Abstract
Rbfox3, a neuron-specific RNA-binding protein, plays an important role in neuronal differentiation during development. An isoform Rbfox3-d31, which excludes the 93-nucleotide cassette exon within the RNA recognition motif of chicken Rbfox3, has been previously identified. However, the cellular functions of Rbfox3-d31 remain largely unknown. Here we find that Rbfox3-d31 mRNA is highly expressed during the early developmental stages of the chicken embryo, while Rbfox3-d31 protein is barely detected during the same stage due to its rapid degradation mediated by the ubiquitin-proteasome pathway. Importantly, this degradation is specific to the Rbfox3-d31 isoform and it does not occur with full-length Rbfox3. Furthermore, suppression of Rbfox3-d31 protein degradation with the proteasome inhibitor MG132 attenuates the splicing activity of another Rbfox family member Rbfox2 by altering the subcellular localization of Rbfox2. These results suggest that Rbfox3-d31 functions as a repressor for the splicing activity of the Rbfox family and its protein level is regulated in an isoform-specific manner in vivo.
Keywords: Rbfox3, Rbfox family, alternative splicing, RNA recognition motif, proteasome degradation, neural development
1. Introduction
Alternative pre-mRNA splicing is a mechanism by which multiple mRNA variants are produced from a single gene, and has a critical role in generating proteome diversity. It is a highly flexible and dynamic process that impacts almost every aspect of eukaryotic cell biology. Alternative pre-mRNA splicing is regulated by both cis-regulatory elements and RNA-binding proteins (RBPs) according to cell type, developmental stage, gender, or response to external stimuli [1].
Among many RBPs, the RNA-binding Fox (Rbfox) family has extensively been investigated as a critical splicing regulator in the development and physiology of the central nervous system [2,3,4]. Rbfox protein contains a single RNA-recognition motif (RRM) in the middle of the molecule and binds the (U)GCAUG RNA element [5,6]. Rbfox protein functions as either an enhancer or a repressor for inclusion of an alternative exon depending on binding location. The binding of Rbfox protein to the upstream intronic region of an alternative exon represses exon inclusion, while exon inclusion is enhanced by the binding of Rbfox protein to the downstream intronic region of an alternative exon [5,7].
The Rbfox family has three members in mammals, Rbfox1, Rbfox2, and Rbfox3. Rbfox2 has a broad expression profile in diverse tissues from the stem cell stage, while Rbfox1 is expressed in heart, skeletal muscle, and neural tissues [5,7,8,9,10]. Rbfox3, which has been identified as the antigen of the anti-NeuN antibody, is exclusively expressed in postmitotic neurons [11,12,13,14]. Each Rbfox protein has various mRNA variants through alternative splicing [11,15,16,17,18]. The variants are differentially expressed in tissues and show different cellular properties. All Rbfox proteins, including chicken Rbfox3 have a 93-nucleotide (nt) alternative exon within the RRM, and alternative exon exclusion is regulated in a tissue specific manner [2,16,18]. Rbfox isoforms lacking the RRM function have a dominant negative effect on the splicing activities of intact Rbfox proteins [15,16]. However, the mechanism by which RRM-truncated Rbfox isoforms inhibit the splicing activity of intact Rbfox protein is largely unknown.
Here, we characterize the chicken Rbfox3 isoform lacking the RRM, Rbfox3-d31. We show that the protein stability of Rbfox3-d31 is dynamically regulated through the ubiquitin-proteasome degradation pathway, and also reveal a novel mechanism by which Rbfox3-d31 regulates subcellular localization of Rbfox2 protein and its splicing activity.
2. Materials and methods
2.1. Cell culture, transfection, and treatment
Human embryonic kidney HEK-293 cells and human neuroblastoma SK-N-SH cells were maintained in DMEM supplemented with 10% FBS and MEMα supplemented with 10% FBS, respectively. Amaxa Nucleofector (Amaxa Biosystems) was used for transfection of the plasmid constructs. Proteasome inhibitor MG132 was purchased from Calbiochem.
2.2. Construction of expression plasmids
The expression constructs encoding myc-Rbfox3-T1-full and myc-Rbfox3-T1-d31 in the pCS3+MT plasmid were previously described as myc-Rbfox3-full and myc-Rbfox3-d31, respectively [2]. The latter two names were used here except in Fig. 1. The expression constructs for myc-Rbfox3-T2-full and myc-Rbfox3-T2-d31 were obtained by the same protocol using the primers 5′-cccggggaattcCATGACCCTCTACACACCAGCACA-3′ and 5′-cccgggtctagaCAGAAGGAAAACGGCTGCGTGTTCA-3′. For construction of GFP-tagged Rbfox3-full and Rbfox3-d31 expression plasmids, the cDNA inserts (T1-full and T1-d31) cut from the pCS3+MT plasmids by EcoRI and SacII were transferred into pEGFP-C1, which contains the GFP coding sequence. The expression construct encoding myc-Rbfox2 was previously described as F011 [15].
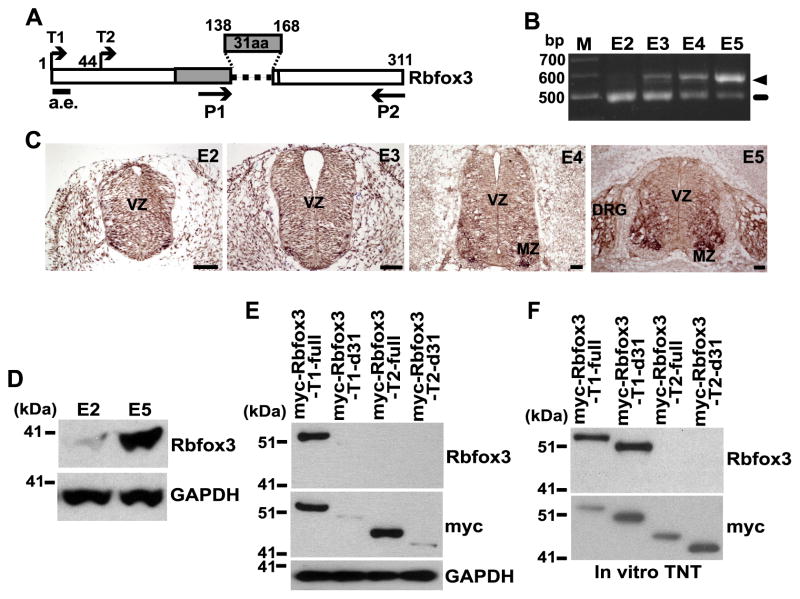
Fig. 1.
The Rbfox3-d31 protein is susceptible to degradation in cells. (A) Diagram of chicken Rbfox3 isoforms. The 31aa are encoded by the alternative exon. The N-terminal residue for the T1 and T2 constructs are shown. P1 and P2 indicate the primers for RT-PCR. a.e. indicates the epitope region for the anti-Rbfox3 antibody. Numbers represent amino acids. RNA-recognition motif (RRM) is indicated by gray bar. (B) Differential mRNA expression of Rbfox3 isoforms during chicken embryo development. RNA extracted from the embryo at the indicated embryonic day was analyzed by RT-PCR using the P1 and P2 primers indicated in panel A. Ethidium bromide stained agarose gel following electrophoresis of Rbfox3-PCR products is shown. The arrow head and line indicate the full-length Rbfox3 and Rbfox3-d31 mRNAs, respectively. M, size marker. (C) Rbfox3 mRNA expression in the developing chicken neural tube. In situ hybridization detecting Rbfox3 mRNA is shown. In situ-positive signals are shown as dark brown staining. Scale bars 50 μm. VZ, ventral zone; MZ, marginal zone; DRG, dorsal root ganglion. (D) Expression of endogenous Rbfox3 protein during development. Chicken embryo extracts harvested at the E2 and E5 were subjected to immunoblotting for the Rbfox3 and GAPDH proteins. (E) Differential protein stability of Rbfox3 isoforms in intact cells. Expression constructs indicated at the top of the blots were transfected into HEK-293 cells. Cell lysates were subjected to immunoblotting for the indicated proteins. (F) Similar protein stability of Rbfox3 isoforms in a cell-free system. Myc-Rbfox3 protein was synthesized in vitro from the indicated constructs at the top of the blots using the TNT Coupled Reticulocyte Lysate System followed by immunoblotting for the indicated proteins.
2.3. RNA preparation and RT-PCR
Total RNA was isolated from chicken embryos and cultured cells using an RNeasy mini kit (Qiagen). RT-PCR was performed using Superscript III reverse transcriptase (Invitrogen) with a random hexamer and FastStart PCR Master (Roche). The PCR primers used for analysis of the chicken Rbfox3 mRNAs surrounding the 93-nt RRM exon region were 5′-CATTTTCAATGAGCGGGGCTCCAA-3′ (P1) and 5′-CATGACCCTCTACACACCAGCACA-3′ (P2). The PCR primers used for the minigene mRNAs analysis were described previously [15].
2.4. Preparation of extracts, immunoprecipitation, and immunoblot analysis
Cell and chicken embryo extracts were prepared using a radioimmunoprecipitation assay buffer (Sigma-Aldrich) supplemented with a protease inhibitor cocktail (Roche). Myc-Rbfox3 protein was synthesized in vitro from the pCS3+MT constructs using the TNT Coupled Reticulocyte Lysate System (Promega). Polyacrylamide gel electrophoresis and immunoblotting were performed as described previously [9]. Primary antibodies used were mouse anti-Rbfox3 (Millipore), mouse anti-myc (Invitrogen), mouse anti-GAPDH (Biodesign, Saco, ME), mouse anti-Tuj1 (Covance, Emeryville, CA), and rabbit anti-Rbfox2 [9]. For immunoprecipitation of Rbfox3 protein for ubiquitination analysis, the GFP-Rbfox3-full and GFP-Rbfox3-d31 expression plasmids were transfected into HEK-293 cells for 24 h. After 2 h of 20 μM MG132 treatment, cells were subjected to immunoprecipitation with anti-Rbfox3 antibody as described previously [11]. The immunocomplexes were collected by incubation with Dynabeads Protein G (Invitrogen) and then subjected to immunoblot analysis by rabbit anti-ubiquitin antibody (Thermo Scientific). For studies on embryos (Fig. 3), embryos removed from eggs were soaked in PBS containing 1 μM MG132 for approximately 1 h during embryo cleaning to prevent Rbfox3-d31 protein degradation in situ. Then embryos were subjected to preparation of extracts or cryosection in the continuous presence of 1 μM MG132.
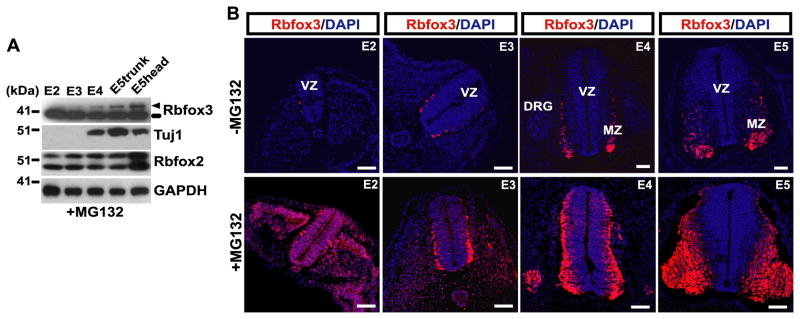
Fig. 3.
Proteasome inhibitor stabilizes the Rbfox3-d31 protein in chicken embryo. (A) Embryos at the indicated embryonic days were soaked in PBS containing 1 μM MG132 for 1 h. Then lysates were prepared in the continuous presence of 1 μM MG132 and subjected to immunoblotting for indicated proteins. The arrow head and line indicate the full-length Rbfox3 and Rbfox3-d31 proteins, respectively. (B) Transverse sections of the neural tube at the indicated embryonic days were prepared with (lower row) or without (upper row) continual treatment with 1 μM MG132, and the sections were stained with anti-Rbfox3 antibody (red) and DAPI for nuclei (blue). Scale bars, 50 μm.
2.5. In situ hybridization
The myc-Rbfox3-full in pCS3+MT plasmid was used as template to prepare an antisense RNA probe. The antisense probe was synthesized using a DIG RNA labeling kit (Roche) and the hybridized probe was detected using a BCIP/NBT Alkaline Phosphatase Substrate kit (Vector Laboratories, Burlingame, CA).
2.6. Immunofluorescence microscopy
Live-cell images of HEK-293 cells transfected with an expression plasmid of GFP-tagged Rbfox3-d31 (for Fig. 2A) were captured using an X-Cite 120 Fluorescence Illumination System equipped Nikon Eclipse TS100. Cryosections of chicken embryos were prepared as described previously [2]. SK-N-SH cells transfected with Rbfox expression plasmids were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in PBS. The sections were stained with the following primary antibodies: mouse anti-Rbfox3 (Millipore), mouse anti-myc (Invitrogen), and rabbit anti-GFP antibodies. Rhodamine Phalloidin (Life technology) staining was performed for F-actin. Alexa Fluor 488- and 594-conjugated goat antibodies against mouse IgG and rabbit IgG (Molecular Probes) were used as secondary antibodies. Nuclear DNA was stained with DAPI (Sigma-Aldrich). Images were captured using an LSM510 Meta confocal laser-scanning microscope (Carl Zeiss).
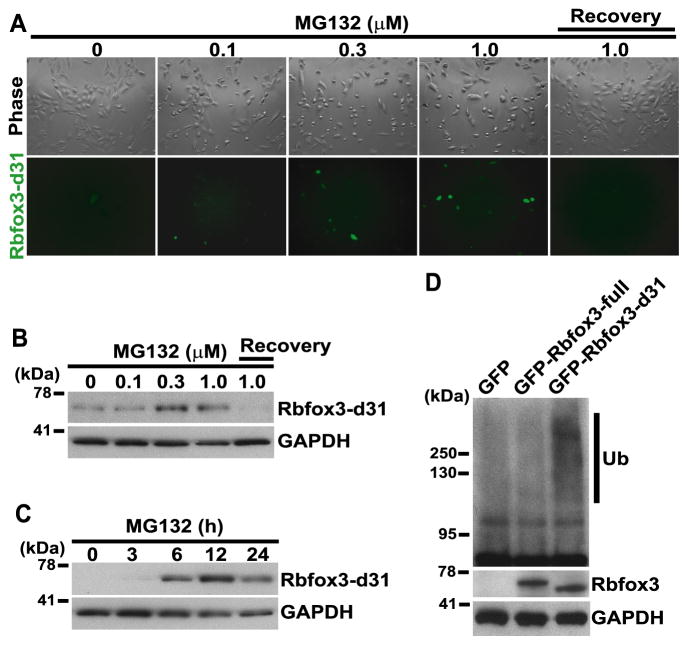
Fig. 2.
The ubiquitin-proteasome pathway is involved in degradation of Rbfox3-d31 protein. (A–C) Proteasome inhibitor treatment increases the Rbfox3-d31 protein level. Expression construct of GFP-tagged Rbfox3-d31 was transfected into HEK-293 cells. 24 h after transfection, cells were treated with the concentrations of MG132 indicated at the top of the panels for 16 h. For the recovery experiment, cells were washed and allowed to recover in complete growth medium for 8 h after the 16 h treatment with 1 μM MG132. Phase-contrast images and GFP signals from GFP-tagged Rbfox3-d31 are shown in panel A. GFP-tagged Rbfox3-d31-expressing HEK-293 cells were treated with different concentrations of MG132 for 16 h (B) or treated with 0.5 μM MG132 for the time periods (C) indicated at the top of the blots. Then cell extracts were subjected to immunoblotting for the indicated proteins. (D) Differential ubiquitination of Rbfox3 isoforms. Expression constructs indicated at the top of the blots were transfected into HEK-293 cells. 24 h after transfection, cells were treated with 20 μM MG132 for 2 h. Cell lysates were subjected to immunoprecipitation with anti-Rbfox3 antibody, and the precipitates were subjected to immunoblotting for the indicated proteins. Ub, ubiquitin.
3. Results and discussion
3.1. Rbfox3-d31 mRNA, but not its protein product, is highly expressed during the early developmental stage of the chicken embryo
We have previously identified two splice variants for the chicken Rbfox3 coding sequence, full-length Rbfox3-full and Rbfox3-d31 with a 93-nt alternative exon exclusion [2]. 31 amino acids (aa) encoded by the 93-nt alternative exon are a part of the RRM and critical for RNA binding, suggesting that Rbfox3-d31 is incapable of binding to target RNAs [16]. To characterize Rbfox3-d31, we examined the expression of Rbfox3-d31 mRNA in chicken embryos at different embryonic developmental stages. RT-PCR analysis showed that Rbfox3-d31 is highly expressed from embryonic day 2 (E2) but its expression then decreases and switches to full-length Rbfox3 expression at E5 (Fig. 1B). Since it is well known that Rbfox3 protein expression is confined to late postmitotic neurons [2,11,13], Rbfox3 mRNA expression during the early embryonic developmental stages is unexpected. To define Rbfox3 mRNA expressing cells, we examined Rbfox3 mRNA expression in chicken embryos at different embryonic developmental stages using in situ hybridization. Rbfox3 mRNA expression is detected in proliferative neural progenitor cells in the ventricular zone (VZ) and its surrounding cells at E2 (Fig. 1C). However, because full-length Rbfox3 mRNA is hardly detected at E2 by RT-PCR, the in situ hybridization signal at E2 must originate from the Rbfox3-d31 mRNA. At E4 and E5, Rbfox3 mRNA expression is confined to most postmitotic neurons in the expanded marginal zone (MZ) of the spinal cord. These results suggest that Rbfox3-d31 is a major splicing variant and its mRNA expression is not restricted to postmitotic neurons during the early embryonic developmental stages.
We next analyzed the protein expression level of Rbfox3 at E2 and E5. Immunoblot analysis using anti-Rbfox3 detected a protein band only at E5 (Fig. 1D). Rbfox-3-d31 mRNA is highly expressed at E2, while its protein product is hardly detected. This result prompted us to investigate the protein stability of Rbfox3-d31. To exclude the possibility that the discrepancy between mRNA and protein expressions of Rbfox3-d31 at E2 is caused by exclusion of the exon which encodes the epitope of anti-Rbfox3 antibody (a.e. in Fig. 1A) [2,12], two different N-terminals (T1 and T2 in Fig. 1A) were used to construct expression plasmids of myc-tagged Rbfox3 with combinations of inclusion or exclusion of the 93-nt alternative exon. These constructs were transfected into human embryonic kidney (HEK)-293 cells, and the exogenously expressed proteins were analyzed by immunoblotting with anti-myc and anti-Rbfox3 antibodies. Though the same amounts of the four constructs were transfected into the cells, immunoblot analyses were unable to detect the protein products from both the myc-Rbfox3-T1-d31 and myc-Rbfox3-T2-d31 constructs that exclude the 93-nt alternative exon, while myc-Rbfox3-T1-full and myc-Rbfox3-T2-full were detected by anti-myc antibody (Fig. 1E, middle panel). Because the epitope of the anti-Rbfox3 antibody used is absent in myc-Rbfox3-T2-full, the anti-Rbfox3 antibody detected only the protein band for myc-Rbfox3-T1-full (hereafter designated Rbfox3-full). We hypothesized that the myc-Rbfox3-T1-d31 and myc-Rbfox3-T2-d31 protein might undergo degradation in the cells. To examine protein stability in a cell-free system, in vitro synthesized proteins using in vitro transcription and translation in reticulocyte lysates were analyzed. Interestingly, myc-Rbfox3-T1-d31 (hereafter designed Rbfox3-d31) and myc-Rbfox3-T2-d31 proteins were strongly detected at the level compatible to full-length Rbfox3 (Fig. 1F). These results suggest that Rbfox3-d31 protein is highly susceptible to degradation in cells and that the discrepancy between mRNA and protein levels of Rbfox3-d31 results from its decreased protein stability in cells.
3.2. Regulation of Rbfox3-d31 protein stability via ubiquitin-proteasome degradation pathway
Two major pathways, the ubiquitin-proteasome pathway and lysosomal proteolysis, mediate protein degradation [19]. The major pathway of selective protein degradation in eukaryotic cells uses ubiquitin as a marker that targets proteins for rapid degradation [20]. To test whether this pathway regulates Rbfox3-d31 protein levels, HEK-293 cells were transfected with GFP-tagged Rbfox3-d31 expression plasmid. 24 h after transfection, cells were treated with different concentrations of the proteasome inhibitor, MG132, for 16 h. MG132 treatment increased the expression of GFP-tagged Rbfox3-d31 protein in a concentration-dependent manner (Fig. 2A). To test whether the increase of GFP-Rbfox3-d31 is reversible upon removal of MG132, cells were washed and allowed to recover in complete growth medium for 8 h. GFP-tagged Rbfox3-d31 was no longer detectable after 8 h recovery time, showing that ubiquitin-proteasome pathway-mediated degradation of Rbfox3-d31 is highly dynamic. The increase in Rbfox3-d31 protein by MG132 treatment was also confirmed by immunoblotting using anti-Rbfox3 antibody (Fig. 2B and C). In addition, we monitored the Rbfox3-d31 protein ubiquitination status in MG132-treated cells. HEK-293 cells were transfected with either GFP-tagged Rbfox3-full or GFP-tagged Rbfox3-d31 expression plasmid. 24 h after transfection, cells were treated with MG132 (20 μM) for 2 h. Total cell extracts from MG132-treated cells were immunoprecipitated with anti-Rbfox3 antibody followed by immunoblotting for ubiquitin. As shown in Fig. 2D, MG132 treatment caused a substantial increase in Rbfox3-d31 ubiquitination but not in Rbfox3-full ubiquitination. These results show that the ubiquitin-proteasome pathway is the central mechanism of Rbfox3-d31 degradation.
To examine whether Rbfox3-d31 protein levels can be increased by proteasome inhibitor treatment in chicken embryos, embryos at different embryonic developmental stages were soaked in PBS containing MG132 for 1 h to prevent Rbfox3-d31 protein degradation in situ. As shown in Fig. 3A, Rbfox3-d31 was abundantly detected during all developmental stages, although it was not detected in untreated embryos (see Fig. 1D). On the other hand, stage-dependent increases in the expression of full-length Rbfox3 and neuron-specific class III β-tubulin (Tuj1) and constitutive expression of Rbfox2 were observed similarly to the previous report where embryos were untreated with MG132 [2]. Furthermore, immunofluorescence analysis revealed that proteasome inhibitor treatment restored Rbfox3-d31 protein in Rbfox3 mRNA positive regions during the early embryonic developmental stages (Fig. 3B). Detection of Rbfox3-d31 in the VZ and its surrounding regions at E2 and E3 in MG132 treated embryos are in contrast to the absence of Rbfox3 in the control embryos. In agreement with the increased expression of Rbfox3-d31 at E4 and E5 on immunoblot analysis, total signals for Rbfox3 in MG132 treated embryos are much higher compared to the control embryos, although its expression is restricted to the MZ and the dorsal root ganglia.
3.3. Rbfox3-d31 regulates the splicing activity of Rbfox2 by altering the subcellular localization of Rbfox2
As Rbfox2 plays an important role in stem cells, embryonic development, and adult physiology [4,7,10,21] and is highly expressed during the early developmental stage of the chicken embryo (Fig. 3A, [2]) we questioned whether Rbfox3-d31 protein influences the splicing activity of Rbfox2. The alternative splicing of exon N30 in nonmuscle myosin heavy chain II-B gene was used as a model [22]. The canonical binding motif of Rbfox family proteins, the UGCAUG element, is located in the downstream intron of N30 (Fig. 4A). Exogenous Rbfox2 expression increased N30 inclusion in human neuroblastoma SK-N-SH cells (Fig. 4B, lane 2), whereas Rbfox3-d31 expression with or without MG132 treatment failed to increase N30 inclusion (Fig. 4B, lanes 3, 4 and 6). Rbfox2-induced N30 inclusion was inhibited by MG132-stabilized Rbfox3-d31 protein (Fig. 4B, lane 8), suggesting that Rbfox3-d31 protein inhibits the Rbfox2 splicing activity.
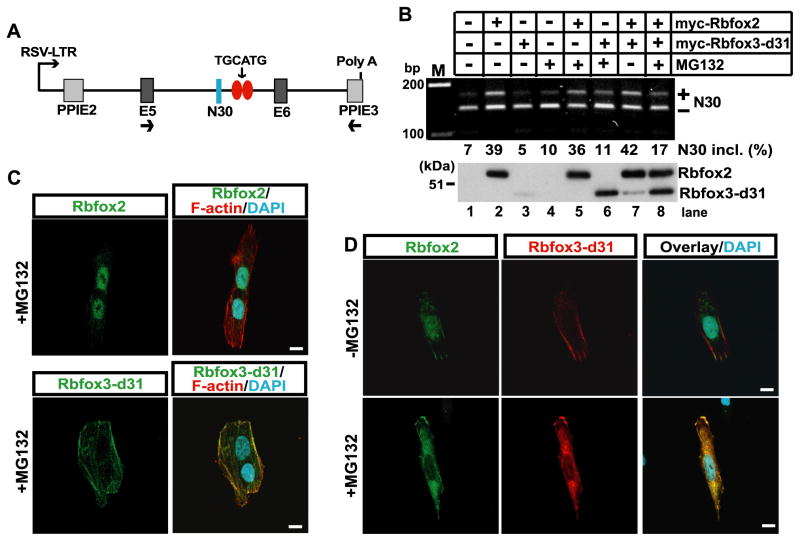
Fig. 4.
Rbfox3-d31 protein regulates the splicing activity of Rbfox2. (A) Diagram of NMHC II-B minigene. N30 is an alternative exon. E5 and E6 indicate constitutive exons from the human NMHC II-B gene. PPIE2 and PPIE3 indicate constitutive exons from the rat pre-proinsulin gene. Two copies of the canonical Rbfox binding motif TGCATG in the downstream intronic region of N30 are drawn as red ovals. Transcription of the minigene is driven by the Rous sarcoma virus long terminal repeat RSV-LTR. Arrows under E5 and PPIE3 indicate the locations of the primers used for RT-PCR. (B) Rbfox3-d31 protein decreases the splicing activity of Rbfox2. Minigene and the expression constructs for the indicated Rbfox2 and Rbfox3-d31 were cotransfected into SK-N-SH cells. After incubation with or without 1 μM MG132 for 12 h, cells were subject to RT-PCR analysis. The plus and minus symbols indicate inclusion and exclusion of N30, respectively (middle row). The myc-tagged Rbfox2 and myc-tagged Rbfox3-d31 expression levels were verified by immunoblotting using anti-myc antibody (bottom row). (C) Rbfox3-d31 protein shows cytoplasmic localization. SK-N-SH cells were transfected with either GFP-tagged Rbfox2 or myc-tagged Rbfox3-d31 expression constructs. After incubation with or without 1 μM MG132 for 12 h, cells were subjected to immunofluorescence analysis using anti-myc antibody for Rbfox3-d31(green), anti-GFP for Rbfox2 (green), F-actin (Rhodamine Phalloidin; red), and DAPI for nuclei (cyan). (D) Rbfox3-d31 protein alters subcellular localization of Rbfox2 protein. The expression constructs for GFP-tagged Rbfox2 and myc-tagged Rbfox3-d31expression constructs were cotransfected into SK-N-SH cells. After incubation with or without f 1 μM MG132 for 12 h, cells were subjected to immunofluorescence analysis using anti-Rbfox3 antibody (red), anti-GFP antibody (green), and DAPI for nuclei (cyan). Scale bars in C and D, 10 μm.
Next, we tried to establish the mechanism by which Rbfox3-d31 can inhibit the splicing activity of Rbfox2. Because Rbfox family proteins function in the nucleus as a splicing regulator, we investigated their subcellular localization to see whether localization was affected by Rbfox3-d31 expression. Either GFP-tagged Rbfox2 or myc-tagged Rbfox3-d31 expression plasmid was transfected into SK-N-SH cells. 24 h after transfection, cells were treated with MG132 for 16 h. Immunofluorescence analysis confirmed strong nuclear localization of Rbfox2, whereas Rbfox3-d31 localized to the cytoplasm (Fig. 4C). We next examined whether Rbfox3-d31 protein affected the subcellular localization of Rbfox2 protein. SK-N-SH cells were co-transfected with both GFP-tagged Rbfox2 and myc-tagged Rbfox3-d31 expression plasmids. Nuclear localization of Rbfox2 protein was not changed by Rbfox3-d31 co-expression without MG132 treatment (Fig. 4D, top panels). Strikingly, MG132-stabilized Rbfox3-d31 protein inhibited the nuclear localization of Rbfox2 (Fig. 4D, bottom panels). These results suggest that Rbfox3-d31 regulates the splicing activity of Rbfox2 by altering the subcellular localization of Rbfox2.
In this report, we have shown that the ubiquitin-proteasome pathway dynamically regulates the protein stability of chicken Rbfox3-d31 at early developmental stages of the chicken embryo. The protein degradation by the ubiquitin-proteasome pathway is essential in diverse aspects of cell physiology and development, and is controlled by multiple mechanisms [19,20]. Although Rbfox3-d31 protein is barely detected without MG132-mediated inhibition of protein degradation in early chicken embryos, the protein degradation of Rbfox3-d31 might be attenuated under certain in vivo conditions. To date, several studies have shown that Rbfox family protein isoforms lacking the RRM function as inhibitors for the splicing activities of full-length proteins in a dominant-negative fashion [15,16]. Our study reveals the cellular mechanism by which Rbfox3-d31 protein influences function of Rbfox family protein. The finding that Rbfox3-d31 protein controls the subcellular localization of its family Rbfox2 protein helps to understand the mechanism of inhibitory activity of Rbfox protein lacking the RRM. Our observations also raise the possibility that Rbfox3-d31 protein might directly or indirectly interact with Rbfox2 in the cytoplasm to inhibit nuclear transport of Rbfox2. Although the detailed molecular mechanism by which Rbfox3-d31 protein alters subcellular localization of Rbfox2 protein remains to be elucidated, Rbfox2 splicing activity regulated by Rbfox3-d31 could be critical in achieving a fine balance of splice isoforms of target genes that allows for proper embryonic development.
Highlights.
Protein stability of Rbfox3 splice isoforms is differentially regulated.
Rbfox3-d31, an Rbfox3 isoform lacking the RRM, is highly susceptible to degradation.
The protein stability of Rbfox3-d31 is regulated by the ubiquitin-proteasome pathway.
Rbfox3-d31 inhibits the nuclear localization of Rbfox2.
Rbfox3-d31 inhibits the splicing activity of Rbfox2.
Acknowledgments
We thank Yoh-suke Mukouyama for helpful discussions and Christian A. Combs (Light Microscope Core Facility, NHLBI) for professional advice on confocal microscopy. We also thank Antoine F. Smith for technical assistance and Jacqueline Parilla for critical reading of the manuscript.
This work was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim KK, Nam J, Mukouyama YS, Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. 2013;200:443–458. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares M, Jr, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gehman LT, Meera P, Stoilov P, Shiue L, O’Brien JE, Meisler MH, Ares M, Jr, Otis TS, Black DL. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKee AE, Minet E, Stern C, Riahi S, Stiles CD, Silver PA. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 2011;39:3064–3078. doi: 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, Conboy JG. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol. 2011;359:251–261. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dredge BK, Jensen KB. NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS One. 2011;6:e21585. doi: 10.1371/journal.pone.0021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- 14.Maxeiner S, Glassmann A, Kao HT, Schilling K. The molecular basis of the specificity and cross-reactivity of the NeuN epitope of the neuron-specific splicing regulator, Rbfox3. Histochem Cell Biol. 2014;141:43–55. doi: 10.1007/s00418-013-1159-9. [DOI] [PubMed] [Google Scholar]
- 15.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 20.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 21.Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, Abou Elela S, Roux P, Lemaitre JM, Tazi J. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013;4:2480. doi: 10.1038/ncomms3480. [DOI] [PubMed] [Google Scholar]
- 22.Kawamoto S. Neuron-specific alternative splicing of nonmuscle myosin II heavy chain-B pre-mRNA requires a cis-acting intron sequence. J Biol Chem. 1996;271:17613–17616. [PubMed] [Google Scholar]