Abstract
Sesame is an important oil crop for the high oil content and quality. The seed oil and protein contents are two important traits in sesame. To identify the molecular markers associated with the seed oil and protein contents in sesame, we systematically performed the association mapping among 369 worldwide germplasm accessions under 5 environments using 112 polymorphic SSR markers. The general linear model (GLM) was applied with the criteria of logP≥3.0 and high stability under all 5 environments. Among the 369 sesame accessions, the oil content ranged from 27.89%–58.73% and the protein content ranged from 16.72%–27.79%. A significant negative correlation of the oil content with the protein content was found in the population. A total of 19 markers for oil content were detected with a R2 value range from 4% to 29%; 24 markers for protein content were detected with a R2 value range from 3% to 29%, of which 19 markers were associated with both traits. Moreover, partial markers were confirmed using mixed linear model (MLM) method, which suggested that the oil and protein contents are controlled mostly by major genes. Allele effect analysis showed that the allele associated with high oil content was always associated with low protein content, and vice versa. Of the 19 markers associated with oil content, 17 presented near the locations of the plant lipid pathway genes and 2 were located just next to a fatty acid elongation gene and a gene encoding Stearoyl-ACP Desaturase, respectively. The findings provided a valuable foundation for oil synthesis gene identification and molecular marker assistant selection (MAS) breeding in sesame.
Introduction
Sesame (Sesamum indicum L.) is an ancient and important oilseed crop and is cultivated mainly in the tropical and subtropical regions of Asia, Africa and Southern America. The harvested area of world sesame reaches to 7.3×107 hm2, and the total product per year is roughly 3.7×107 ton from 2001 to 2010 (FAO data). Compared with other main oil crops, e.g., soybean (18% of average oil content) [1], oilseed rape (41%) [2], sunflower (40–44%) [3] and peanut (51%) [4], sesame is one of the few oil crops with the highest oil content and quality. Sesame seeds contain 55–58% oil and almost 18% proteins. Among the fatty acid compositions in sesame seeds, oleic acid (18∶1) (39.6%) and linoleic acid (18∶2) (46.0%) are the two main components with the ideal ratio of almost 1∶1 [5], [6]. Apart from the seed yield, the content of seed storage oil and proteins is the highlight agronomic trait in sesame breeding [7].
In the past two decades, in order to clarify the high quality of sesame oil and protein, many researchers focused on exploring seed development and fatty acid and storage protein synthesis processes, as well as identifying the lipid synthesis related genes and molecular markers in sesame [8]–[12]. Of all the three available cDNA libraries, two libraries are constructed for seed development analysis [13], [14]. However, oil and protein contents are complex quantitative traits and always are affected by genotype and environment [15]. At present, the mechanism of high oil content and quality in sesame seeds is still unclear. No loci of oil and protein content traits have ever been found in the sesame linkage maps. Even though Wei et al. [12] performed the association analysis of seed oil and protein content and fatty acid composition within 216 Chinese sesame accessions using 79 molecular primer pairs (including SSRs, SRAPs and AFLPs), only one association marker (M15E10-3) was identified under two environments. Therefore, in order to precisely detect the genes or markers associated with oil and protein content traits and to improve the sesame breeding, more efficient markers and germplasm resources with larger phenotypic variation need to be applied [16], [17].
Currently, linkage analysis (QTL mapping) and association mapping are two main and common analysis tools for dissecting complex phenotypic variation. Compared with the traditional linkage analysis based on mapping populations, association mapping offers higher precision for locating QTLs and selecting molecular markers [16], [18]. Till now, association mapping has been extensively used for analyzing important agronomic and quantity traits in wheat, maize, cotton, oilseed rape and other crops [18]–[22]. In the past several years, vast simple sequence repeat (SSR) or microsatellite markers with the high polymorphism are developed in sesame [7], [23], [24]. Accordingly, the association mapping is getting reliable and powerful for detecting the genes or markers associated with key traits and improving the molecular marker-assisted selection (MAS) in sesame breeding programs.
The objectives of this study are: (1) to perform the association mapping of seed oil content (OC) and protein content (PC) traits in worldwide sesame accessions using the GLM and MLM models, (2) to reflect the characteristics of sesame oil and protein contents under various environments, and (3) to determine the key SSR markers associated with seed quality. In this report, a natural population covering 369 worldwide accessions from China and other 15 countries and 112 pairs of polymorphic SSR markers were applied. The results give a foundation for investigating the seed development-related genes and seed quality in sesame.
Results
Seed oil and protein content variation in the natural population
Both the seed oil content (OC) and the protein content (PC) are often influenced by environment. To decrease the environmental effect, we collected the phenotypic data of the 369 sesame accessions under 5 environments of Pingyu and Yuanyang locations in 2011, and Pingyu, Yuanyang and Xinyang locations in 2012, The descriptive parameters under each environment were calculated (Table 1). The results showed that the OC and PC significantly varied among the 369 accessions. In total, the OC of the natural population ranged from 27.89%–58.73%, with an average of 49.59%–53.14%; meanwhile, the PC ranged from 16.72%–27.79% with an average of 20.28%–22.51%. All the datasets showed a normal or nearly normal distribution. To determine the heritability of the phenotypes, we performed the variance analysis of oil and protein contents (Table 2). Results indicated that the OC and PC traits were significantly influenced by genotype and environments (i.e., year and location). No significant interactions between variety and environmental factors (year and location) were detected. Moreover, the OC and PC traits presented the significant negative correlation under the 5 environments, as the correlation coefficient (r) between OC and PC varied from −0.66∼−0.72 (P<0.01) in 2011 and from −0.52∼−0.74 (P<0.01) in 2012, respectively (Data not listed).
Table 1. Phenotypic variation of the seed oil and protein contents in the 369 accessions under 5 environments.
| Trait | Year | Place | Min. (%) | Max. (%) | Mean (%) | Std. ev. | Skewness | Kurtosis |
| OC | 2011 | Pingyu | 27.89 | 55.03 | 49.59 | 4.61 | −2.19 | 1.69 |
| 2011 | Yuanyang | 34.28 | 57.53 | 51.94 | 3.72 | −2.26 | 2.85 | |
| 2012 | Pingyu | 31.95 | 54.80 | 49.82 | 3.43 | −2.54 | 5.13 | |
| 2012 | Yuanyang | 30.77 | 57.88 | 52.23 | 4.22 | −2.53 | 3.98 | |
| 2012 | Xinyang | 32.74 | 58.73 | 53.14 | 4.59 | −2.32 | 2.67 | |
| PC | 2011 | Pingyu | 18.74 | 26.49 | 21.65 | 1.30 | 0.90 | 1.28 |
| 2011 | Yuanyang | 17.60 | 25.10 | 20.28 | 1.16 | 0.97 | 1.55 | |
| 2012 | Pingyu | 20.22 | 27.79 | 22.51 | 1.04 | 0.78 | 1.68 | |
| 2012 | Yuanyang | 16.72 | 27.44 | 20.94 | 1.25 | 1.05 | 5.27 | |
| 2012 | Xinyang | 17.24 | 26.84 | 20.59 | 1.57 | 0.85 | 1.30 |
Table 2. Analysis of Variance of the oil and protein contents in the population under 5 environments.
| Source of variation | DF | OC | PC | ||
| MS | F value | MS | F value | ||
| Year | 1 | 27.33 | 7.46** | 211.90 | 412.83** |
| Location | 2 | 1564.58 | 427.02** | 551.56 | 1074.55** |
| Variety | 368 | 62.78 | 17.13** | 5.23 | 10.19** |
| Year×Variety | 368 | 3.19 | 0.87 | 0.55 | 1.07 |
| Location×Variety | 736 | 2.94 | 0.80 | 0.65 | 1.27 |
| Residual | 367 | 3.66 | 0.51 | ||
** The significance at P<0.01.
DF denotes degree of freedom; MS denotes mean square.
Linkage disequilibrium
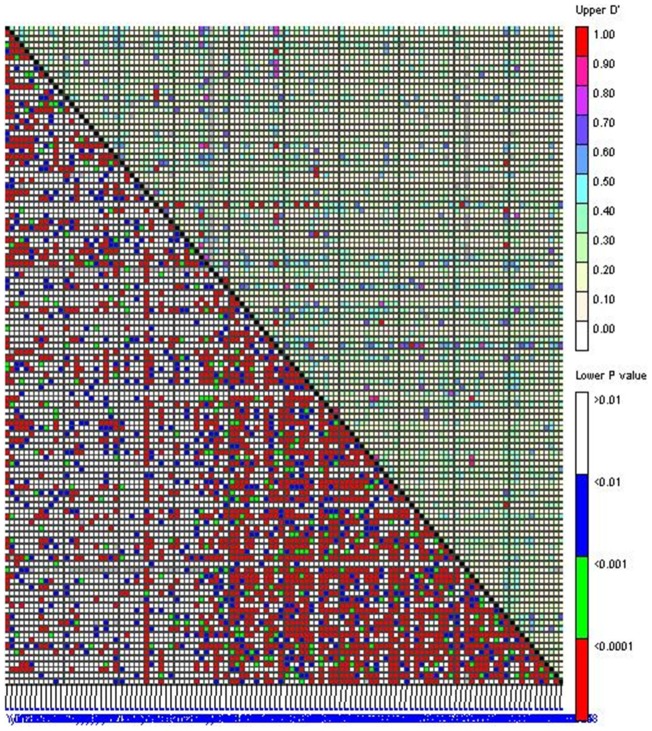
Linkage disequilibrium (LD) refers to the non-random association of alleles between the genetic loci. A total of 112 SSR markers were used for estimating the LD level among the 369 sesame germplasm accessions (Table S2). These SSR markers distributed in 33 contigs/scaffolds with a total length of 180.86 Mb, which approximately represented 67 percentage of the assembly genome size (270 Mb) and 50 percentage of the estimated genome size (360Mb). The average SSR density was 1 SSR per 1.6 Mb. To reflect the associations between the polymorphic loci of the 112 SSR markers, LD P-values were determined among the 6,216 locus pairs (i.e., 112*(112-1)/2) using Fisher's exact test and two indexes of D′ and r 2 (Figure 1). The average values of D′ and r 2 for the 6,216 pairs were 0.1649 and 0.0173, respectively. Of the 6216 pairs, 2584 pairs (41.57%) showed a significant linkage disequilibrium (P<0.01), 363 pairs (5.84%) showed a higher linkage disequilibrium of D′>0.5, and 33 pairs (0.5%) gave a D′ value of 1.0 (i.e., complete linkage). The data indicated that linkage disequilibrium existed among the sesame accessions.
Figure 1. Disequilibrium matrix of 112 SSR polymorphic sites with both the X-axis and Y-axis.
The matrix is divided into two regions by the diagonal line. The upper right region indicates the D′ value of each SSR couples, and the corresponding blocks in the lower left region indicates the significance of D′. Various value intervals of the D′ or P values are shown in different colors according to the right color columns.
SSR marker diversity and population structure
Before analyzing the association, the polymorphism of the 112 SSR markers within the 369 germplasms was investigated (Table S2). Results showed that the number of alleles ranged from 2–5, with an average of 2.47 per locus. The PIC values of the markers ranged from 0.028 (Hs373) to 0.669 (Y1994) with an average value of 0.302. The percentage of heterozygotes per marker varied from 0.27% (Hs373) to 23.12% (Hs4325), with an average of 10.14%. The data indicated that the natural population had the high heterozygosity and was suitable for association mapping analysis. Subsequently, we estimated the population structure, as admixture of subpopulation could result in LD and produce the false-positive results. The most likely number (K) of subgroups in the 369 germplasm accessions was estimated using the 112 SSR markers (Figure 2). As K values increased from 1 to 10, the value of LnP(D) elevated directly; meanwhile, the ΔK reduced straightly with a clear peak value of 643.5 at K = 2. The results indicated that the population was roughly composed of two divergent subgroups according to the Bayesian posterior probability analysis (Figure 3). Of the 369 accessions, 126 accessions were grouped into subgroup 1 (the green ones in Figure 3), 243 accessions into subgroup 2 (the red ones in Figure 3). Most (47 out of 51) of the foreign lines were located in subgroup 2. The two subgroups in the population were considered having the complex ‘admixture’ relationship, and no significant correlation of geographic origin with the subgroups in the Chinese lines were found.
Figure 2. Estimated (a) LnP(D) and (b) ΔK values for a given K.
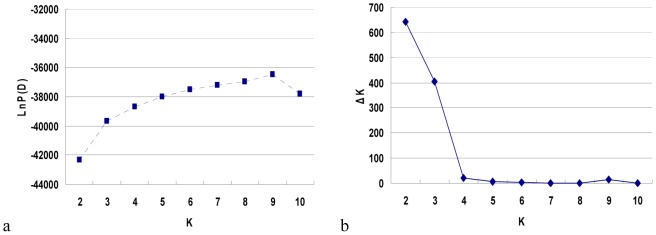
Figure 3. Genetic composition of individuals based on Bayesian posterior probability.
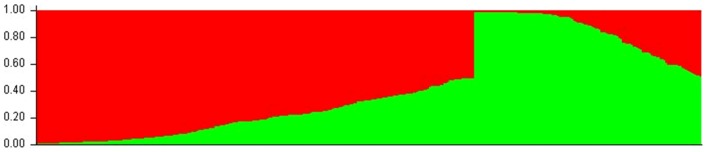
According to the Bayesian posterior probability, the natural population of 369 worldwide sesame lines is divided into two groups in green and red colors, respectively.
Marker-trait associations
In this study, the association analysis was performed using general liner model (GLM) method. The stringent criterion of logP≥3.0 under 5 environments was used for determining the association significance between the OC and PC traits. For the OC trait, 19 markers were detected and the R2 values ranged from 4%–29% (Table 3 & Table S2). According to the primer locations in the sesame genome, 19 markers were mapped in 11 contigs/scaffolds. Of the 19 markers, Hs485 and Hs586 are located in scaffold00001, Hs4381, Hs02 and Hs4082 are located in C01, and Hs4061, Hs345 and Hs19563 are in C04. The distance between the markers that located in the same contig/scaffold was short than 2 Mb (Table S2). For the PC trait, 24 markers were detected with the R2 value range of 3%–29% and mapped in 15 contigs/scaffolds (Table 4 & Table S2). Comparison results indicated that 19 of the above 24 markers were associated with OC trait at the same time; the other 5 markers of Hs425 (in C08), Hs560 (in C11), Hs4265 (in C12), and Hs4089 and Hs1514 (in C19) were unique to PC trait. Simultaneously, in order to assay the stability, we performed association mapping using mixed liner model (MLM) method, with the criterion of logP≥3.0 under at least 3 environments.
Table 3. Association mapping of the OC trait using GLM method under the 5 environments.
| Marker | 2011 | 2012 | |||||||||||||
| Pingyu | Yuanyang | Pingyu | Yuanyang | Xinyang | |||||||||||
| Fmaker | P marker | R2 | Fmaker | P marker | R2 | Fmaker | P marker | R2 | Fmaker | P marker | R2 | Fmaker | P marker | R2 | |
| Hs345 | 35.93 | 3.05E-25 | 0.28 | 26.22 | 4.37E-19 | 0.22 | 13.41 | 3.41E-10 | 0.13 | 26.44 | 3.11E-19 | 0.22 | 37.34 | 4.08E-26 | 0.29 |
| Hs4381 | 61.72 | 8.30E-24 | 0.25 | 43.17 | 1.48E-17 | 0.19 | 40.31 | 1.53E-16 | 0.18 | 46.18 | 1.32E-18 | 0.20 | 75.46 | 3.73E-28 | 0.29 |
| Hs1956 | 47.51 | 7.46E-19 | 0.22 | 39.98 | 2.83E-16 | 0.19 | 16.15 | 2.05E-07 | 0.09 | 24.86 | 8.80E-11 | 0.13 | 40.25 | 2.29E-16 | 0.19 |
| Hs485 | 31.50 | 4.06E-18 | 0.20 | 20.63 | 2.29E-12 | 0.14 | 21.10 | 1.27E-12 | 0.15 | 21.48 | 7.82E-13 | 0.15 | 36.07 | 1.98E-20 | 0.23 |
| Hs1036 | 41.73 | 6.75E-17 | 0.20 | 25.69 | 4.19E-11 | 0.13 | 17.87 | 4.24E-08 | 0.10 | 26.00 | 3.19E-11 | 0.13 | 43.84 | 1.24E-17 | 0.20 |
| Hs205 | 17.18 | 3.11E-15 | 0.19 | 10.41 | 2.42E-09 | 0.12 | 10.37 | 2.68E-09 | 0.13 | 12.46 | 3.65E-11 | 0.15 | 16.29 | 1.75E-14 | 0.18 |
| Hs4209 | 14.91 | 2.67E-13 | 0.17 | 8.15 | 2.64E-07 | 0.10 | 9.28 | 2.55E-08 | 0.12 | 12.37 | 4.41E-11 | 0.14 | 23.92 | 9.21E-21 | 0.25 |
| Hs672 | 16.81 | 1.19E-12 | 0.16 | 11.35 | 1.09E-08 | 0.11 | 16.82 | 1.16E-12 | 0.16 | 20.27 | 4.38E-15 | 0.18 | 26.53 | 2.58E-19 | 0.22 |
| Hs393 | 17.90 | 2.01E-13 | 0.16 | 10.10 | 9.18E-08 | 0.10 | 12.19 | 2.61E-09 | 0.12 | 16.74 | 1.32E-12 | 0.15 | 29.18 | 4.76E-21 | 0.24 |
| Hs4061 | 30.93 | 3.93E-13 | 0.14 | 19.32 | 1.06E-08 | 0.09 | 21.42 | 1.60E-09 | 0.11 | 23.28 | 3.05E-10 | 0.11 | 30.72 | 4.66E-13 | 0.14 |
| Y2129 | 22.99 | 4.73E-10 | 0.13 | 13.89 | 1.64E-06 | 0.08 | 15.29 | 4.57E-07 | 0.09 | 24.86 | 9.25E-11 | 0.13 | 30.65 | 6.69E-13 | 0.16 |
| Hs618 | 25.22 | 5.54E-11 | 0.12 | 12.90 | 3.86E-06 | 0.06 | 7.54 | 6.19E-04 | 0.04 | 17.66 | 4.78E-08 | 0.09 | 22.91 | 4.26E-10 | 0.11 |
| Hs377 | 4.14 | 2.01E-05 | 0.11 | 3.16 | 6.87E-04 | 0.08 | 3.92 | 4.49E-05 | 0.10 | 6.26 | 7.83E-09 | 0.15 | 7.09 | 3.63E-10 | 0.17 |
| Hs635 | 22.15 | 8.38E-10 | 0.11 | 17.23 | 7.11E-08 | 0.08 | 10.34 | 4.30E-05 | 0.05 | 18.79 | 1.71E-08 | 0.09 | 31.73 | 1.98E-13 | 0.15 |
| Hs02 | 20.98 | 2.38E-09 | 0.10 | 17.19 | 7.34E-08 | 0.08 | 10.31 | 4.43E-05 | 0.05 | 12.41 | 6.09E-06 | 0.06 | 36.20 | 4.55E-15 | 0.16 |
| Hs376 | 19.25 | 1.15E-08 | 0.10 | 11.49 | 1.45E-05 | 0.06 | 12.53 | 5.52E-06 | 0.07 | 16.27 | 1.72E-07 | 0.08 | 21.26 | 1.89E-09 | 0.10 |
| Hs1638 | 17.45 | 6.23E-08 | 0.09 | 7.29 | 7.96E-04 | 0.04 | 18.27 | 2.97E-08 | 0.10 | 19.25 | 1.23E-08 | 0.10 | 24.86 | 8.66E-11 | 0.13 |
| Hs586 | 16.84 | 1.01E-07 | 0.08 | 13.48 | 2.25E-06 | 0.07 | 7.30 | 7.82E-04 | 0.04 | 10.90 | 2.52E-05 | 0.06 | 20.14 | 5.06E-09 | 0.10 |
| Hs4082 | 8.18 | 3.36E-04 | 0.04 | 7.56 | 6.08E-04 | 0.04 | 18.30 | 2.69E-08 | 0.09 | 15.51 | 3.44E-07 | 0.08 | 20.61 | 3.33E-09 | 0.10 |
The markers associated with the OC trait under 5 environments are listed in the table.
Table 4. Association mapping of PC trait using GLM method under the 5 environments.
| Marker | 2011 | 2012 | |||||||||||||
| Pingyu | Yuanyang | Pingyu | Yuanyang | Xinyang | |||||||||||
| Fmaker | P marker | R2 | Fmaker | P marker | R2 | Fmaker | P marker | R2 | Fmaker | P marker | R2 | Fmaker | P marker | R2 | |
| Hs345 | 25.46 | 1.41E-18 | 0.21 | 21.52 | 6.24E-16 | 0.19 | 13.41 | 3.41E-10 | 0.13 | 26.44 | 3.11E-19 | 0.22 | 37.34 | 4.08E-26 | 0.29 |
| Hs4381 | 50.30 | 5.14E-20 | 0.21 | 33.86 | 3.25E-14 | 0.15 | 40.31 | 1.53E-16 | 0.18 | 46.18 | 1.32E-18 | 0.20 | 75.46 | 3.73E-28 | 0.29 |
| Hs485 | 26.93 | 9.51E-16 | 0.18 | 21.07 | 1.32E-12 | 0.14 | 21.10 | 1.27E-12 | 0.15 | 21.48 | 7.82E-13 | 0.15 | 36.07 | 1.98E-20 | 0.23 |
| Hs205 | 16.43 | 1.33E-14 | 0.18 | 7.90 | 4.52E-07 | 0.10 | 10.37 | 2.68E-09 | 0.13 | 12.46 | 3.65E-11 | 0.15 | 16.29 | 1.75E-14 | 0.18 |
| Hs1036 | 31.71 | 2.48E-13 | 0.16 | 17.49 | 5.98E-08 | 0.09 | 17.87 | 4.24E-08 | 0.10 | 26.00 | 3.19E-11 | 0.13 | 43.84 | 1.24E-17 | 0.20 |
| Hs393 | 16.01 | 4.43E-12 | 0.15 | 13.34 | 3.73E-10 | 0.12 | 12.19 | 2.61E-09 | 0.12 | 16.74 | 1.32E-12 | 0.15 | 29.18 | 4.76E-21 | 0.24 |
| Hs672 | 17.02 | 8.43E-13 | 0.15 | 11.63 | 6.73E-09 | 0.11 | 16.82 | 1.16E-12 | 0.16 | 20.27 | 4.38E-15 | 0.18 | 26.53 | 2.58E-19 | 0.22 |
| Hs1956 | 26.91 | 1.50E-11 | 0.14 | 22.88 | 4.93E-10 | 0.12 | 16.15 | 2.05E-07 | 0.09 | 24.86 | 8.80E-11 | 0.13 | 40.25 | 2.29E-16 | 0.19 |
| Hs377 | 5.73 | 5.69E-08 | 0.14 | 4.46 | 6.21E-06 | 0.11 | 3.92 | 4.49E-05 | 0.10 | 6.26 | 7.83E-09 | 0.15 | 7.09 | 3.63E-10 | 0.17 |
| Y2129 | 26.65 | 1.99E-11 | 0.14 | 16.79 | 1.17E-07 | 0.09 | 15.29 | 4.57E-07 | 0.09 | 24.86 | 9.25E-11 | 0.13 | 30.65 | 6.69E-13 | 0.16 |
| Hs4209 | 11.18 | 5.03E-10 | 0.13 | 7.07 | 2.56E-06 | 0.09 | 9.28 | 2.55E-08 | 0.12 | 12.37 | 4.41E-11 | 0.14 | 23.92 | 9.21E-21 | 0.25 |
| Hs4061 | 25.55 | 4.14E-11 | 0.12 | 17.49 | 5.59E-08 | 0.08 | 21.42 | 1.60E-09 | 0.11 | 23.28 | 3.05E-10 | 0.11 | 30.72 | 4.66E-13 | 0.14 |
| Hs1638 | 20.72 | 3.30E-09 | 0.11 | 14.13 | 1.29E-06 | 0.08 | 18.27 | 2.97E-08 | 0.10 | 19.25 | 1.23E-08 | 0.10 | 24.86 | 8.66E-11 | 0.13 |
| Hs618 | 22.58 | 5.71E-10 | 0.11 | 12.16 | 7.75E-06 | 0.06 | 7.54 | 6.19E-04 | 0.04 | 17.66 | 4.78E-08 | 0.09 | 22.91 | 4.26E-10 | 0.11 |
| Hs1514 | 19.02 | 1.53E-08 | 0.10 | 15.50 | 3.69E-07 | 0.08 | 6.23 | 2.20E-03 | 0.04 | 14.59 | 8.48E-07 | 0.08 | 14.77 | 7.19E-07 | 0.08 |
| Hs4089 | 19.68 | 7.67E-09 | 0.09 | 18.49 | 2.25E-08 | 0.09 | 6.44 | 1.80E-03 | 0.03 | 17.68 | 4.71E-08 | 0.09 | 13.84 | 1.61E-06 | 0.07 |
| Hs425 | 17.07 | 8.21E-08 | 0.08 | 8.38 | 2.75E-04 | 0.04 | 7.90 | 4.39E-04 | 0.04 | 17.02 | 8.55E-08 | 0.08 | 18.86 | 1.61E-08 | 0.09 |
| Hs560 | 4.53 | 1.96E-04 | 0.07 | 4.98 | 6.48E-05 | 0.08 | 4.69 | 1.33E-04 | 0.07 | 6.43 | 1.91E-06 | 0.10 | 7.36 | 1.97E-07 | 0.11 |
| Hs635 | 13.54 | 2.12E-06 | 0.07 | 13.28 | 2.72E-06 | 0.07 | 10.34 | 4.30E-05 | 0.05 | 18.79 | 1.71E-08 | 0.09 | 31.73 | 1.98E-13 | 0.15 |
| Hs02 | 14.67 | 7.44E-07 | 0.07 | 10.16 | 5.08E-05 | 0.05 | 10.31 | 4.43E-05 | 0.05 | 12.41 | 6.09E-06 | 0.06 | 36.20 | 4.55E-15 | 0.16 |
| Hs4265 | 5.17 | 1.34E-04 | 0.06 | 6.58 | 7.07E-06 | 0.08 | 6.70 | 5.50E-06 | 0.08 | 8.54 | 1.16E-07 | 0.10 | 11.90 | 1.13E-10 | 0.14 |
| Hs376 | 11.29 | 1.76E-05 | 0.06 | 11.04 | 2.22E-05 | 0.06 | 12.53 | 5.52E-06 | 0.07 | 16.27 | 1.72E-07 | 0.08 | 21.26 | 1.89E-09 | 0.10 |
| Hs586 | 11.07 | 2.15E-05 | 0.06 | 11.38 | 1.60E-05 | 0.06 | 7.30 | 7.82E-04 | 0.04 | 10.90 | 2.52E-05 | 0.06 | 20.14 | 5.06E-09 | 0.10 |
| Hs4082 | 12.36 | 6.41E-06 | 0.06 | 8.72 | 2.00E-04 | 0.04 | 18.30 | 2.69E-08 | 0.09 | 15.51 | 3.44E-07 | 0.08 | 20.61 | 3.33E-09 | 0.10 |
The markers associated with the PC trait under 5 environments are listed in the table.
For the OC trait, 9 markers were detected in MLM model and 8 markers of Hs345, Hs4381, Hs485, Hs1036, Hs4061, Hs635, Hs376 and Hs586 were confirmed using GLM and MLM methods. Especially, 4 markers of Hs345, Hs4381, Hs485 and Hs1036 had high R2 values (≥10%) under 5 environments (Table 3 and Table S3). For the PC trait, 9 markers were found and confirmed in both models, of which 7 markers had high R2 values (≥10%) under 5 environments (Table 4 and Table S3). These results suggested that the OC and PC traits are controlled mostly by major genes in sesame.
Marker effect on the phenotypic variation
To explore the association between the above markers and phenotypic variation and the utility potential in sesame MAS breeding program, we performed the allelic effects of the five markers associated with both traits (Table 5). For each marker, the effects estimated were in accordance with the allele character under the 5 environments. Hs345 had the largest effect on the variation of seed oil and protein content. The Hs345-1∶1 and Hs345-2∶2 of Hs345 showed the different variation effects on OC trait, as the average oil content in the accessions ranged from 52.05%–42.82%. In the genotypes carrying the Hs4381-1∶1 allele, oil and protein contents were 43.89% and 23.25%, respectively, while the samples with the Hs485-2∶2 contained 52.00% oil and 21.00% proteins. Furthermore, the specific allele indicated the negative or positive effect to a large extent on the OC or PC trait. For example, the allele effect of Hs345-2∶2 on OC trait ranged from −8.13 to −12.18, whereas the effect on PC trait ranged from 2.05 to 3.82. Comparison results suggested that the alleles of all 5 markers give the opposite effects on OC and PC traits, respectively. The allele that increased the oil content certainly gave the negative effect on protein content, and vice versa. Therefore, these markers could be used for screening sesame lines with either high oil or protein content.
Table 5. Allele effects of 5 markers simutaneously associated with OC and PC traits under 5 environments.
| Marker | Allelea | 2011 | 2012 | ||||||||
| Pingyu | Yuanyang | Pingyu | Yuanyang | Xinyang | |||||||
| OC | PC | OC | PC | OC | PC | OC | PC | OC | PC | ||
| Hs345 | 1∶1 | −0.04 | −0.17 | −0.59 | 0.25 | −1.17 | 0.44 | −1.61 | 0.57 | −0.93 | 0.39 |
| 2∶2 | −10.74 | 2.30 | −8.13 | 2.12 | −8.87 | 2.05 | −12.18 | 2.96 | −11.73 | 3.82 | |
| 1∶2 | −5.53 | 1.38 | −4.59 | 1.67 | −5.13 | 1.29 | −5.84 | 2.08 | −6.66 | 2.58 | |
| Hs4061 | 1∶1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 2∶2 | 8.78 | −2.41 | 4.46 | −1.95 | 6.37 | −1.84 | 8.54 | −2.33 | 7.94 | −2.96 | |
| 1∶2 | 2.58 | −0.98 | −0.18 | −1.06 | 1.78 | −0.80 | 2.58 | −1.14 | 1.60 | −0.86 | |
| Hs1956 | 1∶1 | 1.12 | −0.36 | 1.01 | −0.49 | 0.85 | −0.32 | 1.18 | −0.39 | 1.67 | −0.81 |
| 2∶2 | −5.89 | 1.21 | −4.13 | 0.79 | −4.03 | 0.63 | −5.03 | 0.95 | −5.15 | 1.34 | |
| 1∶2 | −0.74 | 0.15 | 0.11 | −0.28 | −0.86 | 0.26 | −0.60 | 0.49 | −0.85 | 0.02 | |
| Hs4381 | 1∶1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 2∶2 | 9.75 | −2.45 | 6.33 | −1.73 | 6.97 | −1.82 | 9.06 | −2.17 | 9.80 | −3.41 | |
| 1∶2 | 5.02 | −1.05 | 2.34 | −0.49 | 3.16 | −0.90 | 5.12 | −0.75 | 4.58 | −1.51 | |
| Hs485 | 1∶1 | −4.80 | 0.47 | −3.23 | 0.25 | −3.87 | 0.51 | −6.88 | 1.62 | −7.19 | 2.24 |
| 2∶2 | 2.04 | −1.28 | 0.82 | −1.21 | 1.71 | −0.96 | −0.02 | 0.02 | 0.20 | −0.29 | |
| 1∶2 | −3.99 | 0.33 | −3.85 | −0.03 | −2.59 | −0.12 | −4.64 | 1.38 | −5.76 | 1.74 | |
Note: a two of the most common alleles are listed.
Comparative genome analysis
To clarify the distribution and more information of the above associated markers, we performed the comparative genome analysis of the 19 SSR markers associated with the OC trait (Table 6). All genes closest to the markers were explored. Of the 19 markers, 11 are located in gene regions and 8 are in intergenic regions. The flanking genes had various functions, such as ligase (C01.560), transcription factor (C04.26, C13.438) and kinase (C14.22). Moreover, we found that the markers of Hs4082 and Hs345 were just located next to C01.883 (ABC transporter G family member 3 gene) and C04.786 (Stearoyl-ACP Desaturase gene), respectively, which were proved involved in plant lipid biosynthesis. We also screened the upstream and downstream sequences of 500 Kb far from each marker. 17 (out of 19) markers were proved close to plant lipid pathway genes. These result further confirmed our association mapping conclusions.
Table 6. Information of candidate genes related to the markers associated with OC traits.
| Marker | Related genea | Annotation | Nearby lipid genesb | Arabidopsis thaliana homologous genes | Ref. | |
| Locic | Annotation | |||||
| Hs4381 | C01.460 | Thioredoxin M3 | C01.526 | At1G55260 | Lipid Transfer Protein type 5 | [25] |
| Hs02 | C01.560 | E3 ubiquitin-protei | C01.548 | At3G25110 | Acyl-ACP Thioesterase A | [26] |
| ligase RHF2A | C01.575 | At3G07400 | Lipid Acylhydrolase-like | |||
| C01.561 | Unknown | C01.601 | At2G30720 | Acyl-CoA Thioesterase | [27] | |
| Hs4082 | C01.882 | Cell cycle protein | C01.873 | At1G53390 | ABC Transporter | [28] |
| C01.883 | ABC transporter G | C01.883 | At2G28070 | ABC Transporter | [28] | |
| family member 3 | C01.928 | At3G44830 | Diacylglycerol Acyltransferase | [29] | ||
| Hs635 | C02.696 | RAS-related protein Rab11A | C02.739 | At1G47620 | Midchain Alkane Hydroxylase | 30] |
| Hs4209 | C04.25 | Transmembrane domain | C04.38 | At1G31770 | ABC Transporter | [31] |
| containing protein | C04.56 | At1G71010 | Phosphatidylinositol-Phosphate Kinase | [32] | ||
| C04.26 | TCP transcription factor 12 | C04.81 | At1G10900 | Phosphatidylinositol-Phosphate Kinase | [32] | |
| C04.96 | At1G77660 | Phosphatidylinositol-Phosphate Kinase | [32] | |||
| Hs4061 | C04.681 | Inositol trisphosphate 5-phosphatase 1 | — | |||
| Hs345 | C04.785 | Unknown | C04.767 | At1G05630 | Phosphoinositide 5-Phosphatase | — |
| C04.786 | Stearoyl-ACP desaturase | C04.786 | At2G43710 | Stearoyl-ACP Desaturase | [33] | |
| Hs1956 | C04.845 | FAR1-related sequence | The same as Hs345 | |||
| Hs393 | C13.438 | bZIP transcription | C13.388 | At1G10900 | Phosphatidylinositol-Phosphate Kinase | [32] |
| factor 40 | C13.471 | At5G10160 | Hydroxyacyl-ACP Dehydrase | [34] | ||
| C13.504 | At1G17840 | ABC Transporter | [35] | |||
| C13.514 | At4G36480 | Subunit of Serine Palmitoyltransferase | [36] | |||
| Hs205 | C14.21 | Thylakoid membrane | C14.49 | At4G33550 | Lipid Transfer Protein type 3 | [37] |
| phosphoprotein | C14.66 | At1G49430 | Long-Chain Acyl-CoA Synthetase | [38] | ||
| C14.22 | Serine/threonine-protein | C14.111 | At2G46210 | Sphingobase-D8 Desaturase | [39] | |
| kinase WNK4 | C14.132 | At2G46090 | Diacylglycerol Kinase | — | ||
| Hs672 | C14.367 | Cyclin-P3.1 F-box | C14.359 | At2G45150 | CDP-DAG Synthase | [40] |
| protein | C14.413 | At4G33355 | Lipid Transfer Protein type 1 | — | ||
| C14.368 | Unknown | C14.428 | At2G44810 | Acylhydrolase (DAD1-like) | — | |
| Y2129 | C15.825 | Unknown | C15.92 | At2G29980 | Linoleate Desaturase | [41] |
| Hs1638 | C15.840 | Unknown | The same as Y2129 | |||
| Hs618 | C25.41 | IAA-alanine resistance | C25.69 | At5G08415 | Lipoate Synthase | [42] |
| protein 1 | ||||||
| C25.42 | Beta-D-xylosidase | |||||
| Hs376 | C26.474 | ADP-ribosylation factor | C26.417 | At1G15110 | Base-Exchange-type Phosphatidylserine | [43] |
| Synthase | ||||||
| C26.454 | At1G71010 | Phosphatidylinositol-Phosphate Kinase | — | |||
| C26.515 | At1G31812 | Acyl CoA Binding Protein | [44] | |||
| Hs377 | C26.475 | Unknown | The same as Hs376 | |||
| Hs485 | sf00001.885 | Unknown | sf00001.95 | At4G04930 | Dihydrosphingosine Delta-4 Desaturase | [45] |
| sf00001.886 | Unknown | |||||
| Hs586 | sf00001.754 | ADP-ribosylation factor | — | |||
| Hs1036 | sf00044.12 | ABC transporter I | sf00044.12 | At3G20320 | Acid-Binding Protein | [46] |
| family member 15 | sf00044.41 | At2G25170 | Chromatin remodeling factor | [47] | ||
| sf00044.61 | At4G19860 | Acyl acceptor Acyltransferase | — | |||
| sf00044.90 | At1G13210 | Translocase | — | |||
| sf00044.113 | At1G74320 | Choline Kinase | [48] | |||
Note: aRelated genes are the genes containing or close to the screened markers.
Nearby lipid genes refer to the genes located in the upstream and downstream of 500 Kb far from the marker.
Only one of the homologous genes is listed in the table.
— refers to no known lipid genes in the location.
Discussion
To clarify the genetic mechanisms of fatty acid and protein synthesis in sesame seeds, we herein performed the association mapping analysis of the OC and PC traits among 369 sesame accessions using 112 genic-SSR markers. These accessions were collected from 19 provinces of China and 15 other countries, and represented the genetic diversity of sesame germplasm for association mapping study. These accessions included many released Chinese and foreign cultivars. Compared with the traditional linkage analysis (QTL mapping), the association analysis based on linkage disequilibrium (LD) has been applied for the quantitative trait loci (QTLs) detection and location in many crops. Meanwhile, GLM and MLM models are applied individually for evaluating the marker association. In this article, 19 SSR markers associated with the OC trait were detected under each 5 environments in GLM model, while 24 markers were determined and associated with the protein content.
Sesame genetic diversity and the population structure
Sesame is a diploid and self-pollinated oil crop with the karyotype of 2n = 2x = 26. As all cultivars are originated from the sole cultivated species, Sesamum indicum L, the narrow genetic diversity presents in regional sesame resources to a large extent [24], [49]–[51]. In addition, many reports reflect that there is no clear association between genotype and geographical origin, as many sesame accessions from the different geographic locations are clustered into the same group in the dendrogram [51]–[53]. Apart from the natural history of predomesticated ancestors, the diversity pattern of domestic species could be influenced by the breeding practice, germplasm collection and human activity [53]–[55]. In this article, in order to guarantee the broad genetic variation, we selected the 369 worldwide accessions for seed nutrition genetic analysis according to the geographical origin, molecular clustering and the morphologic diversity (Table S1). The heterozygosity ranged from 0.27% (Hs373)-23.12% (Hs4325). The result proved that the natural population could be used as the core germplasm for association mapping (Table S2). Population structure analysis showed that many sesame accessions collected from the same geographic locations were not grouped together, which further proved the unclear association between genotype and geographical origin in sesame germplasm resource [51]–[53].
During performing the association mapping in a population, LD patterns between the functional loci and markers should be analyzed at first [56]. We analyzed the P-values of linkage disequilibrium between the polymorphisms of the 112 SSR marker loci using Fisher's exact test (Figure 1). As 363 (5.84%) pairwise comparisons had the high LDs (D′>0.5), the linkage disequilibrium existed in 369 sesame accessions. Accordingly, we believed that the natural population is suitable for association analysis of the oil and protein contents.
Oil and protein content variation and associated SSR markers
Among large-scale sesame germplasm resources, the oil and protein contents varied significantly. Yermanos et al. [57] evaluated 721 sesame samples collected from more than 19 countries, and found that the oil content varied from 40.4–59.8% with the mean of 53.1%. The protein content ranged from 19–31% with an average of about 25% [58]. In our population, the oil content varied from 27.89%–58.73% with an average of 51.34%, while the protein content varied from 16.72%–27.79% with an average of 21.19% (Table 1). The data reflected the great variation of sesame seed compositions in germplasm [57], [59], [60].
Comparison analysis proved that there is a strong negative correlation of the oil content with protein content. Interestingly, the stringency relationship was also exhibited in the association analysis. As shown in the GLM analysis in Table 3 and 5, all the 19 markers associated with OC were detected for PC trait; and the alleles exhibited the opposite effects on OC and PC at the same time. These phenomena also present in other oil crops, such as oilseed rape, cotton, soybean and peanut [1], [61]–[63]. Zhao et al. [62] found 6 QTLs with pleiotropic effects on both oil content and protein content in oilseed rape. In the cotton backcross inbred population, of 17 QTLs for oil content and 20 QTLs for protein content, 8 QTLs co-localized in the same chromosome regions and controlled oil and protein contents with opposite additive effects [63]. Hwang et al. (2014) detected 25 SNPs associated with seed oil in 13 different genomic regions through GWAS (genome wide association study), and 7 SNPs were significantly associated with both protein and oil traits. For the six of seven marker loci, a negative relationship existed between the protein effect versus that on oil [64]. Meanwhile, QTLs or markers associated with increased protein and oil contents were also found. For example, Zhao et al. (2006) found that 2 QTLs that controlled oil content were independent from protein content by conditional QTL mapping [65]; Hwang et al. (2014) found a SNP at the 4.92 Mb position on Chr 9 was associated with increased protein and oil contents [64].
In many crops, the seed oil content seems to be controlled mostly by major genes [66]–[69]. In this study, we detected 19 markers associated with OC trait in sesame using GLM method, and the R2 values ranged from 4.0–29.0%. Chen et al. (2010) screened 27 QTLs related to oil content in oilseed rape and the individual explanation was high with the range of 4.20–30.20% [66]. In Arabidopsis thaliana, a single QTL or marker could give an explanation of 17–19% for the seed oil content [68]. In soybean, the explanation reached to 14.3–45.6% [69]. In this report, 112 polymorphic SSR markers were used for association mapping. Compared with other common molecular markers, such as SRAP and AFLP, SSR marker is more suitable for sesame diversity analysis due to the narrow genetic basis [24], [49]–[51]. The marker distribution and density were analyzed using the sesame genome assembly data. The contigs/scaffolds carrying the 112 markers approximately covered 180 Mb of the sesame genome and occupied ∼67% of the assembly size (270 Mb) and 50% of the estimated genome size (∼360 Mb, in which 90 Mb was believed to be repeat sequences) (Table S2) [70], [71]. Therefore, the association mapping using 112 markers is meaningful and reliable, even though some QTLs might be missed. To detect more QTLs, new SSR or SNP markers could be applied in further research.
Candidate genes and oil components
To clarify the marker location and more genome information for OC and PC traits, we screened the genes that were close to the associated markers using the sesame genome assembly data. As a result, 36 candidate genes related to lipid pathway were identified (Table 6) [25]–[48]. We found that most candidate genes were involved in three pathways: (1) fatty acid and TAG (triacylglycerol) synthesis and elongation, e.g., C01.526, C01.548 and C01.928; (2) TAG degradation, e.g., C01.601; and (3) fatty acid dehydrogenation, e.g., C04.786 (Stearoyl-ACP desaturase, determining the ratio of saturated and unsaturated fatty acids) (Table S4). Therefore, the seed oil content in the sesame accessions could be regulated by three factors, i.e., oil synthesis ability, oil degradation ability and oil component ratio (e.g., 18∶1 and 18∶2 fatty acids). In various accessions, any alleles of the genes involved in fatty acid and TAG synthesis, TAG degradation or dehydrogenase genes could give effects on oil content. To confirm this hypothesis, further studies of seed oil synthesis should be performed in the future.
Conclusion
We systematically explored the association mapping of seed oil and protein content traits in 369 worldwide sesame accessions using the 112 SSR markers. A significant negative correlationof the oil content with the protein content existed in the population. 19 SSR markers were associated with the oil content trait with high phenotypic variation explanation, and 24 SSR markers were associated with the protein content trait using GLM method. This association results would provide an efficient platform for seed development research and MAS breeding in sesame.
Methods
Plant materials
A population of 369 core sesame germplasm resources was chosen according to the genetic diversity analysis results and phenotypic variation [51]. These core genotypes comprised 318 lines from 19 provinces of China and 51 worldwide lines from the 15 countries, which were reserved at the Sesame Germplasm Bank of Henan Sesame Research Center (HSRC), HAAS (Henan Academy of Agricultural Sciences) (Table S1). All accessions were grown with three replications at three different experimental stations of Yuanyang (113.96°E, 35.05°N), Pingyu (114.62°E, 32.97°N), and Xinyang (114.08°E, 32.13°N) in 2011 and 2012. Five or six young leaves of individual accessions were collected and reserved at −70°C for DNA extraction.
Oil and protein content analysis
After harvested, ∼20 g of seeds were collected from five plants per line, and the seed OC and PC were measured on infrared determination equipment (Perten DA7250, Sweden) according to the manufactures' instructions. The standard curve for measuring the sesame oil and protein contents had been established according to the chemical analysis results of 300 sesame accessions (Unpublished data, HSRC). Three replications of each samples were assayed for phenotypic analysis. Mean value, broad-sense heritability and correlation coefficient of the phenotypic data were analyzed using the statistical analysis system software (SAS Institute Inc. 2002) [72].
SSR genotyping
The 112 polymorphic SSR pairs were selected from our SSR marker bank [24], [51], [73] (Table S2). DNA extraction, PCR amplification, electrophoresis and SSR genotyping analysis were performed according to the methods described by Zhang et al. [24]. The total number of polymorphic alleles at each SSR locus was calculated according to the results of all 369 lines. The polymorphic SSR alleles presented only within 4 (1%) or fewer accessions were recorded as rare alleles.
Linkage disequilibrium (LD)
As the population structure could result in the spurious associations between phenotypes and marker loci, we analyzed the extent and structure of LD within the population before selecting the appropriate association mapping strategy. To assay whether the 112 polymorphic SSR markers were segregated independently or not, LD analysis was conducted according to the dedicated procedure of the TASSEL software [49]. Both D′ and r2 were used for quantifying LD values [74], [75]. Significance (P values) of D′ for each SSR pairs was determined with 100,000 permutations.
Population structure and relatedness analysis
The population structure was determined using STRUCTURE 2.2 [76]. The mixture model and the independent allele frequency model were used to analyze the population dataset. Five runs of STRUCTURE were carried out for each number of populations (K) (from 1–10), and each run started with 10,000 burn-ins, followed by 100,000 iterations. While performing the STRUCTURE, we assumed that the inferred population accord with Hardy Weinberg equilibrium (HWE) and the loci are unlinked. To correct the relatedness of individuals in further analyses, the relatedness between individuals (relative kinship) was evaluated using SPAGeDi 1.2 software [77]. The matrix with the relative kinship coefficients (K matrix) was applied for association analysis using the Mixed Linear Model(MLM, Q+K)method.
Association mapping and marker distribution in sesame genome
Associations between the SSR markers and the oil and protein content traits were investigated using both methods of the general linear model (GLM, Q) and the mixed linear model (MLM, Q+K) in TASSEL 2.1 described by Bradbury et al. [49]. The mean value of the markers at P<0.005 was used for determining the significance of marker-trait associations.
To determine distributions of the associated markers in sesame genome, we carried out the alignment of SSR markers and transcripts with the updated sesame genome data [70], [71]. In the present genome assembly, the number of N50 scaffold was 14, and the number of N90 scaffold was 64. 29,798 gene models were identified. Among related scaffolds or contigs, the sesame lipid synthesis related genes were identified according to the homologous comparison using the genes of A. thaliana from the Acyl Lipids pathway database [78] as queries.
Supporting Information
Origins of 369 sesame accessions.
(DOCX)
Diversity statistics of 112 SSR markers in 369 sesame accessions and locations in the sesame draft genome
(DOCX)
Association mapping of the OC and PC traits using MLM method.
(DOCX)
Functions and involved pathways of the candidate genes.
(DOCX)
Funding Statement
This work was supported by China National “973” Project (2011CB109304) and the earmarked fund for the China Agriculture Research System (CARS-15). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brummer EC, Graef GL, Orf J, Wilcox JR, Shoemaker RC (1997) Mapping QTL for seed protein and oil content in eight soybean populations. Crop Sci 37: 370–378. [Google Scholar]
- 2.Guo SX (2011) Genetic and QTL analysis of oil content and other important quality traits on Brassica Napus L.). PhD thesis. Sichuan Agricultural University, Rice Institute.
- 3. Turhan H, Citak N, Pehlivanoglu H, Mengul Z (2010) Effects of ecological and topographic conditions on oil content and fatty acid composition in sunflower. Bulgarian J Agri Sci 16: 553–558. [Google Scholar]
- 4. Chen SL, Li YR, Cheng ZS, Liao BS, Lei Y, et al. (2009) Heterosis and Genetic Analysis of Oil Content in Peanut Using Mixed Model of Major Gene and Polygene. Scientia Agricultura Sinic 42: 3048–3057. [Google Scholar]
- 5. Anilakumar KR, Pal A, Khanum F, Bawa AS (2010) Nutritional, medicinal and industrial uses of sesame (Sesamum indicum L.) seeds- an overview. Agric Conspec Sci 75: 159–168. [Google Scholar]
- 6. Bedigian D, Harlan JR (1986) Evidence for cultivation of sesame in the ancient world. Econ Bot 40: 137–154. [Google Scholar]
- 7.Zhang HY (2013) Sesame genetic and breeding status and development strategy in China. In: Chinese Journal of Oil Crop Sciences Editorial Office, editors. VII China National Oil Crops Conference altar Paper compilations. Wenchang, China. pp. 29–36.
- 8. Jin UH, Lee JW, Chung YS, Lee JH, Yi YB, et al. (2001) Characterization and temporal expression of a ω-6 fatty acid desaturase cDNA from sesame (Sesamum indicum L.) seeds. Plant Sci 161: 935–941. [Google Scholar]
- 9. Chun JA, Jin UH, Lee JW, Yi YB, Hyung NI, et al. (2003) Isolation and characterization of a myo-inositol 1-phosphate synthase cDNA from developing sesame (Sesamum indicum L.) seeds: functional and differential expression, and salt–induced transcription during germination. Planta 216: 874–880. [DOI] [PubMed] [Google Scholar]
- 10. Leduc V, Moneret-Vautrin DA, Tzen JTC, Morisset M, Guerin L, et al. (2006) Identification of oleosins as major allergens in sesame seed allergic patients. Allergy 61: 349–356. [DOI] [PubMed] [Google Scholar]
- 11. Magni C, Ballabio C, Restani P, Fuggetta D. Alessandri C, et al. (2010) Molecular insight into IgE-mediated reactions to sesame (Sesamum indicum L.) seed proteins. Ann Allerg Asthma Im 105: 458–464. [DOI] [PubMed] [Google Scholar]
- 12. Wei W, Zhang Y, Lv H, Li D, Wang L, Zhang X (2013) Association analysis for quality traits in a diverse panel of Chinese sesame (Sesamum indicum L.) germplasm. J Integr Plant Biol 55: 745–758. [DOI] [PubMed] [Google Scholar]
- 13. Suh MC, Kim MJ, Hur CG, Bae JM, Park YI, et al. (2003) Comparative analysis of expressed sequence tags from Sesamum indicum and Arabidopsis thaliana developing seeds. Plant Mol Biol 52: 1107–1123. [DOI] [PubMed] [Google Scholar]
- 14. Ke T, Dong C, Han M, Zhao Y, Chen H, et al. (2011) Analysis of expression sequence tags from a full-length-enriched cDNA library of developing sesame seeds (Sesamum indicum). BMC Plant Biol 11: 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Singkham N, Jogloy S, Kesmala T, Swatsitang P, Jaisil P, et al. (2010) Genotypic variability and genotype by environment interactions in oil and fatty acids in high, intermediate, and low oleic acid peanut genotypes. J Agr Food chem 58: 6257–6263. [DOI] [PubMed] [Google Scholar]
- 16. Zhao J, Artemyeva A, Del Carpio DP, Basnet RK, Zhang N, et al. (2010) Design of a Brassica rapa core collection for association mapping studies. Genome 53: 884–898. [DOI] [PubMed] [Google Scholar]
- 17. Zou J, Jiang C, Cao Z, Li R, Long Y, et al. (2010) Association mapping of seed oil content in Brassica napus and comparison with quantitative trait loci identified from linkage mapping. Genome 53: 908–916. [DOI] [PubMed] [Google Scholar]
- 18. Abdurakhmonov IY, Saha S, Jenkins JN, Buriev ZT, Shermatov SE, et al. (2009) Linkage disequilibrium based association mapping of fiber quality traits in G. hirsutum L. variety germplasm. Genetica 136: 401–417. [DOI] [PubMed] [Google Scholar]
- 19. Ecke W, Clemens R, Honsdorf N, Becker HC (2010) Extent and structure of linkage disequilibrium in canola quality winter rapeseed (Brassica napus L.). Theor Appl Genet 2010 120: 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eleuch L, Jilal A, Grando S, Ceccarelli S, Schmising MVK, et al. (2008) Genetic diversity and association analysis for salinity tolerance, heading date and plant height of barley germplasm using simple sequence repeat markers. J Integr Plant Biol 50: 1004–1014. [DOI] [PubMed] [Google Scholar]
- 21. Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, et al. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7: e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He D, Xing H, Zhao J, Li T, Tang Y, et al. (2011) Population structure inferring and association analysis of fiber quality in cultivated cotton. J Northwest A & F Univ (Nat Sci Ed) 39: 103–112. [Google Scholar]
- 23. Wei W, Qi X, Wang L, Zhang Y, Wei H, et al. (2011) Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired–end sequencing and development of EST–SSR markers. BMC genomics 12: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang H, Wei L, Miao H, Zhang T, Wang C (2012) Development and validation of genic–SSR markers in sesame by RNA-seq. BMC genomics 13: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, et al. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614. [DOI] [PubMed] [Google Scholar]
- 26. Salas JJ, Ohlrogge JB (2002) Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch Biochem Biophys 403: 25–34. [DOI] [PubMed] [Google Scholar]
- 27. Tilton G, Shockey J, Browse J (2000) Two families of acyl-CoA thioesterases in Arabidopsis. Biochem Soc Trans 28: 946–947. [PubMed] [Google Scholar]
- 28. Quilichini TD, Friedmann MC, Samuels AL, Douglas CJ (2010) ATP-Binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol 154: 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang M, Fan JL, Taylor DC, Ohlrogge JB (2009) DGAT1 and PDAT1 Acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheng D, Nelson TC, Chen J, Walker SG, Wardwell-Swanson J, et al. (2003) Identification of acyl coenzyme A:Monoacylglycerol acyltransferase 3, an intestinal specific enzyme implicated in dietary fat absorption. J Biol Chem 278: 13611–13614. [DOI] [PubMed] [Google Scholar]
- 31. Panikashvili D, Shi JX, Schreiber L, Aharoni A (2011) The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytolt 190: 113–124. [DOI] [PubMed] [Google Scholar]
- 32. Xue HW, Hosaka K, Plesch G, Mueller-Roeber B (2000) Cloning of Arabidopsis thaliana phosphatidylinositol synthase and functional expression in the yeast pis mutant. Plant Mol Biol 42: 757–764. [DOI] [PubMed] [Google Scholar]
- 33. Kachroo A, Shanklin J, Whittle E, Lapchyk L, Hildebrand D, Kachroo P (2007) The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Mol Biol 63: 257–271. [DOI] [PubMed] [Google Scholar]
- 34. Brown A, Affleck V, Kroon J, Slabas A (2009) Proof of function of a putative 3-hydroxyacyl-acyl carrier protein dehydratase from higher plants by mass spectrometry of product formation. FEBS lett 583: 363–368. [DOI] [PubMed] [Google Scholar]
- 35. Bird D, Beisson F, Brigham A, Shin J, Greer S, et al. (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498. [DOI] [PubMed] [Google Scholar]
- 36. Chen M, Han GS, Dietrich CR, Dunn TM, Cahoon EB (2006) The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xing S, Zachgo S (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801. [DOI] [PubMed] [Google Scholar]
- 38. Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen M, Markham JE, Cahoon EB (2012) Sphingolipid delta 8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J 69: 769–781. [DOI] [PubMed] [Google Scholar]
- 40. Haselier A, Akbari H, Weth A, Baumgartner W, Frentzen M (2010) Two closely related genes of Arabidopsis encode plastidial cytidinediphosphate diacylglycerol synthases essential for photoautotrophic growth. Plant Physiol 153: 1372–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arondel V, Lemieux B, Hwang I, Gibson S, Goodman HM, et al. (1992) Map-based cloning of a gene controlling omega-3fatty-acid desaturation in Arabidopis. Science 258: 1353–1355. [DOI] [PubMed] [Google Scholar]
- 42. Yasuno R, Wada H (2002) The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett 517: 110–114. [DOI] [PubMed] [Google Scholar]
- 43. Yamaoka Y, Yu Y, Mizoi J, Fujiki Y, Saito K, et al. (2011) PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana . Plant J 67: 648–661. [DOI] [PubMed] [Google Scholar]
- 44. Engeseth NJ, Pacovsky RS, Newman T, Ohlrogge JB (1996) Characterization of an Acyl-CoA-binding protein from Arabidopsis thaliana . Arch Biochem Biophys 331: 55–62. [DOI] [PubMed] [Google Scholar]
- 45. Michaelson LV, Zauner S, Markham JE, Haslam RP, Desikan R, et al. (2009) Functional characterization of a higher plant sphingolipid Delta4-desaturase: Defining the role of sphingosine and sphingosine-1-phosphate in Arabidopsis. Plant Physiol 149: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Awai K, Xu CC, Tamot B, Benning C (2006) A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc Natl Acad Sci 103: 10817–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94. [DOI] [PubMed] [Google Scholar]
- 48. Monks DE, Goode JH, Dewey RE (1996) Characterization of soybean choline kinase cDNAs and their expression in yeast and Escherichia coli. Plant Physiol 11: 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Laurentin HE, Karlovsky P (2006) Genetic relationship and diversity in a sesame (Sesamum indicum L.) germplasm collection using amplified fragment length polymorphism (AFLP). BMC genetics 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang P, Zhang HY, Guo WZ, Zheng YZ, Wei LB, et al. (2007) Genetic diversity analysis of Sesamum indicum L. germplasms using SRAP and EST-SSR markers. Acta Agro Sin 33: 1696–1702. [Google Scholar]
- 51. Yue WD, Wei L, Zhang T, Li C, Miao H, et al. (2012) Genetic diversity and population structure of germplasm resources in sesame (Sesamum indicum L.) by SSR markers. Acta Agro Sin 38: 2286–2296. [Google Scholar]
- 52. Kim DH, Zur G, Danin-Poleg Y, Lee SW, Shim KB, et al. (2002) Genetic relationships of sesame germplasm collection as revealed by inter–simple sequence repeats. Plant Breed 121: 259–262. [Google Scholar]
- 53. Kumar H, Kaur G, Banga S (2012) Molecular characterization and assessment of genetic diversity in sesame (Sesamum indicum L.) germplasm collection using ISSR markers. J Crop Improve 26: 540–557. [Google Scholar]
- 54. Stankiewicz M, Gadamski G, Gawronski SW (2001) Genetic variation and phylogenetic relationships of triazine resistant and triazine susceptible biotypes of Solanum nigrum-analysis using RAPD markers. Weed Res 41: 287–300. [Google Scholar]
- 55. Garris AJ, Tai TH, Coburn J, Kresovich S, McCOUCH S (2005) Genetic structure and diversity in Oryza sativa L. Genetics. 169: 1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ersoz ES, Yu J, Buckler ES (2007) Applications of linkage disequilibrium and association mapping in crop plants. In: Varshney RK, Tuberosa R, editors. Genomics–assisted crop improvement. Volume 1 . Netherlands: Springer. pp. 97–119. [Google Scholar]
- 57. Yermanos DM, Hemstreet S, Saleeb W, Huszar CK (1972) Oil content and composition of the seed in the world collection of sesame introductions. J Am Oil Chem Soc 49: 20–23. [Google Scholar]
- 58. Johnson LA, Suleiman TM, Lusas EW (1979) Sesame protein: A review and prospectus. J Am Oil Chem Soc 56: 463–468. [DOI] [PubMed] [Google Scholar]
- 59. El Tinay AH, Khattab AH, Khidir MO (1976) Protein and oil compositions of sesame seed. J Am Oil Chem Soc 53: 648–653. [Google Scholar]
- 60. Uzun B, Arslan Ç, Furat Ş (2008) Variation in fatty acid compositions, oil content and oil yield in a germplasm collection of sesame (Sesamum indicum L.). J Am Oil Chem Soc 85: 1135–1142. [Google Scholar]
- 61. Sarvamangala C, Gowda MVC, Varshney RK (2011) Identification of quantitative trait loci for protein content, oil content and oil quality for groundnut (Arachis hypogaea L.). Field Crops Res 122: 49–59. [Google Scholar]
- 62. Zhao J, Becker HC, Zhang D, Zhang Y, Ecke W (2006) Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor Appl Genet 113: 33–38. [DOI] [PubMed] [Google Scholar]
- 63. Yu J, Yu S, Fan S, Song M, Zhai H, et al. (2012) Mapping quantitative trait loci for cottonseed oil, protein and gossypol content in a Gossypium hirsutum×Gossypium barbadense backcross inbred line population. Euphytica 187: 191–201. [Google Scholar]
- 64. Hwang EY, Song Q, Jia G, Specht JE, Hyten DL, et al. (2014) A genome-wide association study of seed protein and oil content in soybean. BMC Genomics 15: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao JY, Becker HC, Zhang D, Zhang Y, Ecke W (2006) Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor Appl Genet 113: 33–38. [DOI] [PubMed] [Google Scholar]
- 66. Chen G, Geng J, Rahman M, Liu X, Tu J (2010) Identification of QTL for oil content, seed yield, and flowering time in oilseed rape (Brassica napus). Euphytica 175: 161–174.. [Google Scholar]
- 67. Zheng P, Allen WB, Roesler K, Williams ME, Zhang S, et al. (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet 40: 367–372.. [DOI] [PubMed] [Google Scholar]
- 68. Hobbs DH1, Flintham JE, Hills MJ (2004) Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol 136: 3341–3349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shi A, Chen P, Zhang B, Hou A (2000) Genetic diversity and association analysis of protein and oil content in food-grade soybeans from Asia and the United States. Plant Breeding 129: 250–256.. [Google Scholar]
- 70. Zhang H, Miao HM, Wang L, Qu L, Liu H, et al. (2013) Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biol. 14: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao HM. The Sesame Genome Project and Sesame Genome Sequencing. Plant and Animal Genome XXII Conference. Plant and Animal Genome, 2014.
- 72. Holland JB, Nyquist WE, Cervantes-Martnez CT (2003) Estimating and interpreting heritability for plant breeding: An update. Plant breed reviews 22: 9–112. [Google Scholar]
- 73. Wei LB, Zhang HY, Zheng YZ, Miao HM, Zhang TZ, et al. (2009) A Genetic Linkage Map Construction for Sesame (Sesamum indicum L.). Genes & genomics 31: 199–208. [Google Scholar]
- 74. Hedrick PW (1987) Gametic disequilibrium measures: proceed with caution. Genetics 117: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weir BS (1996) Genetic data analysis 2: methods for discrete population genetic data. Sunderland: Sinauer Associates Inc Publishers. 445 p. [Google Scholar]
- 76. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620. [Google Scholar]
- 78.Acyl Lipid: pathways Available at: http://aralip.plantbiology.msu.edu
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Origins of 369 sesame accessions.
(DOCX)
Diversity statistics of 112 SSR markers in 369 sesame accessions and locations in the sesame draft genome
(DOCX)
Association mapping of the OC and PC traits using MLM method.
(DOCX)
Functions and involved pathways of the candidate genes.
(DOCX)