high molecular weight (MW), surface charges, and vulnerable tertiary structures.2 Thus specific and robust delivery vehicles are needed to facilitate the loading, delivery, and controlled release of proteins.
Numerous protein carrier platforms have been developed for better therapeutic performance. For example, microspheres7 and hydrogels3, 8 have been utilized to solve the sustained release issue. However, these systems have limited applications for intracellular protein delivery due to their large size. For effective intracellular delivery, other systems including liposomes,9 polymer conjugates,10 nanotubes,11 nanogels,12 and nanoparticles (NPs)4, 6 are being extensively investigated. Among them, biodegradable polymeric NPs have aroused great interest as potentially robust protein carriers due to their biocompatibility, biodegradability, amenability to formulation, and capability for intracellular and systemic delivery.2, 13-15 Nevertheless, the major types of proteins and protein analogues are negatively charged and have complicated, sensitive, and fragile 3-D structures and sizes ranging from ∼ 2 - 15 nm. Thus the currently available polymeric NP platforms require further optimization in order to load and encapsulate proteins with high efficiency and release them in a sustained manner while maintaining their bioactivity.2
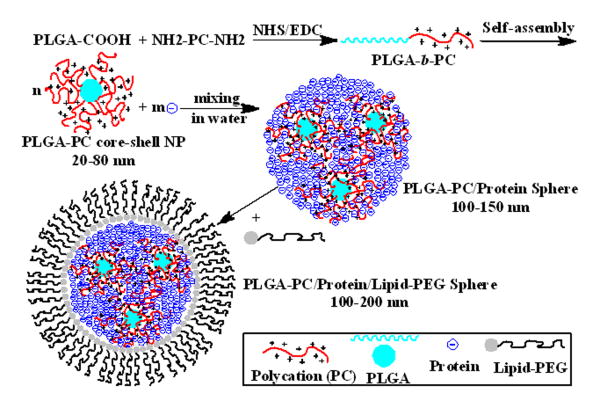
For effective protein delivery, the ideal polymeric NP platform should have: (1) high protein loading (low NP mass to achieve therapeutic dose); (2) “green” protein encapsulation: little or no organic solvents used to prevent protein denaturation; (3) minimal contact with carriers to avoid the low local pH caused by polymer degradation;16, 17 (4) sustainable and controllable protein release with low initial burst. Inspired by the electrostatic interactions between polyelectrolytes, we hypothesized that a NP system with very strong cationic moieties may be ideal to load and release a large quantity of proteins in a safe and controllable manner. A novel protein loading strategy was developed, utilizing very strong cationic NPs to physically adsorb proteins to form a new NP/protein complex nanosphere platform in aqueous solution (Figure 1).
Figure 1.
Illustration of the synthesis of PLGA-b-polycation (PC) copolymer, PLGA-PC NP, PLGA-PC/Protein NP, PLGA-PC/Protein/Lipid-PEG NP, and the multinuclear structure of NP.
To this end, a polymeric NP platform with a cationic shell was first developed from the di-block copolymer of poly(lactide-co-glycolic acid) (PLGA) and L-arginine-based polycation (PC) (Figures 1 and 2). PLGA-based NPs have been widely utilized due to their unique biocompatibility, biodegradability, small particle size, and high drug loading capacity.18-20 During formulation, the hydrophobic PLGA segments form a solid inner core and limit the NP to a small nanosize, while the hydrophilic and water-soluble PC portion forms the outer shell, creating a very strong electrical field for protein adsorption (loading). The final goal was to screen PLGA-PC NPs capable of loading large amounts of proteins (e.g., ≥ 20 wt% of NP) to form new polymer-protein complex NPs, utilizing the electrostatic interaction between proteins and PLGA-PC NPs. The ideal system would require only a simple formulation method (such as mixing in aqueous solution), while the final complex NPs would retain a regular spherical shape and size <200 nm (Figures 1 and 4). The desired structure of the new hybrid NPs would be a nucleus of PLGA-PC NPs surrounded by proteins, which primarily interact with the PC component of PLGA-PC NPs or other proteins.
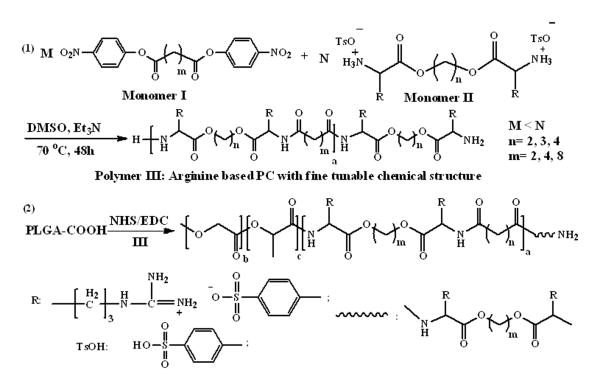
Figure 2.
Synthesis of L-arginine–based PC (1) and PLGA-b-PC copolymer (2)
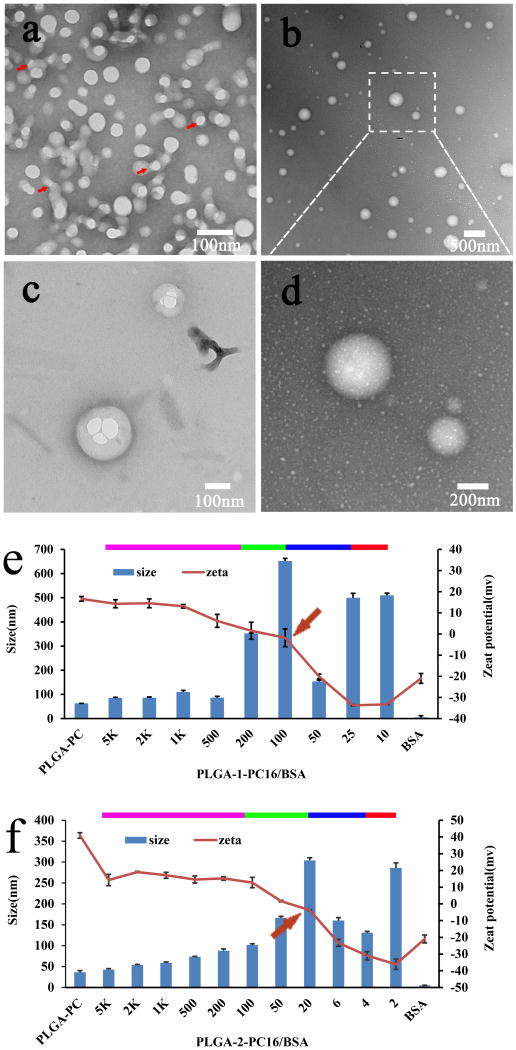
Figure 4.
TEM images of PLGA-PC/Protein nanoparticles (a-d): (a) PLGA2-PC 16 with 2 wt% BSA loading; (b-d) PLGA2-PC16 with 25 wt% BSA loading. Structure-function relationship among the zeta potential, particle size, and weight ratio of PLGA-PC/BSA complex particle (e-f)
First, a water-soluble L-arginine–based PC library (named as PCx) was developed according to a modified synthesis protocol (Figure2 and supporting information).21, 22 L-arginine–based PCs were chosen because arginine remains positively charged in neutral, acidic, and even the most basic environments due to the guanidinium group's high pKa (around 12.5), which enables the PC moiety to interact strongly with the negatively charged proteins and peptides in aqueous solutions. In Figure 2, by setting monomer molar ratios of I/II< 1.0, PCs are obtained with NH2 termini (NH2-PC-NH2), which are needed for further conjugation with PLGA-COOH. To engineer polymeric NPs, di-block copolymers of PLGA-b-PC were prepared by coupling the carboxy terminal of PLGA and the amino functionality of PC (Figure 2 and supporting information). The obtained di-block copolymers of PLGA-b-PC were named PLGAy-PCx (supporting information). Two types of PLGA-COOH with significantly different MWs were used: PLGA1 (Mn = 43.5 kDa) and PLGA2 (Mn = 5.0 kDa). The percentage of PLGA-PC cationic moiety, an important factor for NP protein loading functionality, could be altered by changing the PLGA MW; decreasing the PLGA MW would increase the cationic moiety density of the copolymer and the resulting NP.
The nanoprecipitation method was used to produce small, simple PLGA-PC NPs with the desired core (PLGA)-shell (PC) structure. The particle size and surface charge of PLGA-PC NPs are very important for the interaction with proteins (discussed in detail below). The TEM images and DLS results in Figure 3 show that the formed PLGA-PC NPs were mostly in the range of 20-80 nm, depending on the PLGA-b-PC composition. PLGA MW was the dominating factor in determining NP size: low-MW PLGA2-based PLGA-PC NPs were mainly in the size range of 20-40 nm (Figure 3a); while the high-MW PLGA1-based PLGA-PC NPs were mainly in the size range of 40-80 nm (Figure 3b). Compared to the corresponding PLGA NPs, the PLGA-PC NPs were smaller (Figure 3d). TEM images in Figures 3a and 3b demonstrate that the PLGA-PC NPs were spherical or egg shaped, while the surface morphology was fluffy due to the thick PC shell under dried state in TEM. The zeta potential of the NPs was in the range of +15 to 50 mv and it was not correlated with PLGA or PC structure or MW. The cytotoxicity of PLGA-PC NPs was evaluated via an MTT assay and the NPs were not toxic to either Hela or A549 cells in the concentration range of 1.0 ng/mL to 200 μg/mL.
Figure 3.
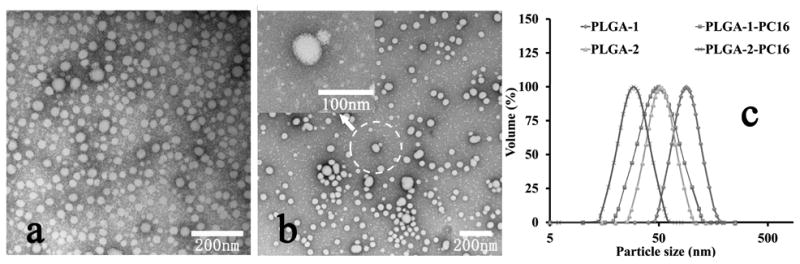
TEM images of PLGA-PC NPs (a)PLGA2-PC16; (b) PLGA1-PC16; (c) illustration of PLGA-PC composition effect (by changing PLGA portion) on nanoparticle size; (d) Particle size of PLGA-PC and PLGA NPs from DLS.
Although many strategies have been developed for loading NPs with proteins, serious challenges remain in easily and rapidly loading a high quantity of proteins into NPs of reasonably small size in a green manner, i.e., without organic solvents. The PLGA-PC NPs were designed with these goals in mind. Body charge density, not surface charge, is believed to determine the protein loading efficiency of a NP in this report. In order to better utilize the electrostatic field of the thick PC shell to strongly adsorb proteins and form new NP complexes with the desired size and protein loading capacity, a few formulation strategies were tried until a very simple method with the lowest contamination was chosen: direct mixing of aqueous solutions containing PLGA-PC NPs and proteins. Bovine serum albumin (BSA) was used as a model protein due to its medium size (MW=66.5 kDa) and suitable charge properties (isoelectric point=4.7). Before the systematic study, a quick experiment was carried out to verify this proof of concept: simply mixing random amounts of PLGA-PC NP solutions with BSA solutions. Mixing produces some changes in terms of zeta potential, particle size, and structure, depending on PLGA-PC type, weight ratio (WR) of PLGA-PC to protein, and other formulation conditions. Figure 4a shows a TEM image example of mixing PLGA2-PC16 NP with 2 wt% BSA in aqueous solution. After mixing, each PLGA-PC NP was loaded with some proteins on its surface. Some NPs connected through bound proteins, but the structure of this complex was random and irregular. We then wanted to investigate the relationships among protein loading efficiency, particle size, structure, and surface charge of the PLGA-PC NP/BSA complex. We sought to determine whether we could produce PLGA-PC/BSA complexes with uniform nanostructure, small size, and high protein loading. To systematically investigate the NP-protein interaction and the structure-function relationship, two types of PLGA-PC NPs (PLGA1-PC16 and PLGA2-PC16) were selected and they had very similar structure: the PC blocks were identical, while PLGA blocks had the same structure but significantly different MWs (43.5kDa and 5.0 kDa, respectively). Since the MW of PC16 is around 4.5 kDa, the PC composition of PLGA1-PC16 and PLGA2-PC16 NPs are about 9.4 wt% and 47.4 wt%, respectively. Therefore, the completely different PC composition (a five-fold difference) made comparison straightforward.
For each PLGA-PC NP system, various PLGA-PC/BSA hybrid complexes were formed by simply mixing aqueous solutions with a series of desired WRs of PLGA to BSA, ranging broadly from 5,000 to 2. Figures 4e and 4f show the particle size and zeta potential of PLGA-PC/BSA particles as a function of the WR of PLGA-PC NP to BSA. In Figures 4e and 4f, the zeta potential trend of can be divided into four regions. In the first region, as the WR of PLGA-PC NP to BSA decreased from 5,000 to a few hundred, the zeta potential of the complex particles also decreased, suggesting that if more proteins were mixed with PLGA-PC NPs, more of the positively charged surface of the PLGA-PC NP was surrounded or covered by negatively charged proteins via electrostatic interaction. In the second region, with more proteins added (WR from a few hundred to a few tens), almost all of the PLGA-PC NP surface was covered with proteins, and the zeta potential changed from positive to zero or slightly negative. At this stage, the corresponding WR to the 0 mv zeta potential is determined by the PC weight percentage. For the third region, a further decrease of the WR of PLGA-PC NP to protein continued to decrease the zeta potential of the complexed particle to around -30 mv. For the fourth region, the zeta potential continued to decrease or plateaued at a value slightly more negative than the zeta potential of pure BSA. The particle size trend in Figures 4e and 4f can also be divided into the same four regions. In the first region, as the WR of PLGA-PC to BSA decreased, the size of the complex particle slightly and steadily increased, suggesting that as more protein was added into the PLGA-PC NP solutions, the PLGA-PC NPs began to adsorb more proteins on their surface. In the second region, as the WR of PLGA-PC NP to protein decreased further, the particle size of PLGA-PC NP/protein complexes significantly increased beyond the expected size range, indicating some form of connections of NPs. For the largest particle size obtained in this region, the corresponding zeta potential was around zero. After the second region, however, a continuing decrease of WR in a narrow range (third region) immediately resulted in a significant reduction of complex particle size. This phenomenon suggests that the PLGA-PC NP/protein complex particle may be stable within this narrow WR range and is likely the optimal formulation. One possible reason for this stable state is that the total positive charge from PLGA-PC NPs is equal to or close to the total negative charge from BSAs. The small particle size had a corresponding zeta potential of approximately -20 mv. In this region, the PLGA2-PC16 NPs had loaded protein in the range of 15-30 wt% with encapsulation efficiency over 95%. However, for the PLGA1-PC16 NPs, the corresponding protein-loading region was in the range of 2-3 wt% due to the low PC density with similar encapsulation efficiency. In the fourth region, the continuously decreasing WR caused significant particle size increases, indicating a lack of stable complexes and heavy aggregation at higher WR of BSA to PLGA-PC NP. These results show that for each type of PLGA-PC16 NP, the maximum effective loading efficiency was achieved at a small particle size with a zeta potential range of -10 to -30mv, in a specific and narrow WR range of PLGA-PC to BSA. Figures 4e and 4f show the same trend for both size and zeta potential, but different corresponding WR windows for the specific small particle size, suggesting that this is a function of polymer structure. These relationships hold true for all other developed PLGA-PC NPs. Briefly, PLGA2-PC NPs have around 10 times the BSA loading capability of the corresponding PLGA1-PC NPs, and the highest BSA loading with small particle size always happened in a narrow WR range. When the order of addition was changed (nanoparticle solution was added into BSA), the complexed particles were larger in the 3rd region.
Based on the above findings regarding zeta potential and particle size, our findings in the third region of the PLGA2-PC system were highly interesting: the complexed particles had small sizes but maintained significant protein loading. TEM was used to study the complexed particle structure in this region for the PLGA2-PC system. To acquire better image quality and resolve more details, the formulation protocol was slightly modified (supporting information) so that NPs with larger size could be obtained without affecting structure. Figure 4b shows PLGA2-PC/BSA NPs with relatively large size at a WR of 4. It was surprising to find that all of the new NPs formed by simple mixing were uniform and spherical complexed NPs. Figure 4d shows a magnified image of two such nanospheres from Figure 4b. The structure and surface morphology of the complexed NPs were different from the pure PLGA-PC NPs in Figure 3. Based on the NP size and protein loading amount, it was predicted that multiple PLGA-PC NPs would be contained in a single complex NP (size ≥80 nm). Through TEM, the inner structure of some NP complexes was examined, confirming multiple PLGA-PC NPs inside, acting as nucleus. Figure 4c shows an example of complexed PLGA2-PC16/BSA, with the size of the internal, smaller PLGA2-PC16 NPs consistent with those in Figure 3a.
Other proteins/peptides, including ovalbumin, TNF-α, insulin, and Ac2-26 peptide, were evaluated for interaction with PLGA-PC NPs. The results and relationships were similar to those found above: complex nanoparticles were formed with PLGA2-PC16 with ≥20 wt% loading and ≤200 nm size. The MWs of proteins/peptides did not significantly affect interactions between PLGA-PC NPs and proteins. Additionally, multiple types of proteins could be loaded simultaneously without making obvious sacrifices in loading efficiency of each protein (supporting information). The in vitro and in vivo bioactivity of loaded proteins was evaluated by testing the released TNF-α and insulin, respectively. The results indicated that this loading strategy did not significantly affect the bioactivity of released proteins (supporting information). Other types of PLGA-b-PCs with similar MW and PLGA composition were developed for comparison. NPs around or <50-60 nm formed, but they could not effectively load large amounts of proteins (supporting information).
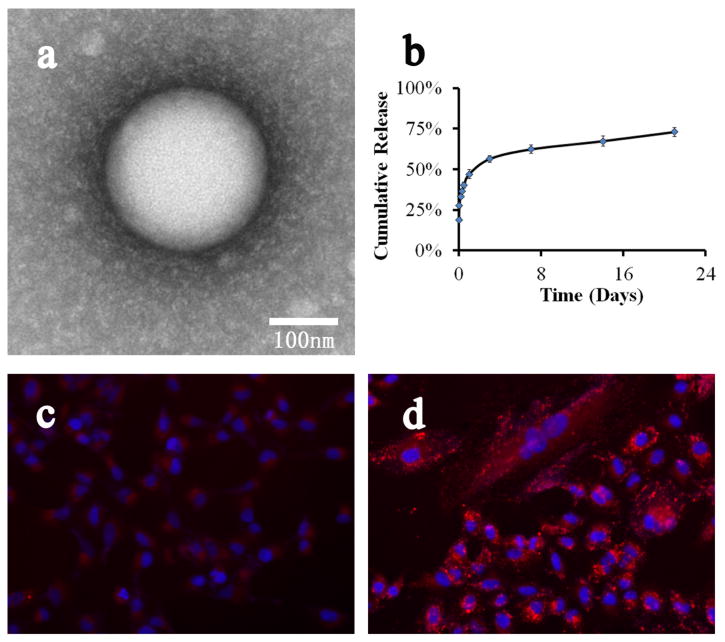
The specific complexed PLGA2-PC16/Protein nanospheres formed in the third region were unstable in buffered solutions. After optimization, it was found that lipid/lipid-PEG (such as DSPE-PEG(MW≥2,000)) effectively stabilized the negatively charged nanospheres without causing obvious size change, with only a 5-10 wt% loss in overall protein loading (supporting information). The stabilization could be due to self-assembly of cationic and hydrophobic lipid/lipid-PEG on the surface of PLGA2-PC16/protein nanospheres, which was confirmed by the TEM, the changes of particle size and surface charge and small protein loss. Figure 5a shows a TEM image of a PLGA2-PC/protein nanosphere coated with DSPE–PEG3000/Lecithin. Study of the release profile of these nanospheres in PBS buffer (Figure 5b) revealed some burst release (around 25 wt%) after coating of lipid-PEG and addition of buffers. Fast release occurred over the first 1-2 days, especially in the initial hours. After that, release of BSA was sustained and steady for at least three weeks. Finally, BSAs labeled with red fluorescent dye RITC were loaded to study the cellular interaction of the PLGA2-PC/Protein nanospheres (supporting information). Figures 5c and 5d show the cellular uptake of pure RITC-BSA compared to the complex of PLGA2-PC NP/RITC-BSA/DSPE-PEG3000 after four hours of incubation. After four hours, large numbers of NPs entered the cells, and the proteins began to be released in a sustained fashion.
Figure 5.
(A) and (B), TEM image and release profile of PLGA2-PC16 NP/BSA/DSPE–PEG3000; (C) and (D), the cellular uptake of fluorescence BSA (c) and PLGA2-PC16 NP/fluorescence BSA / DSPE–PEG3000 (d) after 4 hours.
In summary, a new PLGA-PC NP platform with a core-shell structure and a new “green” protein-loading strategy were developed and evaluated for protein delivery as well as structure–function relationships. Two issues were investigated: 1) the effect of PLGA-PC structure on the resulting NP structure and protein encapsulation capability, and 2) the relationship between the size of PLGA-PC/Protein NPs, their zeta potential, and protein loading efficiency. The results indicated that PLGA-PC polymer structure primarily dictates NP size and interaction with proteins. And the particle size, zeta potential, and weight ratio of PLGA-PC NP/Protein are closely related. Some screened PLGA-PC NPs were able to interact with proteins, forming new multi-nuclear spherical NPs with high protein loading (20-40 wt%) and 100-200 nm size by simple mixing in aqueous solutions. These findings could facilitate the development of new robust protein-delivery platforms or the modification of currently existing platforms for green protein loading.
Supplementary Material
Footnotes
This research was supported by the National Institutes of Health under grant number CA151884, EB015419-01 and the David Koch-Prostate Cancer Foundation Program in Cancer Nanotherapeutics. O.C.F. has financial interest in BIND Therapeutics, Selecta Biosciences, and Blend Therapeutics, biopharmaceutical companies that are developing therapeutic nanoparticles. J.S. acknowledges the financial support from NCI R00CA160350 and PCF Young Investigator Award.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx.
Contributor Information
Dr. Jun Wu, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801; MIT-Harvard Center for Cancer Nanotechnology Excellence, Massachusetts Institute of Technology, Cambridge, MA, 02139 (USA)
Dr. Nazila Kamaly, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801; MIT-Harvard Center for Cancer Nanotechnology Excellence, Massachusetts Institute of Technology, Cambridge, MA, 02139 (USA)
Dr. Jinjun Shi, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801
Lili Zhao, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801; School of Life Science, Nanjing University, Nanjing, Jiangsu, 210093, P.R. China.
Dr. Zeyu Xiao, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801
Geoffrey Hollett, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801.
Rohit John, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801.
Shaunak Ray, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801.
Dr. Xiaoyang Xu, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801; MIT-Harvard Center for Cancer Nanotechnology Excellence, Massachusetts Institute of Technology, Cambridge, MA, 02139 (USA)
Dr. Xueqing Zhang, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801
Prof. Philip W. Kantoff, Lank Center for Genitourinary Oncology, Danan-Farber Cancer Institute, Harvard Medical School, Boston, MA, 02115, USA
Prof. Omid C. Farokhzad, Email: ofarokhzad@zeus.bwh.harvard.edu, Laboratory of Nanomedicine and Biomaterials, Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, 02115 (USA); Fax: (+1) 617-730-2801; MIT-Harvard Center for Cancer Nanotechnology Excellence, Massachusetts Institute of Technology, Cambridge, MA, 02139 (USA); King Abdulaziz University, Jeddah, Saudi Arabia
References
- 1.Langer R, Tirrell DA. Nature. 2004;428:487. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 2.Gu Z, Biswas A, Zhao M, Tang Y. Chemical Society Reviews. 2011;40:3638. doi: 10.1039/c0cs00227e. [DOI] [PubMed] [Google Scholar]
- 3.Vermonden T, Censi R, Hennink WE. Chemical Reviews. 2012;112:2853. doi: 10.1021/cr200157d. [DOI] [PubMed] [Google Scholar]
- 4.Kamaly N, Fredman G, Subramanian M, Gadde S, Pesic A, Cheung L, Fayad ZA, Langer R, Tabas I, Cameron Farokhzad O. Proceedings of the National Academy of Sciences. 2013;110:6506. doi: 10.1073/pnas.1303377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W, Priceman S, Wu L, Zhou ZH, Liu Z, Segura T, Tang Y, Lu Y. Nat Nano. 2010;5:48. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- 6.Pridgen EM, Alexis F, Kuo TT, Levy-Nissenbaum E, Karnik R, Blumberg RS, Langer R, Farokhzad OC. Science Translational Medicine. 2013;5:213ra167. doi: 10.1126/scitranslmed.3007049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang HC, Lee JE, Bae YH. Journal of controlled release : official journal of the Controlled Release Society. 2012;160:440. doi: 10.1016/j.jconrel.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peppas NA, Wood KM, Blanchette JO. Expert Opinion on Biological Therapy. 2004;4:881. doi: 10.1517/14712598.4.6.881. [DOI] [PubMed] [Google Scholar]
- 9.Yan W, Huang L. Polymer Reviews. 2007;47:329. [Google Scholar]
- 10.Duncan R. Nat Rev Drug Discov. 2003;2:347. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 11.Brahmachari S, Das D, Shome A, Das PK. Angewandte Chemie International Edition. 2011;50:11243. doi: 10.1002/anie.201105622. [DOI] [PubMed] [Google Scholar]
- 12.Ayame H, Morimoto N, Akiyoshi K. Bioconjugate Chemistry. 2008;19:882. doi: 10.1021/bc700422s. [DOI] [PubMed] [Google Scholar]
- 13.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Chemical Society Reviews. 2012;41:2971. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, Xiao Z, Kamaly N, Farokhzad OC. Accounts of chemical research. 2011;44:1123. doi: 10.1021/ar200054n. [DOI] [PubMed] [Google Scholar]
- 15.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Advanced Drug Delivery Reviews. 2014;66:2. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park TG, Lu W, Crotts G. Journal of Controlled Release. 1995;33:211. [Google Scholar]
- 17.Zolnik BS, Burgess DJ. Journal of Controlled Release. 2008;127:137. doi: 10.1016/j.jconrel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Farokhzad OC, Jon S, Khademhosseini A, Tran TNT, LaVan DA, Langer R. Cancer Research. 2004;64:7668. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, Xiao Z, Votruba AR, Vilos C, Farokhzad OC. Angewandte Chemie-International Edition. 2011;50:7027. doi: 10.1002/anie.201101554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Chan JM, Gu FX, Rhee JW, Wang AZ, Radovic-Moreno AF, Alexis F, Langer R, Farokhzad OC. ACS Nano. 2008;2:1696. doi: 10.1021/nn800275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J, Wu D, Mutschler MA, Chu CC. Advanced Functional Materials. 2012;22:3815. [Google Scholar]
- 22.Wu J, Yamanouchi D, Liu B, Chu CC. Journal of Materials Chemistry. 2012;22:18983. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.