Gene delivery has the potential to treat a range of inherited and acquired diseases. Research has primarily focused on the use of viral vectors for this purpose, due to efficient infection of cells with viruses as well as long-term gene expression. However, the use of viral vectors for gene therapy is limited by safety concerns, production/manufacturing challenges, and limited nucleic acid carrying capacity [1, 2]. Thus, increased attention has been focused on biomaterials including cationic polymers as gene transfection agents [3-5] due totheir electrostatic interactions with plasmid DNA to form cationic nanoparticles. Polymers, including polyethylenimine (PEI), are useful in a variety of gene therapy applications [6-10]. One promising class of polymers, poly(β-amino ester)s (PBAEs), are degradable and have enhanced delivery compared to PEI [11-13]. Recent studies show that the amine-terminated polymers are generally more effective at promoting cellular uptake and DNA delivery than their acrylate-capped counterparts [14, 15].
We hypothesized that small molecules conjugated to the ends of the linear polymers could modify the gene delivery efficacy of these polymers in a cell-type specific manner. We initially screened a large library of poly(β-amino ester)s for gene delivery [16]. The base polymer with the highest gene transfer efficiency, C32, is synthesized by mixing 1,4-butanediol diacrylate (C) with 5-amino-1-pentanol (32) at a 1.2:1.0 amine to diacrylate molar ratio. The C32 polymer has excellent biocompatibility in-vivo, effectiveness as a gene delivery vector for the treatment of prostate cancer via intra-tumoral injection in mice, and is promising as a delivery system for pancreatic cancer [17, 18].
Using a combinatorial approach, an initial library of end-modified C32 polymers was synthesized and tested [19-21]. Beyond increasing efficacy in an easy-to-transfect cell line, we wanted to explore whether small molecule end groups could change gene delivery efficacy differentially among varying cell types and in difficult to transfect conditions such as 100% serum. Therefore, we synthesized a new library of end-modified C32 polymers using structural information gained from previous studies, where we noted that diamine end groups that contain a three-carbon spacer between amine functionalities were most effective [13]. We chose flow cytometry (and GFP expression) to quantify transfection efficacy so that we could analyze cell populations, which is not possible with luminescence assays. We have previously seen good correlation between these two methods [16]. The synthesis, biophysical characterization, and delivery efficacy of the nanoparticles formed with these new polymers is presented here.
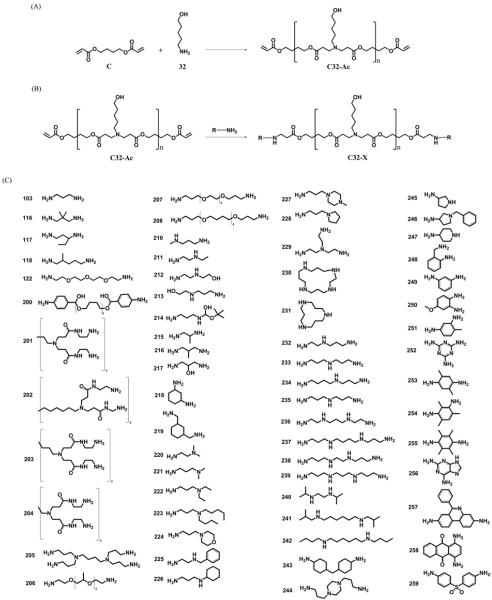
A library of polymers was synthesized from a base polymerization of (C) and (32) to form acrylate terminated C32 (Fig. 1a) and then this polymer was end-capped (Fig. 1b) with small molecules that contain amine groups (Fig. 1c). Polymers were characterized by 1H-NMR and gel permeation chromatography (see methods and Figure S1 and S8). The effect of modifying the ends of the same base polymer with different amine end groups is dramatic. Figures 2 and 3 demonstrate that the end group modulates gene delivery from very low (<1% positive) to very high (>90% positive) and does so in a cell specific manner.
Figure 1.
(A) Synthesis of acrylate-terminated C32 polymer (C32-Ac). (B) Synthesis of end-modified C32 polymers (C32-X). (C) Amine capping molecules.
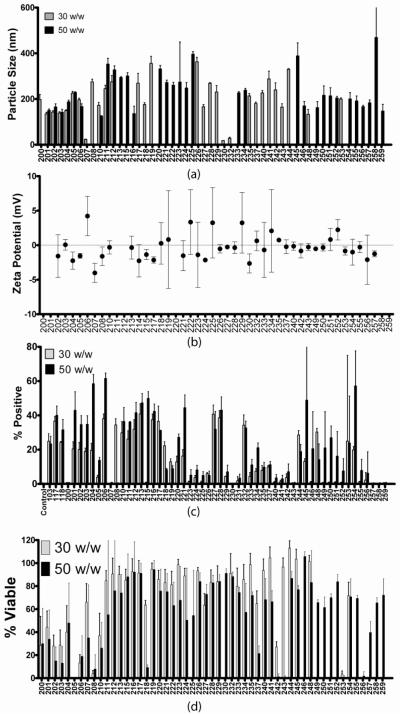
Figure 2.
Biophysical and gene delivery characterization of small molecule modified polymers. (a) Particle size (b) Zeta potential (c) Gene delivery efficacy in COS-7 (d) Viability in COS-7. Biophysical characterization shown for each polymer at its optimal formulation (Mean+SD).
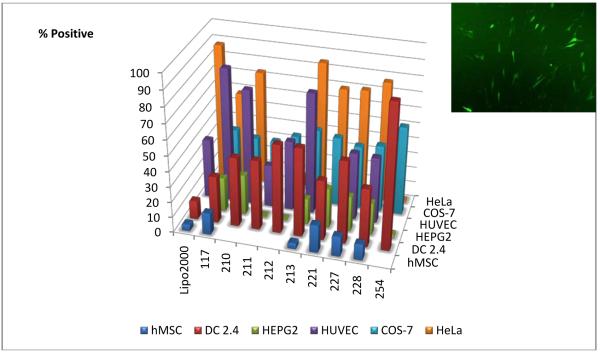
Figure 3.
Gene delivery efficacy of lead small molecule end-capped polymers across six cell types. Each bar represents the optimal formulation for each polymer and ranges from 30-75 Weight Polymer:Weight DNA. Inset: Fluorescence micrograph showing gene delivery efficacy of 221 to human mesenchymal stem cells (~18% positive cells).
Through electrostatic interactions, these polymers bind DNA and form nanoparticles. We examined the particle size and zeta potential of these particles by dynamic light scattering at the same concentrations and media conditions as used during transfection as has been previously described [22]. We found that for most polymers, 30 and 50 weight polymer:weight DNA (wt/wt) formulations formed particles of similar size. However, changes to polymer end-group could considerably vary particle size from no particle formation to larger than 400 nm particles (Figure 2a). End-modification had minimal effects on particle zeta potential (Figure 2b) and both 30 w/w and 50 w/w formulations had the same neutral zeta potential. Most polymers formed nanoparticles that were ~200 nm in size and had neutral zeta potential.
We initially screened our library of end-modified C32 polymers in COS-7 monkey kidney fibroblasts in 10% serum with a 600 ng DNA dose/well (Fig. 2c). At a 30 w/w ratio (polymer:DNA), 5 new polymers (206, 213, 216, 227, 228) achieved a nominally higher transfection rate than the previous best polymer, C32-117. At a 50 w/w ratio, 11 new polymers achieved a nominally higher transfection rate than the previous best polymer. Almost all of the new polymers showed normal viability at the concentrations and wt/wt ratio tested (Figure 2d), although a few of the best polymers showed significant toxicity (206, 208, 210, and 253). The four cytotoxic polymers may be useful at lower polymer doses, but are less suitable for transfection that the other high-performing polymers that also have high viability (such as 213, 216, 227, 228, 245, 254, etc). Interestingly, biophysical properties of the nanoparticles (Figure 2) or molecular weight (Figure S9) did not significantly correlate to transfection efficacy. This suggests that the functionality of the small molecule polymer end-groups does not come from tuning the physical properties of the nanoparticle, but instead from changing the interaction between the complex and the cell, perhaps by playing a role in cellular uptake.
To investigate the biological effects of end-modification, we compared gene delivery of the nanoparticles across additional cell lines including HeLa, a human cervical cancer cell line (Figure S2), HepG2, a human hepatocellular carcinoma (Figure S3), HUVEC, human primary endothelial cells (Figure S4), DC 2.4, murine dendritic cells (Figure S5), and hMSC, human mesenchymal stem cells (Figure S6). All experiments were conducted in standard cell growth media that includes 10-12% serum proteins. Several of the polymers (206, 208, 210, 213, 221, 227, and 228) were effective at transfecting HeLa cells and were comparable to the commercially available reagent, Lipofectamine 2000. In HepG2 cells, a different set of lead polymers (210, 218, 221, 227, and 228) was most effective. In HUVECs, a hard to transfect human primary cell type, several polymers including 117, 210, and 213 were highly effective, delivering exogenous genes to over twice as many cells as Lipofectamine 2000 (Figure S5). Polymer C32-210 is particularly interesting as it is both more efficient relative to the other similarly structured PBAEs and effective, meaning that less than half the amount of polymer 210 (30 wt/wt) is needed to achieve similarly high transfection (~75% positive) compared to polymer 213 (75 wt/wt). Many other polymers were unable to achieve this high level of efficacy at any formulation. The fact that such small changes to polymer structure can make the polymer more relatively efficient (or in the case of C32-206 or 211 relatively inefficient) is surprising (Figure S4). End-modified polymers were found to be particularly effective for gene delivery to DC 2.4 cells (Figure S5). Lipofectamine 2000, a commercially available non-viral transfection reagent, transfects 10% of these antigen-presenting immune cells. Polymer C32-254, on the other hand, is able to transfect 90% of these cells. This result suggests the potential utility of these biodegradable particles as efficient genetic vaccines. A final difficult-to-transfect cell type that we are interested in is hMSC. Here, polymer C32-221 is the lead biomaterial, and while transfection is not as high as in other cell types, it remains significantly higher than levels from Lipofectamine 2000 (Figures S6a, S6b, S6e). The polymeric nanoparticles are also shown to be minimally cytotoxic to these fragile cells (Fig. S6c-d), similar to the COS-7 findings. To further increase gene delivery to hMSCs, we increased the DNA dose which enhanced gene expression for some polymers up to 5-fold while not having any adverse effects on cell viability. Some of these formulations also retain effectiveness in the presence of 100% serum during transfection, with C32-206 and C32-204 best for COS-7s and HUVECs respectively (Figure S7). This suggests that some of these materials may retain their effectiveness following intravenous administration. As compared to Lipofectamine 2000, these polymers require a higher dose of biomaterial (w/w polymer) to form optimized gene delivery particles.
A few polymers have high transfection across most cell types and point to some general features of highly effective of polymeric gene delivery particles. Polymers C32-213, C32-227, and C32-228 are especially effective and structurally, are very similar. They, like all polymers in this library, contain the same polymer backbone and hydroxyl-terminated side chains. They also each have an end-group that is composed of two amines separated by exactly three carbons and then terminated in a slightly differing arrangement of a few atoms. Unlike many end-groups in the library (such as 103-208 and 232-239), these three polymers are not terminated by primary amines. Polymers C32-213, C32-227, and C32-228 have molecular weights between 8 – 16.7 kDa, form nanoparticles that are 170-290 nm in size (slightly larger than average), and have a zeta potential of ~ 0 mV (most neutral) in the presence of cell media.
While some polymers are strong transfection agents overall, one polymer is not optimal for all cell types (Figure 3). Rather, for each cell type there is a particular polymer that is most effective (COS-7:C32-206, HeLa:C32-213, HepG2:C32-210, HUVEC:C32-117, DC2.4:C32-254, hMSC:C32-221). Some polymers are also highly effective for delivery to one cell type, while simultaneously being poor for delivery to another (for example, C32-254 is especially effective for delivery to DC2.4s rather than HepG2s or HeLas, whereas C32-228 transfects 80% of HeLas, but only 10-40% in the other five cell types tested). When a double DNA dose is used and multiple cell types are transfected side by side in the same plate, the same cell specificity trends are observed, although the transfection of HeLas increases overall (Figure S10). Future work is needed to elucidate the mechanisms for this cell specificity, which may be due to changes to particle uptake kinetics and/or polymer/DNA binding interactions that regulate DNA release. Combined with other modalities for cell targeting such as targeting ligands [23, 24] or transcriptional targeting [17], proper biomaterial selection may further improve the safety/efficacy window for nanomedicine. In conclusion, our approach has revealed materials that are promising for the non-viral delivery of genes to cancer cells, immune cells, and human stem cells and points to polymer end-group as a regulator for cell-type specificity.
Experimental Section
Polymer Synthesis
Acrylate-terminated poly(β-amino ester) C32-Ac was synthesized by mixing 3532 mg of 1,4-butanediol diacrylate (17.8 mmol) with 1533 mg of 5-amino-1-pentanol (14.8 mmol) for 24 hr at 90°C in the dark. End chain capping reactions were performed by mixing 160 μL of C32-Ac/DMSO solution (500 mg/mL polymer in DMSO) with 640 μL of 0.25 M amine solution. Excess amine is used to fully end-modify the polymer without causing detectable cross-linking or aminolysis. Reactions were performed in 1.5 mL eppendorf tubes with constant agitation for 24 hours at room temperature. Following initial screening, lead polymers 117, 210, 211, 212, 213, 221, 227, 228, 254 were scaled up by adding 9.1 g of THF to 5 g of C32-Ac, vortexing, and then transferring to a 100 mL flask with a stir-bar. Forty mL of 0.25 M end-capping amine solution was then added, and the mixture was left stirring at room temperature in the dark for 24 hrs. End-modified polymers were precipitated by the addition of 10 volumes of diethyl ether and centrifugation at 2,500 rpm for 2 minutes. Polymers were washed twice with diethyl ether and dried in a vacuum desiccator.
Polymer Characterization
Polymers were characterized by 1H-NMR (300 MHz, CDCl3) on a Varian mercury spectrometer. C32-Ac: δ (ppm) 1.3–1.6 (m, -NCH2(CH2)3CH2OH), 1.7 (bs, -N(CH2)2COOCH2CH2- and CH2CHCOOCH2CH2-), 2.3–2.5 (m, -COOCH2CH2N- and -NC H2(CH2)4OH), 2.7–2.8 (m, -COOCH2CH2N-), 3.6 (bs, -N(CH2)4CH2OH), 4.1 (bs, -N(CH2)2-COOCH2CH2-), 4.2 (m, CH2CHCOOCH2CH2-), 4.4 (bs, -N(CH2)5OH), 5.9 (m, CH2CHCOOCH2CH2-), 6.1–6.2 (m, CH2-CHCOOCH2CH2-), 6.4 (m, CH2CHCOOCH2CH2-). End-modification of polymers was confirmed by the disappearance of the acrylate peaks at 5.9, 6.1-6.2, and 6.4. Organic phase Gel Permeation Chromatography (GPC) was performed using a solution of 0.1 M piperidine in 95% THF/5% DMSO (v/v) as the eluent at a flow rate of 1.0 mL/min in a Waters GPC system equipped with an autosampler (Waters Corporation, Milford, MA). A Phenogel (Phenomenex, Torrance, CA) MXL column (5 μm, 300 × 7.8 mm) and a Waters Styragel HR4 column were used in series and the molecular weights of the polymers are reported relative to monodisperse poly(2-vinylpyridine) standards. Unmodified C32-Ac and most end-modified polymers synthesized generally had Mw ~ 8,500 Da. However, four polymers sythesized in larger scale, C32-221, −227, −228, and −254, had higher molecular weights ~14-20 kDa, approximately twice as large as the others, indicating low levels of crosslinking of the C32 oligomers by these diamines (Figure S8). The small variance in molecular weight among the polymers does not appear to be correlated to particle size, zeta potential, or transfection efficacy (see Figures 2, S8, and S9) among the cell types tested. Polymers were stored at −20°C with desiccant.
Particle Sizing and Zeta-Potential
Particle size and Ζ-potential measurements were measured by using a ZetaPALS dynamic light scattering detector as previously described [24].
Cell Culture
COS-7 (ATCC, Manassas, VA), HeLa (ATCC), HepG2 (ATCC), and HUVECs (Lonza, Walkswille, MD) were grown in accordance with manufacturer instructions. DC2.4s were generously given to us by Herman Eisen (MIT) and cultured according to published procedures [25]. Bone marrow-derived hMSCs (Lonza, Walkswille, MD) were grown in MSC growth medium consisting of Dulbeccos modified Eagle Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (Gibco), 100 mM sodium pyruvate and 100 units of penicillin and streptomycin. Cells were subcultured upon confluence until passage 5 before use.
Cell Transfections
Cells were plated in 96-well plates at 15,000 cells/well and allowed to adhere overnight. EGFP-1 DNA (Elim Biopharmaceuticals, Hayward, CA)) was diluted in 25 mM sodium acetate (NaAc, pH=5) to 0.04 mg/mL. Polymers at 100 mg/mL in DMSO were diluted into NaAc buffer to yield the different polymer to DNA weight ratios. One hundred microliters of diluted polymer solution was mixed vigorously with 100 μL of DNA solution in 500 μL eppendorf tubes. After 10 minutes wait time, 180 μL of each was added to 720 μL of media in a deep-well polypropylene plate. The media over the cells was then removed with a 12-channel aspirator wand and followed by the addition of 150 μL/well of polymer-DNA complex solution in media. Complexes were incubated over the cells for four hours, then aspirated off and replaced with 100 μL/well of fresh media. Cells were allowed to grow for two days at 37°C, 5% CO2 and then analyzed by FACS for GFP expression. For transfections in 100% serum, we prepared the particles in 25 mM sodium acetate buffer as all our formulations are prepared. We then added these particles to 100% serum instead of regular complete cell medium. These diluted particle mixtures are then added to freshly aspirated cells. While the particle diluent was initially 100% serum rather than cell medium, due to the presence of residual buffer, the final concentration of serum is 80% serum during the transfection. No cell culture medium or any other additives were present during transfection. For hMSC experiments, all volumes were scaled up 5-fold and performed in 24-well plates. 75,000 cells were seeded 24 hours prior to transfection and particle doses of 3 μg and 6 μg DNA were added to the cells for four hours, after which they were aspirated off and replaced with 500 μL/well of fresh media.
FACS
GFP expression was measured using Fluorescence Activated Cell Sorting (FACS) on a FACSCalibur (Becton Dickinson, San Jose, CA, USA) or BD LSR II. Cells were aspirated, washed, aspirated, trypsonized, and then suspended in FACS buffer (98% PBS, 2% FBS, 1:200 propidium iodide) while in 96-well plates. Propidium iodide staining was used to exclude dead cells from the analysis and at least 5,000 live cells per sample were acquired. Two-dimensional gating was used to separate increased auto-fluorescence signal from increased GFP signal to more accurately count positively-expressing cells. Gating and analysis was performed using FlowJo 6.3 software (TreeStar, Ashland, OR, USA).
CellTiter Assay for Cell Viability
Ninety six-well plates were seeded with COS-7 cells in DMEM (10% FBS, 1% Pen/Strep) to 15,000 cells/well and allowed to attach overnight. Transfection was performed as described previously. The media + complexes were aspirated off 4 hours post-transfection and replaced with fresh media. 24 hours post-transfection, 20 uL of CellTiter 96® AQueous One Solution Reagent was added to each well. Absorbance measurements were taken following manufacturer instructions. Viability is reported as normalized cell metabolic activity compared to untreated controls. hMSC viability measurements were performed similarly.
Supplementary Material
Acknowledgements
This work was supported by NIH Grant EB000244.
Footnotes
J.S. and J.J.G. contributed equally to this manuscript. This work was supported by NIH Grant EB000244.
Additional figures are available in the supporting information online free of charge.
References
- [1].Kootstra NA, Verma IM. Annu Rev Pharmacol Toxicol. 2003;43:413. doi: 10.1146/annurev.pharmtox.43.100901.140257. [DOI] [PubMed] [Google Scholar]
- [2].Thomas CE, Ehrhardt A, Kay MA. Nat Rev Genet. 2003;4:346. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- [3].Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discov. 2005;4:581. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- [4].Putnam D. Nat Mater. 2006;5:439. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- [5].Wagner E, Kloeckner J. Adv Polym Sci. 2006;192:135. [Google Scholar]
- [6].Khalil IA, Kogure K, Akita H, Harashima H. Pharmacol Rev. 2006;58:32. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- [7].Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci U S A. 1995;92:7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Neu M, Fischer D, Kissel T. J Gene Med. 2005;7:992. doi: 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- [9].Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. Mol Ther. 2005;11:990. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- [10].Varga CM, Tedford NC, Thomas M, Klibanov AM, Griffith LG, Lauffenburger DA. Gene Ther. 2005;12:1023. doi: 10.1038/sj.gt.3302495. [DOI] [PubMed] [Google Scholar]
- [11].Lynn DM, Anderson DG, Putnam D, Langer R. J Am Chem Soc. 2001;123:8155. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- [12].Lynn DM, Langer R. J Am Chem Soc. 2000;122:10761. [Google Scholar]
- [13].Green JJ, Anderson DG, Langer R. Accounts of Chemical Research. 2007;41:749. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Akinc A, Anderson DG, Lynn DM, Langer R. Bioconjug Chem. 2003;14:979. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- [15].Anderson DG, Akinc A, Hossain N, Langer R. Mol Ther. 2005;11:426. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- [16].Anderson DG, Lynn DM, Langer R. Angew Chem Int Ed Engl. 2003;42:3153. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- [17].Anderson DG, Peng W, Akinc A, Hossain N, Kohn A, Padera R, Langer R, Sawicki JA. Proc Natl Acad Sci U S A. 2004;101:16028. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Showalter SL, Huang YH, Witkiewicz A, Costantino CL, Yeo CJ, Green JJ, Langer R, Anderson DG, Sawicki JA, Brody JR. Cancer Biol Ther. 2008;7:1584. doi: 10.4161/cbt.7.10.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Green JJ, Zugates GT, Tedford NC, Huang YH, Griffith LG, Lauffenburger DA, Sawicki JA, Anderson DG. Adv Materials. 2007;19:2836. [Google Scholar]
- [20].Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, Sawicki JA, Anderson DG. Mol Ther. 2007;15:1306. doi: 10.1038/sj.mt.6300132. [DOI] [PubMed] [Google Scholar]
- [21].Green JJ, Zhou BY, Mitalipova MM, Beard C, Langer R, Jaenisch R, Anderson DG. Nano Lett. 2008;8:3126. doi: 10.1021/nl8012665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Bioconjugate Chemistry. 2006;17:1162. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- [23].Ogris M, Walker G, Blessing T, Kircheis R, Wolschek M, Wagner E. J. Controlled Release. 2003;91:173. doi: 10.1016/s0168-3659(03)00230-x. [DOI] [PubMed] [Google Scholar]
- [24].Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Nano Letters. 2007;7:874. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- [25].Shen Z, Reznikoff G, Dranoff G, Rock KL. Journal of Immunology. 1997;158:2723. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.