Abstract
Vigna reflexo-pilosa, which includes a neglected crop, is the only one tetraploid species in genus Vigna. The ancestral species that make up this allotetraploid species have not conclusively been identified, although previous studies suggested that a donor genome of V. reflexo-pilosa is V. trinervia. In this study, 1,429 azuki bean EST-SSR markers were developed of which 38 EST-SSR primer pairs that amplified one product in diploid species and two discrete products in tetraploid species were selected to analyze 268 accessions from eight taxa of seven Asian Vigna species including V. reflexo-pilosa var. glabra, V. reflexo-pilosa var. reflexo-pilosa, V. exilis, V. hirtella, V. minima, V. radiata var. sublobata, V. tenuicaulis and V. trinervia to identify genome donor of V. reflexo-pilosa. Since both diploid and tetraploid species were analyzed and each SSR primer pair detected two loci in the tetraploid species, we separated genomes of the tetraploid species into two different diploid types, viz. A and B. In total, 445 alleles were detected by 38 EST-SSR markers. The highest gene diversity was observed in V. hirtella. By assigning the discrete PCR products of V. reflexo-pilosa into two distinguished genomes, we were able to identify the two genome donor parents of créole bean. Phylogenetic and principal coordinate analyses suggested that V. hirtella is a species complex and may be composed of at least three distinct taxa. Both analyses also clearly demonstrated that V. trinervia and one taxon of V. hirtella are the genome donors of V. reflexo-pilosa. Gene diversity indicates that the evolution rate of EST-SSRs on genome B of créole bean might be faster than that on genome A. Species relationship among the Vigna species in relation to genetic data, morphology and geographical distribution are presented.
Introduction
The Leguminosae genus Vigna comprises about 100 species. These species are morphologically diverse and geographically widespread and mainly found in Africa (African Vigna; subgenus Vigna) and Asia (Asian Vigna; subgenus Ceratotropis) [1]. The Asian Vigna comprises 21 species in which seven are domesticated and/or cultivated in various geographical and climatic regions, and cropping systems in Asia [2]. The seven domesticated/cultivated species include V. aconitifolia (moth bean), V. angularis (azuki bean), V. mungo (black gram), V. radiata (mungbean), V. reflexo-pilosa (créole bean), V. stipulacea (jungli bean) and V. umbellata (rice bean). All Asian Vigna species are diploid having 11 haploid chromosomes (2n = 2x = 22) with the exception for créole bean which is a tetraploid species with number of haploid chromosome of 22 (2n = 4x = 44). In fact, créole bean is the only natural amphidiploid in the subtribe Phaseolinae [3]. Cytogenetic analyses of V. reflexo-pilosa showed that the species formed 22 bivalents without multivalent at meiosis and was, therefor considered to be an amphidiploid [4] [5].
Cultivated and wild forms of créole bean are classified as V. reflexo-pilosa var. glabra and V. reflexo-pilosa var. reflexo-pilosa, respectively [6]. Wild créole bean is widely distributed in East, Southeast and South Asia, and across the islands from the west to the north Pacific islands. It is also found in Papua New Guinea and northern Australia [7] [8] [9]. The cultivated créole bean was formerly recognized as a glabrous variety of mungbean, V. radiata var. glabra [1]. Then, it was treated as a distinct species, V. glabrescens [3]. It is differentiated from its wild progenitor principally by thick glabrous stem and erect growth habit and reported to be cultivated as pulse in Vietnam and the Philippines or as forage in India, Mauritius and Tanzania [2]. This crop shows resistance to several insect pests and diseases such as bruchids, bean fly, powdery mildew, and cucumber mosaic virus [10], and is partially cross-compatible with mungbean [11]. Thus, créole bean has potential to be a gene source for breeding other Vigna crops. In addition, cultivated créole bean could be considered as a novel crop for the future and wild créole bean as a wider genepool to improve cultivated créole bean.
The origin and genome donors of créole bean have been the subject of debate. Based on the study on isozyme banding patterns and interspecific hybridization among the Ceratotropis species, Egawa et al [10] [12] proposed that V. reflexo-pilosa var. reflexo-pilosa is the descendant of natural interspecific hybridization between V. trinervia and V. minima followed by spontaneous chromosome doubling. An accession used as V. minima in Egawa et al. [10] [12] was correctly identified as V. hirtella by Konarev et al. [13], and was used in the present study (No. 21, JP108851, from Malaysia). Proteinase inhibitors polymorphism study in Asian Vigna by Konarev et al. [13] supported that one genome donor of créole bean is V. trinervia and suggested that the other genome donor is most likely V. hirtella or its closely related species. The phylogenetic studies based on plastid sequence data also supported that V. trinervia is a genome donor of créole bean [14] [15]. In contrast, rDNA-ITS sequence variation showed high similarity between créole bean and V. exilis, V. hirtella and V. umbellata [16], suggesting that one of these species is the genome donor of V. reflexo-pilosa.
The objective of this study was to determine the putative genome donor species of tetraploid créole bean using EST-SSR markers. To do so, we developed EST-SSR from azuki bean and used them to analyze accessions of créole bean and other Vigna species that are candidate genome donor species.
Materials and Methods
Plant Materials
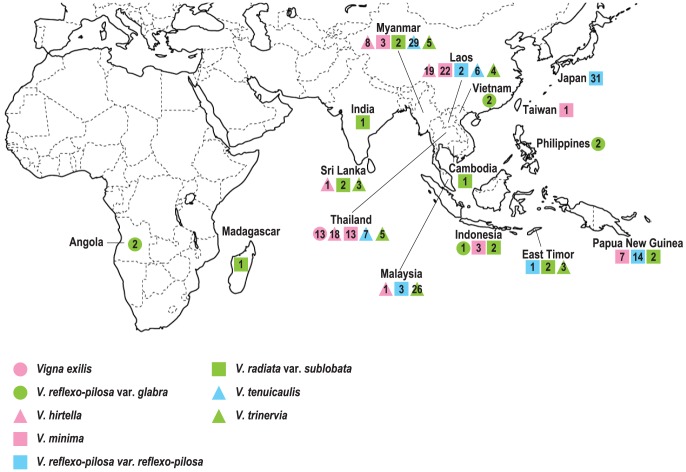
A Japanese azuki bean cultivar ‘Erimo-shouzu’ (V. angularis var. angularis, accession no. JP37752) obtained from the Genebank, National Institute of Agrobiological Sciences (NIAS), Tsukuba, Japan was used for development of EST-SSR markers. Eight accessions consisting of four major Vigna crop species; azuki bean, rice bean, black gram and mungbean were used for assessing transferability and polymorphisms of the EST-SSR markers (Table 1). Two hundred and sixty-eight accessions from eight taxa of seven Asian Vigna species (Figure 1, Table S1) including 7 of V. reflexo-pilosa var. glabra, 51 of V. reflexo-pilosa var. reflexo-pilosa, 13 of V. exilis, 47 of V. hirtella, 49 of V. minima, 13 of V. radiata var. sublobata, 42 of V. tenuicaulis, and 46 of V. trinervia were used to analyze the genome origin of V. reflexo-pilosa.
Table 1. A summary of transferability and polymorphism of 1,429 azuki bean EST-SSR markers in the four Asian Vigna species.
| Code | Species | Domestication status | Common name | Genebank acc. no | Origin | Linkage map | PCR products amplified | % | Simple banding pattern | % | Complex banding pattern | Exceeded of expected size | Number of markers compared between cultivated and wild | Polymorphic between cultivated and wild | % |
| VAC | V. angularis var. angularis | Cultivated | Azuki bean | JP81481 | Japan | Han et al. (2005) | 1327 | 92.9 | 1100 | 82.9 | 46 | 181 | 1307 | 236 | 18.1 |
| VAW | V. nepalensis | Wild | JP107881 | Nepal | Han et al. (2005) | 1312 | 91.8 | 1090 | 83.1 | 45 | 177 | ||||
| VMC | V. mungo var. mungo | Cultivated | Black gram | JP219132 | Thailand | Chaitieng et al. (2006) | 1216 | 85.1 | 1007 | 82.8 | 39 | 170 | 1196 | 164 | 13.7 |
| VMW | V. mungo var. silvestris | Wild | JP107873 | India | Chaitieng et al. (2006) | 1194 | 83.6 | 998 | 83.6 | 36 | 160 | ||||
| VUC | V. umbellata | Cultivated | Rice bean | JP217439 | Myanmar | Isemura et al. (2010) | 1324 | 92.7 | 1091 | 82.4 | 48 | 185 | 1296 | 187 | 14.4 |
| VUW | V. umbellata | Wild | JP210639 | Thailand | Isemura et al. (2010) | 1304 | 91.3 | 1073 | 82.3 | 48 | 183 | ||||
| VRC | V. radiata var. radiata | Cultivated | Mungbean | JP229096 | Thailand | Isemura et al. (2012) | 1197 | 83.8 | 987 | 82.5 | 44 | 166 | 1198 | 277 | 23.1 |
| VRW | V. radiata var. sublobata | Wild | JP211874 | Myanmar | Isemura et al. (2012) | 1204 | 84.3 | 985 | 81.8 | 42 | 177 |
Figure 1. Distribution of 286 accessions of 8 taxa of the genus Vigna subgenus Ceratotropis.
Numbers of accessions analyzed are shown within circles, triangles and squares.
Development of Azuki Bean EST-SSR Markers
Total RNA was extracted from seedlings and young pods of Erimo-shouzu using Plant RNA Purification Reagent (Invitrogen, CA, USA). Purification of polyadenylated RNA and conversion to cDNA were performed as described by [17]. Synthesized cDNA was resolved by 1% agarose gel electrophoresis, and fragments ranging from 1 to 3 kb were recovered. The recovered fragments were cloned into the Eco RI-Xho I site of the pBluescript II SK- plasmid vector (Stratagene, CA, USA) and introduced into the E. coli ElectroTen-Blue strain (Stratagene, CA, USA) by electroporation. To generate ESTs, plasmid DNAs were amplified from the colonies using TempliPhi (GE Healthcare UK Ltd, Buckinghamshire, England) and subjected to sequencing using a BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, CA, USA). The reaction mixtures were run on an automated ABI PRISM 3730 DNA Analyzer (Applied Biosystems).
Sequencing chromatograms were converted into nucleotide bases with Phred [18] [19], and the sequences derived from the vector and linkers were removed with CROSSMATCH [19]. The EST reads were quality-trimmed with TRIM2 [20] using the Phred quality score ≥20, and ambiguous regions including more than ten X or N bases were trimmed. Contiguous, high-quality reads ≥100 bp were submitted to the DDBJ/EMBL/GenBank databases under the accession numbers HX939204 to HX950377 (11,174 entries). The PHRAP program with default parameters was used to cluster and identify non-redundant azuki bean ESTs [19].
Simple sequence repeats (SSRs) ≥15 nucleotides in length, which contained all possible combinations of di-nucleotide (NN), tri-nucleotide (NNN), and tetra-nucleotide (NNNN) repeats, were identified from the non-redundant azuki bean ESTs using fuzznuc program in EMBOSS [21] for SSRs within two mismatches. Primer pairs for amplification of SSR-containing regions were designed based on the flanking sequences of each SSR with the aid of the Primer3 [22] so that the amplified fragment sizes were between 90 bp and 300 bp in length. The newly developed markers were designated as VES (Vigna EST-derived SSR) markers.
EST-SSR Marker Analysis
Total genomic DNA was extracted from young leaves of each Vigna accession using the method described by [23] with a slight modification. The DNA was quantified against a lambda DNA on 1.0% agarose gel stained with ethidium bromide and diluted to 5 ng/µl for PCR amplification.
Amplification, transferability and polymorphisms of all the EST-SSR markers were confirmed using eight accessions, two each from azuki bean complex, blackgram, rice bean and mungbean (Table 1). Five microliters of PCR reaction mixture including 0.5 ng of total genomic DNA, 0.02 U BIOTAQ DNA polymerase (BIOLINE, UK), 1x PCR buffer (BIOLINE, UK), 3 mM MgCl2, 0.2 mM dNTPs and 2 pmol of the forward and reverse EST-SSR primers. The PCR thermal cycling was performed as follows: 94°C for 1 min; 3 cycles of 94°C for 30 s and 70°C for 30 s, followed by 3 rounds of the same program in which the annealing and extension temperatures were decreased by 2°C every 3 cycles; 3 cycles of 94°C for 30 s, 62°C for 30 s and 72°C for 30 s, followed by 2 rounds of the same program in which the annealing temperatures were decreased by 2°C every 2 cycles; 30 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s, and a final cycle at 72°C for 10 min. The PCR products were separated by 10% polyacrylamide gel electrophoresis in tris-borate-ethylene diamine tetraacetic acid (TBE) buffer according to the standard protocol, and banding patterns in the gel were stored as pictures. The characteristics of PCR products such as intensity, banding pattern and size range were recorded for each accession and EST-SSR markers. Polymorphism between wild and cultivated accessions in each species was recorded.
Out of 1,429 azuki bean EST-SSR markers, 175 markers with good transferability in all the four major Vigna crop species were labeled with fluorescent dyes and further examined for polymorphism in a panel of eight taxa of Asian Vigna consisting of 16 accessions, two accessions for each species randomly chosen from the 268 accessions (Table S1). The 5′-end of the reverse primers were labeled with one of the following fluorescent dyes; 6-FAM (blue), HEX (green), and NED (yellow) (Applied Biosystems). Five microliters of PCR reaction mixture contained 5 ng of genomic DNA, 1× QIAGEN Multiplex PCR Master Mix, and 5 pmol of the forward and reverse primers. PCR reactions were performed in a GeneAmp PCR System 9700 (Applied Biosystems). The PCR thermal cycling was programmed as follows: 95°C for 15 min followed by 40 cycles of 94°C for 30 s, 55°C for 90 s, 72°C for 60 s, and a final cycle at 72°C for 10 min. One microliter of 10 times dilution PCR product was mixed with 8.5 µL of Hi-Di formamide and 0.125 µL of ROX size standard (Applied Biosystems). The mixture was denatured at 95°C for 5 min and then run on an ABI PRISM 3130xl DNA Analyzer (Applied Biosystems). Allele size for the highest stutter peak with the height ranging between 500 and 10,000 RFU were recorded. The genotyping was performed using GeneMapper 4.0 (Applied Biosystems).
Thirty-eight EST-SSR primer pairs amplified one product in diploid species and two discrete products in tetraploid species with height ranging RFU (Table S2) were selected for further analysis in the 268 Vigna accessions. Three primers with different labels and size products were mixed into a single PCR reaction mixture and amplified as multiplex PCR. After genotyping, a single allele size was scored for each marker in each accession corresponding to the strongest peak.
Data Analysis
Since both diploid (1 genome set) and tetraploid (2 genome set) species were analyzed together in this study and each SSR primer pair detected 2 loci in the tetraploid species, we separated the genome of V. reflexo-pilosa, into 2 different diploid types, viz. A and B. Therefore 10 genomes were recognized from the 8 taxa of 7 Vigna species.
Genetic distance (D A) [24] for all possible pairs of accessions was calculated using software POPULATIONS 1.2.28 (available at www.cnrs-gif.fr/pge/bioinfo/populations). D A among the 10 genomes was also calculated using the same software. A phylogenic tree was constructed to reveal relationships among accessions based on D A using neighbor-joining clustering method by software MEGA 5.05 [25] with bootstrap support (1,000 replicates) obtained by re-sampling the allelic frequency data. In order to confirm the results from neighbor-joining clustering, principal coordinate analysis (PCoA) was also performed using software PAST [26] to reveal the relationship among different accessions. In addition, based on sources of V. hirtella, Seehalak et al. (2006) divided V. hirtella in to 3 subgroups (a1, a2 and b), thus the relationship of individuals based on PCoA analysis enabled 12 sub-genome groups to be determined. Using average D A genetic distance between 12 sub-genomes, an unrooted dendrogram showing relationships between these genomes was constructed by neighbor-joining method using software POPULATIONS 1.2.32. (available at www.cnrs-gif.fr/pge/bioinfo/populations) with bootstrap support (400 replicates) obtained by re-sampling the allelic frequency data. The trees were visualized with MEGA ver. 5.05 [25].
Results
Azuki Bean EST-SSR Markers
A total of 11,167 cDNA clones were sequenced consisting of 7,534 clones from a seedling library and 3,633 clones from a young pod library. After clustering, 4,896 potential non-redundant EST sequences, including 2,350 contigs and 2,546 singletons, were generated with a total of 4,284,693 qualified bases (Table S3). By using the fuzznuc program in EMBOSS, a total of 1,188 SSRs were identified in the 4,896 non-redundant EST sequences. Of the 1,188 SSRs, di-, tri-, and tetra-nucleotide SSRs accounted for 71.6%, 26.7%, and 1.7%, respectively (Table S4). Assuming that total length of the non-redundant azuki bean EST sequences is 4.3 Mbp, the frequency of occurrence of the SSRs in transcribed regions was estimated to be one in every 3.6 kb. Altogether105 primer pairs were initially designed on the flanking regions of the 132 perfect SSR motifs (Table S4). To increase the number of candidate EST-SSR markers, additional 1,324 primer pairs were designed on the flanking regions of 1,545 imperfect SSR motifs allowing one or two base mismatching. As a result, a total of 1,429 EST-SSR markers were designed (Table S4), of which 149, 28, 5 and 5 markers identified 2, 3, 4 and ≤5 SSRs in the regions between the primer pairs, respectively. Thus the total number of identified SSRs by the 1,429 EST-SSR markers was 1,677 that consisted of 137 (8.2%) di-nucleotide repeats, 1,400 (83.5%) tri-nucleotide repeats and 140 (8.3%) tetra-nucleotide repeats (Table S4). Among the di-nucleotide repeats, poly(AG)n (n = 88, 5.2% of total) were most frequently observed, followed by poly(AT)n (n = 35, 2.1%) and poly (AC)n (n = 14, 0.8%). Among the ten types of tri-nucleotide repeats observed, poly(AAG)n (n = 322, 19.2%) were the most abundant, followed by poly(GGA)n (n = 195, 11.6%) and poly(ATC)n (n = 180, 10.7%). Among the thirteen tetra-nucleotide repeats, poly(AAAG)n (n = 45, 2.7%) were the most frequently observed, followed by poly(AAAT)n (n = 30, 1.8%), and poly(AAAC)n (n = 20, 1.2%). The details of the designed azuki bean EST-SSR primers, along with the corresponding SSR motif, product size, and primer sequence, are available at http://marker.kazusa.or.jp/Azuki and in Table S5.
Amplification and Transferability of Azuki Bean EST-SSR Markers
Transferability and polymorphism of the 1,429 azuki bean EST-SSR markers were initially examined by polyacrylamide gel electrophoresis using eight accessions consisting of four major Vigna crop species; azuki bean complex, black gram, rice bean and mungbean (Table 1, Table S1). These wild and cultivated parental accessions had been used for a linkage map construction in each species ([27] [28] [29] [30]. Amplification of azuki bean EST-SSR markers in the same species, V. angularis, and the closely related species, V. umbellata, were 91.3 and 92.9% which were higher than those in V. radiata and V. mungo (Table 1). However, the size and pattern of PCR products of EST-SSR markers did not always reveal simple banding pattern with expected size, even in the same species. Among the amplified markers, 81.8 to 83.6% of them revealed simple banding pattern suitable for further application. When polymorphism between cultivated and wild accessions was examined, V. radiata possessed the highest polymorphism in which 277 markers (23.1% of amplified markers) were polymorphic. Further, 175 markers with good transferability in the four Vigna species were tested for polymorphism in a panel of 16 accessions from 8 taxa of Asian Vigna (Table S1). The test revealed that 22 markers (12.6%) failed to amplify, 36 (20.6%) amplified some accessions, whereas 117 (66.8%) successfully amplified all the 16 accessions (Table 2). Among the 117 amplifiable markers, 2 (1.1%) gave product size larger than 500 bp, 4 (2.3%) amplified multiple products, 45 (25.7%) amplified one product in tetraploid species, 10 (5.7%) were monomorphic, and 56 (32%) were polymorphic, amplifying one product in diploid species and two discrete products in tetraploid species and were considered to be suitable for analyzing origin of genome donor of the tetraploid species.
Table 2. Characteristics of 196 EST-SSR primers developed in this study.
| Type | Description | Number of primers (%) | |
| 1 | Not amplified in all accessions | 22 (12.6) | |
| Amplified in some accessions | 36 (20.6) | ||
| 2–1) | 1) Amplified in 1 to 8 accession(s) | 5 (2.9) | |
| 2–2) | 2) Amplified in 9 to 15 accessions | 31 (17.7) | |
| Amplified in all 16 accessions | 117 (66.8) | ||
| 3–1) | 1) PCR product of more than 500 bp | 2 (1.1) | |
| 3–2) | 2) Multiple PCR product | 4 (2.3) | |
| 3–3) | 3) Monomorphic PCR product | 10 (5.7) | |
| 3–4) | 4) Single PCR product in tetraploid | 45 (25.7) | |
| 3–5) | 5) Two PCR products in tetraploid | 56 (32) | |
| Total | 175 (100.0) | ||
EST-SSR Polymorphism and Genetic Diversity
Thirty-eight primer pairs of azuki bean EST-SSR were analyzed in 268 accessions of 10 genomes from eight taxa of Asian Vigna. Based on discrimination of the diploid species we distinguished two discrete products detected by each marker of V. reflexo-pilosa into two types, viz. A and B. The product size that is the same or very similar to V. trinervia was scored as type A, while the other product size was scored as type B. In total, 445 alleles were detected in 10 genomes by the 38 EST-SSR loci (Tables 3, 4). The number of alleles detected per locus ranged between 3 (VES0777 and VES1271) and 34 (VES1172) with a mean of 11.7 alleles per locus (Table 4). The PIC values ranged from 0.36 (VES0116) to 0.94 (VES1172) with a mean of 0.67. None of the EST-SSR markers had PIC values lower than 0.3, while 20 markers had PIC values higher than 0.7. The markers showed high PIC value in V. hirtella (0.44) and V. minima (0.42), but low value in V. reflexo-pilosa (0.04-0.09). In general, each marker showed a PIC value of 0 in both populations of V. reflexo-pilosa. The allelic richness was between 2.0 for marker VES0777 and 9.9 for marker VES1172 with a mean of 4.7. Fifteen primers had allelic richness higher than 5.0 (Table 4).
Table 3. Genome number assigned in this study with number of alleles, gene diversity and observed heterozygosity analyzed by 38 EST-SSR markers.
| Genome | No. of accessions | No. of loci typed | No. of alleles | Gene diversity | Observed heterozygosity |
| 1 (Vigna exilis) | 13 | 38 | 100 | 0.305 | 0.012 |
| 2 [V. reflexo-pilosa var. glabra (a)] | 7 | 38 | 44 | 0.053 | 0.000 |
| 3 [V. reflexo-pilosa var. glabra (b)] | 7 | 38 | 44 | 0.052 | 0.000 |
| 4 (V. hirtella) | 47 | 38 | 171 | 0.478 | 0.019 |
| 5 (V. minima) | 49 | 38 | 203 | 0.458 | 0.017 |
| 6 [V. reflexo-pilosa var. reflexo-pilosa (a)] | 51 | 38 | 56 | 0.058 | 0.000 |
| 7 [V. reflexo-pilosa var. reflexo-pilosa (b)] | 51 | 38 | 76 | 0.094 | 0.000 |
| 8 (V. radiata var. sublobata) | 13 | 38 | 101 | 0.357 | 0.008 |
| 9 (V. tenuicaulis) | 42 | 38 | 122 | 0.309 | 0.022 |
| 10 (V. trinervia) | 46 | 38 | 98 | 0.306 | 0.028 |
| Total | 326 | 38 | 445 | 0.711 | 0.013 |
Table 4. EST-SSR primers used, number of alleles per locus, allele size range, polymorphic information content (PIC) and allelic richness for each genome.
| PIC | Allelic richness | |||||||||||||||||||||||
| Genome number | Genome number | |||||||||||||||||||||||
| Primer | No. of alleles | Allele size range (bp)† | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Overall | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Overall |
| VES0019 | 16 | 253–283 (30) | 0.45 | 0.00 | 0.21 | 0.77 | 0.81 | 0.00 | 0.07 | 0.40 | 0.85 | 0.61 | 0.77 | 3.6 | 1.0 | 2.0 | 5.9 | 6.2 | 1.0 | 1.5 | 2.8 | 7.2 | 3.6 | 5.8 |
| VES0021 | 18 | 217–289 (72) | 0.46 | 0.00 | 0.00 | 0.51 | 0.59 | 0.00 | 0.00 | 0.13 | 0.37 | 0.04 | 0.75 | 3.0 | 1.0 | 1.0 | 3.6 | 4.1 | 1.0 | 1.0 | 1.8 | 3.0 | 1.3 | 5.3 |
| VES0070 | 7 | 265–275 (10) | 0.00 | 0.00 | 0.00 | 0.50 | 0.46 | 0.00 | 0.07 | 0.64 | 0.17 | 0.00 | 0.61 | 1.0 | 1.0 | 1.0 | 3.3 | 2.7 | 1.0 | 1.5 | 3.8 | 2.0 | 1.0 | 3.5 |
| VES0093 | 11 | 183–199 (16) | 0.00 | 0.00 | 0.00 | 0.51 | 0.15 | 0.00 | 0.00 | 0.66 | 0.05 | 0.40 | 0.75 | 1.0 | 1.0 | 1.0 | 3.3 | 1.9 | 1.0 | 1.0 | 3.9 | 1.3 | 2.3 | 5.0 |
| VES0116 | 4 | 278–284 (6) | 0.13 | 0.37 | 0.00 | 0.00 | 0.00 | 0.13 | 0.00 | 0.13 | 0.00 | 0.22 | 0.36 | 1.8 | 2.0 | 1.0 | 1.0 | 1.0 | 1.7 | 1.0 | 1.8 | 1.0 | 1.9 | 2.2 |
| VES0120 | 16 | 245–293 (48) | 0.13 | 0.00 | 0.00 | 0.16 | 0.52 | 0.00 | 0.10 | 0.73 | 0.00 | 0.00 | 0.51 | 1.8 | 1.0 | 1.0 | 2.0 | 4.2 | 1.0 | 1.6 | 4.8 | 1.0 | 1.0 | 3.5 |
| VES0202 | 15 | 218–254 (36) | 0.55 | 0.00 | 0.00 | 0.58 | 0.24 | 0.00 | 0.00 | 0.43 | 0.46 | 0.00 | 0.71 | 4.4 | 1.0 | 1.0 | 3.4 | 2.1 | 1.0 | 1.0 | 2.9 | 2.9 | 1.0 | 4.6 |
| VES0204 | 20 | 310–366 (56) | 0.61 | 0.00 | 0.00 | 0.69 | 0.70 | 0.10 | 0.24 | 0.44 | 0.21 | 0.31 | 0.71 | 4.5 | 1.0 | 1.0 | 4.7 | 5.4 | 1.6 | 2.6 | 2.8 | 2.4 | 2.5 | 5.0 |
| VES0335 | 15 | 265–284 (19) | 0.44 | 0.00 | 0.21 | 0.78 | 0.29 | 0.04 | 0.40 | 0.00 | 0.60 | 0.55 | 0.81 | 2.8 | 1.0 | 2.0 | 6.4 | 2.3 | 1.3 | 2.7 | 1.0 | 3.7 | 3.6 | 6.5 |
| VES0427 | 9 | 316–329 (13) | 0.50 | 0.32 | 0.00 | 0.26 | 0.47 | 0.00 | 0.00 | 0.34 | 0.16 | 0.31 | 0.71 | 3.0 | 2.0 | 1.0 | 2.1 | 3.3 | 1.0 | 1.0 | 2.8 | 1.8 | 2.4 | 4.5 |
| VES0478 | 11 | 298–309 (11) | 0.72 | 0.00 | 0.00 | 0.22 | 0.21 | 0.13 | 0.13 | 0.50 | 0.09 | 0.08 | 0.66 | 5.4 | 1.0 | 1.0 | 1.9 | 1.9 | 1.7 | 1.7 | 3.3 | 1.6 | 1.5 | 4.3 |
| VES0546 | 26 | 431–476 (45) | 0.63 | 0.00 | 0.00 | 0.32 | 0.84 | 0.00 | 0.37 | 0.48 | 0.58 | 0.48 | 0.85 | 4.4 | 1.0 | 1.0 | 2.9 | 7.2 | 1.0 | 3.2 | 3.0 | 3.0 | 2.9 | 7.5 |
| VES0624 | 4 | 265–271 (6) | 0.00 | 0.00 | 0.00 | 0.00 | 0.15 | 0.04 | 0.00 | 0.00 | 0.00 | 0.00 | 0.42 | 1.0 | 1.0 | 1.0 | 1.0 | 1.9 | 1.3 | 1.0 | 1.0 | 1.0 | 1.0 | 2.6 |
| VES0665 | 9 | 195–213 (18) | 0.26 | 0.00 | 0.00 | 0.31 | 0.35 | 0.00 | 0.00 | 0.67 | 0.13 | 0.24 | 0.68 | 2.6 | 1.0 | 1.0 | 2.7 | 2.8 | 1.0 | 1.0 | 5.0 | 1.9 | 2.3 | 4.2 |
| VES0670 | 6 | 99–114 (15) | 0.44 | 0.00 | 0.00 | 0.56 | 0.20 | 0.00 | 0.00 | 0.00 | 0.38 | 0.08 | 0.71 | 2.8 | 1.0 | 1.0 | 3.8 | 2.1 | 1.0 | 1.0 | 1.0 | 3.1 | 1.5 | 4.4 |
| VES0678 | 7 | 293–318 (25) | 0.45 | 0.00 | 0.00 | 0.40 | 0.54 | 0.00 | 0.00 | 0.36 | 0.28 | 0.41 | 0.70 | 2.9 | 1.0 | 1.0 | 2.3 | 3.3 | 1.0 | 1.0 | 2.0 | 2.0 | 2.5 | 4.5 |
| VES0679 | 18 | 310–365 (55) | 0.44 | 0.00 | 0.00 | 0.40 | 0.78 | 0.31 | 0.14 | 0.63 | 0.61 | 0.74 | 0.81 | 2.8 | 1.0 | 1.0 | 3.2 | 5.6 | 2.0 | 1.9 | 4.4 | 3.9 | 5.2 | 7.8 |
| VES0749 | 15 | 208–236 (28) | 0.29 | 0.00 | 0.00 | 0.60 | 0.27 | 0.00 | 0.00 | 0.62 | 0.09 | 0.43 | 0.79 | 2.0 | 1.0 | 1.0 | 3.8 | 2.3 | 1.0 | 1.0 | 4.4 | 1.6 | 2.5 | 5.8 |
| VES0762 | 7 | 242–266 (24) | 0.13 | 0.00 | 0.21 | 0.62 | 0.08 | 0.00 | 0.04 | 0.37 | 0.00 | 0.00 | 0.43 | 1.8 | 1.0 | 2.0 | 3.6 | 1.5 | 1.0 | 1.3 | 2.0 | 1.0 | 1.0 | 3.0 |
| VES0777 | 3 | 166–175 (9) | 0.00 | 0.00 | 0.00 | 0.36 | 0.40 | 0.00 | 0.10 | 0.18 | 0.14 | 0.00 | 0.37 | 1.0 | 1.0 | 1.0 | 2.0 | 2.3 | 1.0 | 1.6 | 1.9 | 1.7 | 1.0 | 2.0 |
| VES0803 | 12 | 292–308 (16) | 0.00 | 0.00 | 0.00 | 0.64 | 0.71 | 0.00 | 0.10 | 0.29 | 0.41 | 0.00 | 0.80 | 1.0 | 1.0 | 1.0 | 4.8 | 4.6 | 1.0 | 1.6 | 2.0 | 2.6 | 1.0 | 6.3 |
| VES0868 | 12 | 213–288 (75) | 0.13 | 0.00 | 0.00 | 0.53 | 0.27 | 0.00 | 0.00 | 0.26 | 0.35 | 0.00 | 0.62 | 1.8 | 1.0 | 1.0 | 4.1 | 2.2 | 1.0 | 1.0 | 2.6 | 2.0 | 1.0 | 4.2 |
| VES0987 | 9 | 298–310 (12) | 0.26 | 0.21 | 0.00 | 0.55 | 0.36 | 0.00 | 0.00 | 0.13 | 0.31 | 0.59 | 0.73 | 2.6 | 2.0 | 1.0 | 3.4 | 3.0 | 1.0 | 1.0 | 1.8 | 2.0 | 3.0 | 5.0 |
| VES1001 | 10 | 227–250 (23) | 0.29 | 0.00 | 0.00 | 0.39 | 0.55 | 0.00 | 0.00 | 0.64 | 0.00 | 0.00 | 0.65 | 2.5 | 1.0 | 1.0 | 2.7 | 3.6 | 1.0 | 1.0 | 3.9 | 1.0 | 1.0 | 3.9 |
| VES1020 | 6 | 170–183 (13) | 0.26 | 0.00 | 0.00 | 0.48 | 0.51 | 0.07 | 0.07 | 0.00 | 0.00 | 0.37 | 0.73 | 2.6 | 1.0 | 1.0 | 2.9 | 3.3 | 1.5 | 1.5 | 1.0 | 1.0 | 2.0 | 4.6 |
| VES1023 | 7 | 140–157 (17) | 0.00 | 0.00 | 0.00 | 0.28 | 0.56 | 0.00 | 0.10 | 0.00 | 0.37 | 0.00 | 0.59 | 1.0 | 1.0 | 1.0 | 2.2 | 3.7 | 1.0 | 1.6 | 1.0 | 2.2 | 1.0 | 3.4 |
| VES1029 | 9 | 111–128 (17) | 0.58 | 0.00 | 0.00 | 0.62 | 0.04 | 0.00 | 0.00 | 0.00 | 0.52 | 0.00 | 0.61 | 3.8 | 1.0 | 1.0 | 3.8 | 1.3 | 1.0 | 1.0 | 1.0 | 2.9 | 1.0 | 3.9 |
| VES1067 | 14 | 447–477 (30) | 0.13 | 0.00 | 0.37 | 0.68 | 0.60 | 0.00 | 0.32 | 0.36 | 0.17 | 0.37 | 0.83 | 1.8 | 1.0 | 2.0 | 4.5 | 3.9 | 1.0 | 2.8 | 2.0 | 2.1 | 2.0 | 6.7 |
| VES1082 | 14 | 281–317 (36) | 0.40 | 0.00 | 0.00 | 0.51 | 0.70 | 0.00 | 0.00 | 0.34 | 0.13 | 0.46 | 0.70 | 2.8 | 1.0 | 1.0 | 3.1 | 4.8 | 1.0 | 1.0 | 2.8 | 1.9 | 2.8 | 5.0 |
| VES1085 | 13 | 390–403 (13) | 0.00 | 0.00 | 0.00 | 0.33 | 0.60 | 0.31 | 0.13 | 0.00 | 0.50 | 0.74 | 0.82 | 1.0 | 1.0 | 1.0 | 2.6 | 4.1 | 2.5 | 1.7 | 1.0 | 2.8 | 4.7 | 6.6 |
| VES1172 | 34 | 217–268 (51) | 0.71 | 0.53 | 0.37 | 0.85 | 0.89 | 0.58 | 0.82 | 0.60 | 0.41 | 0.59 | 0.94 | 6.0 | 3.0 | 2.0 | 7.4 | 9.0 | 3.8 | 6.5 | 3.8 | 2.9 | 4.3 | 9.9 |
| VES1196 | 5 | 203–215 (12) | 0.00 | 0.00 | 0.00 | 0.30 | 0.15 | 0.00 | 0.00 | 0.00 | 0.09 | 0.00 | 0.38 | 1.0 | 1.0 | 1.0 | 2.5 | 1.9 | 1.0 | 1.0 | 1.0 | 1.5 | 1.0 | 2.1 |
| VES1231 | 6 | 120–139 (19) | 0.23 | 0.00 | 0.00 | 0.53 | 0.08 | 0.00 | 0.00 | 0.00 | 0.34 | 0.00 | 0.52 | 2.0 | 1.0 | 1.0 | 2.9 | 1.5 | 1.0 | 1.0 | 1.0 | 2.6 | 1.0 | 3.5 |
| VES1258 | 10 | 369–398 (29) | 0.54 | 0.00 | 0.00 | 0.22 | 0.59 | 0.04 | 0.00 | 0.00 | 0.50 | 0.48 | 0.76 | 3.0 | 1.0 | 1.0 | 1.9 | 3.6 | 1.3 | 1.0 | 1.0 | 2.9 | 3.0 | 5.5 |
| VES1263 | 8 | 305–320 (15) | 0.13 | 0.00 | 0.00 | 0.00 | 0.43 | 0.07 | 0.00 | 0.56 | 0.00 | 0.30 | 0.68 | 1.8 | 1.0 | 1.0 | 1.0 | 2.7 | 1.5 | 1.0 | 3.6 | 1.0 | 2.0 | 4.4 |
| VES1271 | 3 | 307–310 (3) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.05 | 0.37 | 0.42 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.3 | 2.0 | 2.6 |
| VES1310 | 7 | 288–309 (21) | 0.23 | 0.00 | 0.00 | 0.37 | 0.25 | 0.00 | 0.10 | 0.60 | 0.60 | 0.30 | 0.55 | 2.0 | 1.0 | 1.0 | 3.0 | 2.5 | 1.0 | 1.6 | 3.8 | 3.6 | 2.0 | 3.3 |
| VES1469 | 29 | 128–178 (50) | 0.00 | 0.21 | 0.21 | 0.82 | 0.79 | 0.17 | 0.04 | 0.61 | 0.64 | 0.66 | 0.86 | 1.0 | 2.0 | 2.0 | 6.5 | 6.1 | 2.0 | 1.3 | 3.8 | 5.3 | 5.0 | 7.7 |
| Total | 445 | |||||||||||||||||||||||
| Average | 11.7 | 0.28 | 0.04 | 0.04 | 0.44 | 0.42 | 0.05 | 0.09 | 0.32 | 0.28 | 0.27 | 0.67 | 2.43 | 1.16 | 1.16 | 3.24 | 3.34 | 1.27 | 1.53 | 2.49 | 2.33 | 2.15 | 4.75 | |
Difference between the largest and smallest fragments amplified by each primer is shown in parentheses.
See Table 3 for the abbreviations of genome number.
V. hirtella possessed the highest gene diversity (0.478), while V. reflexo-pilosa showed very low gene diversity (0.053 to 0.094) (Table 3). All genomes of V. reflexo-pilosa showed no observed heterozygosity, while the other genomes showed very low observed heterozygosity from 0.008 to 0.028 (Table 3).
Genetic Relationship among Genomes
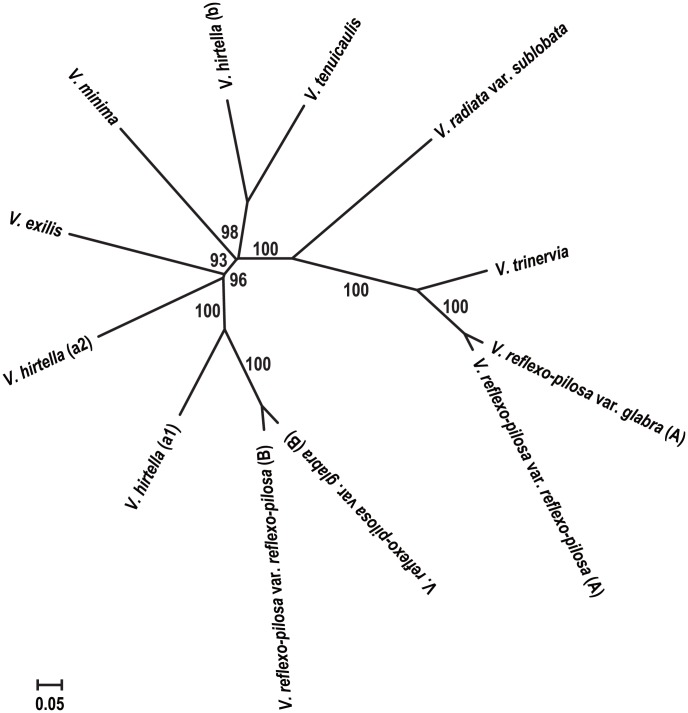
Genetic distance (D A) among the ten genomes is shown in Table 5a. In most cases D A among genomes was high (>0.6). Nonetheless, D A between V. trinervia and V. reflexo-pilosa var. glabra (A) or V. reflexo-pilosa var. reflexo-pilosa (A) was low being 0.333 and 0.313, respectively. While, both V. reflexo-pilosa var. glabra (B) and V. reflexo-pilosa var. reflexo-pilosa (B) showed lowest D A with V. hirtella being 0.585 and 0.589 (Table 5a) or with V. hirtella (a1), being 0.429 and 0.422 (Table 6), respectively. Interestingly, V. trinervia, which belongs to section Angulares showed lower D A with V. radiata var. sublobata (section Ceratotropis) than with those species in the section Angulares.
Table 5. Genetic distance(D A) within and among 10 genomes with (Vigna hirtella is not divided).
| Genome | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| 1 | 0.326 | |||||||||
| 2 | 0.900 | 0.061 | ||||||||
| 3 | 0.695 | 0.985 | 0.060 | |||||||
| 4 | 0.649 | 0.927 | 0.585 | 0.482 | ||||||
| 5 | 0.740 | 0.881 | 0.740 | 0.693 | 0.464 | |||||
| 6 | 0.901 | 0.079 | 0.987 | 0.922 | 0.870 | 0.059 | ||||
| 7 | 0.690 | 0.989 | 0.098 | 0.589 | 0.740 | 0.990 | 0.097 | |||
| 8 | 0.848 | 0.817 | 0.875 | 0.838 | 0.884 | 0.817 | 0.878 | 0.385 | ||
| 9 | 0.712 | 0.895 | 0.716 | 0.652 | 0.716 | 0.894 | 0.720 | 0.829 | 0.310 | |
| 10 | 0.891 | 0.333 | 0.963 | 0.890 | 0.886 | 0.313 | 0.965 | 0.799 | 0.849 | 0.304 |
Table 6. Genetic distance(D A) within and among 10 genomes (Vigna hirtella is divided into three genomes).
| Genome | 1 | 2 | 3 | 4 (a1) | 4 (a2) | 4 (b) | 5 | 6 | 7 | 8 | 9 | 10 |
| 1 | 0.326 | |||||||||||
| 2 | 0.900 | 0.061 | ||||||||||
| 3 | 0.695 | 0.985 | 0.060 | |||||||||
| 4 (a1) | 0.625 | 0.911 | 0.429 | 0.377 | ||||||||
| 4 (a2) | 0.626 | 0.931 | 0.600 | 0.609 | 0.298 | |||||||
| 4 (b) | 0.749 | 0.926 | 0.672 | 0.672 | 0.646 | 0.321 | ||||||
| 5 | 0.740 | 0.881 | 0.740 | 0.704 | 0.689 | 0.700 | 0.464 | |||||
| 6 | 0.901 | 0.079 | 0.987 | 0.908 | 0.925 | 0.922 | 0.870 | 0.059 | ||||
| 7 | 0.690 | 0.989 | 0.098 | 0.422 | 0.606 | 0.680 | 0.740 | 0.990 | 0.097 | |||
| 8 | 0.848 | 0.817 | 0.875 | 0.899 | 0.824 | 0.831 | 0.884 | 0.817 | 0.878 | 0.385 | ||
| 9 | 0.712 | 0.895 | 0.716 | 0.713 | 0.697 | 0.446 | 0.716 | 0.894 | 0.720 | 0.829 | 0.310 | |
| 10 | 0.891 | 0.333 | 0.963 | 0.892 | 0.895 | 0.874 | 0.886 | 0.313 | 0.965 | 0.799 | 0.849 | 0.304 |
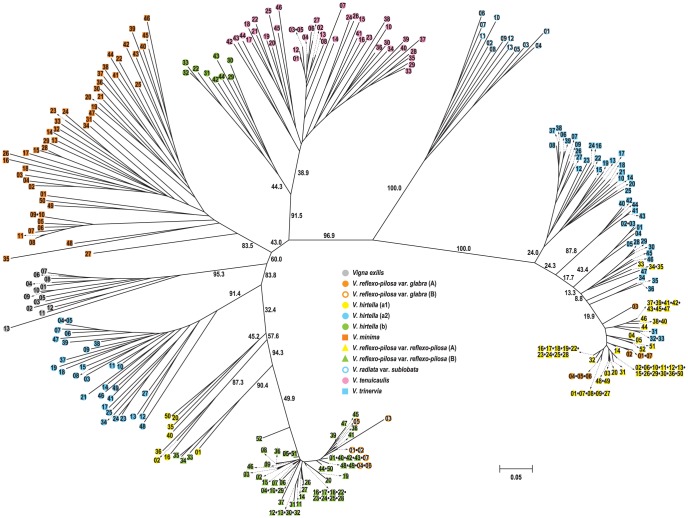
A neighbor-joining tree was constructed based on the genetic distances for all possible pairs of 268 accessions. The tree showed two major groups with 96.9% bootstrap value, namely azuki bean group and mungbean group (Figure 2). V. exilis, V. minima, V. tenuicaulis, V. hirtella, V. reflexo-pilosa var. glabra (B), and V. reflexo-pilosa var. reflexo-pilosa (B) were clustered in the azuki bean group. While V. radiata var. sublobata, V. trinervia, V. reflexo-pilosa var. glabra (A) and V. reflexo-pilosa var. reflexo-pilosa (A) were clustered in the mungbean group. In the azuki bean group, 13 accessions of V. exilis and 49 accessions of V. minima were independent clusters with 95.3% and 83.5% bootstrap values, respectively. V. hirtella showed high divergence in which 9 accessions in the population of V. hirtella (b) clustered strongly with V. tenuicaulis with 91.5% bootstrap value, while the other 38 accessions in V. hirtella (a) clustered with V. reflexo-pilosa var. glabra (B) and V. reflexo-pilosa var. reflexo-pilosa (B) with 83.8% bootstrap value. Those 38 accessions of V. hirtella also showed two subclusters a1 and a2 (Figure 2 and 3). In the mungbean group, V. trinervia clustered with V. reflexo-pilosa var. glabra (A) and V. reflexo-pilosa var. reflexo-pilosa (A) with a 100% bootstrap support. Five accessions from Myanmar (40–44) formed a distinct branch with a bootstrap value of 87.8% and were separated from the other accessions (Figure 2). Three accessions from Laos (31–33) were differentiated from the other accessions and were clustered together with V. reflexo-pilosa (A) accessions.
Figure 2. A phylogenetic tree showing relationship among 286 accessions in 12 sub-genome groups of the genus Vigna subgenus Ceratotropis based on variation at 38 EST-SSR loci.
Figure 3. A phylogenetic tree showing relationship among 12 sub-genome groups of the genus Vigna subgenus Ceratotropis constructed based on the genetic distance shown in Table 6.
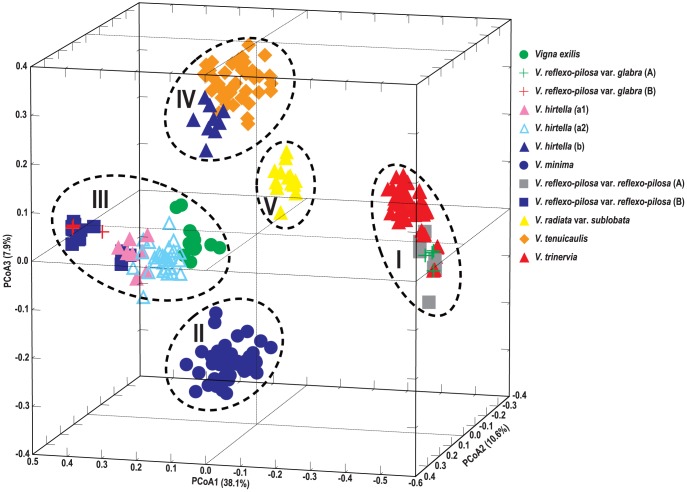
The principle coordinate analysis (PCoA) was also conducted to confirm species relationship based on the neighbor-joining tree using the same genetic distance estimates. The first three PCs together accounted up to 56.6% of the total variation. The first, second and third PCs accounted for 38.1%, 10.6% and 7.9%, respectively (Figure 4). A three-dimensional PC plot (PC1, PC2 and PC3) of the twelve genome groups is shown in Figure 4, which clearly separated the genomes into 5 distinct groups. Group I comprised V. reflexo-pilosa var. reflexo-pilosa (A), V. reflexo-pilosa var. glabra (A), and V. trinervia. Group II comprised V. minima only. Group III comprised V. reflexo-pilosa var. glabra (B), V. reflexo-pilosa var. reflexo-pilosa (B), V. exilis and V. hirtella (a1, a2). Group IV comprised V. hirtella (b), and V. tenuicaulis. Group V comprised solely V. radiata var. sublobata. Distribution range of V. reflexo-pilosa var. glabra (A) and V. reflexo-pilosa var. reflexo-pilosa (A) in Group I was narrower than V. reflexo-pilosa var. glabra (B) and V. reflexo-pilosa var. reflexo-pilosa (B) in Group III. Of all species used in this study, only V. hirtella was unambiguously distinguished into two sub-groups, a and b. Subgroup a showed close relationship with populations of V. reflexo-pilosa var. reflexo-pilosa (B); V. reflexo-pilosa var. glabra (B) and V. exilis, while subgroup b showed close relationship with V. tenuicaulis (Figure 3).
Figure 4. PCA scattered plot depicting relationship among 286 accessions in 12 sub-genome groups of the genus Vigna subgenus Ceratotropis based on variation at 38 EST-SSR loci.
Discussion
Transferability of Azuki Bean EST-SSRs
One of the advantages of SSRs derived from EST (EST-SSR markers) to other DNA marker systems is the high transferability across species and genera because of primer pairs are designed from conserved transcribed regions, although they are generally less polymorphic than genomic SSR markers (gSSR). In this study, more than 83% of the azuki bean EST-SSR markers were able to amplify DNA from 8 Asian Vigna species including four major crop species, viz. V. angularis, V. umbellata, V. radiata and V. mungo of the subgenus Ceratotropis. This result is in agreement with that of [31] who reported that more than 80% of mungbean EST-SSR markers were transferable to other species in the subgenus Ceratotropis. Transferability of the EST-SSR markers from azuki bean (in this study) and mungbean [31] is greater than that of gSSR markers from azuki bean (>67% transferable; [28]) and mungbean (>59% transferable; [32]). The high transferability of azuki bean EST-SSRs and of mungbean EST-SSRs to the related Vigna species indicates high genome conservation among the species in the subgenus Ceratotropis. Therefore, the azuki bean EST-SSR markers developed in this study will be useful for comparative genomic and genetic diversity studies in Vigna species. Nonetheless, as expected, polymorphism rate of the azuki bean EST-SSR markers detected between cultivated and wild accessions of azuki bean, rice bean, mungbean and black gram is low (14% in black gram and 23% in mungbean). In the same plant materials, Chaitieng et al. [28] reported that polymorphism rate of the genomic azuki bean SSRs was as high as 50% in black gram to 63% in mungbean. Thus low polymorphism rate of the azuki bean EST-SSRs can be an undesirable character for the use of these markers in comparative genome mapping in the genus Vigna. However, EST-SSRs represent true genetic diversity which may be more directly associated with traits of interest in breeding as compared to gSSRs [33], and they may reflect better relationships among the related species in genetic diversity study.
Evolution and Domestication of Tetraploid V. reflexo-pilosa
V. reflexo-pilosa is the only tetraploid species of the genus Vigna. Although previous studies on Vigna species using DNA markers and plastid sequences clearly demonstrated that V. trinervia is a genome donor of V. reflexo-pilosa, those studies were unable to identify the other genome donor. In the present study, five putative genome donor species of V. reflexo-pilosa belong to the section Angulares and one species representing the section Ceratotropis were analyzed with azuki bean EST-SSRs. By assigning the discrete PCR products of V. reflexo-pilosa into two distinguished genomes (designated as genome A and B), we were able to identify the two genome donor parents of créole bean. Phylogenetic tree (Figure 2) and PCoA plot (Figure 4) clearly demonstrated that genome A of V. reflexo-pilosa is received from V. trinervia. This result confirms the previous results reported by [7] [10] [13] [14] [15] that V. trinervia is a diploid genome donor. V. trinervia and V. reflexo-pilosa shares several similar morphological characters such as seed shape (rectangular), non-protruded hilum and large golden flower. The phylogenetic tree and the PCoA plot also unambiguously demonstrated that the other genome (genome B) of V. reflexo-pilosa is originated from V. hirtella. This result is in agreement with the result reported by Tateishi [7] [10] [13] [16] that V. hirtella is one of candidate genome donor of créole bean. Results from phylogenetic analysis of plastid DNA sequences of [14] [15] suggested that V. trinervia is the maternal genome donor of V. reflexo-pilosa. Since the previous and our results demonstrated close genetic relationship between tetraploid V. reflexo-pilosa and diploid V. trinervia/V. hirtella, we propose that créole bean evolved from interspecific hybridization between V. trinervia as female parent and V. hirtella as male parent, followed by genome duplication.
Phylogenetic tree (Figure 2) and PCoA plot (Figure 4) showed lower divergence of genome A of V. reflexo-pilosa compared to genome B. Very low gene diversity within both genomes A and B suggested that this tetraploid species has a monophyletic (single) origin and only evolved recently. Gene diversity within the genome A in both V. reflexo-pilosa var. reflexo-pilosa (wild form) and V. reflexo-pilosa var. glabra (cultivated form) is very similar (0.058 vs. 0.053), while gene diversity of the genome B in wild créole bean is about twice of that in the cultivated one (0.94 vs. 0.52) (Table 3). This indicates that the evolution rate of EST-SSRs on genome B of créole bean might be faster than that on genome A. The marked morphological differences between the cultivated and wild créole beans are thicker and erect glabrous stem in the former. Moreover, cultivated V. reflexo-pilosa still possesses a relatively high degree of pod shattering (Somta and Chankaew, personal observation), an important domestication trait for legume crops [34]. Therefore, the cultivated V. reflexo-pilosa can be treated as a semi-domesticated form. Based on cultivation, utilization and distribution of diploid genome donor species, créole bean appears to have been domesticated in Southeast Asia. The crop is now very rarely cultivated, although it is produced in northern mountainous villages in Vietnam under the same name and consumed in the same way as mungbean [2].
Genetic Structure of Diploid Asian Vigna Species Detected by EST-SSR
We found that V. hirtella possessed greater genetic variation than V. exilis, V. minima, and V. tenuicaulis. A previous study based on AFLP suggested that V. hirtella germplasm collected from Thailand, Malaysia and Myammar is a species complex consisting of two taxa (types), called V. hirtella (a) and V. hirtella (b) [35]. Although the study used only 3 accessions for V. hirtella (a) and 5 accessions for V. hirtella (b), all the 3 V. hirtella (a) accessions were included in V. hirtella (a1) group, and 5 V. hirtella (b) accessions were included in V. hirtella (a2) group in the present study. In addition, our EST-SSR results detected the existence of another genetically distinct type of materials designated as V. hirtella (b) which is closely related to V. tenuicaulis (Figure 2, 3, 4). These accessions should be re-examined morphologically whether they can be included within a morphological variation of V. tenuicaulis.
V. trinervia is widely distributed across Asia and also found in Papua New Guinea, Madagascar and East Africa. It is the second most widely distributed species in the subgenus Ceratotropis after V. radiata var. sublobata [2]. Nevertheless, gene diversity of V. trinervia was not high as compared to the other related species (Table 3). In Southeast Asia, especially in Thailand and the peninsular Malaysia, V. trinervia is often found on rural roadside habitats and in or near to rubber or oil palm plantations. It seems that some of the populations in those areas were introduced recently as a cover plant in the plantations [2]. This may account for the low diversity of V. trinervia.
V. tenuicaulis distributes in open wet habitats in northern Southeast Asia, and appeared to be closely related to V. hirtella (b) and V. angularis [13] [35] [6] [36]. Tomooka et al. suggested that there are no major barriers to hybridization between V. tenuicaulis and V. hirtella (b) [2]. Our results confirmed the close relationship between these two species (Figure 2, 4). Previous studies based on molecular, biochemical and morphological variations all showed high level of distinctness and variation within V. tenuicaulis. EST-SSR marker variation in our study revealed contrasting results. Although we used as many as 30 accessions of V. tenuicaulis from three countries, viz. Laos, Myanmar and Thailand. (Figure 1, Table S1), the species showed relatively low gene diversity (0.309).
V. exilis has been reported only in Thailand and Myanmar [2] [14]. In Thailand, this species is restricted to rocky limestone mountains. Its habitats suggest that it may be useful as gene sources for resistant to alkaline soil and drought conditions. SSR analysis revealed high level of intra-specific diversity in V. exilis [37]. Our results also supported this despite only 13 accessions from narrow geographical origin (west of Thailand) of this species were used (Figure 1, Table S1), the species showed similar level of gene diversity to V. tenuicaulis and V. trinervia (Table 3).
V. minima has broad environments adaptation and grows well in shaded deciduous forest floors and open-wet habitats in East and Southeast Asia [2]. It is the only species in section Angulares that is found on the forest floor. Among the Asian Vigna species analyzed in this study, V. minima is the second most diverse species after V. hirtella (Table 3). The high differentiation of V. minima is due to wide geographical distribution of the analyzed accessions. In Southeast Asia, isolation of V. minima in forests in different mountainous regions across Thailand and Myanmar and its sporadic occurrence in patches of forests in those regions may account for high level of population divergence [35]. Accessions of V. minima in the present study were most closely related with V. hirtella, especially V. hirtella (b), followed by V. exilis (Figure 2, 4). Similar finding was reported by [35]. This agrees with their morphological appearance [2] that V. minima is sometimes confused with V. hirtella. V. minima can be distinguished from V. hirtella by smaller bracteole and more protruding hilum with well-developed rim-aril [2].
In summary, one of the advantages of EST-SSR markers to other DNA marker systems is their co-dominant nature and high transferability to closely related species. In this study, we used EST-SSR primer pairs, which amplified one PCR product in diploid species and two discrete PCR products in tetraploid species to separate genomes of V. reflexo-pilosa into 2 different diploid types, viz. A and B, that could successfully determine different sets of the donor genomes. Phylogenetic analyses revealed that V. trinervia and V. hirtella are the genome donors of V. reflexo-pilosa. Both genomes of cultivated and wild créole bean accessions showed low divergence suggesting that domestication of V. reflexo-pilosa is a relatively recent event.
Supporting Information
Origin of 268 accessions used to analyze origin of genome of V. reflexo-pilosa .
(XLSX)
Transferability and polymorphisms of azuki bean EST-SSR markers in the other Vigna species.
(XLSX)
EST composition in non-redundant azuki bean EST sequences.
(XLSX)
Summary of SSRs in the non-redundant azuki bean ESTs and designed EST-SSR primers.
(XLSX)
Primer sequences, SSR motifs, expected sizes and EST sequences of the designed EST-SSR markers.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All azuki bean EST-SSR primers, along with the corresponding SSR motif, product size, and primer sequence, are available at http://marker.kazusa.or.jp/Azuki. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by the Thailand Research Fund (TRF). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Verdcourt B (1970) Studies in the Leguminosae–Papilionoideae for the flora of tropical East Africa, IV. Kew Bull 24: 507–569. [Google Scholar]
- 2.Tomooka N, Vaughan D A, Moss H, Maxted N (2002) The Asian Vigna: genus Vigna subgenus Ceratotropis genetic resources. Kluwer Academic Press. 270 pp. [Google Scholar]
- 3. Maréchal R, Mascherpa J M, Stainer F (1978) Etude taxonomique d'un groupe complexe d'espe′ces des genres Phaseolus et Vigna (Papilionaceae) sur la base de donne′es morphologiques et polliniques, traite′es par l'analyse informatique. Boissiera 28: 1–273. [Google Scholar]
- 4. Swindell RE, Watt EE, Evans GM (1973) A natural tetraploid mungbean of suspected amphidiploid origin. J Hered 64: 107. [Google Scholar]
- 5. Egawa Y, Siriwardhane D, Yagasaki K, Hayashi H, Takamatsu M, et al. (1990) Collection of millets and grain legume in Shimonai district of Nagano Prefecture, 1989. Annual report on exploration and introduction of plant genetic resources (NIAS, Tsukuba, Japan) 6: 1–22. [Google Scholar]
- 6. Tomooka N, Maxted N, Thavarasook C, Jayasuriya AHM (2002) Two new species, new species combinations and sectional designations in Vigna subgenus Ceratotropis (Piper) Verdcourt (Leguminosae, Phaseoleae). Kew Bull 57: 613–624. [Google Scholar]
- 7. Tateishi Y (1985) A revision of the Azuki bean group, the subgenus Ceratotropis of the genus Vigna (Leguminosae). Ph. D. Thesis, Tohoku University, Japan [Google Scholar]
- 8. Tateishi Y, Ohashi H (1990) Systematics of the azuki bean group in the genus Vigna . In Bruchid and Legumes: Ecology and Coevolution eds Fujii, K., et al Kluwer Akademic Publishers, Netherlands, pp 189–199. [Google Scholar]
- 9. Tomooka N, Kobayashi N, Kamuou RN, Risimeri J, Poafa J, et al. (2005) Ecological survey and conservation of legume symbiotic rhizobia genetic diversity in Papua New Guinea, 2004. Annual Report on Exploration and Introduction of Plant Genetic Resources NIAS. Vol. 21: 135–143. [Google Scholar]
- 10. Egawa Y, Bujang IB, Chotechuen S, Tomooka N (1996) TateishiY (1996) Phylogenetic differentiation of tetraploid Vigna species, V. glabra and V. reflexo-pilosa. . JIRCAS J 3: 49–58. [Google Scholar]
- 11. Chen HK, Mok MC, Shanmugasundaram S, Mok DWS (1989) Interspecific hybridization between Vigna radiata L. Wilczek and V. glabra . Theor Appl Genet 78: 641–647. [DOI] [PubMed] [Google Scholar]
- 12.Egawa Y, Chotechuen S, Tomooka N, Thavarasook C, Kitbamroong C (1996) Cross-compatability among the subgenus Ceratotropis of the genus Vigna. In Egawa Y, Chotechuen S (eds.) Phylogenetic differentiation of mungbean germplasm (subgenus Ceratotropis of the genus Vigna) and evaluation for breeding program. Japan International Research Center for Agricultural Science, Japan, pp. 19–30.
- 13. Konarev AV, Tomooka N, Vaughan DA (2002) Proteinase inhibitor polymorphism in the genus Vigna subgenus Ceratotropis and its biosystematic implications. Euphytica 123: 165–177. [Google Scholar]
- 14. Ye Tun Tun, Yamaguchi H (2007) Phylogenetic relationship of wild and cultivated Vigna (Subgenus Ceratotropis, Fabaceae) from Myanmar based on sequence variations in non-coding regions of trnT-F . Breed Sci 57: 271–280. [Google Scholar]
- 15. Javadi F, Ye Tun Tun, Kawase M, Guan K, Yamaguchi H (2011) Molecular phylogeny of the subgenus Ceratotropis (genus Vigna, Leguminosae) reveals three eco-geographical groups and Late Pliocene–Pleistocene diversification: evidence from four plastid DNA region sequences. Ann Bot 108: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Doi K, Kaga A, Tomooka N, Vaughan DA (2002) Molecular phylogeny of genus Vigna subgenus Ceratotropis based on rDNA-ITS and atpB-rbcL intergenic spacer region of cpDNA sequences. Genetica 114: 129–145. [DOI] [PubMed] [Google Scholar]
- 17. Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S (1999) A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res 6: 369–373. [DOI] [PubMed] [Google Scholar]
- 18. Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185. [DOI] [PubMed] [Google Scholar]
- 19. Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8: 186–194. [PubMed] [Google Scholar]
- 20. Huang X, Wang J, Aluru S, Yang SP, Hillier L (2003) PCAP: a whole-genome assembly program. Genome Res13: 2164–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386. [DOI] [PubMed] [Google Scholar]
- 23. Lodhi MA, Ye GN, Weeden NF, Reisch BI (1994) A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol Biol Rep 12: 6–13. [Google Scholar]
- 24. Nei M, Tajima F (1983) TatenoY (1983) Accuracy of estimated phylogenetic trees from molecular data. J Mol Evol 19: 153–170. [DOI] [PubMed] [Google Scholar]
- 25. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4(1): 9 pp. [Google Scholar]
- 27. Han OK, Kaga A, Isemura T, Wang XW, Tomooka N, et al. (2005) A genetic linkage map for azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi] Theor Appl. Genet111: 1278–1287. [DOI] [PubMed] [Google Scholar]
- 28. Chaitieng B, Kaga A, Tomooka N, Isemura T, Vaughan DA (2006) Development of a black gram [Vigna mungo (L.) Hepper] linkage map and its comparison with an azuki bean [Vigna angularis (Willd.) Ohwi and Ohashi] linkage map. Theor Appl Genet 113: 1261–1269. [DOI] [PubMed] [Google Scholar]
- 29. Isemura T, Kaga A, Tomooka N, Shimizu T, Vaughan DA (2010) The genetics of domestication of rice bean, Vigna umbellate. . Ann Bot 106: 927–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Isemura T, Kaga A, Tabata S, Somta P, Srinives P, et al. (2012) Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 7(8): e41304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Somta P, Seehalak W, Srinives P (2009) Development, characterization and cross-species amplification of mungbean (Vigna radiata) genic microsatellite markers. Conserv Genet 10: 1939–1943. [Google Scholar]
- 32. Tangphatsornruang S, Somta P, Uthaipaisanwong P, Chanprasert J, Sangsrakru D, et al. (2009) Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vigna radiata (L.) Wilczek). BMC Plant Biol 9: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choudhary S, Sethy NK, Shokeen B, Bhatia S (2009) Development of chickpea EST-SSR markers and analysis of allelic variation across related species. Theor Appl Genet 118: 591–608. [DOI] [PubMed] [Google Scholar]
- 34. Harlan JR (1992) Crops and Man. 2nd ed. Am. Soc. Agronomy, Madison, WI.. [Google Scholar]
- 35. Seehalak W, Tomooka N, Waranyuwat A, Thipyapong P, Laosuwan P, et al. (2006) Genetic diversity of the Vigna germplasm from Thailand and neighboring regions revealed by AFLP analysis. Genet Resour Crop Evol 53: 1043–1059. [Google Scholar]
- 36. Tomooka N, Yoon MS, Doi K, Kaga A, Vaughan DA (2002) AFLP analysis of a Vigna subgenus Ceratotropis core collection. Genet Resour Crop Evol 49: 521–530. [Google Scholar]
- 37. Kaewwongwal A, Jetsadu A, Somta P, Chankaew S, Srinives P (2013) Genetic diversity and population structure of Vigna exilis and Vigna grandiflora (Phaseoleae, Fabaceae) from Thailand based on microsatellite variation. Botany 91: 653–661. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Origin of 268 accessions used to analyze origin of genome of V. reflexo-pilosa .
(XLSX)
Transferability and polymorphisms of azuki bean EST-SSR markers in the other Vigna species.
(XLSX)
EST composition in non-redundant azuki bean EST sequences.
(XLSX)
Summary of SSRs in the non-redundant azuki bean ESTs and designed EST-SSR primers.
(XLSX)
Primer sequences, SSR motifs, expected sizes and EST sequences of the designed EST-SSR markers.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All azuki bean EST-SSR primers, along with the corresponding SSR motif, product size, and primer sequence, are available at http://marker.kazusa.or.jp/Azuki. All other relevant data are within the paper and its Supporting Information files.