Abstract
Mutation specific immunohistochemistry (IHC) is a promising new technique to detect the presence of the BRAFV600E mutation in colorectal carcinoma (CRC). When performed in conjunction with mismatch repair (MMR) IHC, BRAFV600E IHC can help to further triage genetic testing for Lynch Syndrome. In a cohort of 1426 patients undergoing surgery from 2004 to 2009 we recently demonstrated that the combination of MMR and BRAFV600E IHC holds promise as a prognostic marker in CRC, particularly because of its ability to identify the poor prognosis MMR proficient (MMRp) BRAFV600E mutant subgroup. We attempted to validate combined MMR and BRAFV600E IHC as a prognostic indicator in a separate cohort comprising consecutive CRC patients undergoing surgery from 1998 to 2003. IHC was performed on a tissue microarray containing tissue from 1109 patients with CRC. The 5 year survivals stratified by staining patterns were: MMRd/BRAFwt 64%, MMRd/BRAFV600E 64%, MMRp/BRAFwt 60% and MMRp/BRAFV600E 53%. Using the poor prognosis MMRp/BRAFV600E phenotype as baseline, univariate Cox regression modelling demonstrated the following hazard ratios for death: MMRd/BRAFwt HR = 0.71 (95%CI = 0.40–1.27), p = 0.31; MMRd/BRAFV600E HR = 0.74 (95%CI = 0.51–1.07), p = 0.11 and MMRp/BRAFwt HR = 0.79 (95%CI = 0.60–1.04), p = 0.09. Although the findings did not reach statistical significance, this study supports the potential role of combined MMR and BRAF IHC as prognostic markers in CRC.
Introduction
The development of biomarkers to predict outcome after definitive treatment of malignancy is an area of active research. Despite literally thousands of biomarkers having been explored in various cohorts, [1] very few have entered routine clinical practice. Reasons for the failure to translate into clinical care include cost, impracticality, lack of availability and failure of validation in different cohorts or across diverse populations. [1] An ideal biomarker would be inexpensive, readily deployable in the routine clinical setting and add genuine prognostic information in addition to that already provided by simple measures such as age, stage and grade.
In many institutions patients with colorectal carcinoma (CRC) undergoing surgery with curative intent are routinely offered reflex immunohistochemistry (IHC) for the DNA mismatch repair (MMR) proteins MLH1, PMS2, MSH2 and MSH6 in order to triage formal molecular testing for Lynch Syndrome. [2] We recently demonstrated, in a single institution cohort of patients undergoing surgery for CRC at Royal North Shore Hospital between calendar years 2004 and 2009, that the addition of BRAFV600E mutation specific IHC to MMR IHC holds promise as a biomarker for all cause survival. [3] This approach identifies the poor prognostic group of mismatch repair proficient (MMRp) BRAFV600E mutant CRC which accounted for 6.4% of CRC in our previous study. [3] Because the presence of BRAFV600E determination by either molecular means or IHC virtually excludes Lynch Syndrome in mismatch repair deficient (MMRd) CRC and is therefore commonly performed in many institutions in MMRd CRC, [2] this approach requires minimal extra expense and fits well into routine laboratory workflow.
In this study we sought to validate the combination of MMR and BRAFV600E IHC as a prognostic marker in CRC by examining its prognostic power in a different cohort – namely all patients undergoing surgery for CRC at the same institution from June 1998 to 2003.
Materials and Methods
Patients
We searched the database of the Department of Anatomical Pathology, Royal North Shore Hospital, for all patients who underwent surgery for CRC with curative intent from June 1998 to the end of calendar year 2003. During this period this department provided a centralized pathology service for 2 major quaternary centres with dedicated colorectal surgery units as well as four community hospitals with general surgery units. Patients treated endoluminally, with histologies other than adenocarcinoma or with tissue blocks unavailable for review were excluded. The pathology reports of all cases were reviewed (and if necessary the slides from cases were retrieved and reassessed) in order to stage the tumours according to the AJCC 7th edition 2009 staging system [4].
Immunohistochemistry
Tissue microarrays (TMAs) containing two 1 mm cores of carcinoma were created. IHC for the MMR associated proteins MLH1, PMS2, MSH2 and MSH6 was performed and interpreted using standard and previously described methods. [6] BRAFV600E mutation specific IHC was performed using a commercially available mouse monoclonal antibody (clone VE1, SpringBioscience, Pleasonton CA) using the same methods we have previously described. [2], [3], [5] Briefly VE1 IHC was performed using the Leica BondIII autostainer (Leica Microsystems, Mount Waverley, VIC, Australia) used according to the manufacturer’s protocol with alkaline antigen retrieval (solution ER2, VBS part no: AR9640, Leica Microsystems) with the primary antibody being used at a dilution of 1 in 80. BRAFV600E staining was interpreted as positive if >20% of neoplastic cells stained positively. The presence of definitive negative staining for any one of the four MMR markers was interpreted as evidence of mismatch repair deficiency (MMRd). MMR and BRAFV600E IHC was interpreted by observers who were blinded to all other clinical and pathological data.
Survival Data
Follow up data was obtained by examination of hospital medical records and the hospital pathology database, assessment of records from surgeons’ private rooms and examination from publicly available death notices up to January 2014. Overall survival was defined as the duration alive from time of definitive surgery. In patients with metachronous CRCs, survival was taken from the time of surgery for the first CRC with subsequent tumors (either recurrences or second primary tumors) being excluded from survival analysis.
Statistical analysis
Single variable p-values were computed using either the chi-square test for categorical variables or the Mann-Whitney-U test for scalar variables such as age at diagnosis. Five year survival values were obtained via Kaplan Meier analysis for each of the four MMR/BRAF IHC tumour phenotypes.
The effect of MMR/BRAF tumour IHC phenotype on overall survival was explored using Cox regression proportional hazards analysis, including a final model adjusted for gender, age at diagnosis, anatomic location, histologic grade and overall stage.
A p-value of <0.05 was taken as significant. All analysis was performed using IBM SPSS statistics for MAC, Version 21.0 (IBM Corp, Armonk NY USA, released 2012).
This study was approved by the Northern Sydney Local Health District Human Research Ethics Committee under protocol 1201-035 M. The ethics committee waived the need for consent to use the archived formalin fixed paraffin embedded tissue blocks and to access medical records on the basis that the study was only performed on archived formalin fixed paraffin embedded tissue removed during routine care many years previously. All patient information was anonymized and de-identified prior to analysis.
Results
A total of 1109 colorectal carcinomas met inclusion criteria and had cores available in the TMA sections. The clinical and pathological details are presented in Table 1. Briefly, the median age at diagnosis was 72 years, 49.2% were female, and 75% had stage 2 or 3 disease. 856 patients were MMRp (85.9%) of which 133 (13.4% of the total) were MMRp/BRAFV600E mutant and 720 (72.5% of the total) were MMRp/BRAFwt. 144 were MMRd (14.1%), of which 108 (10.9% of the total) were MMRd/BRAFV600E and 32 (3.2% of the total) were MMR-deficient/BRAF wild type.
Table 1. Clinical and pathological features of 1109 consecutive patients with CRC.
| Variable | Count (%) | SingleVariablep-value | Univariateanalysis HR(95%CI),p-value | Multivariateanalysis HR(95%CI),p-value |
| Gender | 0.63 | |||
| female | 546 (49.2) | 1.00 | 1.00 | |
| male | 563 (50.8) | 1.15 (0.96–1.37), 0.13 | 1.22 (1.00–1.50), 0.05 | |
| Age at diagnosis | 72 (28–100) | N/A | 1.05 (1.04–1.05), <0.01 | 1.05 (1.04–1.06), <0.01 |
| Anatomic location | <0.01 | |||
| rectum | 278 (25.5) | 1.00 | 1.00 | |
| caecum | 156 (14.3) | 1.17 (0.86–1.59), 0.32 | 0.85 (0.60–1.21), 0.37 | |
| ascending colon | 218 (20.0) | 1.16 (0.89–1.50), 0.28 | 0.87 (0.64–1.18), 0.37 | |
| transverse colon | 130 (11.9) | 1.23 (0.91–1.67), 0.18 | 0.80 (0.55–1.15), 0.23 | |
| descending colon | 48 (4.4) | 0.99 (0.63–1.55), 0.97 | 0.90 (0.54–1.48), 0.67 | |
| sigmoid colon | 260 (23.9) | 1.19 (0.93–1.54), 0.17 | 1.04 (0.79–1.38), 0.78 | |
| Histologic grade | <0.01 | |||
| low | 835 (80.9) | 1.00 | 1.00 | |
| high | 197 (19.1) | 1.39 (1.11–1.75), <0.01 | 1.13 (0.87–1.47), 0.36 | |
| AJCC Stage | <0.01 | |||
| I | 207 (18.7) | 1.00 | 1.00 | |
| IIA | 295 (26.6) | 0.28 (0.17–0.46), <0.01 | 0.10 (0.05–0.20), <0.01 | |
| IIB | 54 (4.9) | 0.35 (0.21–0.57), <0.01 | 0.13 (0.06–0.25), <0.01 | |
| IIC | 15 (1.4) | 0.95 (0.53–1.69), 0.86 | 0.36 (0.17–0.79), 0.01 | |
| IIIA | 40 (3.6) | 1.06 (0.48–2.33), 0.90 | 0.32 (0.12–0.81), 0.02 | |
| IIIB | 321 (28.9) | 0.31 (0.15–0.61), <0.01 | 0.15 (0.06–0.34), <0.01 | |
| IIIC | 107 (9.6) | 0.63 (0.39–1.01), 0.06 | 0.22 (0.11–0.44), <0.01 | |
| IVA | 39 (3.5) | 1.49 (0.89–2.50), 0.13 | 0.59 (0.28–1.22), 0.16 | |
| IVB | 5 (0.5) | 2.23 (1.26–3.98), <0.01 | 1.30 (0.60–2.83), 0.50 | |
| MMR IHC status | <0.01 | N/A | ||
| proficient (MMRp) | 856 (85.9) | 1.00 | ||
| deficient (MMRd) | 140 (14.1) | 0.90 (0.69–1.17), 0.42 | ||
| BRAF IHC status | <0.01 | N/A | ||
| wild type (BRAFwt) | 774 (76.2) | 1.00 | ||
| mutant (BRAFV600E) | 242 (23.8) | 1.12 (0.91–1.38), 0.30 | ||
| MMR/BRAF IHC phenotype | <0.01 | |||
| MMRp/BRAFV600E | 133 (13.4) | 1.00 | 1.00 | |
| MMRd/BRAFwt | 32 (3.2) | 0.71 (0.40–1.27), 0.25 | 1.12 (0.61–2.06), 0.72 | |
| MMRd/BRAFV600E | 108 (10.90 | 0.74 (0.51–1.07), 0.11 | 0.87 (0.58–1.31), 0.51 | |
| MMRp/BRAFwt | 720 (72.5) | 0.79 (0.60–1.04), 0.09 | 0.80 (0.60–1.08), 0.15 | |
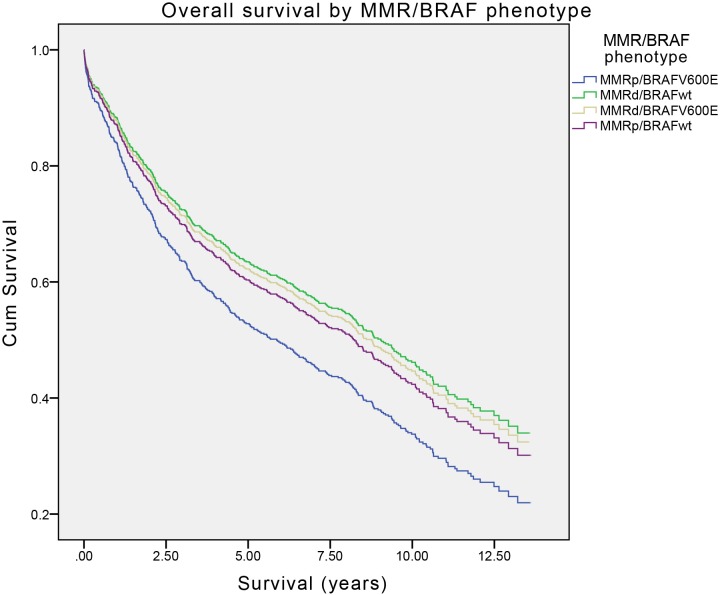
Table 2 presents the overall survival figures for each of the four MMR/BRAF phenotypes as determined by the Kaplan Meier analysis. The 5-year survivals were 52.6% for MMRp/BRAFV600E, 64.2% for MMRd/BRAFwt, 64.1% for MMRd/BRAFV600E and 60.1% for MMRp/BRAFwt CRCs.
Table 2. Overall survivals of each of the four MMR/BRAF phenotypes by Kaplan Meier actuarial analysis.
| MMR/BRAF phenotype | 5-year survival | Mean survival |
| MMRp/BRAFV600E | 52.6% | 7.12 years (95%CI = 5.87–8.37) |
| MMRd/BRAFwt | 64.2% | 8.36 years (95%CI = 6.16–10.56) |
| MMRd/BRAFV600E | 64.1% | 8.08 years (96%CI = 7.04–9.40) |
| MMRp/BRAFwt | 60.1% | 8.06 years (95%CI = 7.62–8.50) |
The univariate Cox regression survival function demonstrating the crude (unadjusted) relationship between survival and MMR/BRAF status is presented in Table 1 and Figure 1. Tumours segregated according to their MMR/BRAF phenotype in a consistent trend throughout the period of follow-up, with the MMRp/BRAFV600E trending towards worse prognosis compared to the other three phenotypes. Compared to the MMRp/BRAFV600E phenotype, tumours displaying the MMRp/BRAFwt phenotype tended towards significantly improved survival with a hazard ratio of 0.79 (95%CI = 0.60–1.04, p = 0.09). This effect was markedly diminished in the multivariate model due to the dominant effects of gender, age at diagnosis and tumour stage on overall survival (adjusted effect).
Figure 1. Overall survival of patients with CRC stratified by MMR and BRAF status (Cox regression modelling).
Discussion
The determination of BRAF mutation status by immunohistochemistry has the significant advantages over molecular techniques of being both inexpensive and fitting easily into standard surgical pathology workflow. In laboratories where all CRCs routinely undergo screening for Lynch Syndrome with MMR IHC, the addition of BRAFV600E mutation specific immunohistochemistry would simply be a matter of performing IHC for 5 rather than 4 markers entailing minimal extra labour or handling costs and would have the added advantage of further triaging molecular testing for Lynch Syndrome in MMRd CRC. Therefore, if the addition of BRAFV600E mutation specific IHC to all CRCs can be validated as a biomarker, there is real potential that it may become the first prognostic biomarker for CRC deployed into routine clinical practice.
When we previously investigated the prognostic power of combined MMR and BRAFV600E IHC in a group of 1426 CRC from 2004 to 2009 [3], univariate analysis demonstrated that MMRp/BRAFV600E CRCs had a statistically significantly worse outcome compared to the other phenotypes (hazard ratio of 1.79 (95%CI = 1.24–2.60), (p)<0.01). This result was negated in multivariate analysis (hazard ratio of 1.10 (95%CI = 0.69–1.76), (p) = 0.68) primarily due to the dominant effect of stage and age on overall survival. In the current study, comprising CRCs from 1109 from the same institution resected from 1998 to 2003, MMRp/BRAFV600E CRCs trended towards a worse prognosis compared to all other tumour groups but failed to gain statistical significance (MMRd/BRAFwt p = 0.31, MMRd/BRAFV600E p = 0.11 and MMRp/BRAFwt p = 0.09). Whilst our findings support the prognostic utility of the combination of MMR and BRAFV600E IHC, the failure to achieve statistical significance indicates that further studies in larger cohorts will be needed to validate this approach. Ideally such further studies should be in truly independent external cohorts (that is from other institutions) rather than merely representing a preceding cohort from the same institution as in this case.
To date 13 studies have directly compared the accuracy of BRAF mutation status determination by IHC with molecular techniques. In 11 studies BRAFV600E mutation specific IHC has either outperformed or performed comparably to molecular techniques [2], [7], [8], [9], [10], [11], [12], [13], [14],[15],[16] whereas in two studies mutation specific IHC was found to be less reliable. [17], [18] A fair reading of the literature would support the approach taken by Kuan et al, [12] that mutation specific IHC is reliable but requires rigorous technical optimization and ongoing quality assurance including the performance of molecular testing in equivocal cases. Whilst this study was not intended or designed to assess the accuracy of BRAFV600E mutation specific IHC we note that the antibody has previously been proven to be extremely reliable in our hands. [2] The overall rate of BRAF mutation as determined by IHC in this study (23.8%) is in keeping with the 18.4% incidence we reported in a similar cohort of consecutiveCRCs from 2011 from the same institution tested by molecular means alone [2].
Although there are limitations to this study, most importantly that it did not represent a true external validation cohort but rather a validation cohort from the same institution, our finding of a trend towards survival differences amongst CRC when stratified by BRAFV600E and MMR IHC status is very similar to that which we have previously reported. [3] This provides cautious support to the use of a combination of BRAFV600E and MMR IHC as prognostic biomarkers in CRC. Ultimately similar studies will need to be performed in large truly independent cohorts before this approach can be considered validated. In the interim, the combination of BRAFV600E and MMR IHC still has a clear role in the triaging of patients with CRC encountered in routine clinical practice for formal molecular testing for Lynch Syndrome. [2], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16] The strong likelihood that this approach can also have the added benefit of predicting outcome can be considered a likely downstream benefit of universal screening for Lynch Syndrome by immunohistochemistry.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by funding from the Cancer Institute NSW as part of a translational research centre grant and internally by the department of anatomical pathology Royal North Shore Hospital. No other funding support was received. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ludwig JA, Weinstein JN (2005) Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer 5: 845–856. [DOI] [PubMed] [Google Scholar]
- 2. Toon CW, Walsh MD, Chou A, Capper D, Clarkson A, et al. (2013) BRAFV600E Immunohistochemistry facilitates universal screening of colorectal cancers for Lynch Syndrome. Am J Surg Pathol 37: 1592–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toon CW, Chou A, DeSilva K, Chan J, Patterson J, et al. (2014) BRAFV600E immunohistochemistry in conjunction with mismatch repair status predicts survival in patients with colorectal cancer. Modern Pathology 27: 644–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edge SE, Byrd DR, Compton CC (2009) AJCC Cancer Staging Manual. 7th ed. New York, NY: Springer.
- 5. Hall G, Clarkson A, Shi A, Langford E, Leung H, et al. (2010) Immunohistochemistry for PMS2 and MSH6 alone can replace a four antibody panel for mismatch repair deficiency screening in colorectal adenocarcinoma. Pathology 42: 409–413. [DOI] [PubMed] [Google Scholar]
- 6. Bullock M, O’Neill C, Chou A, Clarkson A, Dodds T, et al. (2012) Utilisation of a Monoclonal Antibody for BRAFV600E Detection in Papillary Thyroid Carcinoma. Endocrine Related Cancer 19: 779–784. [DOI] [PubMed] [Google Scholar]
- 7. Affolter K, Samowitz W, Tripp S, Bronner MP (2013) BRAF V600E mutation detection by immunohistochemistry in colorectal carcinoma. Genes Chromosomes Cancer 52: 748–52. [DOI] [PubMed] [Google Scholar]
- 8. Capper D, Voigt A, Bozukova G, Ahadova A, Kickingereder P, et al. (2013) BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. Int J Cancer 133: 1624–30. [DOI] [PubMed] [Google Scholar]
- 9. Thiel A, Heinonen M, Kantonen J, Gylling A, Lahtinen L, et al. (2013) BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Archiv 463: 613–621. [DOI] [PubMed] [Google Scholar]
- 10. Routhier CA, Mochel MC, Lynch K, Dias-Santagat D, Louis DN, et al. (2013) Comparison of 2 monoclonal antibodies for immunohistochemical detection of BRAF V600E mutation in malignant melanoma, pulmonary carcinoma, gastrointestinal carcinoma, thyroid carcinoma, and gliomas. Hum Pathol. 44: 2563–70. [DOI] [PubMed] [Google Scholar]
- 11. Rössle M, Sigg M, Rüschoff JH, Wild PJ, Moch H, et al. (2013) Ultra-deep sequencing confirms immunohistochemistry as a highly sensitive and specific method for detecting BRAF V600E mutations in colorectal carcinoma. Virchows Arch 463: 623–31. [DOI] [PubMed] [Google Scholar]
- 12. Kuan SF, Navina S, Cressman KL, Pai RK (2014) Immunohistochemical detection of BRAF V600E mutant protein using the VE1 antibody in colorectal carcinoma is highly concordant with molecular testing but requires rigorous antibody optimization. Hum Pathol. 45: 464–72. [DOI] [PubMed] [Google Scholar]
- 13. Sajanti S, Sirniö P, Väyrynen JP, Tuomisto A, Klintup K, et al. (2014) VE1 immunohistochemistry accurately detects BRAF V600E mutations in colorectal carcinoma and can be utilized in the detection of poorly differentiated colorectal serrated adenocarcinoma. Virchows Arch. 464: 637–643. [DOI] [PubMed] [Google Scholar]
- 14. Ilie MI, Long-Mira E, Hofman V, Mouroux J, Vignaud JM, et al. (2014) BRAFV600E mutation analysis by immunohistochemistry in patients with thoracic metastases from colorectal cancer. Pathology 46: 311–5. [DOI] [PubMed] [Google Scholar]
- 15. Nolan S, Arnason T, Drucker A, Huang WY (2014) The Utility of BRAFV600E Mutation-specific Antibody for Colon Cancers With Microsatellite Instability. Appl Immunohistochem Mol Morphol 22: e8–e13. [DOI] [PubMed] [Google Scholar]
- 16. Sinicrope FA, Smyrk TC, Tougeron D, Thibodeau SN, Singh S, et al. (2013) Mutation-specific antibody detects mutant BRAFV600E protein expression in human colon carcinomas. Cancer 119: 2765–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adackapara CA, Sholl LM, Barletta JA, Hornick JL (2013) Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology 63: 187–93. [DOI] [PubMed] [Google Scholar]
- 18.Lasota J, Kowalik A, Wasag B, Wang ZF, Felisiak-Golabek A, et al.. (2014) Detection of the BRAF V600E Mutation in Colon Carcinoma: Critical Evaluation of the Imunohistochemical Approach. Am J Surg Pathol [epub ahead of print, May 14]. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.