Abstract
Mammalian immune system development depends on instruction from resident commensal microorganisms. Diseases associated with abnormal immune responses towards environmental and self antigens have been rapidly increasing over the last 50 years. These diseases include inflammatory bowel disease (IBD), multiple sclerosis (MS), type I diabetes (T1D), allergies and asthma. The observation that people with immune mediated diseases house a different microbial community when compared to healthy individuals suggests that pathogenesis arises from improper training of the immune system by the microbiota. However, with hundreds of different microorganisms on our bodies it is hard to know which of these contribute to health and more importantly how? Microbiologists studying pathogenic organisms have long adhered to Koch's postulates to directly relate a certain disease to a specific microbe, raising the question of whether this might be true of commensal–host relationships as well. Emerging evidence supports that rather than one or two dominant organisms inducing host health, the composition of the entire community of microbial residents influences a balanced immune response. Thus, perturbations to the structure of complex commensal communities (referred to as dysbiosis) can lead to deficient education of the host immune system and subsequent development of immune mediated diseases. Here we will overview the literature that describes the causes of dysbiosis and the mechanisms evolved by the host to prevent these changes to community structure. Building off these studies, we will categorize the different types of dysbiosis and define how collections of microorganisms can influence the host response. This research has broad implications for future therapies that go beyond the introduction of a single organism to induce health. We propose that identifying mechanisms to re-establish a healthy complex microbiota after dysbiosis has occurred, a process we will refer to as rebiosis, will be fundamental to treating complex immune diseases.
What is dysbiosis?
Our current knowledge of the architecture of a healthy microbiota comes from multiple studies in individuals with no overt signs of disease (Huttenhower et al., 2012). This structure includes Bacteroidetes and Firmicutes as the dominant bacterial phyla present in stool samples and Proteobacteria and Actinobacteria being a small but consistent presence in most people. Broadly defined, dysbiosis is any change to the composition of resident commensal communities relative to the community found in healthy individuals. In the last decade, a number of studies have documented significant changes in the structure of microbial communities in patients and mouse models of inflammatory bowel diseases (IBD) such as Crohn's and ulcerative colitis (UC) (Frank et al., 2007), diabetes (Karlsson et al., 2013), asthma (Abrahamsson et al., 2013), allergies and even autism (Parracho et al., 2005). Given the emerging importance of the microbiota to host development, it is speculated that these observed changes in microbial composition are contributing factors to the initiation and/or persistence of many of these diseases.
There are multiple ways that the structure of the microbial community can be influenced. This includes the genetics of the host, diet, infection, or medical interventions (such as antibiotics). The hygiene hypothesis originally proposed that antibiotic usage and lifestyle alterations that limit microbial exposure were predisposing populations of people in developed countries to autoimmune disease (Strachan, 2000). Therefore it was suggested that, due to the inability to differentiate between pathogen and commensal, antibiotics disturb microbiota structure and, subsequently, the co-evolutionary relationship between our immune system and the symbionts we host. Since then a significant amount of research has gone into exploring the functional importance of the microbiota and the influence antibiotic treatment has on its architecture (Ubeda et al., 2010; Buffie et al., 2012; Cho and Blaser, 2012). Interestingly, antibiotics commonly used in the clinic have significant impacts on our microbial residents (Table 1). Importantly, many antibiotics have long lasting effects on the microbiota, leading to the permanent loss of some organisms, while others outgrow and persist. While these studies have documented dysbiosis in the context of disease or antibiotic use, recent investigations have begun to identify causality between dysbiosis and disease progression. Collectively, these studies now allow us to better understand what dysbiosis is and begin to categorize it into three types that will be discussed in depth here. These include (i) loss of beneficial microbial organisms, (ii) expansion of pathobionts or potentially harmful microorganisms and (iii) loss of overall microbial diversity (Fig. 1). These three types of dysbiosis are not mutually exclusively and may all occur concurrently. This review is intended to help synthesize the current data to allow a more defined understanding of what dysbiosis is and how it influences health, immune system development and disease.
Table 1.
Antibiotic induced alterations to the microbiota
|
Antibiotic |
Alterations to the microbiota |
|||
|---|---|---|---|---|
| Name | Activity/spectrum | Reduced diversity | Community loss | Community expansion |
| Amoxicillin |
|
|
|
|
| Vancomycin |
|
|
|
|
| Ciprofloxacin |
|
|
|
|
| Cephalosporin (Cephazolin) |
|
|
|
|
| Streptomycin |
|
|
|
|
Antibiotics commonly used in the clinic lead to alterations in the composition of the microbiota. This table describes how various antibiotics influence specific bacterial communities. M, mouse; NM, neonatal mouse; NR, neonatal rat; C, clinical.
Mangin et al. (2010) Anaerobe 16: 433–438.
Schumann et al. (2005) Physiol Genomics.
Sekirov et al. (2008) Infect Immun.
Russell et al. (2012) EMBO Rep.
Vreize et al. (2013) J Hepatol.
Perez-Cobas et al. (2013) Gut.
Dethlefsen et al. (2008) PLoS Biol.
Figure 1.
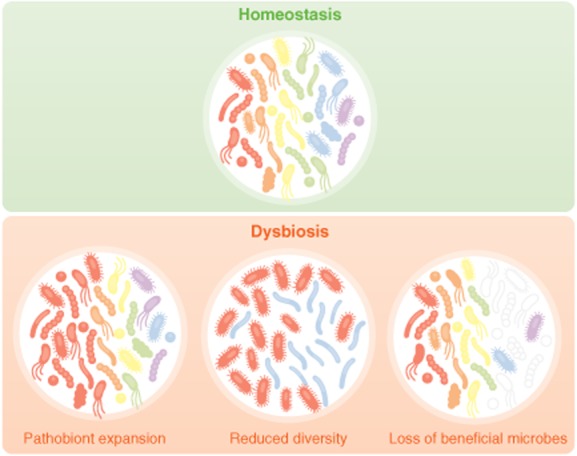
A loss of beneficial microbes, expansion of pathobionts, and loss of diversity are events that encompass dysbiosis. During healthy, homeostatic conditions the microbiota is composed of a diversity organisms that are known to benefit host development and health. However, environmental insults, such as antibiotic use or diet can lead to disruptions in the structure of the microbial community. These disruptions can lead to a loss of organisms that are beneficial to the host and a subsequent overgrowth of commensals that have the potential to cause harm, termed pathobionts. Domination of the microbiota by pathobionts can lead to inflammation and pathology. Additionally, multiple studies have described the diversity of contributions made by the various members of the microbiota. Oftentimes, these are non-redundant influences on host health, thus a total loss of diversity in the microbiota can also influence disease progression or severity and thus also represents a dysbiosis event.
Loss of beneficial microbial organisms
It has become well established that the microbiota is important for the maturation and development of appropriate intestinal immune responses (Round and Mazmanian, 2009; Cho and Blaser, 2012; Hooper et al., 2012). Recent studies have begun to identify the organisms and their mechanisms responsible for eliciting immune development within the host. The immune response must be carefully balanced between the inflammation required for pathogen eradication and tolerant reactions that prevent unwanted immune responses towards self tissue and commensals. Tolerant mechanisms evolved by the host include production of mucus and antimicrobial peptides to create a barrier between host tissue and microbes (Johansson et al., 2008; 2011,; Vaishnava et al., 2011), modification of bacterial products to make them less immunogenic (Bates et al., 2007), production of antibodies that can directly bind to and influence microbial function (Fagarasan and Honjo, 2003; Peterson et al., 2007; Slack et al., 2012) and anti-inflammatory T cells. Interestingly, specific members from the microbiota have been identified that can directly co-ordinate many aspects of the host immune response.
Tolerance to self tissue and resident commensals is, in part, governed by a specialized subset of T lymphocytes termed T regulatory cells (Tregs) (Josefowicz et al., 2012). These cells are marked by the expression of a transcription factor, Foxp3 (Fontenot et al., 2003). Mutations in the Foxp3 gene result in a loss of Treg development in both mouse and humans and subsequent inflammation and autoimmunity at multiple organ sites, demonstrating the importance of Treg populations to host health. Several recent investigations have identified resident microorganisms that are capable of inducing these cell types within the intestine of the host (Round and Mazmanian, 2010; Atarashi et al., 2011). Initial experiments demonstrated that Treg function is compromised in germfree mice and can be restored by either mono-association with the human commensal Bacteriodes fragilis or with a defined mix of Clostridium strains from groups IV and XIVa. Colonization of animals with either of these organisms protects animals from colitis in a Treg-dependent manner through distinct mechanisms. The Clostridium strains induce TGF-β expression from intestinal epithelial cells (IECs) to enhance the differentiation of inducible colonic Tregs (Atarashi et al., 2013); while B. fragilis utilizes a capsular polysaccharide, PSA, that binds to TLR2 on both dendritic cells and T cells to induce IL-10 and enhance the suppressive capability of Tregs (Round et al., 2011; Shen et al., 2012). Other Bacteriodes strains have also been reported to increase the total numbers of Tregs within the colon, including B. caccae, B. thetaiotaomicron, and B. vulgaris, although this is not seen in all systems (Faith et al., 2014). Several other strains of commensal bacteria have evolved the capacity to induce Tregs and confer protection from inflammatory disease. These include multiple strains of Lactobacillus such as L. acidophilus and several strains of Bifidobacterium, including B. breve, likely making induction of Tregs a common mechanism employed by bacteria to induce tolerance in the gut. The mechanisms by which these many of these organisms influence Treg biology remain untested but likely include bacterial derived products such as short-chain fatty acids (SCFAs) (Schmidt et al., 2010). SCFAs are microbial fermentation products from dietary fibre and include propionate, acetate, butyrate, and formate. These products have been shown by multiple groups to be responsible for regulating the Treg pool within the colon and provide protection from experimental colitis (Arpaia et al., 2013; Furusawa et al., 2013; Smith et al., 2013). Butyrate and propionate, but not acetate, seem to be the most prominent SCFAs capable of driving Treg responses. Butyrate, in particular, is capable of blocking histone deacetylases that promote DNA condensation (HDACs) and can thereby regulate gene expression. Interestingly, if T cells are treated with butyrate under Treg inducing conditions in vitro, there is a marked increase in acetylation of the Foxp3 promoter region as well as enhancer elements thereby allowing Foxp3 to be expressed. SCFAs are sensed by the host through a variety of receptors, including G-protein coupled receptors GPCR43 and GPR109a (Smith et al., 2013; Singh et al., 2014). Not surprisingly, GPCR43 expression is increased on Tregs specifically within intestinal tissues (Smith et al., 2013). Consequently, loss of these receptors leads to susceptibility of animals to colonic inflammation and colon cancer.
While Tregs are a key component of tolerance induction within the intestine, commensal organisms have evolved other mechanisms to suppress inflammation. Patients with IBD have elevated levels of inflammatory cytokines such as TNF-α and IP-10. As discussed above, the levels of these cytokines are often opposed by the induction of Tregs, and this is the mechanism utilized by the host to maintain homeostasis within the gut. Recently, however, a group uncovered a mechanism by which commensal bacteria can directly reduce inflammation by targeting the cytokines themselves. Lactobacillus paracasei and L. casei both encode a protease called lactoceptin that specifically degrades the inflammatory cytokine IP-10 (von Schillde et al., 2012). Degradation of secreted and cell-associated IP-10 lead to reduced lymphocyte recruitment during an animal model of ileitis. Thus, members of the microbiota have evolved ways to directly antagonize the effects of the host inflammatory response and therefore directly aid in the maintenance of intestinal homeostasis.
Unlike T cells that recognize protein antigens, invariant NKT cells (iNKT) recognize lipid antigens and play important roles in innate and adaptive inflammation. Interestingly, the development of these cell types is dynamically regulated by the microbiota such that GF mice have elevated numbers of these cells within the gut (Olszak et al., 2012). Animals containing increased numbers of iNKT cells are more susceptible to colitis, thus the identification of how the microbiota functions to suppress iNKT cell numbers in the gut could lead to an important therapeutic intervention. A recent report demonstrated that mono-association of germfree mice with B. fragilis is sufficient to suppress the development of this population (An et al., 2014). B. fragilis contains a gene with high homology to enzymes involved in sphinoglipid biogenesis. Deletion of this gene in B. fragilis prevents the suppression of iNKT cell development, indicating that B. fragilis utilizes these lipids to influence iNKT cell biology. Indeed, these investigators purified sphingolipids from B. fragilis and showed that they are capable of reducing iNKT cell activation and protecting animals from disease. Importantly, early exposure to B. fragilis is required to inhibit iNKT cell development, and mono-colonization of adult germfree mice is insufficient to protect against colitis. As discussed earlier, B. fragilis is also able to modulate Treg responses (Ochoa-Reparaz et al., 2010; Round and Mazmanian, 2010). These studies reveal that a single commensal organism can possess several mechanisms to positively influence host biology and highlights the complexity and intimacy of host-commensal relationships.
Clinical investigations provide support for these animal studies. Culture-independent analyses of patients with CD and UC have revealed a distinct loss of symbionts residing within the colonic mucosa as compared to healthy individuals. Specifically, loss of Clostridium groups XIVa and IV is observed within faecal and mucosal samples in IBD patients (Gophna et al., 2006; Frank et al., 2007). These observed reductions were seen consistently whether patients were actively experiencing clinical symptoms or within remission and were not correlated with recent antibiotics. This argues that the loss of beneficial microbes is an underlying issue and not simply a reflection of increased inflammation or treatment. Conflicting results have been reported for the phyla Bacteroidetes. Using mucosal tissues from small intestines and colons from 190 patients that included equal numbers of CD, UC, and healthy controls, Frank et al. identified approximately 800–1600 OTUs and observed a significant decrease in the OTUs and relative abundance of Bacteroidetes within IBD patients. Other studies including microbial identification from faecal samples have observed increases in Bacteroidetes within IBD patients (Mangin et al., 2004; Gophna et al., 2006). This, however, may reflect differences in culture-dependent versus -independent techniques, the depth of microbial sequence sampling, or tissue versus faecal sampling differences. Collectively, these studies identify important ways in which commensal bacteria positively influence mammalian biology, thus loss of these organisms represents an important aspect of dysbiosis.
Expansion of pathobionts
The microbiota also contains members that have the capacity to cause harm to the host. These organisms have been termed pathobionts to indicate their potential to cause pathology (Chow and Mazmanian, 2010). Pathobionts are typically kept at low levels within a healthy gut and do not cause problems in immune-competent hosts; however, there are several examples that outgrowth of these organisms can contribute to disease. Thus, expansion of pathobionts represents a second category of dysbiosis.
The most often reported incidence of pathobiont expansion is that of Proteobacteria and in particular the family Enterobacteriaceae, which contains members such as Escherichia coli, Shigella and Klebsiella (Lupp et al., 2007). This is true in multiple mouse models of colitis, including animal models carrying mutations within genes that are associated with IBD (Ayres et al., 2012). In particular, the deletion of TLR5, a Toll-like receptor (TLR) responsible for recognizing flagellar proteins, results in low grade inflammation, colitis and metabolic syndrome with incomplete penetrance (Vijay-Kumar et al., 2010; Carvalho et al., 2012; Cullender et al., 2013). These disease phenotypes are lost when animals are re-derived with a microbiota from Jackson labs, suggesting a role for the composition of the microbiota in driving disease. To identify microbial communities associated with colitis in TLR5 deficient animals, the microbiota was compared between colitic and non-colitic TLR5−/− animals. Interestingly, animals that developed spontaneous colitis also had a threefold increase in Proteobacteria. More importantly, members of Enterobacteriaceae, such as E. coli, were found to be preferentially associated with the mucosa and able to penetrate the tissue. To show the requirement for E. coli for colitis induction in TLR5−/− animals, they antibiotic treated TLR5−/− and WT animals and then treated the mice with an adherent invasive E. coli (AIEC) strain. TLR5−/− animals were unable to manage levels of these flagellated bacteria and harboured a 10-fold larger bacterial load. Moreover, all TLR5−/− animals receiving this strain succumbed to colitis, demonstrating that E. coli were sufficient to cause colitis in the absence of TLR5.
Klebsiella pneumonia and Proteus mirabilis, both members of the Enterobacteriaceae family, also expanded to induce colitis within T-bet−/− X Rag2−/− mice (Garrett et al., 2007). Rag deficient animals lack any T or B cells and therefore crossing these mice causes a lack of Tbet in cells of the innate immune system. Interestingly, these species required an intact microbiota for disease induction and were not sufficient to induce colitis on their own. As a final example, when animals are treated with antibiotics and subsequently induced for colitis using DSS, expansion of a multi-drug-resistant E. coli strain occurs that is able to penetrate the intestinal mucosa and cause sepsis (Ayres et al., 2012). Multiple studies have demonstrated how these organisms are able to exploit the inflamed environment and expand their numbers, and these mechanisms have been reviewed elsewhere (Spees et al., 2013). Importantly, the expansion of Enterobacteriaceae is also seen in patients suffering from CD and UC in both tissue and faecal samples (Frank et al., 2007), consistent with what is seen in mouse models; however, much work needs to be done to better understand whether these organisms truly initiate disease or simply exacerbate disease progression.
Other types of organisms have been reported to expand in other animal models of immune deficiency. NLRP6 is a member of the nucleotide-binding oligomerization domain protein like receptors (NLRs), a class of cytoplasmic receptors that recognize bacterial products. NLRP6−/− mice have an increased abundance of Prevotella spp. and TM7 and are highly susceptible to colitis induced by dextran sodium sulfate (DSS) (Elinav et al., 2011). The colitis is transmissible as co-housing NLRP6−/− and WT mice results in increased susceptibility to disease in the WT animals. Thus, the NLRP6 deficiency is not required for pathology, but rather allows for the expansion of a subset of organisms that promote inflammation.
Finally, while the vast majority of our microbial inhabitants are bacteria, mammals are also colonized by fungal and viral members. In general, yeast are not susceptible to antibiotic treatment, and one common side-effect of antibiotic use is the outgrowth of yeast strains such as Candida albicans. A recent study highlighted the importance of controlling fungal overgrowth to intestinal health by investigating the role of Dectin-1 within the gut (Iliev et al., 2012). Dectin-1 is a receptor that recognizes alpha-mannans from the cell wall of yeast, and mice deficient in this receptor are more susceptible to DSS colitis that can be treated with fluconazole, an antifungal drug that does not target bacteria. These data indicate that yeast outgrowth can worsen disease and our immune system has evolved elegant mechanisms to regulate yeast populations in the gut. Similarly, a study showed that antibiotic treatment of animals allowed overgrowth of C. albicans, which increased plasma concentrations of prostaglandin E2 (PGE2) (Kim et al., 2014). The increase in PGE2 caused susceptibility to allergies in these animals that could be suppressed when PGE2 synthesis was blocked. Thus, antibiotic treatment can allow for outgrowth of fungal species in the gut that can influence extra-intestinal disease within the host. Viral members of our microbiota are only just beginning to be identified, presenting new players in the complex relationship between genetic and environmental factors that lead to disease. Polymorphisms within the gene ATG16L1 confer a twofold higher risk to the development of IBD. When mice contain a hypomorphic mutation within ATG16L1, they experience histological abnormalities and increased colitis similar to Crohn's disease patients carrying polymorphisms within the same gene (Cadwell et al., 2010). Interestingly, chronic infection with a specific strain of mouse norovirus is required for the observed phenotype within mice. Although this is not an example of expansion, it highlights the likelihood of unknown viral pathobionts playing a role in the development of dysbiosis associated diseases.
It is unlikely that pathobiont expansion alone would cause disease, as decades of research have been dedicated to identifying a single organism capable of inducing IBD and none have been found. Supporting this, subclinical twins and immediate family members posses a microbiota that resembles that of a related IBD patient; however, none of these infectious agents had the ability to induce IBD within a healthy individual. Therefore, it is possible that concomitant reductions in beneficial microbes or a loss of diversity must also occur to promote disease. Collectively, these studies investigate both well-characterized and newly discovered pathobionts, capable of inciting harmful effects on the host when given the opportunity to expand.
Loss of diversity: more is better
There is evidence that members of the microbiota have diverse and non-redundant contributions to host health. For instance, some organisms promote the development of anti-inflammatory networks while others induce protective inflammatory responses. Additionally, while some host–microbe interactions can be initiated by multiple species, others involve a more unique relationship with exclusive community members (Faith et al., 2014).Therefore, gaining the maximum health benefits from the microbiota may require a more complex and diverse collection of organisms, and, indeed, several recent experiments support this notion. Thus, an important aspect of dysbiosis can also include a loss of total microbial diversity.
As discussed above, Treg function is compromised in germfree animals. In order to identify organisms that complement this deficiency, diluted human stool samples enriched for chloroform-resistant bacteria were used to colonize germfree animals. Interestingly, these Clostridia species, containing over 30 different strains, induced a threefold expansion of Tregs within the gut over un-colonized controls. Importantly, Treg induction within the host was diminished when mice were colonized with only a single strain from this Clostridia collection, as compared to animals colonized with over 15 different Clostridia strains. These data suggest that a greater diversity of organisms, even within the same family, can maximally induce host cellular development (Atarashi et al., 2011; 2013,). Whether these strains amplify Treg activity through the same pathway or work through distinct mechanisms remains unclear, but will be important to our understanding of how these organisms influence host health.
Early exposure to these diverse communities of microbes appears to be an additional facet to maintaining a healthy host–microbiota relationship (Cho and Blaser, 2012; Cho et al., 2012). The antibody isotype, IgE, is associated with allergic responses, and animals with increased concentrations of IgE develop worsened allergic disease. Despite a reduction in all other isotypes, IgE is greatly elevated in the serum of germ-free mice, indicating a specific requirement for the microbiota in suppressing this antibody (Cahenzli et al., 2013). Colonization of germfree mice early in life with a complex microbiota reduces IgE levels and prevents allergy; however, if germfree animals are colonized during adult stages, suppression of IgE levels does not occur. To better understand the minimal microbial diversity required for IgE suppression, germfree mice were first colonized by a single species, E. coli, which was incapable of suppressing IgE levels. Bi-colonization with two species, Lactobacillus murinus and Parabacteriodes distasonis, also failed to suppress IgE levels. When germfree animals were colonized with eight different organisms that included L. murinus and P. distasonis half of the animals still displayed hyper-IgE production. Suppression of IgE was only evident when the animals were colonized with up to 40 different strains of bacteria. Taken together, these studies suggest that a greater diversity of microbial organisms is required for maximal benefits to the host.
While many studies have found correlations with changes in the microbiota abundance and allergy, few studies have been able to look at the microbiota early in life and then follow the same children out to see if they develop disease. However, a recent study was able to collect microbiota samples from children during there first week of life and again at 1 month and 12 months after birth (Abrahamsson et al., 2013). These same children were then assessed for allergic disease and asthma at age seven. Interestingly, the children that developed asthma all had lower microbial diversities early in life when compared to the non-asthmatic children, suggesting that early life microbial diversity might protect from disease later in development. Rigorous studies still need to be performed; however, these studies suggest that increased microbial diversity is better. While we have discussed three distinct types of dysbiosis, it is likely that dysbiosis encompasses all three of these manifestations concomitantly to influence disease.
Rebiosis: establishing a microbial community back to a healthy state
A handful of studies have now demonstrated that replacement of beneficial bacterial species can protect and even cure diseases in animal model systems and human patients (Mazmanian et al., 2008; Sokol et al., 2008; Ochoa-Reparaz et al., 2010; Round and Mazmanian, 2010). In humans, this has been best established for Clostridium difficile infections (Aas et al., 2003). C. difficile infection is a major cause of antibiotic-associated diarrhoea and becoming an increasing healthcare burden (Kachrimanidou and Malisiovas, 2011). C. difficile infections can cause a wide range of diseases that include severe diarrhoea, pseudomembranous colitis, and toxic megacolon. Infection is usually treated with vancomycin or metronidazole; however, in 25–30% of cases a recurrent disease follows, with each relapse increasing the likelihood of another recurrence. Interestingly, when a more narrow spectrum antibiotic is used that does less damage to the microbiota, there are lower recurrence rates of C. difficile, suggesting that the microbiota may play a protective role during infection. It has now been shown in both mouse models and, more importantly, humans that a faecal microbiota transplant (FMT) from a healthy donor can help displace C. difficile infection and prevent recurrences. After a large number of uncontrolled trials demonstrated high success rates for microbiota transplantation in resolving C. difficile infections, a randomized study recently revealed an 81% success rate following a single transplant with a 98% success rate after a second (van Nood et al., 2013; Vrieze et al., 2013; Pamer, 2014). These studies highlight the potential therapeutic value of identifying ways to restore a ‘healthy’ microbiota, a process we will refer to as rebiosis.
There are several potential candidate approaches for rebiosis aside from FMT described above. Multiple investigations have sought to identify functional members of the microbiota that can be cultured and therefore used as a probiotic either in the form of a pill or embedded in food items. Given the importance of microbial diversity it is likely that no one single organism will work most effectively, but rather a complex assortment of organisms will provide the maximum benefit. Indeed, yogurts and probiotics are beginning to incorporate multiple strains of organisms. The downside to this therapy is that it relies on the ability to culture these organisms within the lab, yet it is estimated that only 20–30% of these organisms are culturable, meaning that a large, likely very important constituent of the microbiota will not be sampled. Another method to maintain or restore a healthy microbiota could potentially be the use of pre-biotics, which are non-digestible ingredients that stimulate the growth and activity of certain bacterial groups. There are a few known substances that meet these criteria, such as inulin, but much work is needed to identify the best nutritional sources for the microbiota. A better understanding of how host immune pathways influence the structure of the microbial community might also open up new avenues for improved therapies for manipulating microbial communities. Mammalian immune systems have acquired elegant mechanisms for mounting antigen-specific immunity against pathogens and therefore might also use similar mechanisms to control the microbiota. If true, then engineered therapies that target specific microbes while keeping the rest of the microbiota intact would be possible. The microbiota represents an important aspect of human health that we are only beginning to understand; therefore, much work lies ahead to tap into the therapeutic value of these microbial symbionts.
Acknowledgments
These authors have no conflict of interest.
We would like to thank members of the Round and O'Connell labs for their critical review of the manuscript. C.P. is supported by a T32 fellowship in microbial pathogenesis (AI-055434). Work in the lab is supported by the Edward Mallinckrodt Jr. Foundation, Pew Scholars Program, NSF CAREER award (IOS-1253278), Packard Fellowship in Science and Engineering, NIAID (AI95375), NIAID (AI107090), and NIAID (AI95375) to J.L.R.
References
- Aas J, Gessert CE. Bakken JS. Recurrent Clostridium difficile colitis: case series involving 18 patients treated with donor stool administered via a nasogastric tube. Clin Infect Dis. 2003;36:580–585. doi: 10.1086/367657. [DOI] [PubMed] [Google Scholar]
- Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L. Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2013 doi: 10.1111/cea.12253. doi: 10.1111/cea.12253 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, DeRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Ayres JS, Trinidad NJ. Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JM, Akerlund J, Mittge E. Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahenzli J, Koller Y, Wyss M, Geuking MB. McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I. Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J. Mazmanian SK. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe. 2010;7:265–276. doi: 10.1016/j.chom.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML. Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasan S. Honjo T. Intestinal IgA synthesis: regulation of front-line body defences. Nat Rev Immunol. 2003;3:63–72. doi: 10.1038/nri982. [DOI] [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J. Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra211. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA. Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N. Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna U, Sommerfeld K, Gophna S, Doolittle WF. Veldhuyzen van Zanten SJ. Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J Clin Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR. Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger J, Chinwalla A, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L. Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Larsson JM. Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl. 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF. Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachrimanidou M. Malisiovas N. Clostridium difficile infection: a comprehensive review. Crit Rev Microbiol. 2011;37:178–187. doi: 10.3109/1040841X.2011.556598. [DOI] [PubMed] [Google Scholar]
- Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- Kim YG, Udayanga KG, Totsuka N, Weinberg JB, Nunez G. Shibuya A. Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE(2) Cell Host Microbe. 2014;15:95–102. doi: 10.1016/j.chom.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC. Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Mangin I, Bonnet R, Seksik P, Rigottier-Gois L, Sutren M, Bouhnik Y, et al. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Microbiol Ecol. 2004;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Mangin I, Suau A, Gotteland M, Brunser O. Pochart P. Amoxicillin treatment modifies the composition of Bifidobacterium species in infant intestinal microbiota. Anaerobe. 2010;16:433–438. doi: 10.1016/j.anaerobe.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL. Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- van Nood E, Dijkgraaf MG. Keller JJ. Duodenal infusion of feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:2145. doi: 10.1056/NEJMc1303919. [DOI] [PubMed] [Google Scholar]
- Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL. Kasper LH. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–495. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamer EG. Fecal microbiota transplantation: effectiveness, complexities, and lingering concerns. Mucosal Immunol. 2014;7:210–214. doi: 10.1038/mi.2013.117. [DOI] [PubMed] [Google Scholar]
- Parracho HM, Bingham MO, Gibson GR. McCartney AL. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- Perez-Cobas AE, Artacho A, Knecht H, Ferrus ML, Friedrichs A, Ott SJ, Moya A, Latorre A. Gosalbes MJ. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. 2013;8:e80201. doi: 10.1371/journal.pone.0080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL. Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Round JL. Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL. Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA. Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM. Finlay BB. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schillde MA, Hormannsperger G, Weiher M, Alpert CA, Hahne H, Bauerl C, et al. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe. 2012;11:387–396. doi: 10.1016/j.chom.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Schumann A, Nutten S, Donnicola D, Comelli EM, Mansourian R, Cherbut C, Corthesy-Theulaz I. Garcia-Rodenas C. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genomics. 2005;23:235–245. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- Schmidt EG, Claesson MH, Jensen SS, Ravn P. Kristensen NN. Antigen-presenting cells exposed to Lactobacillus acidophilus NCFM, Bifidobacterium bifidum BI-98, and BI-504 reduce regulatory T cell activity. Inflamm Bowel Dis. 2010;16:390–400. doi: 10.1002/ibd.21068. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C. Finlay BB. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD. Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack E, Balmer ML, Fritz JH. Hapfelmeier S. Functional flexibility of intestinal IgA – broadening the fine line. Front Immunol. 2012;3:100. doi: 10.3389/fimmu.2012.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees AM, Lopez CA, Kingsbury DD, Winter SE. Baumler AJ. Colonization resistance: battle of the bugs or Menage a Trois with the host? PLoS Pathog. 2013;9:e1003730. doi: 10.1371/journal.ppat.1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan DP. Family size, infection and atopy: the first decade of the ‘hygiene hypothesis’. Thorax. 2000;55(Suppl. 1):S2–S10. doi: 10.1136/thorax.55.suppl_1.s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, de Groot PF, Kootte RS, Knaapen M, van Nood E. Nieuwdorp M. Fecal transplant: a safe and sustainable clinical therapy for restoring intestinal microbial balance in human disease? Best Pract Res Clin Gastroenterol. 2013;27:127–137. doi: 10.1016/j.bpg.2013.03.003. [DOI] [PubMed] [Google Scholar]


