Abstract
WWOX, the gene that maps to common chromosomal fragile site FRA16D, is frequently affected by aberrations in multiple types of cancers. WWOX encodes a 46 kDa protein that contains two WW domains and a short-chain oxidoreductase (SDR) domain. We recently demonstrated that ectopic expression of WWOX inhibits xenograft tumor growth of tumorigenic breast cancer cells. Little is known of the biochemical function(s) of WWOX. The SDR domain is predicted to be involved in sex-steroid metabolism and the WW domains are likely involved in protein–protein interactions. In this report, we identify the specific proline-rich ligand for WWOX as PPXY and show that the amino-terminal WW domain is responsible for this interaction. Using the WWOX WW domains as a probe, we screened high-density protein arrays and identified five candidate-binding partners. The binding to one of these candidates, small membrane protein of the lysosome/late endosome (SIMPLE), was further analysed, and we observed that a specific PPSY motif in the SIMPLE amino-acid sequence was required to interact with the amino-terminal WW domain of WWOX. In addition, immunofluorescence staining demonstrated that endogenous WWOX and SIMPLE co-localize to peri-nuclear compartments of MCF-7 human breast cancer cells. These studies demonstrate that WWOX contains a Group I WW domain that binds known cellular proteins containing the specific ligand PPXY. Identification and characterization of WWOX interacting proteins will lead to an understanding of the biological functions of WWOX in normal and tumor cells.
Keywords: tumor suppressors, WW domain, protein–protein interaction
Introduction
We recently cloned WW domain oxidoreductase (WWOX), the gene encoded by the common chromosomal fragile site FRA16D (Bednarek et al., 2000; Ludes-Meyers et al., 2003). This region is frequently affected by LOH in breast, ovarian, esophageal, prostate and hepatocellular cancers. We demonstrated that ectopic WWOX expression inhibited tumor growth of breast cancer cells, suggesting that WWOX behaves as a potential tumor-suppressor gene (Bednarek et al., 2001).
The WWOX gene spans a region over 1 Mb in size, which is composed of nine exons coding for a protein of 414 amino acids. The WWOX amino-acid sequence predicts two protein structural motifs consisting of two WW domains and a short-chain dehydrogenase/reductase domain (SDR) (Bednarek et al., 2000). The SDR region of WWOX may be involved in sex-steroid metabolism due to its high amino-acid sequence homology to specific oxidoreductases (e.g. 17beta-hydroxysteroid reductase 3; Kallberg et al., 2002).
Although the biological function of WWOX remains to be determined, the biochemical function of WW domain motifs is known to be involved in protein–protein interactions. WW domains are grouped according to their binding preference to a diverse set of proline-rich ligands (Bedford et al., 2000). There are four groups of WW domains: Group I binds PPXY ligands (Chen and Sudol, 1995), Group II binds PPLP (Chan et al., 1996), Group III binds polyproline sequences flanked by Arg or Lys (Bedford et al., 1998) and Group IV binds phospho-Ser-Pro or phospho-Thr-Pro (Lu et al., 1999).
In this study, we determined that WWOX contains a Group I WW domain that specifically binds the PPXY ligand in vivo and in vitro, and we identified five potential WWOX interacting proteins. One of these proteins is the small membrane protein of the lysosome/ late endosome (SIMPLE) (also called PIG7 or LITAF; Moriwaki et al., 2001) a p53 inducible protein involved in the cellular response to TNFα signaling. To better understand the biochemical function of the WWOX WW domains, we characterized the WWOX–SIMPLE interaction in vitro and in vivo. Furthermore, we demonstrated that endogenously expressed WWOX and SIMPLE partially co-localize to the Golgi apparatus.
Results
WWOX contains a group I WW domain
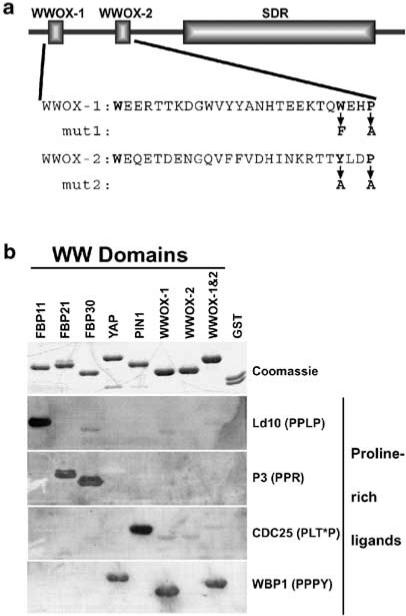
The WWOX protein consists of two structural domains. The N-terminal 88 amino acids contain two regions that are homologous to the WW protein interaction motif (Figure 1a) (Bednarek et al., 2000). To determine the specificity of WWOX WW domain binding, we generated GST-fusion proteins, each containing individual WW domain (GST–WWOX-1 and GST–WWOX-2) and both WW domains (GST–WWOX-1 and -2). The GST-fusion proteins were expressed and purified from bacteria, separated by SDS–PAGE and transferred to a nitrocellulose membrane. A representative set of other WW domains was included. The immobilized proteins were probed with four different proline-rich ligands to identify specific ligand–WWOX interactions (Figure 1b). GST–WWOX-1 and -2 and GST–WWOX-1 specifically bound the peptide containing the PPPY motif. Interestingly, GST–WWOX-2 did not interact with any of the ligands tested. These results show that the WWOX WW domain binds the ligand containing the PPXY motif, defining it as a class I WW-domain (Chen and Sudol, 1995). It also raises the possibility that WWOX-2 recognizes a novel motif or contributes to the structural integrity of the domain.
Figure 1.
WWOX WW domain I binds the PPXY ligand. (a) Full-length WWOX protein is shown schematically, highlighting the WW domains and the SDR domain. Below the schematic are the amino-acid sequences of the WW domains. Mut1 and mut2 show the amino-acid changes made in GST–WWOX-1 and -2 to inactivate the WWOX-1 and WWOX-2 domains, respectively. (b) Purified GST-fusion proteins (top panel) containing the WW domains from several different proteins were probed with each of four proline-rich ligands (bottom four panels). Equal amounts of each GST-fusion protein and GST alone were separated by SDS–PAGE and stained with Coomassie blue to visualize protein bands (Coomassie) or transferred to PVDF membrane for ligand binding. The biotinylated peptide ligands (PPLP, PPR, PLT*P, and PPPY) were bound to streptavidin-conjugated HRP prior to incubation with the membrane-bound proteins. Ligands bound by GST-fusion proteins were visualized by ECL. The complete amino-acid sequences of the peptide ligands are given in Materials and methods. T* = phosphothreonine
Identification of potential WWOX ligand proteins
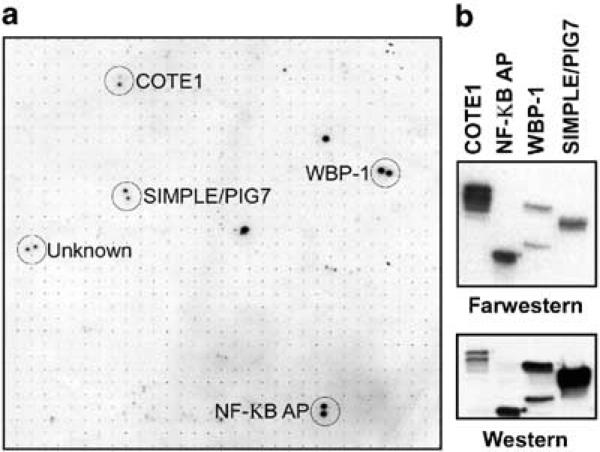
In order to identify potential WWOX binding partners, we probed a high-density protein array (Bussow et al., 1998) with a radiolabeled probe containing both WWOX WW domains. The high-density protein filters used in these experiments represent a novel product for high-throughput proteomics, and were generated by the German Human Genome Project (www.rzpd.de). The filters are PVDF membranes that contain 27 648 human brain cDNA clones arrayed and spotted in duplicate. Each cDNA clone expresses a histidine-tagged fusion protein. We performed a far-Western analysis of these membranes using a phospho-labeled GST-fusion protein as probe. Using this procedure, we identified five bacterial clones that express candidate WWOX-binding proteins (Figure 2). The ‘address’ of the candidate proteins was determined and the corresponding bacterial stocks were obtained from the German Genome Center.
Figure 2.
Identification of candidate WWOX interacting proteins. (a) High-density protein arrays were probed with a 32P radiolabeled WW domain. Proteins bound to the WWOX WW domains were visualized by autoradiography and were identified by mapping their coordinates on the array grid. A partial view of the array is shown with duplicate positive colonies circled. The name of each protein identified by BLAST search is shown. (b) The interaction of the WWOX WW domains with the identified proteins was confirmed by far-Western analysis of bacterial expressed candidate proteins. Each candidate protein was bound to nickel agarose beads under denaturing conditions, separated by SDS–PAGE and transferred to PVDF membranes. Membranes were analysed by Western blotting (bottom panel) using antibodies that recognized the histidine tag, and by far-Western analysis (top panel) using radiolabeled WWOX WW domains as a probe
The cDNA clones of all the candidate His-tagged fusion proteins were obtained and their identities confirmed by DNA sequencing. Four of the encoded proteins have been previously described (Table 1) and are (1) putative NF-κB activating protein (cDNA clone 120) (Matsuda et al., 2003), (2) COTE1 (Winfield et al., 1997), (3) SIMPLE (Moriwaki et al., 2001) and (4) WW domain-binding protein-1 (WBP-1) (Chen and Sudol, 1995). One clone has not been identified as a cellular encoded protein and contains no recognizable protein structural motifs in current protein databases. All of the protein amino-acid sequences have at least one PPXY WWOX-binding motif (Table 1); in fact, WBP-1 contains the PPXY motif used to originally determine the ligand specificity of WW domains (Figure 1).
Table 1.
Identity of WWOX candidate binding partners
Each of the identified proteins was expressed in bacteria and partially purified with nickel agarose beads. Confirmation of WWOX binding to the specific proteins encoded by the cDNA clones was carried out by far-Western analysis using the WW domains as a probe (Figure 2b, top panel). Expression of the tagged cDNA proteins was determined by Western blotting using an antibody that recognizes the histidine tag (Figure 2b, bottom panel). WW domain binding to this set of four proline-rich ligands was thus confirmed.
The candidate binding protein SIMPLE was first identified as a protein encoded by a differentially expressed transcript in BCG-CWS-treated human monocytes (Moriwaki et al., 2001). Most importantly, the SIMPLE protein was observed to be located in a perinuclear region of the cell (Moriwaki et al., 2001), similar to what we observed for WWOX subcellular localization (Bednarek et al., 2001). Little is known about the biological functions, such as subcellular localization, of the other candidate proteins. This potential co-localization with WWOX made SIMPLE the strongest candidate to be a bona fide in vivo WWOX partner. Therefore, we further characterized the WWOX WW domain interaction with SIMPLE.
WWOX WW domain binding specificity
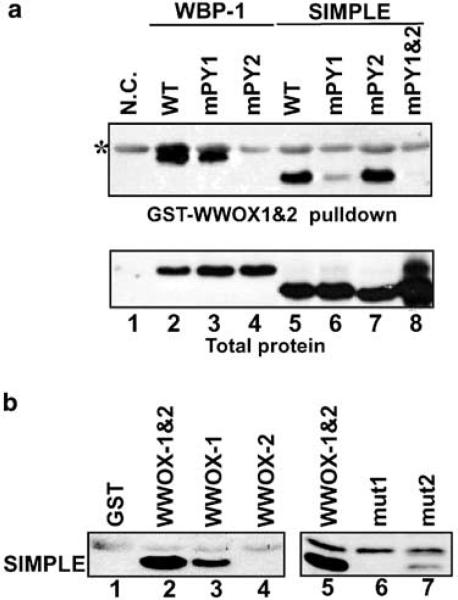
Both WBP-1 and SIMPLE contain two PPXY motifs (Table 1). It is well documented that loss of the ligand's tyrosine residue abolishes the interaction with a group I WW domain (Chen and Sudol, 1995). Therefore, to determine the specificity of the WWOX–SIMPLE interaction, we performed site-directed mutagenesis of the PPXY motifs of SIMPLE and WBP-1. The mutant proteins were expressed in bacteria and GST pull-down assays were used to determine whether the mutant proteins could bind to the WWOX WW domain. Histidine-tagged proteins that interacted with GST–WWOX-1 and -2 were visualized by Western blotting using anti-RGS antibody. Interaction of the WWOX WW domains to SIMPLE was clearly observed (Figure 3a, lane 5, top panel), validating the previous far-Westerns. Mutation of the first SIMPLE PPSY motif significantly reduced the interaction (Figure 3a, lane 6), while mutation of the second PPSY motif had no observable effect (Figure 3a, lane 7). Mutation of both PPSY motifs showed no detectable binding (Figure 3a, lane 8), even though more total protein was present in the cell extract.
Figure 3.
(a) WWOX interacts with specific PPXY motifs of candidate interacting proteins. Site-directed mutagenesis was used to change the tyrosine of PPXY to alanine of SIMPLE (lanes 5–8) and WBP-1 (lanes 2–4). Additionally, a double mutant of SIMPLE was created (lane 8). To determine protein interactions, total protein extracts from bacteria expressing the wild-type and mutant proteins were used for GST pull-down assays. Proteins pulled down by GST–WWOX-1 and -2 were separated by SDS–PAGE, transferred to PVDF membranes and visualized by Western blotting using antibody to the histidine tag (top panel). Equal amounts of expressed proteins were determined by Western blotting of total protein extracts (bottom panel). Total protein extracts from the parental bacterial strain that does not express any histidine-tagged proteins were used as the negative control (lane 1). *Band due to nonspecific binding of the antibody to the GST protein. (b) The first WW domain is required for interaction with endogenously expressed SIMPLE. Whole-cell extracts from the breast cancer cell line MCF-7 were used for GST pull-down assays using GST fused to each individual WWOX WW domain and to GST–WWOX-1 and -2 containing altered amino acids in each of the WW domains. Proteins pulled down by the indicated GST-fusion proteins were separated by SDS–PAGE, transferred to PVDF and analysed by Western blotting with antibody that specifically recognizes SIMPLE. Left panel: Binding to the complete WWOX WW domain (GST–WWOX-1 and -2, lane 2) and to the individual WWOX WW domains (GST–WWOX-1 and GST–WWOX-2, lanes 3–4, respectively). The observed binding by endogenous SIMPLE was specific for WWOX, as shown by the GST pull-down with GST alone (lane 1). Right panel: Binding to the complete WW domain (GST–WWOX-1 and -2, lane 5) and to the mutant WW domains (mut1 and mut2, lanes 6 and 7, respectively). Each WW domain had two amino acids changed in the context of GST–WWOX-1 and -2 (Figure 1)
Binding to WBP-1 was used as a positive control (Figure 3a, lanes 2–4) because the nine amino-acid sequence (gtpPPPYtv) from WBP-1 was used in the peptide ligand-binding assay (Figure 1). Mutagenesis of this PPXY motif demonstrated that it is the target of WWOX WW domain binding in the context of the full-length WBP-1 protein. These analyses demonstrated that the WWOX WW domains specifically interact with the PPXY ligand in different protein contexts.
We then determined whether the WWOX WW domains could interact with the SIMPLE protein endogenously expressed by mammalian cells. GST pull-down assays from total cellular extracts from the breast cancer cell line MCF7 demonstrated that the WWOX WW domains bind endogenous SIMPLE (Figure 3b, lanes 2 and 5). The interaction was specific for the WW domains as no SIMPLE was pulled down by GST itself (Figure 3b, lane 1). To determine whether the interaction is specific for WWOX-1 or WWOX-2, GST pull-downs with GST fused to each of the WW domains was performed. In addition, mutations were made in GST–WWOX-1 and -2 to inactivate either the WWOX-1 or WWOX-2 domains (shown in Figure 1a). When GST was fused to WWOX-1 alone efficient binding to SIMPLE was maintained; however, no detectable binding was observed to WWOX-2 (Figure 3b, lanes 3 and 4, respectively). In agreement with these results, mutation of the first WW domain eliminated the interaction, while the mutated second WW domain retained binding activity (Figure 3b, lanes 6 and 7, respectively). The interaction with the individual WW domains and with the mutated WW domains both showed reduced binding to the first WW domain. This may indicate that the second WW domain functions to stabilize the interaction or is involved in maintaining the structural integrity of the domain. These results are in perfect agreement with the peptide-binding studies (Figure 1b) that identified WWOX-1 as a Group I WW domain.
WWOX and SIMPLE partially co-localize to the Golgi
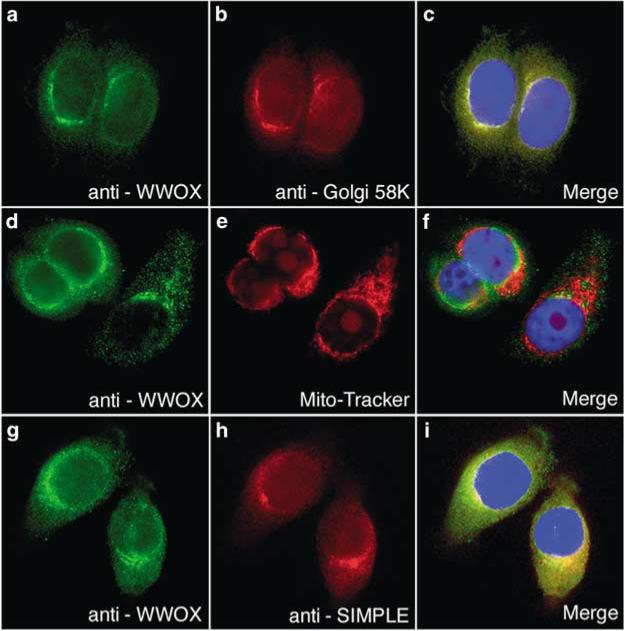
SIMPLE was chosen for further analysis as a WWOX-binding candidate, because it was previously shown that SIMPLE resides in a perinuclear compartment of the cell (Moriwaki et al., 2001) similar to WWOX. If WWOX and SIMPLE are protein interaction partners, they have to exist, at least transiently, in the same region of the cell. Therefore, we performed immunofluorescence staining of MCF-7 cells (Figure 4) that express both WWOX and SIMPLE. First, we determined the subcellular localization of endogenous WWOX in the MCF-7 human breast cancer cell line using affinitypurified anti-WWOX antibody generated in our laboratory (Figure 4a–f). MCF-7 cells were co-stained with anti-WWOX antiserum (Figure 4a) and with an antibody that recognizes the 58 kDa Golgi protein (Figure 4b). Both proteins showed perinuclear staining with significant overlap when the images were merged (Figure 4c). This is consistent with our original observation using a GFP–WWOX fusion protein that identified WWOX as a Golgi protein (Bednarek et al., 2001). However, contrary to our published observations, and to the results presented here, it has been reported that endogenous WWOX and GFP–WWOX fusion proteins localize to the mitochondria (Chang et al., 2001). To determine whether our anti-WWOX antibodies recognized WWOX localized in the mitochondria, we stained MCF-7 cell mitochondria with MitoTracker Orange CMTMRos (Figure 4e) prior to immunofluorescence using anti-WWOX antibodies. Consistent with our previous experiments, the WWOX antibodies stained a perinuclear region of the cell (Figure 4d); however, there was little, if any, overlap with the MitoTracker stain. Again, our immunofluorescence results were consistent with our previous experiments using a GFP–WWOX fusion protein (Bednarek et al., 2001). Next, we determined whether WWOX and SIMPLE co-localized to the same sub-cellular region. Co-immunofluorescence staining with our anti-WWOX antibodies (Figure 4g) and antibodies that recognize SIMPLE (Figure 4h) showed a similar cytoplasmic pattern of staining that partially co-localized to the perinuclear region of the cell (Figure 4i). Each protein is also observed to reside in smaller distinctive partially nonoverlapping areas of the cell, which may indicate that SIMPLE may transiently exist in the same subcellular location as WWOX while moving through the trans-Golgi network to the final destination of the lyosome/late endosome.
Figure 4.
WWOX and SIMPLE partially co-localize to the Golgi apparatus. Panels a–c: Endogeous WWOX co-localizes with the 58 kDa Golgi marker in MCF-7 cells. Note full overlap of the fluorescence signals (panel c, yellow color). Panels d–f: Immunofluorescence detection of WWOX and fluorescent staining of mitochondria. As can be observed, no overlap is evident in the merge image (panel f). Panels g–i: Endogenous WWOX partially co-localizes with SIMPLE. Immunofluorescence detection of WWOX (panels a, d and g) was carried out using affinity-purified anti-WWOX rabbit polyclonal antiserum and Alexafluor 488 conjugated anti-rabbit secondary antibody. Immunofluorescence detection of 58 kDa Golgi marker (panel b) was carried out using mouse monoclonal anti-58 kDa and Alexafluor 647-conjugated anti-mouse secondary antibody. Immunofluorescence detection of SIMPLE (panel h) was carried out using mouse monoclonal anti-SIMPLE antibody and Alexafluor 647 secondary antibody
Discussion
We have demonstrated that the amino-terminal WWOX WW domain behaves as a Group I WW domain that specifically binds the ligand PPXY. Using the powerful tool of high-density protein arrays, we identified five proteins as candidate WWOX interaction partners. Importantly, each interacting protein contained at least one PPXY sequence. The identities of these proteins are: (1) putative NF-κB activating protein (cDNA clone 120) (Matsuda et al., 2003), (2) COTE1 (Winfield et al., 1997), (3) SIMPLE (Moriwaki et al., 2001) and (4) WBP-1 (Chen and Sudol, 1995). One clone has not been identified to date.
One candidate protein, COTE1, is encoded by the gene COTE1, first identified by virtue of its close proximity to the glucocerebrosidase gene on chromo-some 1q21 (Winfield et al., 1997). Currently, no known function for COTE1 has been determined. COTE1 contains a predicted transmembrane region, making it possible that it would reside in a membrane-bound subcellular organelle such as the Golgi. This remains to be determined.
The candidate partner protein SIMPLE was first identified as a differentially expressed transcript in BCG-CWS-treated human monocytes (Moriwaki et al., 2001). The SIMPLE cDNA sequence was identical to the p53 inducible transcript LITAF/PIG7, except for one guanine nucleotide that was missing at the exon 3–4 junction that changes the translation-reading frame of the two cDNAs. The PIG7/LITAF mRNA codes for a 228 AA nuclear protein, whereas the SIMPLE mRNA codes for a 161 AA protein integral membrane protein located in the lysosome/late endosome. Owing to the subcellular localization of SIMPLE and the potential involvement of SIMPLE in TNFα signaling, we further characterized the WWOX–SIMPLE interaction.
We demonstrated that the WWOX WW domain specifically interacted with the SIMPLE amino-terminal PPSY motif using in vitro binding assays (Figure 3a). Interestingly, SIMPLE has been identified as the binding partner for the WW domain of the ubiquitin-protein ligase NEDD4 and that interaction was shown to depend on the same PPSY motif (Jolliffe et al., 2000). In addition, we demonstrated that WWOX binding to endogenously expressed SIMPLE was dependent on the amino-terminal WWOX WW domain, consistent with our in vitro ligand-binding experiments.
Using immunofluorescence staining, we observed that our anti-WWOX antibody consistently recognized endogenous WWOX in the Golgi apparatus with little, if any, detectable staining in the mitochondria (Figure 4a– f). Although this is in agreement with our previous findings using a GFP–WWOX fusion protein (Bednarek et al., 2001), it is inconsistent with the observations of others (Chang et al., 2001). Although the reasons for this discrepancy are not clear, it may be partially explained due to the use of different cell types and different antibodies. Most importantly, we observed that WWOX and SIMPLE partially co-localize to a perinuclear region of the cell. Since SIMPLE was originally identified as a late endosome/lysosomal protein (Moriwaki et al., 2001) and because the late endosomes/lysosomes are continuations of the Golgi apparatus, the observed partial co-localization could be a representation of the dynamics of this organelle. Moreover, this also emphasizes a possible transient nature of the WWOX–SIMPLE interaction in the Golgi.
WWOX has been reported to directly interact with two other proteins, the tumor suppressor p53 and JNK1 (Chang et al., 2001, 2003). In addition, mapping studies using the yeast two-hybrid system identified WWOX-1 as the WW domain responsible for the interaction with p53, and the binding site was mapped to the p53 proline-rich region (AA 66–110). The fact that neither p53 nor JNK was identified in our experiments may indicate that the cDNA clones encoding these proteins are not represented in the high-density protein array. It is worth noting, however, that the p53 proline-rich region does not contain any PPXY motifs, and it has been well documented that ligand preference by the various known WW domains is ‘highly specific’ (Kay et al., 2000; Sudol and Hunter, 2000; Kasanov et al., 2001; Otte et al., 2003). Indeed, we have presented evidence that the WWOX WW domain specifically binds the PPXY motif. Moreover, WWOX binding to two candidate interacting proteins was demonstrated to have preference for one of two PPXY motifs within their sequence, demonstrating that a higher level of specificity also exists, as has been observed for many other Group I WW domains (Kasanov et al., 2001). It remains to be determined which p53 and JNK1 amino acids directly interact with WWOX as they may represent novel WW domain interaction motifs.
The biochemical functions of WWOX and SIMPLE are poorly understood. Moriwaki et al. (2001) speculated that SIMPLE may be involved in the apoptotic response for intracellular killing of mycobacteria. WWOX also has been implicated as a proapoptotic protein involved in the enhancement of TNFα cytotoxi-city synergistically with p53 (Chang et al., 2001). Since SIMPLE is induced by both p53 and TNFα, the reported apoptotic effects may, in part, be due to a WWOX–SIMPLE functional interaction.
Recently, SIMPLE was identified as a protein that activated NF-κB-dependent transcription. Remarkably, in the same report (Matsuda et al., 2003), the putative NF-κB activating protein clone 120 was also identified, a protein we describe here as a WWOX-binding candidate. The identification of these two WWOX-binding partners may indicate that WWOX is involved in the regulation of NF-κB transcriptional activation.
Future studies on WWOX protein–protein interactions will help elucidate the biological function(s) of WWOX and its role in normal and diseased states.
Materials and methods
Construction, purification and labeling GST-fusion proteins
GST-fusion proteins containing WWOX-1 and -2 domains, and the individual WWOX-1 and WWOX-2 domains were constructed by insertion of PCR-amplified fragments of the WWOX cDNA into the BamH1 and EcoR1 sites of the bacterial expression vector pGEX-2TK (Pharmacia). The resulting fusion proteins contain the WWOX amino acids: AA 16-94, GST–WWOX-1 and -2; AA 16–53, GST–WWOX-1; AA 53–94, GST–WWOX-2 (Figure 1). Site-directed mutagenesis was performed using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene).
GST-fusion proteins were expressed in Escherichia coli strain BL21 and purified essentially as described by the manufacturer. Cleared bacterial lysates from 100 ml cultures were made by sonication in PBS containing protease inhibitors (Roche). GST-fusion proteins were purified using Glutathione Sepharose 4B (Pharmacia).
The WW domain was radioactively labeled as described in Sambrook and Russell (2001). GST–WWOX-1 and -2 bound to glutathione sepharose 4B beads was incubated with protein kinase (Sigma) and [γ-32P]ATP, followed by digestion with thrombin protease (Pharmacia) to release the labeled WW domain from GST.
For far-Western ligand-binding studies, GST-fusion proteins were separated by SDS–PAGE and transferred to Immobilon-P (Millipore). Membranes were blocked for at least 4 h at 4°C with PBS containing 3% non-fat dry milk and 0.1% Tween-20. Peptides were prebound to horseradish peroxidase (HRP)-conjugated streptavidin (Sigma Chemical Co.), then incubated with membrane-bound GST-fusion proteins overnight at 4°C in blocking buffer. Membranes were washed four times at 4°C and then developed using ECL (Amersham).
Peptides used in this study were: Ld1D (PPLP) biotin-SGSGAPPTPPLPP; P3(PPR) biotin-GVSVRGRGAAPPP PPVPRGRGVGP; CDC25 (PLT*P) biotin-SGSGEQP LT*PVTDL (T* = phosphorylated threonine); WBP-1 (PPPY) biotin-SGSGGTPPPPYTVG.
For GST pull-down assays, bacterial lysates were made as described above, followed by the addition of NP-40 to a final concentration of 0.1%. Cleared total cell lysates from MCF-7 cells were prepared using the lysis buffer (50 mM Tris–Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 10% glycerol). GST pull-down assays were performed by addition of 10 μg of purified GST-fusion protein and 20 μl of Glutathione Sepharose 4B (50% slurry) to equal amounts of total cell extracts. After incubation overnight at 4°C with mixing, the beads were washed three times with cold lysis buffer and once with cold PBS. Bound proteins were eluted by boiling in 1 SDS–PAGE loading buffer.
Immunodetection was performed using KPL Protein Detector™ chemiluminescence detection reagents. RGShis monoclonal antibody (1 : 1000 dilution), SIMPLE specific monoclonal antibody (1 : 1000) and HRP-conjugated anti-mouse antibody (1 : 5000 dilution) were incubated with membrane for 1 h at room temperature.
Fluorescence microscopy
MCF-7 cells were cultured in IMEM containing 5% FBS and gentamicin. Mitochondria were stained by incubating cells with complete medium containing 100 nM MitoTracker Orange CMTMRos for 20 min in a 37°C humidified CO2 incubator. The cells were fixed for 10 min with PBS containing 10% formaldehyde and permeabilized by washing three times with PBS, 0.1% IGEPAL. Primary and secondary antibodies were diluted in PBS, 0.1% IGEPAL, 3% NGS and incubated with the cells for 1 h each at room temperature, followed by three washes with PBS, 0.1% IGEPAL. The stained cells were mounted using Vectashield mounting medium containing DAPI stain (Vector Labs). The primary antibodies used were rabbit anti-WWOX at 1 : 25 (Ludes-Meyers et al., 2003), mouse anti-SIMPLE antibody (BD transduction labs.) at 1 : 50. The secondary antibodies used were goat anti-rabbit AlexaFluor™ 488 (Molecular Probes, Inc.) at 1 : 50 and goat anti-mouse AlexaFluort 647™ (Molecular Probes, Inc.) at 1 : 50.
Acknowledgements
This work was supported by NCI RO1 CA 102 444, a NIEHS center grant ES07784 and a Pharmacia award.
Abbreviations
- BCG-CWS
Mycobacterium bovis Bacillus Calmette-Guerin
- ECL
enhanced chemiluminescence
- JNK
c-Jun N-terminal kinase
- LITAF
lipopolysaccharide-induced transcription factor
- LOH
loss of heterozygosity
- NEDD4
neuronal precursor cells expressed developmentally downregulated protein 4
- NF-κB
nuclear factor-kappa B
- NGS
normal goat serum
- PIG7
p53 inducible gene 7
- SDR
short-chain dehydrogenase/reductase
- TNF
tumor necrosis factor
- WBP-1
WW domain-binding protein-1
- WWOX
WW domain oxidoreductase
References
- Bedford MT, Reed R, Leder P. Proc. Natl. Acad. Sci. USA. 1998;95:10602–10607. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford MT, Sarbassova D, Xu J, Leder P, Yaffe MB. J. Biol. Chem. 2000;275:10359–10369. doi: 10.1074/jbc.275.14.10359. [DOI] [PubMed] [Google Scholar]
- Bednarek AK, Keck-Waggoner CL, Daniel RL, Laflin KJ, Bergsagel PL, Kiguchi K, Brenner AJ, Aldaz CM. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- Bednarek AK, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. Cancer Res. 2000;60:2140–2145. [PubMed] [Google Scholar]
- Bussow K, Cahill D, Nietfeld W, Bancroft D, Scherzinger E, Lehrach H, Walter G. Nucleic Acids Res. 1998;26:5007–5008. doi: 10.1093/nar/26.21.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Bedford MT, Leder P. EMBO J. 1996;15:1045–1054. [PMC free article] [PubMed] [Google Scholar]
- Chang N-S, Doherty J, Ensign A. J. Biol. Chem. 2003;278:9195–9202. doi: 10.1074/jbc.M208373200. [DOI] [PubMed] [Google Scholar]
- Chang NS, Pratt N, Heath J, Schultz L, Sleve D, Carey GB, Zevotek N. J. Biol. Chem. 2001;276:3361–3370. doi: 10.1074/jbc.M007140200. [DOI] [PubMed] [Google Scholar]
- Chen HI, Sudol M. Proc. Natl. Acad. Sci. USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe CN, Harvey KF, Haines BP, Parasivam G, Kumar S. Biochem. J. 2000;351(Part 3):557–565. [PMC free article] [PubMed] [Google Scholar]
- Kallberg Y, Oppermann U, Jornvall H, Persson B. Protein Sci. 2002;11:636–641. doi: 10.1110/ps.26902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasanov J, Pirozzi G, Uveges AJ, Kay BK. Chem. Biol. 2001;8:231–241. doi: 10.1016/s1074-5521(01)00005-9. [DOI] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- Ludes-Meyers JH, Bednarek AK, Popescu NC, Bedford MT, Aldaz CM. Cytogenet. Genome Res. 2003;100:101–110. doi: 10.1159/000072844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, Doi T, Shimotohno K, Harada T, Nishida E, Hayashi H, Sugano S. Oncogene. 2003;22:3307–3318. doi: 10.1038/sj.onc.1206406. [DOI] [PubMed] [Google Scholar]
- Moriwaki Y, Begum NA, Kobayashi M, Matsumoto M, Toyoshima K, Seya T. J. Biol. Chem. 2001;276:23065–23076. doi: 10.1074/jbc.M011660200. [DOI] [PubMed] [Google Scholar]
- Otte L, Wiedemann U, Schlegel B, Pires JR, Beyermann M, Schmieder P, Krause G, Volkmer-Engert R, Schneider-Mergener J, Oschkinat H. Protein Sci. 2003;12:491–500. doi: 10.1110/ps.0233203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DA. Molecular Cloning: A Laboratory Manual. Third edn Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY.: 2001. [Google Scholar]
- Sudol M, Hunter T. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- Winfield SL, Tayebi N, Martin BM, Ginns EI, Sidransky E. Genome Res. 1997;7:1020–1026. doi: 10.1101/gr.7.10.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]