Abstract
Just as the Petri dish has been invaluable to the evolution of biomedical science in the last 100 years, microfluidic cell assay platforms have the potential to change significantly the way modern biology and clinical science are performed. However, an evolutionary process of creating an efficient microfluidic array for many different bioassays is necessary. Specifically for a complete view of a cell response it is essential to incorporate cytotoxic, protein and gene analysis on a single system. Here we present a novel cellular and molecular analysis platform, which allows access to gene expression, protein immunoassay, and cytotoxicity information in parallel. It is realized by an integrated microfluidic array plate (iMAP). The iMAP enables sample processing of cells, perfusion based cell culture, effective perturbation of biologic molecules or drugs, and simultaneous, real-time optical analysis for different bioassays. The key features of the iMAP design are the interface of on-board gravity driven flow, the open access input fluid exchange and the highly efficient sedimentation based cell capture mechanism (~100% capture rates). The operation of the device is straightforward (tube and pump free) and capable of handling dilute samples (5-cells per experiment), low reagent volumes (50 nL per reaction), and performing single cell protein and gene expression measurements. We believe that the unique low cell number and triple analysis capabilities of the iMAP platform can enable novel dynamic studies of scarce cells.
Introduction
Harnessing the inherent characteristics of operating at the micrometre scale has provided a variety of novel assays previously not possible using conventional macroscale techniques. Microscale operations hold the promise of integrating complete protocols onto a single chip (i.e., lab-on-a-chip)1–4 and the potential of significantly enhancing assay reproducibility and quantitation5 relative to macroscale techniques.
Microscale techniques for cell-based analysis have been developed for a myriad of applications ranging from single cell analyses and cytometry-like techniques,6 to gradient generating devices for cell treatment,7 in vivo like 3-dimensional culture,8 to miniaturizations of traditional perfusion9 and static10 cell cultures. However most of the microscale cell culture research has focused on high throughput assays.11–15
An important impediment to the adoption of microscale cell culture systems is the relative lack of methods for quantitative analysis of more complex cellular functions such as protein expressions or genetic transcriptions. While a strength of microfluidic devices is the small sample size, this is also a limiting factor when it comes to using many of the traditional off-chip bio-analytical assays such as flow cytometry and polymerase chain reaction. Without the ability to probe more complex aspects of cellular responses beyond cell viability or proliferation assays on a microfluidic chip, the adoption of these microscale assays to cell biology is very limited.
In an attempt to resolve these issues, many of the “stand alone” microfluidic components for performing bio-analytical functions such as cell lysis,10,17,18 immunoassays,19,20 capillary electrophoresis (CE)21,22 and polymerase chain reaction23,24 will have to be directly integrated with microscale cell culturing and assaying. Recent progress in that regard has been shown by Lindstrom et al.25 where approximately 5 stem cells per well (500 nL) were loaded in a microfluidic array. The cells were cultured on the chip for several days before subjecting them to lysis and PCR analysis within the same chip. The authors demonstrated that they could detect single-base mutations in the p53 gene. Also in the same direction Paguirigan and Beebe26 have demonstrated an antibody based In Cell Western (ICW) technique that can accurately quantify protein expression changes in microfluidic cell cultures without removing the cells from the microscale culture chamber. Even though these developments have been important advancements, for a complete view of a cell response it is essential to incorporate protein and gene readout on a single system and be able to combine these with cytotoxic and stimulation analysis.
In addition, current miniaturized cell-based assay platforms struggle with low cell capture efficiencies.16 Improvement of cell capture methods will open new ways of research on primary cells where low passage numbers need to be maintained to avoid genetic variation.27,28 In particular primary endothelial cells have a limited lifespan and display characteristics that differ from batch to batch.29 Circulating tumor cells (CTCs) are another example of low occurrence (generally less than 50 cells mL−1) heterogeneous5 cells that require highly efficient cell micro-culture and analysis systems. Development of a highly efficient cell capture devices will therefore improve the culture and analysis of rare clinical isolates.
Here we present a novel integrated microfluidic array plate (iMAP) for low cell number, dynamic studies with several readout capabilities such as real-time Nucleic Acid Sequence-Based Amplification (NASBA) and immunofluorescent (IF) protein expression detection. The microfluidic system is capable of performing 512 dynamically configurable integrated cell based assays coupled with real-time NASBA and IF analysis. It has been designed for simple, nearly 100% efficient and shear stress free microfluidic loading of cells and reagents that cannot be done with existing laboratory tools. The device has an on-board gravity driven flow control system making it independent of tubing, cumbersome syringe pumps or other active macro-scale flow control components such as pressure sources and solenoid valves. The device can therefore be self-contained, mobile and directly used in standard laboratory cell culture incubation chambers and microscope systems.
Results and discussion
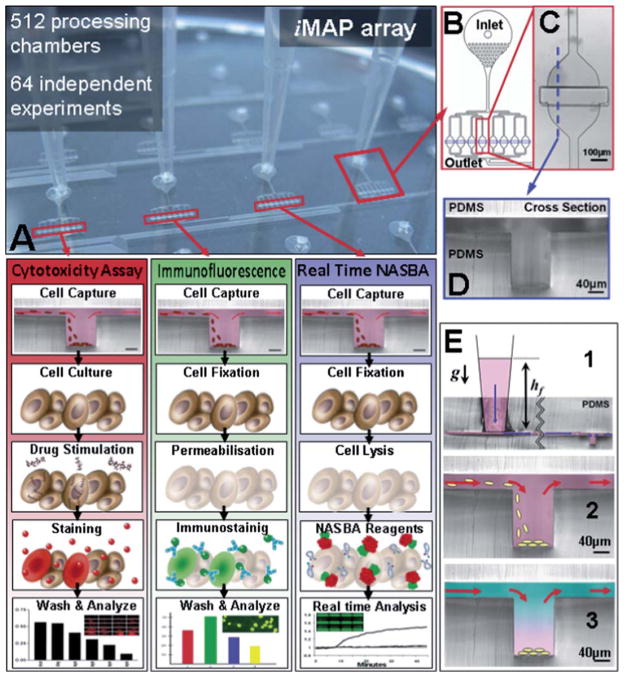
The iMAP (Fig. 1) is a novel microfluidic plate design that allows for simple cell loading, cell capture, reagent loading, reagent mixing and parallel processing operations. These unit laboratory operations can be performed in any order and repetition to achieve more complex experimental procedures. Fig. 1A demonstrates how these unit operations can be combined to perform cytotoxicity, immunofluorescence and gene-expression assays on a single chip. Assays are performed in the basic processing chamber structure (Fig. 1C) which consists of a chamber with a micro-trench (Fig. 1D). The micro-trench allows the capture of cells and particles through gravitational sedimentation. In addition, a controlled flow over the top of the trench allows successive, diffusion-driven loading, mixing and replacement of reagent while retaining the cells free of hydrodynamic shear stress within the trench. The required flow control is achieved by setting the height of a liquid in an attached vessel, e.g. a pipette tip (Fig. 1E), which generates a hydrostatic pressure head at the inlet. The column is open on the top, which allows for loading of cells and the replacement of the fluid being loaded into the device. The operation paradigm of this device is different to the more established “connection of specialized blocks” paradigm used in electronic integrated circuits and high density fluidic circuits (e.g. as in fluidic devices shown by Quake and co-workers30). Instead, the device operates by using a single multipurpose structure that can retain cells or particles and delay the diffusion of molecules of interest in the same space where different reactions are sequentially performed. This approach has several advantages. First, higher assay sensitivities are achieved because the molecules of interest do not have to be transported. Hence, they contact less surface area, thereby reducing the (unspecific) loss of target “molecules”. Second, the purpose or function of the structure is completely determined by the fluid flow into it and its previous contents. Thus, by selecting the sequence of input fluids, the same multipurpose structure can be ‘programmed’ to perform a certain repertoire of complex assays. Thirdly, the small footprint of the multipurpose trench structure allows it to be easily multiplexed into an array that can perform a large number of simultaneous integrated assays.
Fig. 1. iMAP array.
(A) consists of 64 processing modules (B) that can perform 64 independent simultaneous integrated assays. The integrated function of each processing module can be flexibly selected by the user and depends entirely on the sequence and timing of the fluid inputs. The basic steps required for three different cell based assays are illustrated (A). This versatility allows the user to combine cell stimulation and gene and protein expression analysis on a single microfluidic platform. For statistical replication purposes each module consists of 8 parallel and equally distributed (C) processing chambers that contain a central trench structure (D). Each processing chamber can execute any sequence laboratory unit operations (E), gravity driven flow (E1), cell capture (E2) and reagent loading and mixing (E3).
Micro-trench fluidic simulation
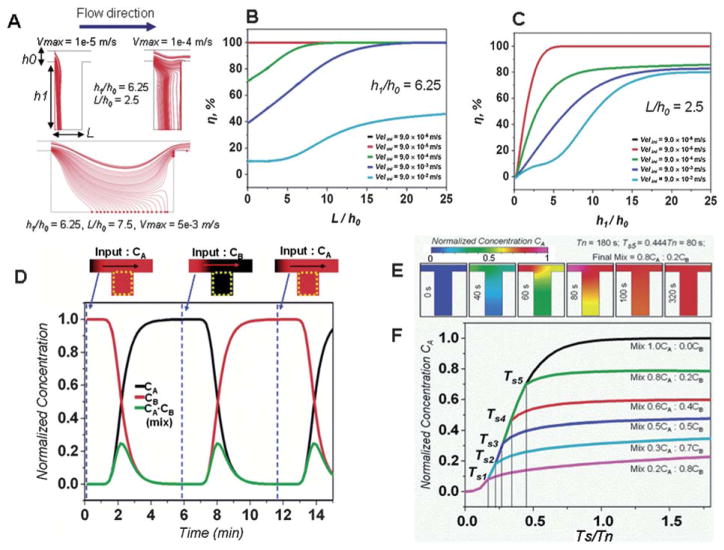
Computational fluid dynamics were used to characterize two basic laboratory unit operations; (1) Cell capture and (2) Fluid loading and diffusive mixing (Fig. 2). These simulations help to predict the effect of the main design parameters impacting the performance of these basic laboratory unit operations. Gravitational sedimentation allows the loading and retention of cells or particles in the processing chamber which are more dense than the medium. The particle trajectories were calculated by solving the ballistic equation of motion for each particle. A spherical particle with a mass m and a radius rp immersed in a flowing liquid (density ρ, dynamic viscosity η) is subjected to gravitational forces, the hydrodynamic Stokes drag as well as the related buoyancy. The Khan and Richardson force
Fig. 2.
Effect of the main design parameters on the performance of the basic laboratory unit operations using computational fluid dynamics simulation. (A) Particle trajectories for different inlet velocities and trench geometries. (B) Trench length and inlet velocity effect on the particle capture efficiency η. (C) Trench depth and inlet velocity effect on the particle capture efficiency η. Please note that the results from the 9 × 10−6 m s−1 velocity (black line) and the 9 × 10−6 m s−1 (red line) are identical. (D) Sequential loading (inlet velocity 10−4 m s−1) of species CA (D ≈ 10 −10 m2 s−1) into a trench structure h1/h0 = 6.25, L/h0 = 2.5 with species CB. (E) Time lapse of the CA concentration within the trench during the creation of a 0.8CA : 0.2CB mixture, flow is stopped at Ts = 80 s or at Tn = 0.444. Within 40 s the CA concentration (D ≈ 10−10 m2 s−1) homogenizes and stabilizes within the trench. (F) Creation of several mixture ratios of the initial species CB and the new added species CA in the trench structure, the mixture ratio is controlled by the flow stopping time TS which is less than the refreshment time Tn.
| (1) |
is an empirical estimate of the fluid drag force that is valid for a wide range of Reynolds numbers, (including Re < 1). The fluid velocity is represented by vector u, while up is the particle velocity.
Since the Khan and Richardson force is directly dependent on the fluid velocity field, the incompressible Navier–Stokes equations
| (2) |
were used to predict the fluid velocity field within the processing chamber. The channel lengths (70 mm) within the device are significantly greater than the channel height (40 μm), thus the system can be successfully modeled using a 2D simulation.
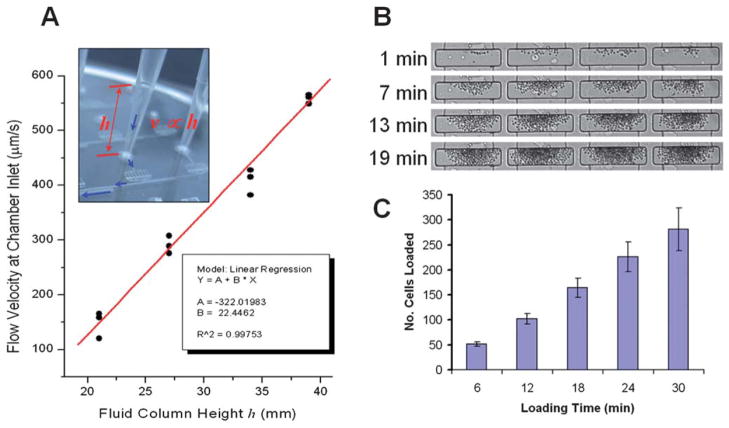
The particle capture efficiency within the micro-trench was evaluated for different inlet velocities, trench geometries and particles sizes. The calculations shown in Fig. 2A–C demonstrate particle capture efficiencies close to 100% which can be achieved as long as fluid velocity at the inlet is kept below 100 μm s−1 and the ratio of the channel to trench depth (h1/h0) is above 5. As shown by Manbachi et al. 2008,31 once the particles reach the bottom of the processing chamber, i.e., the trench, they are shielded from shear stress. This is due to fluid velocities of at least three orders of magnitude smaller at the bottom of a micro-trench than on the top for channel to trench depth (h1/h0) ratios exceeding 5. This also means that once cells or particles reach the bottom of the trench they are effectively captured. This was confirmed in experiments with 1.5 μm silica beads and input flow velocities of 20 μm s−1, showing that nearly 100% of the beads were captured (Fig. S4†, ESI2). Experiments with adherent cells (HeLa, MCF7) and non-adherent cells (plasma cells U266, macrophage J774) also demonstrated very high capture and retention efficiencies (Movie S8, ESI†). The total number of cells retained in each processing chamber can be controlled by the loading time (Fig. 3B and C) and the concentration of cells in suspension. In addition, we were also able to capture and retain bacterial cells (E. coli) at the bottom of the micro-trench, however; the capture efficiency was lower due to the lower density of bacterial cells (Fig. S2† and Movies S3 and S4).
Fig. 3.
Laboratory unit operations: (A) gravity driven fluid control that depends on the height of the fluid column. The height of the fluid column can be regulated by controlling the amount of fluid inserted into the tip. (B and C) Cell capture is accomplished by allowing cells in the flow to sediment to the bottom of the central trench structure. Flow velocities at the bottom of the trench structure are 3 orders of magnitude lower than in the rest of the device allowing the cells to be effectively retained. Due to the high capture efficiency the number of loaded cells in the trenches can be controlled with the cell loading time.
Species loading and mixing within the processing chamber and the trench geometries was predicted by calculating the transient convection and diffusion mass transport model
| (3) |
for a transient incompressible Navier–Stokes velocity field (eqn (2)).
The calculations (Fig. 2D–E) demonstrate that the average concentration of a species within the trench can be controlled by the liquid flow over the trench. The removal of the initial species by a new species is simply implemented by flowing the new species over the trench, and allowing for the new species to diffuse into the trench while the initial one diffuses out of the trench (see Movie S7†). This can be repeated in a cyclic manner to produce a sequential exposure of the cells in the trench to different reagents (Fig. 2D). Intermediate mixtures of species can be achieved by stopping the flow at a time Ts < Tn where Tn is the time to complete species replacement within the micro-trench as shown in Fig. 2E and F (also see CFD ESI†). Once the flow has been stopped, the two species continue to mix through diffusion and the concentration within the trench is homogenized (Fig. 2E and F). Species form within the trench can diffuse out and into the surrounding channels but the 40 μm height (40 × 50 μm cross-sectional area) of the connecting channels significantly delays that process. This way, the average concentration within the trench can be kept constant within a ± 10% variation for over 150 min (Fig. S10, ESI†) which is sufficient for most enzymatic reactions such as PCR or NASBA.32 This is a similar strategy to the one used by Du et al. 200933 to maintain stable gradients within microfluidic channels for several hours. Experimental results with a fluorescent dye confirmed that when a new solution is flowed into the processing chamber it gradually mixes with and finally replaces the previous contents of the chamber (Fig. S8A–B, ESI†). Furthermore, the time to complete species replacement within the trench Tn is dependent on the flow velocity over the trench.
Gravity driven flow
Gravity driven fluid control is accomplished by inserting a standard pipette tip into a primed device inlet and then loading it with the input fluid (Fig. 3A). The flow velocity is simply regulated by the height of the fluid column, i.e. the pressure head. By using standard 200 μL pipette tips, velocities exceeding 600 μm s−1 can be achieved at the inlet of the processing chamber. The hydrostatic pressure head of small inlet volumes can be increased by stacking a second (immiscible and inert) liquid such as mineral oil on top. This allows volumes as low as 50 nL to be readily loaded and driven into each processing chamber (Fig. S3, ESI†). This method makes the device ideal for applications where sample or reagent volumes are very scarce or expensive.
Full experimental protocols
Laboratory unit operations can be dynamically and seamlessly combined into sequences that perform more complex, integrated assays. We can perform a wide range (Fig. 1) of cell-based assays in parallel on the same platform. Fig. 1 illustrates the combination of individual operation units required for a cytotoxicity assay, real-time gene expression analysis (NASBA) and immunofluorescent protein analysis. Priming of the chip, loading with the cell suspension and cell capture are common unit operations for all three assays. The user has now the option to grow the cells within the chip by continuously perfusing the cells with fresh medium or to perform molecular analysis directly on the captured cells. Cells can be stimulated with drugs and fixed, permeabilized or lysed for the consecutive analysis. In this study we implemented on-chip a tmRNA based E. coli diagnostics assay, a Paclitaxel cytotoxicity assay, a gene expression analysis on mRNA level of the estrogen receptor α (ESR1 which is associated with breast cancer) and the protein expression analysis of the ESR1 nuclear receptors. In order to further demonstrate the versatility of our chip, we performed a drug treatment study for the down regulation of the ESR1 nuclear protein.
Micro-trench cell culture
The initial steps for on-chip experiments are often shared by a number of different cell-based assays (Fig. S7†). The priming of the chip and cell capture is usually followed by a sequence of unit operations such as cell culture, fluorescent staining and lysis (Fig. S8D–E†). On chip cell culture showed quick adoption of cells to the new microenvironment and exponential cell growth until reaching of confluence in three days. Cells were cultured under continuous perfusion with a constant feed of nutrients with fresh medium and constant removal of cell growth byproducts with the outgoing feed. Moreover, cells captured in the trench structure are protected from the shear stress of the flowing medium due to minimal velocities in the bottom of the trench. Trapped cells were exposed to Calcein AM staining and Propidium iodide (PI) showing cell viability after five days of cell culture (Fig. S9D, ESI†). Finally, fluorescently stained cells were lysed using a commercial buffer compatible with nucleic acid amplification for 30 min and the efficiency of the cell lysis was monitored in real-time by fluorescent microscopy. Fig. S9E† demonstrates the process of cell lysis which initiates within five minutes and it is almost complete within less than ten minutes from the loading of the lysis reagent.
On-chip real-time NASBA
Nucleic acid analysis based amplification of RNA and real-time detection can be readily performed within the device. Real-time NASBA is used for the nucleic acid detection on-chip. NASBA is an enzymatic transcription-based RNA amplification system developed by Compton in 1991.32 NASBA is an isothermal (41 °C) process that can exponentially produce more than 109 RNA copies in 90 min. Compared to other in vitro amplification methods such polymerase chain reaction (PCR),34 strand-displacement amplification (SDA)35 or rolling-circle amplification (RCA),36 NASBA has the unique characteristic that it can, in a single step, amplify specific RNA sequences. Furthermore, NASBA has the distinctive ability to specifically amplify RNA in a background of DNA of comparable sequence,37 which alleviates the requirements on sample purification. Also, since NASBA operates at 41 °C, i.e. a much lower temperature than the 95 °C required in PCR cycling, problems with on-chip sample drying or thermal displacement are reduced. This eliminates the need for valves or other active components to encapsulate the contents of the reaction chamber and allows the microfluidic channels to limit the diffusion of molecules out of the reaction. It also reduces the thermal design requirements of the chip and makes the chip operation simpler.
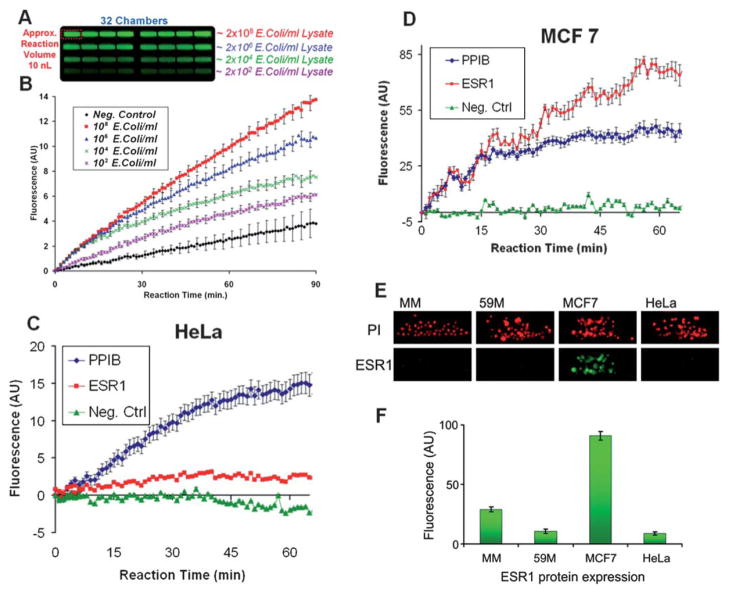
We demonstrate NASBA (Fig. 4, SM6) by performing an on-chip E. coli diagnostic assay. Based on custom designed Ribo-SEQ targeting primers38 and molecular beacons we were able to detect down to 10 E. coli bacteria in 100 μL within 7 minutes (Fig. 4B), giving a sensitivity of approximately a single target copy. Note that each E. coli contains approximately 500–1000 copies39 of the tmRNA RiboSEQ target fragments resulting in approximately a single target copy within a 10 nL volume. tmRNA also has a relatively high stability compared to mRNA which also increases the detection robustness. This sensitivity is also reinforced by the increased depth of the trench geometry within the processing chamber compared to other conventional microfluidic chambers. The trench insures that more fluorescent signal is collected in a single exposure. This is also evident (Fig. S9A–B, ESI†) where at constant fluorophore concentration and excitation, the intensity of the fluorescent signal collected from the trench area is 8 fold higher than one from the inlet channel. The negative control was implemented by amplifying a deionized water sample. Evidently there is also a gradual increase in the negative control signal; we believe that could be due to non-specific amplification or E. coli tmRNA contamination that might be present in the commercial recombinant enzyme reagents.
Fig. 4.
Gene and protein expression analysis on the iMAP array. (A) End-point fluorescence within the processing chambers after the (B) real-time NASBA E. coli diagnostic assay. This demonstrates that rapid and sensitive NASBA can be performed within the iMAP array chip. (C and F) Oncogene: Estrogen Receptor α (ESR1) integrated single-chip cell based assays. Multiplexed real-time NASBA assays detect the expression of the ESR1 gene in comparison to the house keeping gene PPIB in (C) HeLa and (D) MCF7 cells. As expected the ESR1 gene is highly expressed in the MCF7 cells. (E) This was further verified with the ESR1 nuclear receptor protein detection immunofluorescent assay. U266 (MM), 59M, MCF7 and HeLa cells on-chip, fixed, permeabilised and nuclear stained using Propidium iodide (PI) and anti-ESR1 (green) antibody. (F) MCF7 cells show specific nuclear staining for ESR1. All standard error bars are based on the eight replicates (n = 8) in each processing module.
Next, we performed NASBA on eukaryotic cells captured in the iMAP array. Since estrogen receptor alpha (ESR1) is a major oncogene in breast cancer,40 we used ESR1 positive MCF7 breast cancer cells to demonstrate multiplexed, real-time NASBA-based gene expression analysis. Furthermore, this analysis was complemented with an ESR1 protein detection based on immunofluorescent staining (Fig. S5†). Using primers and molecular beacons specific to the ESR1 gene and to a house keeping gene encoding for the peptidylprolyl isomerase B (PPIB),41 the device performed a multiplexed real-time NASBA protocol (Fig. 4C and D) from as few as 10 cells. The ESR1 signals are more noisy compared to the E. coli reaction. This was due to the fact that we increased the camera gain in order to compensate for a weaker fluorescent signal strength. This increased the signal noise level.
On-chip immunofluorescent analysis
We performed an immunostaining assay on-chip to detect the nuclear expression of ESR1 protein. Since a high level of ESR1 gene expression was shown in MCF7 cells and ESR1 was not expressed in HeLa cells in NASBA experiments, we used the same cells for ESR1 immunostaining. ESR1 protein is known to be localized in the nucleus of MCF7 cells4 and absent in HeLa cells.42 Additionally, we used other cell lines, the ovarian cancer cell line 59M and non-adherent plasma cells U-266 (MM) to investigate the protein expression. Cells were captured, fixed, permeabilized and immunostained on the chip (Fig. 4E and F). Immunofluorescent staining with the anti-ESR1 antibody showed an approximately ten fold increase in signal from the, ESR-1 positive, MCF7 cells over the negative control cells HeLa, 59M and U-266. This demonstrates the specificity of the stain. Please note that as in conventional assays to achieve successful protein detection, the antibodies used should bind to accessible target epitopes. Thus the antibodies have to be compatible with the specific fixation and permeabilization protocol.
Drug dosage analysis
The previously mentioned procedures can be further combined into a complete drug study experimental protocol. We demonstrated this (Fig. 5) with a cytotoxicity analysis of the anti-cancer drug paclitaxel43 on HeLa cells and a study of the effects of the As2O3 drug on ESR1 expression in breast cancer cells MCF7.
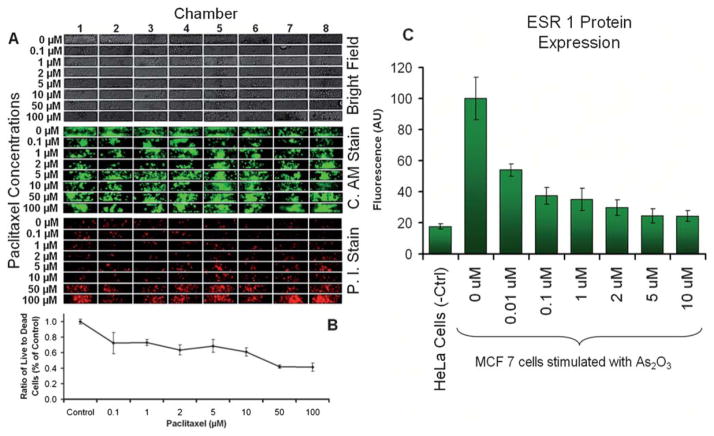
Fig. 5.
Complete drug study experimental protocol execution on the iMAP array. (A) HeLa based cytotoxicity assay validating the effect of paclitaxel, a well know anti-cancer agent that (B) reduces the growth and proliferation of cancer cells. (C) Study of the down regulation effects of the As2O3 drug on ESR1 in breast cancer cells MCF7. In a fully integrated on-chip assay the effects of arsenic trioxide on the protein expression of ESR1 in MCF7 cells were studied by loading 128 processing chambers with MCF7 and HeLa cells and stimulating the cells with several concentrations of As2O3 over a period of 72 h, after which the protein expression was analyzed on chip with immunofluorescent nuclear staining and detection. All standard error bars are based on the eight replicates (n = 8) in each processing module.
Paclitaxel is a well know anti-cancer agent that reduces the growth and proliferation of cancer cells. To perform the dynamic cytotoxicity experiments we loaded 64 processing chambers with approximately 50 HeLa cells and cultured them for one day after which we stimulated the cells with eight different concentrations of Paclitaxel raging from 0 μM (control) up to 100 μM for 48 hours. Cell viability was quantified through Propidium iodide and Calcine AM fluorescent staining. As expected, the fluorescent imaging revealed (Fig. 5A and B) that the ratio between dead and alive cells increases with the drug concentration.
The effects of As2O3 on ESR1 expression were studied by loading 128 processing chambers with ESR1 positive MCF7 cells and as a negative control we used ESR1 negative HeLa cells. The MCF7 cells were stimulated for 48 hours with seven different concentrations of As2O3 and the ESR1 protein quantified with nuclear immunofluorescent staining. As in previous reports44 the results show (Fig. 5C) that As2O3 has a strong effect of an approximately three-fold down-regulation of the expression of ESR1 in MCF7 cells.
A limitation of the current setup is the available space for cell growth, thus confluence is reached quicker than in conventional dishes. In addition the signals obtained are from fewer cells (5 to 400) which in the case of a heterogeneous response might increase the result variability.
Discussion
The iMAP array is compatible with standard existing laboratory infrastructure. The inlet distribution is the same as that of the standard 96 well plate, so cells and reagents can be loaded using multichannel pipettes and/or pipetting robots (Fig. S6, ESI†). Additionally in contrast to standard microfluidic devices that use syringe pumps, reagent replacement or refilling is done in a simple and bubble free manner, cells are loaded efficiently and with very low shear stress into the device. The micro-trench structure captures and isolates cells from the direct flow which also enables the capturing and processing of bacterial cells (Fig. S2† and Movies S3, S4). Furthermore, the iMAP array integrates flow control and gravity driven pumping which makes it independent of tubes or external pumps or other macroscale active components and allows it to be easily inserted and used within incubation or environment control chambers and standard fluorescent microscope systems or even some plate readers. In essence the iMAP array is a nanolitre processing multi-chamber plate that can perform dynamic flow assays with high sensitivity, high level of integration (experiment to answer), and spatial multiplexing without the need for cumbersome active control components such a pumps, pressure sources, solenoid valves, etc. The fabrication process is a simple, 2-layer soft photolithography that does not require precise alignment of multiple layers since only the 1.3 mm inlets have to be aligned to the fluidics layer. All the processing modules are fluidically independent so in applications that do not require the use of the complete 512 processing chambers, the user can load a subset of modules without having to seal the remaining inlets. This simplifies the iMAP array operation protocols.
Conclusion
In conclusion the iMAP combines parallel microscale cell culture with quantitative gene expression, protein immunoassays, and cytotoxicity readout without requiring any off chip steps providing a truly experiment to answer capability. The gravity-driven perfusion cell culture is an effective mechanism for on-chip cell perturbation with biological molecules or drugs and simultaneous, real-time optical analysis. The small sample size (5 cells per experiment), the highly efficient sedimentation based cell capture mechanism (~100% capture rates) and the single cell protein and gene expression sensitivities make the iMAP array an indispensable tool for studying rare clinical isolates. The open access input fluid exchange and the tube and pump free operation of the device enable its direct incorporation with existing laboratory infrastructure making it a versatile fully integrated microfluidic array platform for dynamic cell studies which can be universally and economically used in any biological laboratory.
Materials and methods
Fabrication
The microfluidic device was fabricated using standard soft lithography replica molding techniques.45 A mould was created through a double-layer process (Fig. SM1, ESI†) first using negative photoresist, SU8-3050 (Microchem USA) and then SU8-2150 (Microchem USA) both layers were deposited onto a clean silicon wafer using a spin coater (P6700 Specialty Coating Systems, Inc., USA). The first layer photoresist (5 mL) was spread onto the wafer at 500 RPM for 10 s, and the rotation rate was then ramped up at an acceleration of 350 RPM s−1 to 4000 RPM, at which rate the sample was spun for 30 s to form a 40 μm thin layer. The wafer was then soft baked at 95 °C for 15 min and UV-exposed for 30 s at 9.5 mW cm−2 using a Karl-Süss KSM MJB-55W mask aligner. The wafer was post-exposure baked for 1 min at 65 °C and 62 min at 95 °C, allowed to cool to room temperature, developed in Microposit EC Solvent (Chestech Ltd., UK) developer and sonicated (Branson 5510, Kell-Storm, USA) for 4 min, and blown dry with nitrogen. The second layer photoresist (5 mL) SU8-3025 (Microchem USA) was spread onto the patterned wafer at 500 RPM for 10 s, and the rotation rate was then ramped up at an acceleration of 350 RPM s−1 to 4000 RPM, at which rate the sample was spun for 3 s. The wafer was then soft baked at 95 °C for 10 min. Then another layer of photoresist (5 mL) SU8-2150 (Microchem USA) was spread onto the wafer at 500 RPM for 5 s, and the rotation rate was then ramped at an acceleration of 200 RPM s−1 to 1500 RPM, at which rate the sample was spun for 30 s to form a 250 μm thick layer. The wafer was then soft baked at 65 °C for 6 min and then at 105 °C for 15 min, then aligned and UV-exposed for 70 s at 9.5 mW cm−2. The wafer was post-exposure baked for 6 min at 65 °C and 12 min at 95 °C, allowed to cool to room temperature, developed in Microposit EC Solvent (Chestech Ltd., UK) developer for 12 min, and blown dry with nitrogen. The two layer SU-8 mould was then coated with perfluorosilane in a vacuum chamber for reducing the PDMS adhesion. PDMS prepolymer (Sylgard 184, Dow Corning) was prepared at 10 : 1 (w/w) ratio, degassed in a vacuum chamber for 30 min, then spin coated (500 RPM for 30 s) on the SU8 mould and cured in a 100 °C oven for 5 min producing a ~500 μm thick fluidic layer. The PDMS was then carefully peeled off the mould. As the upper seal, a second unpatterned wafer was also coated with a 2 mm thick layer of PDMS prepolymer and cured at 80 °C for 4 h. Fluid inlets and outlets were punched into the 2 mm thick layer upper layer. A flat-tip needle was used for making the inlet (1.3 mm OD) and outlet (3 mm OD) holes. The device was assembled by first placing the fluidic layer (with the fluidic structures exposed on the upper side) on a 500 μm thick, 4-inch glass wafer for support. Then the assembly was treated with O2 plasma (PDC-002 Harrick Scientific Corp. USA) for 2 min after which the upper PDMS layer (containing the inlets and outlets) was manually aligned and bonded at 60 °C for 2 h. The entire device was degassed overnight for removing bubbles that may form between the glass and the PDMS.
Device loading and flow control
The device is primed with PBS by placing the empty chip into a vacuum desiccator and degassing it for 10 min. This removes any bubbles formed during the initial fluid loading process (Movie S1, ESI†). Then a 1000 μL Eppendorf pipette loaded with 100 μL of PBS is inserted into each inlet that will be utilized during the experiment. With this pipette the PBS solution is introduced into the device until a droplet is formed in the outlet. Next, the pipette tip is released from the pipette and left inserted into the device inlet. The resulting liquid column generates the pressure head driving the flow. The height of the hydrostatic column and thus the flow velocity through the device can be regulated (Fig. 3A) by inserting or removing fluid from the top of the pipette tip with a second long tip pipette (0.5 mm round tip Corning® gel-loading tips, Sigma Aldrich, USA). Cells or other particles are loaded by first loading the fluid column with PBS and establishing a flow within the chip and then using the long-tip pipette the cells are injected at the bottom of the fluid column. For experiments with eukaryotic cells (MCF7, HeLa, etc.), ~2000 cells suspended in 10 μL of culture media were loaded into each processing module. Cell numbers were quantified using a hemocytometer (BS.748 Hawksley, UK) according to the manufacturer’s instructions. Any additional or non-loaded cells were removed from the inlet by extracting them from the bottom of the fluid column with long tip pipette (0.5 mm round tip Corning® gel-loading tips, Sigma Aldrich, USA). The PBS fluid column is then replaced with culture medium.
Device coating for adherent cell culture
To prepare the device for adherent cell culture and stimulation experiments the processing chambers are coated with a poly-L-lysine layer by allowing the 1% poly-L-lysine solution (Sigma Aldrich, USA) to flow continuously for 2 h at room temperature. Then the poly-L-lysine is replaced at the inlet with 300 μL of PBS (Sigma Aldrich, USA) and left to rinse the device for 2 h. The remaining PBS is replaced with 300 μL of cell culture medium.
Cell culture
MCF7, HeLa, 59M and U266 cells were cultured on and off chip using a CO2-independent medium supplemented with 10% bovine serum albumin (BSA), L-Glutamine and penicillin/ streptomycin. In the on-chip cell culture and stimulation experiments the entire device was incubated at 37 °C in an inverted fluorescence microscope (Olympus IX81) fitted with an incubation chamber (Solent Scientific, UK).
Cell staining and drugs
Cells were stained on chip by perfusion with a PBS solution containing fluorescent agents. For live cell staining, 4 μL mL−1 of Calcin AM (Sigma Aldrich, USA) was used while for dead cells 4 μL mL−1 of PI (Sigma Aldrich, USA) nuclear stain was used. Both stains were dissolved together in PBS and an aliquot of 300 μL loaded into each attached pipette tip. After 20 minutes, fluorescent images were acquired of each processing chamber. In cell stimulation experiments the drug or stimulating agent is dissolved in the cell culture medium according to the required concentration. Cells would receive the stimulation drug or agent after a 24 h drug free cell culture period.
Bacterial cell lysis
For the E. coli experiments cell lysis was performed off chip using MicroLYSIS PLUS buffer (Microzone Ltd., UK) according to the manufacturer’s recommendations. An overnight culture of E. coli (OD600 of 1.36) with a cell density of 3.4 × 108 colony forming units (CFU) per mL−1 (based on plate counts) was centrifuged at 10 000 rpm for 5 min, producing a pellet volume of 5 μL, and mixed with 40 μL of lysis buffer. Temperature cycling was then performed between 65 °C and 96 °C for a total of 23.5 min (65 °C for 15 min, 96 °C for 2 min, 65 °C for 4 min, 96 °C for 1 min, 65 °C for 1 min, 96 °C for 30 s) by transferring the tubes between two heating blocks. This crude cell lysate was either used directly or diluted with PBS to produce required cell concentrations. Samples of the cell lysate were stored at −15 °C until required.
On-chip eukaryotic cell lysis
Cells were loaded onto the chip and captured within the micro-trench structure. On-chip eukaryotic cell lysis was then performed by using a MicroLYSIS PLUS buffer. Briefly, 200 μL of lysis buffer were loaded into the fluid column and allowed to flow into the processing chamber for 5 minutes. The lysis buffer flow was then stopped and the device mounted on a hot plate at 65 °C for 5 min.
Real-time NASBA
All reagents required to perform the NASBA were supplied as part of a NucliSens Basic Kit (bioMérieux, UK), except primers and molecular beacon probes, which were supplied by Sigma Aldrich (UK). The NASBA mix was prepared according to the recommendations of manufacturer; the final concentration of each primer was 5 μM, and 5 μM for the molecular beacon probe. For the E. coli detection the design of both primers and the molecular beacon probe was taken from Dimov et al. 2008.38 For the estrogen receptor α detection, the primer and the molecular beacon probe designs are described in Verjat et al.41 Off-chip real-time NASBA was performed using a LightCycler 480 Real-time PCR System (Roche Applied Science, USA); the reagent mixture, without enzymes, was mixed with the RNA template and heated on a block heater at 65 °C for 5 min and at 41 °C for 5 min. Next, the enzymes were added, the NASBA mixture was heated for 5 min at 41 °C, loaded into a 96-well plate, and placed in the LightCycler system. The fluorescence (530 nm) was measured at 1 minute intervals for 90 min.
For on-chip experiments, the reagent mixture was mixed off chip with the enzyme mixture to produce a master NASBA mixture. This was then flowed into the chip for 15 min at ~200 μm s−1. In the case of the E. coli detection experiments, the reagent mixture contained varying concentrations of E. coli lysate (for the negative control RNA free water replaced the E. coli lysate). After that the device is mounted on a thermally controlled inverted fluorescent microscope and the green and red fluorescence imaged every minute in every processing chamber. Real-time NASBA curves were calculated from the average fluorescence increase over the initial background signal as a function of time from the sets of acquired images.
Cell fixing, permeabilization and immunofluorescent staining
Cells captured within the chip were fixed with 4% paraformaldehyde for 25 min at 100 μm s−1 flow rate. After washing once briefly with PBS-T (Dulbelcco’s PBS containing 0.01% Triton X-100), the cells were treated with 0.05% Triton X-100 in PBS for 10 min for permeabilization at a flow rate of 200 μm s−1. The cells briefly were washed once with PBS-T, followed by blocking in PBS-T containing 5% fetal bovine serum (FBS) for 30 min at room temperature and a flow rate of 200 μm s−1. The cells were then incubated for 1 hour at room temperature with anti-ESR1 antibodies (F10, Santa Cruz, CA, USA) diluted at 1 : 50 in PBS-T containing 1% FBS at a flow rate of 200 μm s−1. The cells were washed with PBS-T, for 20 min at 600 μm s−1. Then the cells were incubated for 15 min at room temperature with Alexa Fluor 488 fluorescent dye-conjugated secondary antibodies (Molecular Probes Inc., Eugene, OR, USA) (1 : 200) in PBST containing 1% FBS at 200 μm s−1. The cells were washed with PBS-T for 1 h at 600 μm s−1 flow rate, before staining with Propidium Iodide. Immunofluorescent staining was observed under Olympus IX81 Fluorescence Microscope with Optronics MagnaFire digital camera (Microscope Image Center, UVM).
Device read out
For readout the device is mounted on a thermally controlled inverted fluorescent microscope. In our case an Olympus IX81 model fitted with an incubation chamber from Solent Scientific (UK) was used. All image acquisitions (bright field, and fluorescent) were done with a CCD sensor (Hamamatsu C4742-80-12AG) and through the 10× objective. For green fluorescence imaging, an excitation filter BP492/18 with a xenon light source, CellR MT20 (Olympus) and a U-MF2 filter cube (Olympus) were used. For red fluorescence imaging was carried out with an excitation filter BP540/10 with the previously mentioned xenon light source and a U-MW1GA3 filter cube (Olympus). When scanning multiple processing chambers, a programmed motorized stage positioned and focused the objective over each processing chamber. The image analysis and motorized stage programming was done using the ImageJ and the Cell R software packages (Olympus).
Acknowledgments
The authors would like to thank Dr Arman Rahman for providing the non-adherent plasma cells U266. This work was supported by the Science Foundation Ireland under Grant No. 05/CE3/B754, Irish Cancer Society Research Fellowship Award CRF10KIJ, U. S. National Cancer Institute (NCI) award CCNE-T U54CA151459 and the Center for Nanostructured Materials Technology (Grant Nos. 2010K000352, 2010K000353, and 2010K000354) under “21st Century Frontier R&D Programs” of the Ministry of Education, Science and Technology, Korea.
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c1lc20105k
References
- 1.Figeys D, Pinto D. Anal Chem. 2000;72:330A–335A. doi: 10.1021/ac002800y. [DOI] [PubMed] [Google Scholar]
- 2.Dittrich PS, Tachikawa K, Manz A. Anal Chem. 2006;78:3887–3908. doi: 10.1021/ac0605602. [DOI] [PubMed] [Google Scholar]
- 3.Khandurina J, McKnight TE, Jacobson SC, Waters LC, Foote RS, Ramsey JM. Anal Chem. 2000;72:2995–3000. doi: 10.1021/ac991471a. [DOI] [PubMed] [Google Scholar]
- 4.Kamei K-i, Ohashi M, Gschweng E, Ho Q, Suh J, Tang J, Yu ZTF, Clark AT, Pyle AD, Teitell MA, Lee K-B, Witte ON, Tseng H-R. Lab Chip. 2010;10:1113–1119. doi: 10.1039/b922884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sims CE, Allbritton NL. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 7.Keenan TM, Folch A. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Folch A, Toner M. Annu Rev Biomed Eng. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 9.Kim L, Toh YC, Voldman J, Yu H. Lab Chip. 2007;7:681–694. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Alexander CM, Beebe DJ. Lab Chip. 2007;7:726–730. doi: 10.1039/b618793e. [DOI] [PubMed] [Google Scholar]
- 11.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, Monuki ES, Jeon NL. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 12.Fujii S-i, Uematsu M, Yabuki S, Abo M, Yoshimura E, Sato K. Anal Sci. 2006;22:87–90. doi: 10.2116/analsci.22.87. [DOI] [PubMed] [Google Scholar]
- 13.Li CW, Yang J, Yang M. Lab Chip. 2006;6:921–929. doi: 10.1039/b600058d. [DOI] [PubMed] [Google Scholar]
- 14.Pihl J, Sinclair J, Sahlin E, Karlsson M, Petterson F, Olofsson J, Orwar O. Anal Chem. 2005;77:3897–3903. doi: 10.1021/ac050218+. [DOI] [PubMed] [Google Scholar]
- 15.Yamada M, Seki M. Anal Chem. 2006;78:1357–1362. doi: 10.1021/ac0520083. [DOI] [PubMed] [Google Scholar]
- 16.Lee PJ, Hung PJ, Rao VM, Lee LP. Biotechnol Bioeng. 2006;94:5–14. doi: 10.1002/bit.20745. [DOI] [PubMed] [Google Scholar]
- 17.Di Carlo D, Ionescu-Zanetti C, Zhang Y, Hung P, Lee LP. Lab Chip. 2005;5:171–178. doi: 10.1039/b413139h. [DOI] [PubMed] [Google Scholar]
- 18.Di Carlo D, Jeong KH, Lee LP. Lab Chip. 2003;3:287–291. doi: 10.1039/b305162e. [DOI] [PubMed] [Google Scholar]
- 19.Verpoorte E, de Rooij NF, Sigrist H, Thormann W, Linder V. Electrophoresis. 2002;23:740–749. doi: 10.1002/1522-2683(200203)23:5<740::AID-ELPS740>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Bernard A, Michel B, Delamarche E. Anal Chem. 2001;73:8–12. doi: 10.1021/ac0008845. [DOI] [PubMed] [Google Scholar]
- 21.Manz A, Harrison DJ, Verpoorte EMJ, Fettinger JC, Paulus A, Lüdi H, Widmer HM. J Chromatogr, A. 1992;593:253–258. [Google Scholar]
- 22.Simpson PC, Roach D, Woolley AT, Thorsen T, Johnston R, Sensabaugh GF, Mathies RA. Proc Natl Acad Sci U S A. 1998;95:2256–2261. doi: 10.1073/pnas.95.5.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, Xu J, Ma W, Zheng W. Biotechnol Adv. 2006;24:243–284. doi: 10.1016/j.biotechadv.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Science. 2006;314:1464–1467. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 25.Lindstrom S, Hammond M, Brismar H, Andersson-Svahn H, Ahmadian A. Lab Chip. 2009;9:3465–3471. doi: 10.1039/b912596e. [DOI] [PubMed] [Google Scholar]
- 26.Paguirigan AL, Beebe DJ. Integr Biol. 2009;1:182–195. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wenger SL, Senft JR, Sargent LM, Bamezai R, Bairwa N, Grant SG. Biosci Rep. 2004;24:631–639. doi: 10.1007/s10540-005-2797-5. [DOI] [PubMed] [Google Scholar]
- 28.Chang-Liu CM, Woloschak GE. Cancer Lett. 1997;113:77–86. doi: 10.1016/s0304-3835(97)04599-0. [DOI] [PubMed] [Google Scholar]
- 29.Bouïs D, Hospers G, Meijer C, Molema G, Mulder N. Angiogenesis. 2001;4:91–102. doi: 10.1023/a:1012259529167. [DOI] [PubMed] [Google Scholar]
- 30.Hong JW, Quake SR. Nat Biotechnol. 2003;21:1179–1183. doi: 10.1038/nbt871. [DOI] [PubMed] [Google Scholar]
- 31.Manbachi A, Shrivastava S, Cioffi M, Chung BG, Moretti M, Demirci U, Yliperttula M, Khademhosseini A. Lab Chip. 2008;8:747–754. doi: 10.1039/b718212k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Compton J. Nature. 1991;350(6313):91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- 33.Du Y, Shim J, Vidula M, Hancock MJ, Lo E, Chung BG, Borenstein JT, Khabiry M, Cropek DM, Khademhosseini A. Lab Chip. 2009;9:761–767. doi: 10.1039/b815990d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullis KB, Faloona FA. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 35.Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fire A, Xu SQ. Proc Natl Acad Sci U S A. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deiman B, van Aarle P, Sillekens P. Mol Biotechnol. 2002;20:163–179. doi: 10.1385/MB:20:2:163. [DOI] [PubMed] [Google Scholar]
- 38.Dimov IK, Garcia-Cordero JL, O’Grady J, Poulsen CR, Viguier C, Kent L, Daly P, Lincoln B, Maher M, O’Kennedy R, Smith TJ, Ricco AJ, Lee LP. Lab Chip. 2008;8:2071–2078. doi: 10.1039/b812515e. [DOI] [PubMed] [Google Scholar]
- 39.Glynn B, Lacey K, Reilly J, Barry T, Smith TJ, Maher M. Res J Biol Sci. 2007;2:564–570. [Google Scholar]
- 40.Pinzone JJ, Stevenson H, Strobl JS, Berg PE. Mol Cell Biol. 2004;24:4605–4612. doi: 10.1128/MCB.24.11.4605-4612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamy PJ, Verjat T, Paye M, Servanton AC, Grenier J, Leissner P, Mougin B. Clin Chem Lab Med. 2006;44:3–12. doi: 10.1515/CCLM.2006.002. [DOI] [PubMed] [Google Scholar]
- 42.Sato H, Nishida S, Tomoyori H, Sato M, Ikeda I, Imaizumi K. Biosci, Biotechnol, Biochem. 2004;68:1790–1793. doi: 10.1271/bbb.68.1790. [DOI] [PubMed] [Google Scholar]
- 43.Jordan MA, Wendell K, Gardiner S, Brent Derry W, Copp H, Wilson L. Cancer Res. 1996;56:816–825. [PubMed] [Google Scholar]
- 44.Stoica A, Pentecost E, Martin MB. Endocrinology. 2000;141:3595–3602. doi: 10.1210/endo.141.10.7704. [DOI] [PubMed] [Google Scholar]
- 45.Younan Xia GMW. Angew Chem, Int Ed. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]