Summary
Background
The PINK1-Parkin pathway is known to play important roles in regulating mitochondria dynamics, motility, and quality control. Activation of this pathway can be triggered by a variety of cellular stressor signals that cause mitochondrial damage. How this pathway senses different levels of mitochondrial damage and mediates cell fate decisions accordingly is incompletely understood.
Results
Here, we present evidence that PINK1-Parkin has both cytoprotective and pro-apoptotic functions. PINK1-Parkin operates as a molecular switch to dictate cell fate decisions in response to different cellular stressors. Cells exposed to severe and irreparable mitochondrial damage agents such as valinomycin can undergo PINK1-Parkin-dependent apoptosis. The proapoptotic response elicited by valinomycin is associated with the degradation of Mcl-1. PINK1 directly phosphorylates Parkin at Ser65 of its Ubl domain and triggers activation of its E3 ligase activity through an autocatalytic mechanism which amplifies its E3 ligase activity towards Mcl-1.
Conclusions
Autocatalytic activation of Parkin bolsters it accumulation on mitochondria and apoptotic response to valinomycin. Our results suggest that PINK1-Parkin constitutes a damage-gated molecular switch that governs cellular context-specific cell fate decisions in response to variable stress stimuli.
Introduction
Mutations in PINK1 and Parkin are associated with early-onset familial autosomal recessive Parkinson's disease (PD) [1, 2]. Although the exact molecular mechanism which causes PD is not clearly understood, genetic studies in model organisms coupled with mechanistic studies in mammalian cells suggest that PINK1 acts upstream of Parkin to regulate mitochondrial integrity, dynamics, and motility [3-5].
The level of PINK1 is low in unperturbed mitochondria due to proteolytic degradation of PINK1 by the protease ParL and subsequent retrotranslocation into the cytosol for proteasomal degradation [6]. Upon the loss of mitochondrial membrane potential by decouplers, such as cyanide, the import and degradation of PINK1 are blocked, allowing it to accumulate on the outer mitochondrial membrane [7-9]. Increased expression and activity of PINK1 lead to phosphorylation of mitofusin 2 [10], Parkin [9, 11], Miro [12], and other substrates. Elevated PINK1 activity promotes translocation of Parkin from the cytosol to the mitochondria. In accordance with the significance of Parkin mitochondrial recruitment, many patient derived Parkin mutants are defective in mitochondrial translocation [13, 14]. Parkin is a member of the RING-IBR-RING (RBR) family of ubiquitin E3 ligases with a conserved catalytic cysteine residue analogous to the HECT domain E3s [15]. The auto-ubiquitination activity of Parkin is abolished if this residue is mutated [16, 17]. A diverse set of protein substrates have been shown be ubiquitinated by Parkin [2], including several proteins localized in the mitochondrial outer membrane such as Mfn1 and Mfn2 [10, 18, 19], Drp1 [20], voltage-dependent anion channel 1 (VDAC1) [21] and Miro [12]. Ubiquitination and degradation of these proteins is linked to mitochondrial fission during mitophagy or mitochondrial motility. Proteomics studies revealed that Parkin may directly or indirectly regulate ubiquitination of more than 100 mitochondrial proteins upon mitochondrial depolarization [22].
Despite these current insights, there are still many outstanding questions that remain to be answered. First, the mechanism by which PINK1 mediates Parkin mitochondrial translocation is still incompletely understood. Second, it remains to be determined how the E3 ligase activity of Parkin is regulated. Biochemical and structure studies revealed that Parkin exists in an autoinhibited conformation due to an interaction between the N-terminal Ubiquitin like domain (Ubl), the repressor element (REP) and the RING1 domain [23]. These observations raise an interesting question about the potential mechanisms that can allosterically activate Parkin in vitro and in vivo. Finally, the diversity of Parkin substrates begs the question whether there is selective ubiquitination of these substrates and whether ubiquitination of these substrates contribute to cell fate decisions in response to distinct cellular stressor stimuli.
The PINK1-Parkin pathway has also been shown to have cytoprotective activity under various stress conditions [2, 5]. There appears to be distinct Parkin-dependent pathways for preventing cell death depending on the severity of mitochondrial damage and the nature of stress signals. Reactive oxygen species (ROS) can induce PINK1-Parkin dependent repairing damaged mitochondria through the mitochondria derived vesicles (MDVs) trafficking pathway [24]. Upregulation of Parkin-dependent NF-kB signaling has a prosurvival role under moderate stress [25]. Mitophagy is the best characterized PINK1-Parkin-dependent pathway for removal of damaged mitochondria [4]. Collectively these results suggest that the PINK1-Parkin nexus can mount different cellular responses depending on the levels of cellular stress signals.
Here we investigated mechanisms by which PINK1-Parkin regulates cell survival. We discovered that beyond its well recognized cytoprotective function, PINK1-Parkin is also required for valinomycin-induced apoptosis. Mechanistically, the pro-apoptotic function of PINK1-Parkin in response to severe apoptotic signal is associated with ubiquitination of anti-apoptotic protein Mcl-1 and activation of the caspase cascade. We reconstituted Parkin-dependent ubiquitination of Mcl-1 in vitro and demonstrated that PINK1 allosterically regulates the Parkin E3 ligase activity through phosphorylation of Ser65 of Parkin. This phosphorylation event sets off autocatalytic activation of Parkin. Finally, our study showed that ubiquitin is also a substrate for PINK1 and phosphorylated ubiquitin promotes Parkin mitochondrial targeting and afford additional elevation of Parkin activity. Our results reveal a new function of the PINK1-Parkin pathway in cell fate decisions and a Parkin activation cascade in response to diverse stress stimuli.
Results
Parkin-dependent mitophagy and apoptotic cellular responses in response to mitochondrial depolarization
It is well established that the PINK1/Parkin pathway is involved in elimination of depolarized mitochondria by mitophagy [26]. As expected, HeLa cells stably expressing fluorescence protein tagged wild type Parkin but not mutant Parkin T240R (seen in Parkinson's patients) undergo mitophagy in the presence of mitochondrial uncoupler protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (Figure 1A). The mitophagic response can be demonstrated by the increased colocalization of Parkin and LC3 with the mitochondria marker RFP-Smac-mts (RFP-Smac) - a fusion of the mitochondrial targeting sequence of Smac with RFP (Supplementary Figure 1AB and Supplementary movie 1 and 2). Under these conditions, no significant apoptosis is seen in either cell lines based on RFP-Smac release (Figure 1B).
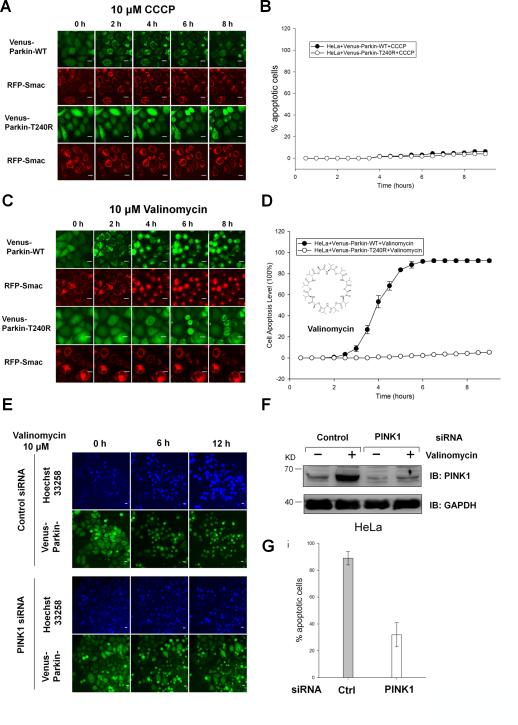
Figure 1. Valinomycin triggers Parkin mitochondrial recruitment and apoptosis in a PINK1/Parkin-dependent manner.
(A-D) HeLa cells expressing RFP-Smac-mts and either Venus-Parkin-WT or Venus-Parkin-T240R were treated with 10μM CCCP (A and B) or 10μM Valinomycin (C and D) in a 9-hour time course. Scale bar, 10 μm
(A) CCCP induced mitochondrial recruitment of Parkin-WT, but not Parkin-T240R. Neither Parkin-WT nor Parkin-T240R triggered RFP-Smac release from the mitochondria. (B) Apoptotic cell death was quantified by counting cells positive for RFP-Smac release. CCCP did not trigger significant RFP-Smac release in either cell lines. (C) Valinomycin induced mitochondrial recruitment of Parkin-WT, but not Parkin-T240R. (D) Quantitation of cell death by counting cells positive for RFP-Smac release in valinomycin treated cells.
(E-G) HeLa cells expressing Venus-Parkin-WT were transfected with Control or PINK1 siRNA and treated with 10μM valinomycin in a 12-hour time course. Apoptotic cell death was visualized by staining with Hoechst 33258 which enters all cells regardless cell health. The average intensity of DNA labeling significantly increases in apoptotic cells as nuclei condense during apoptosis. Quantitation of cell death was performed using automated cell heath application module in MetaXpress software. Representative images are shown. Scale bar, 10 μm.
(E) PINK1-dependent apoptotic cell death was visualized by fluorescent microscopy. Cells were incubated with valinomycin for 1.5h prior to image capture.
(F) WT and PINK1 knockdown cells in response to valinomycin were immunoblotted using an antibody for PINK1.
(G) Apoptotic cell death was quantified by counting cells positive for cells with condensed chromatin at 12h. More than five hundred cells were scored. Error bars represent standard deviation.
To further probe specificity of Parkin-induced mitophagy in response to mitochondrial depolarization, we tested mitophagic responses to a panel of mitochondrial damaging agents including rotenone, dinitrophenol, and valinomycin. Surprisingly, we found only valinomycin, a potassium ionophore known to collapse the potassium gradient across the mitochondrial inner membrane and depolarize mitochondria, induces rapid cellular apoptosis in HeLa cells expressing wild type Parkin but not Parkin T240R mutant (Figure 1CD and Supplementary movie 3). Valinomycin resistance is not due to the expression of the Parkin mutant since wild type HeLa cells display identical apoptosis phenotype upon valinomycin treatment (data not shown). Valinomycin induced cell death can be suppressed by the treatment with the caspase inhibitor Z-VAD-FMK, which is consistent with cell death by apoptosis (Supplementary Figure 1GH). Both CCCP and valinomycin induce rapid Venus-Parkin accumulation on mitochondria with similar kinetics (Supplementary Figure 1CDEF). However, despite similar effects on Parkin mitochondrial accumulation, cells exposed to these two depolarizing agents adopt different cell fates.
PINK1 is required for valinomycin-induced Parkin-dependent apoptosis
Because Parkin-dependent mitophagy requires PINK1 stabilization and accumulation on mitochondria [9, 13, 21, 27], we investigated whether PINK1 is also required for Parkin-dependent apoptosis in response to valinomycin using siRNA-mediated silencing of PINK1. Similar to what we observed for CCCP, valinomycin stabilized PINK1 expression (Figure 1F). As expected, PINK1 knockdown blocks Venus-Parkin accumulation on mitochondria and stabilization of PINK1 (Figure 1EF). The levels of apoptosis are significantly reduced in PINK1 knockdown cells (Figure 1EFG). This result indicates that the PINK1-Parkin pathway is involved in both mitophagy and apoptosis in response to the two different stimuli.
Valinomycin specifically suppresses the expression of anti-apoptotic protein Mcl-1 and activates initiator caspase-8 and caspase-9
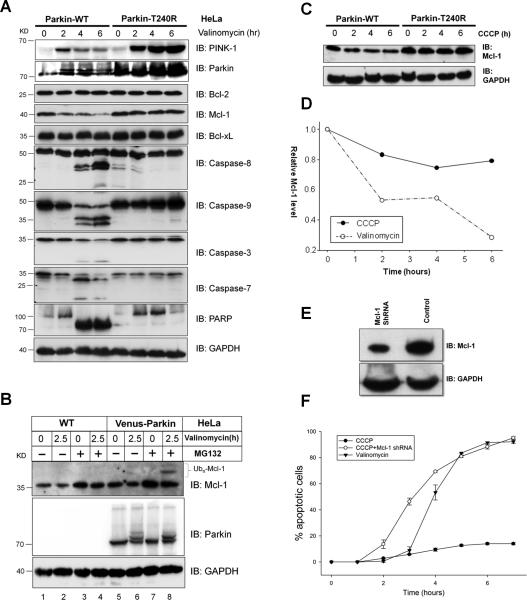
To address the mechanism of valinomycin-induced apoptosis, we initially assessed the expression of anti-apoptotic proteins and the activation status of several apoptotic hallmark proteins in Parkin wild type and Parkin T240R mutant HeLa cells (Figure 2A). Expression of Parkin T240R is higher than wild type Parkin which may be a result of their differential stability in cells. However, this difference is unlikely to be important for interpreting the experimental results since Parkin T240R is effectively null in valinomycin induced apoptotic responses and serves a better control to demonstrate the specificity of PINK1/Parkin-dependent cellular responses. Interestingly, in cells expressing wild type Parkin, valinomycin treatment slightly elevated PINK1 expression at 2 hr, after which levels declined and suppressed. In contrast, valinomycin strongly induced expression of PINK1 in HeLa cells expressing Parkin T240R mutant (Figure 2A, top panel). The pattern of PINK1 induction in the absence of functional Parkin in HeLa cells is reminiscent to CCCP treatment described previously [26]. Among the three well characterized Bcl2 proteins, only Mcl-1 is significantly down regulated by valinomycin treatment in a Parkin-dependent manner. No changes in Bcl-xL or Bcl2 were observed. Valinomycin treatment causes activation of initiator caspase-8 and caspase-9 within 4 hr in HeLa cells expressing the wild type but not in cells expressing the mutant Parkin (Figure 2A). In agreement with strong apoptotic activation by valinomycin, effector caspase-3, caspase-7, and PARP are cleaved in a Parkin-dependent manner (Figure 2A). To determine whether suppression of Mcl-1 is due to Parkin-dependent ubiquitin-mediated degradation, HeLa cells with or without Parkin expression were treated with valinomycin in the presence or absence of proteasome inhibitor MG132. High molecular weight Mcl-1 only presents in cells expressing Parkin and accumulates in the presence of MG132 (Figure 2B, lane 8). This finding suggests Mcl-1 is likely ubiquitinated by Parkin upon treatment with valinomycin. Activation of apoptosis by valinomycin is associated with suppression of Mcl-1 and activation of the intrinsic and extrinsic apoptotic initiator caspases.
Figure 2. Valinomycin induces Parkin-dependent suppression of Mcl-1 and activation of apoptotic initiator caspases.
(A) HeLa cells expressing Parkin-WT or Parkin-T240R were subjected to 10μM valinomycin in a 6-hour time course and collected at 2-hour intervals. Pro-apoptotic and anti-apoptotic proteins were monitored by western blotting using antibodies for their respective proteins. Upon valinomycin treatment, cells expressing Parkin-WT exhibited a depletion of Mcl-1 levels, activation of Caspase-8 and -9, and cleavage of Caspase-3, -7 and PARP. The corresponding apoptotic response was not detected in cells expressing Parkin-T240R.
(B) HeLa cells with or without Parkin expression were treated with 10μM valinomycin and/or 10μM MG132 for 2.5 hr. The response was monitored by western blotting using an antibody for Mcl-1 and Parkin. Cells expressing Parkin degrade Mcl-1 when treated with valinomycin and accumulated Ub-Mcl-1 when treated simultaneously with the proteasome inhibitor MG132.
(C) Effect of CCCP on Mcl-1 levels. The indicated cell lines were treated with 10μM CCCP in a 6-hour time course. Endogenous Mcl-1 was blotted with anti-Mcl-1 with GAPDH as a loading control. (D) Mcl-1 levels from (A) and (C) was plotted.
(E) Suppression of endogenous Mcl-1 expression by shRNA. Stable knockdown HeLa cells expression Venus-Parkin and RFP-Smac were obtained by infection with shRNA for Mcl-1.
(F) CCCP induces apoptosis in HeLa cells with reduced Mcl-1 expression. Apoptotic cell death was quantified by counting cells positive for RFP-Smac release.
If Mcl-1 suppression is linked to differences in cell fates upon exposure to CCCP and valinomycin, we would expect that CCCP should not cause significant Mcl-1 degradation. Immunoblotting analysis revealed that CCCP only causes a slight drop in Mcl-1 and remains stabilized after six hours as opposed to steady decline of Mcl-1 in response to valinomycin (Figure 2C and D). Conversely, if Mcl-1 level is a key determinant of cell fate decision, lowering Mcl-1 should be sufficient to alter the CCCP response. To test this hypothesis, we knocked down expression of Mcl-1 by shRNA (Figure 2E). Shown in Figure 2F, HeLa cells with lower endogenous Mcl-1 undergo apoptosis in response to CCCP. These data suggests that Parkin catalyzes selective degradation of specific substrates in response to specific stimuli. Suppression of Mcl-1 by Parkin is critical for differential cell fate decisions in response to CCCP and valinomycin.
Valinomycin induces PINK1- and Parkin-dependent apoptotic response in MEFs
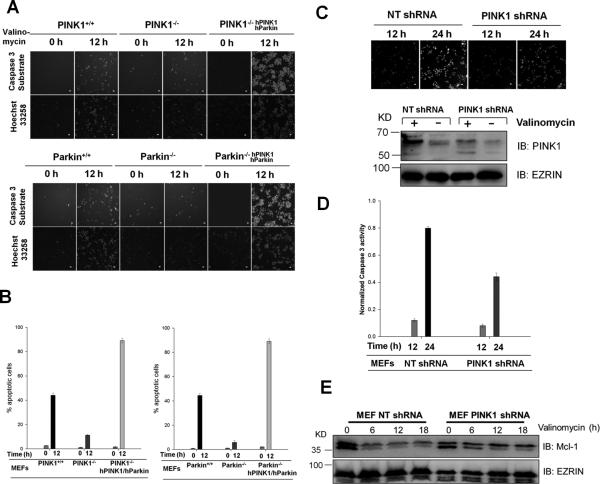
To determine whether the endogenous PINK1 or Parkin is required for valinomycin-induced apoptosis in another cell system, we first investigated the response to valinomycin in MEFs derived from the parental and PINK1 knockout mice as well as MEFs from Parkin knockout mice [28]. Using time-lapse live cell imaging, we monitored cell apoptotic responses during the time course of valinomycin exposure by a caspase-3 activity sensor NucView 488 (Biotium). DEVD-NucView™ 488 caspase-3 substrate is a cell membrane permeable fluorescence sensor. Upon cleavage by active caspase-3, the DNA-binding dye is released and migrates to the cell nucleus to stain the nucleus bright green. The nuclei and their condensation state were also scored independently by Hoechst 33258 fluorescence using an automated MetaXpress application module. As seen in HeLa cells, parental wild type MEFs are more prone to valinomycin induced apoptosis than PINK1 or Parkin null MEFs (Figure 3AB). To further demonstrate that the endogenous PINK1 has a pro-apoptotic role in valinomycin induced apoptotic response, we knocked down the expression of PINK1 in wild type MEFs using shRNA. A mammalian non-targeting shRNA (NT shRNA) was used as a control. Depletion of PINK1 was confirmed by immunoblotting (Figure 3C). Treatment with valinomycin induced stronger apoptosis in control MEFs (Figure 3CD). Treatment with valinomycin also suppresses Mcl-1 expression in the control MEFs (Figure 3E). However, down regulation of Mcl-1 is partially abrogated in PINK1 knockdown MEF cells (Figure 3E). These results suggest that valinomycin induces PINK1 and Parkin-dependent suppression of Mcl-1 and apoptosis in MEFs.
Figure 3. Valinomycin-induced apoptosis requires PINK1 and Parkin.
(A) and (B) Wild type MEFs, PINK1 or Parkin null MEFs or MEF nulls reconstituted with human PINK1 and Parkin were stained with NucView caspase-3 sensor and Hoechst 33258 and treated with 10μM valinomycin for 12 hr. Apoptotic cell death was visualized with fluorescent microscopy (A) and quantified by measuring the percentage of apoptotic nuclei (B). Scale bar, 10 μm. (C) Wild type MEF cells were stably introduced with nontargeting or PINK1 shRNA and treated with 10μM valinomycin for 24 hrs. PINK1 expression in control and knockdown cells was measured by western blot. Apoptosis was monitored by caspase-3 sensor and quantified. (D) and (E) PINK1 knockdown attenuates valinomycin-induced apoptosis and suppressed Mcl-1 degradation.
We tested whether valinomycin sensitivity can be rescued in PINK1 or Parkin null MEFs by stably expressing the two genes. The human versions of these genes were used because commercial antibodies against mouse Parkin are largely ineffective. Nevertheless, stable expression of human PINK1 and Parkin fully restores the defects of valinomycin response in MEFs that are deficient for either gene (Figure 3AB). Immunoblotting confirmed similar pattern of regulation of caspases and Bcl2 proteins seen in HeLa cells (Supplementary Figure 2A) and Z-VAD-FMK sensitivity (Supplementary Figure 2BC). These findings indicate that the PINK1-Parkin pathway controls cell fate determination in response to valinomycin.
PINK1 phosphorylates a highly conserved Ser65 residue in Parkin ubiquitin-like (Ubl) domain and ubiquitin
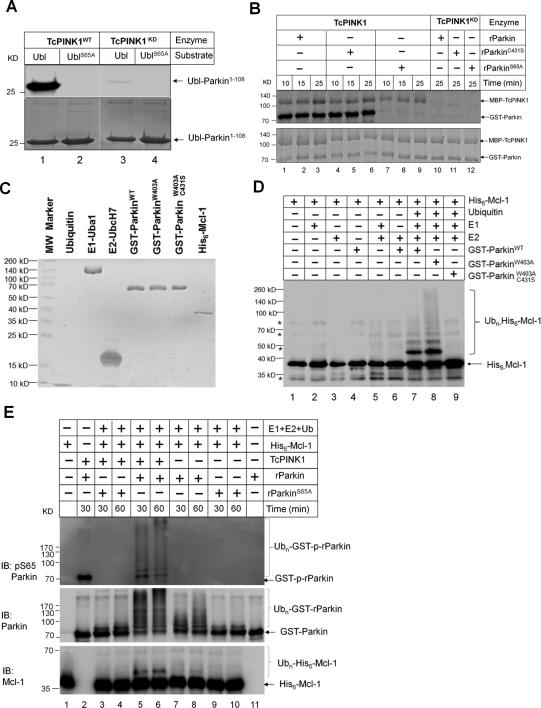
To further probe the biochemical mechanisms by which PINK1 and Parkin regulate cellular apoptosis, we investigated how the PINK1 and Parkin activity controls Mcl-1 ubiquitination in vitro. Our initial attempt was to use recombinant PINK1 to drive the pathway. Despite intensive efforts, we were unable to detect robust kinase activity of human PINK1. Previous studies by Muqit and colleagues showed that an insect orthologue of PINK1 (TcPINK1) from Tribolium castaneum is catalytically active [29]. In agreement with the previous report, we find TcPINK1 can phosphorylate human Parkin1-108 and Ser65 is most likely the site targeted by TcPINK1 as no phosphorylation can be detected when Ser65 was mutated to Ala (Figure 4A, lane 1 versus 2). Despite robust phosphorylation of recombinant human Parkin1-108 by TcPINK1 in vitro, full length human Parkin was reported to be a poor substrate for TcPINK1 [11]. In light of the recent success with the full length rat Parkin (rParkin) protein in structural studies [23], we tested whether recombinant rat Parkin may be a better substrate for TcPINK1 because rParkin is well folded. Shown in Figure 4B, rParkin is robustly phosphorylated by TcPINK1. Mutation of catalytic Cys at 431 to Ser does not affect its phosphorylation. However, mutation of Ser65 to Ala in the Ubl domain of rParkin largely eliminates its phosphorylation (Figure 4B, lanes 7-9). Since the Ubl domain of Parkin shares significant sequence and structure homology with ubiquitin and Ser65 is conserved and solvent exposed (Supplementary Figure 3ABC), we tested whether ubiquitin may also be a substrate for PINK1. As seen with Parkin Ubl, TcPINK1 can phosphorylate wild type ubiquitin and this phosphorylation is significantly reduced when Ser at 65 is mutated to Ala or Asp (Supplementary Figure 4A), suggesting Ser65 is the primary phosphorylation site of ubiquitin. These results indicate that PINK1 phosphorylates Parkin and ubiquitin primarily at a single Ser residue (Ser65).
Figure 4. Full length Parkin and Parkin Ubl domain are PINK1 substrates and in vitro reconstitution of Mcl-1 ubiquitination by Parkin.
(A) and (B) In vitro kinase assay with [γ-32P] ATP. (A) UblWT-Parkin1-108 or UblS65A -Parkin1-108 as the substrate for TcPINK1WT. Ser65 in Parkin Ubl domain is required for PINK1 phosphorylation. (B) Phosphorylation of rParkinWT, rParkinC431S or rParkinS65A by TcPINK1WT and kinase dead TcPINK1D359A. (C) Coomassie blue staining of proteins used in the in vitro ubiquitination reaction. (D) In vitro ubiquitination assay was performed at 37°C for 3 hrs using 1 μM His6-Mcl-1 as the substrate and was monitored by western blotting using an antibody for Mcl-1. rParkin-W403A exhibiting enhanced auto-ubiquitination and rParkinW403AC431S exhibiting no ligase activity were used as controls. 0.7 μM rParkin was used in ubiquitination reactions. (E) Phosphorylation of Parkin Ser65 promotes Parkin auto- and trans-ubiquitination activity. Wild type rParkin and Ser65 rParkin mutant were incubated with TcPINK1 or kinase dead TcPINK1 for 60 min prior to initiating ubiquitination reactions. Phosphorylation of Ser65 of Parkin was monitored by a Ser65 phospho-specific antibody. Ubiquitin ligase activity was monitored by western blotting using an antibody for pSer65 Parkin, Parkin and Mcl-1. Ubiquitination reactions were terminated at indicated time.
Mcl-1 is a substrate for Parkin E3 ubiquitin ligase
To demonstrate that Mcl-1 is a substrate for Parkin, we reconstituted Parkin-dependent Mcl-1 ubiquitination using highly purified ubiquitin, Uba1(E1), UbcH7 (E2), rat Parkin, and Mcl-1 (Figure 4C) and incubated for 3 hr at room temperature (Figure 4D). Polyubiquitination of Mcl-1 is more robust with a ParkinW403A (Figure 4D lane 7 versus 8), a mutant that has been shown to inactivate the REP element and to elevate its autoubiquitination activity [23]. A double mutant with the substitution of catalytic Cys 431 to Ser has no activity (Figure 4D, lane 9). Omission of any component required in the ubiquitin transfer reaction abrogated polyubiquitination of Mcl-1, suggesting this reaction is highly specific (Figure 4D, lanes 1-6). This finding indicates Mcl-1 is a substrate for Parkin in vitro.
Activation of Parkin E3 ubiquitin ligase activity by PINK1 through phosphorylation of Parkin Ubl at Ser65
Given that Parkin Ser65 is phosphorylated by PINK1, we analyzed the effects of Parkin phosphorylation on its auto-ubiquitination and Mcl-1 ubiquitination activity. Shown in Figure 4E, GST-rParkin is phosphorylated by PINK1 and this phosphorylation is absent in GST-rParkinS65A as detected by a Parkin phospho-Ser65 antibody (Figure 4E, lane 2 versus 3). Phosphorylation of Parkin greatly increases its auto- and trans-ubiquitination activity by the early appearance of high molecular weight Parkin conjugates and ubiquitinated Mcl-1 (Figure 4E, lanes 5 -6 versus lanes 7-8). GST-rParkinS65A is inactive in the presence or absence of PINK1 (Figure 4E, lane 3-4 versus 9-10). Since PINK1 also phosphorylates ubiquitin at Ser65 (Supplementary Figure 4A), we also tested the effect of this phosphorylation on Parkin E3 ligase activity. Ubiquitin was first incubated with PINK1 or kinase dead PINK1 for 2 hr to allow it to be phosphorylated. Phosphorylated ubiquitin and control ubiquitin were used as inputs in the reconstituted Mcl-1 ubiquitination reaction by Parkin in vitro. In the reaction with phosphorylated ubiquitin, ubiquitinated Mcl-1 showed up earlier than the control reaction (Supplementary Figure 4B, lanes 1-3 versus lanes 4-6) as well as the auto-ubiquitinated Parkin bands. Consistent with the positive effect of ubiquitin phosphorylation, phosphomimetic ubiquitinS65D stimulates Parkin auto-ubiquitination more effectively than ubiquitinS65A with Ser65 phosphorylated rParkin or unphosphorylated rParkin (Supplementary Figure 4C). These results indicate that phosphorylation of Parkin by PINK1 contributes to its activation in vitro. The effect of ubiquitin Ser65 phosphorylation on Parkin mitochondrial recruitment was tested by comparing HA-Ub and HA-UbS65A in HeLa cells expressing Venus-Parkin. Expression of HA-UbS65A but not HA-Ub caused a significant reduction in Parkin recruitment to mitochondria in response to valinomycin treatment (Supplementary Figure 4D and E). This result points to a potential role for ubiquitin phosphorylation in Parkin recruitment to mitochondria.
Phosphorylation of Parkin Ser65 is required for valinomycin induced apoptotic responses
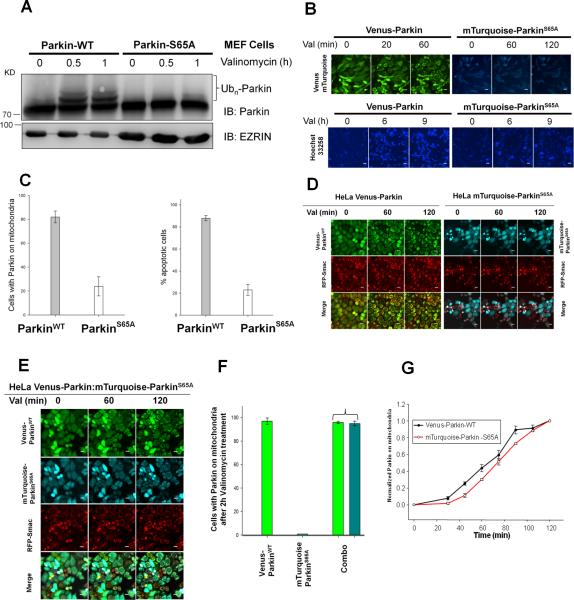
To address the significance of Parkin Ser65 phosphorylation in Parkin pro-apoptotic signaling in response to valinomycin, we stably expressed mTurquoise-ParkinS65A in Parkin null MEF and HeLa cells. The mitochondrial recruitment behavior of mTurquoise-Parkin is identical to Venus-Parkin (data not shown). While valinomycin induces wild type Parkin mobility shift (Figure 5A), ParkinS65A is largely unresponsive to valinomycin treatment. mTurquoise-ParkinS65A is also defective in mitochondrial recruitment in response to valinomycin (Figure 5B and 5C). MEF cells expressing mTurquoise-ParkinS65A are resistant to valinomycin induced cell death (Figure 5B and C), suggesting phosphorylation of Parkin Ser65 is required for initiating Parkin recruitment to mitochondria and transducing apoptotic signal.
Figure 5. Phosphorylation of Parkin Ser65 is required for valinomycin-induced apoptosis.
PINK1−/− Parkin−/− MEFhPINK1 cells expressing Venus-Parkin or mTurqoise-ParkinS65A were treated with valinomycin for indicated time. (A) Immunoblotting analysis of Parkin and ParkinS65A response to valinomycin treatment. (B) Parkin mitochondrial recruitment is perturbed by Ser65 mutation. Cells expressing mTurqoise-ParkinS65A are resistance to valinomycin in apoptosis. (C) Quantitation of (B). (D) Parkin mitochondrial recruitment upon treatment with valinomycin in HeLa cells expressing either Venus-Parkin or mTurqoise-ParkinS65A or (E) both Venus-Parkin and mTurqoise-ParkinS65A. Scale bar, 10 μm. (F) Quantitation of Venus-Parkin or mTurqoise-ParkinS65A accumulation on mitochondria. (G) Time course of Venus-Parkin and mTurqoise-ParkinS65A mitochondrial recruitment in response to valinomycin treatment in double positive HeLa cells.
Autocatalysis is a key mechanism for Parkin activation and mitochondrial recruitment
Inefficient accumulation of ParkinS65A on mitochondria could be due to its inability to associate with mitochondria or lack of catalytic activity to generate a positive feedback that is required for rapid and efficient activation of Parkin. Since Ser65 phosphorylated Parkin has robust auto-ubiquitination activity, we reasoned that the defect of ParkinS65A in mitochondrial recruitment could be rescued by the presence of wild type Parkin in trans. We tested this hypothesis by expressing Venus-Parkin and mTurquoise-ParkinS65A independently or together in the same HeLa cells. As expected, Venus-Parkin is recruited to mitochondria while mTurquoise-ParkinS65A is not when expressed independently upon valinomycin treatment (Figure 5D). In contrast, when both were co-expressed in the same cell, we found that mTurquoise-ParkinS65A is efficiently recruited and indistinguishable from Venus-Parkin at 2 hr (Figure 5E and F). Detailed kinetics analysis revealed that there was an approximately 10 min delay in the early phase of accumulation for mTurquoise-ParkinS65A and both approached similar levels at approximately 110 min (Figure 5G). This result suggests that catalytically active Parkin converts ParkinS65A to the wild type Parkin phenotype in terms of mitochondrial recruitment.
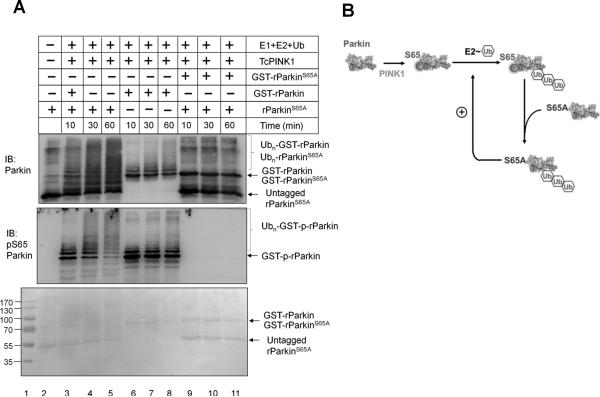
One hypothesis that can explain the rescue of ParkinS65A by wild type Parkin is that Parkin activation is through an autocatalytic mechanism. To test this hypothesis, we set up an in vitro ubiquitination assay using a combination of wild type and mutant Parkin proteins (Figure 6AB). An untagged rParkinS65A was purified to mix with either GST-rParkin or GST- rParkinS65A at 3:1 ratio for the ubiquitination reaction after incubation with TcPINK1 to allow phosphorylation of GST-rParkin to occur. With a three-fold reduction of GST-rParkin, the rate of Parkin auto-ubiquitination slowed down (Figure 6A, lanes 6-8 versus Figure 4E, lanes 5-6). When nonphosphorylatable untagged rParkinS65A was included in the reaction, polyubiquitination of rParkinS65A occurred judging by the disappearance of the substrate (Figure 6A, top panel, lane 3-5). No reaction was seen in the control with GST- rParkinS65A with rParkinS65A (Figure 6A, lane 9-11), suggesting neither protein has catalytic activity in the presence of TcPINK1. With the Parkin pSer65 antibody, we can see the autoubiquitination rate of GST-rParkin increased dramatically in the presence of rParkinS65A (Figure 6A, lanes 3-5 versus 6-8). This is unlikely due to the stimulation of GST-rParkin phosphorylation by rParkinS65A since the overall phosphorylation of GST-rParkin stayed the same (Figure 6A, lane 3-5). This result indicates that Parkin activation likely occurs via an autocatalytic mechanism. PINK1 phosphorylation of Parkin at Ser65 provides the initial spark for the nonlinear amplification of Parkin activity.
Figure 6. Autocatalytic activation of Parkin by PINK1.
(A) In vitro ubiquitination of wild type and mutant Parkin through autocatalysis. 0.15 μM GST-rParkin or GST-rParkinS65A was mixed with 0.45 μM rParkinS65A prior to incubation with 0.5 μM TcPINK1 for phosphorylation at 30oC for 60 min. Ubiquitination reactions were monitored by immunoblotting with anti-Parkin or anti-pSer65 Parkin antibodies. (B) Schematic diagram of the autocatalytic ubiquitination reaction.
Autocatalytic activity but not mitochondrial recruitment of Parkin is required for Parkin-dependent apoptotic response to valinomycin
Previous studies showed that mitochondrial expression of linear ubiquitin chain promotes mitochondrial targeting of ParkinC431S mutant that is defective in ubiquitin E3 ligase activity in response to CCCP[16]. To test whether mitochondrial recruitment of Parkin is sufficient for triggering apoptotic response in the presence of valinomycin, we stably expressed mitochondrial targeted linear ubiquitin chain Tom70-4xUbi in Parkin−/−MEF cells along with Venus-ParkinC431S. Immunoblotting showed that ParkinC431S had an extra high molecular weight band, which was independent of valinomycin treatment or Tom70-4xUbi expression (Supplementary Figure 5AB). Upon treatment with valinomycin, Venus-ParkinC431S accumulates on mitochondria in 40% of cells expressing both proteins compared to less than 5% cells expressing Venus-ParkinC431S alone at 90 min (Supplementary Figure 5CD). Meanwhile, valinomycin induced apoptosis is indistinguishable between these two cell lines similar to the cell line expressing Venus-ParkinS65A whereas Venus-Parkin showed dramatic apoptotic response (Supplementary Figure 5EF). Thus in the absence of catalytic activity, Parkin recruitment to mitochondria does not switch cellular responses. This result suggests that Parkin mitochondrial targeting alone is insufficient to trigger valinomycin-induced apoptosis and in line with the notion that Parkin autocatalytic activation and ubiquitination of Mcl-1 are required for mediating the apoptotic responses.
Discussion
Here we show that the PINK1-Parkin pathway can mediate different cell fates in response to different cell stressor stimuli. We uncovered a new pro-apoptotic function of PINK1 and Parkin when cells are exposed to valinomycin in addition to its well documented role in mitophagy and cytoprotection. The PINK1-Parkin dependent apoptotic response is associated with suppression of Mcl-1. PINK1 phosphorylates Parkin at Ser65 and unleashes the autocatalytic activity of Parkin and ubiquitination of Mcl-1, which contributes to irreversible molecular switches resulting in apoptotic responses. In addition, we reveal that PINK1 can also phosphorylate ubiquitin at Ser65. These two posttranslational events bolster Parkin accumulation on mitochondria and apoptotic response to valinomycin. Our results suggest that PINK1-Parkin constitutes a damage-gated molecular switch that governs cellular context-specific cell fate decisions in response to variable stress stimuli.
PINK1 as a molecular gauge for cellular stress
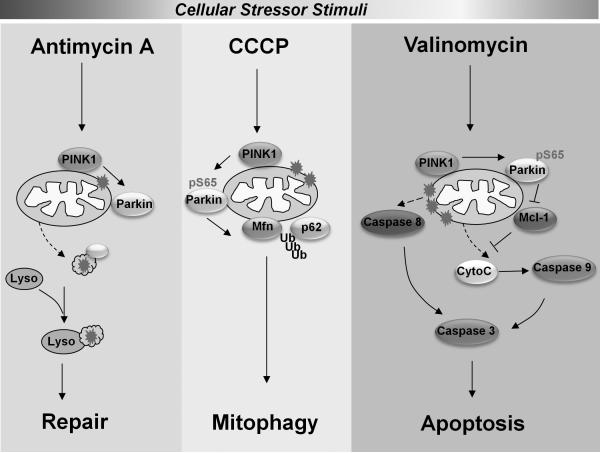
The anti-apoptotic and cytoprotective function of PINK1 against cell stressors has been well recognized although the exact mechanism is still debatable. For example, PINK1 is known to antagonize MG132-induced cell death [30-32]. The pro-survival function of PINK1 has been attributed to the inhibition of BAX translocation by cytoplasmic PINK1 activity [32] or phosphorylation of Bcl-xL by mitochondrial PINK1 [30, 31]. Some of the protective activity of PINK1 is independent of Parkin or its mitochondria recruitment as PINK1without mitochondria targeting signal retains anti-apoptotic activity in response to MG132 or MPTP [32, 33]. Here we demonstrate that PINK1 can also mediate pro-apoptotic signals and this function requires Parkin. To reconcile this apparent paradoxical role of PINK1 in cell death regulation, we have to consider the type of cellular damage each cellular stressor inflicts. McLelland et al. recently showed that excessive production of reactive oxygen species (ROS) by antimycin A causes mild oxidative damage which generates Mitochondria-derived vesicles (MDVs) in PINK1 and Parkin-dependent manner. These vesicles contain oxidized cargo that is transported to lysosomes for disposal [24] (Figure 7). This pathway is distinct from PINK1-Parkin dependent mitophagy which is triggered by the dissipation of ∆ψm as a result of exposure to decouplers such as CCCP and irreversible damage of mitochondria. In many cellular contexts, mitophagy or autophagy may act as a rescue mechanism that the cell uses to escape from cell death [34]. With the treatment of valinomycin, the severity of mitochondria or organelle damage is likely heightened to a threshold beyond repair due to severe stress. Under these conditions, the PINK1-Parkin pathway executes apoptotic program. Hence the cell fate determination dictated by PINK1 or Parkin can be stimulus specific.
Figure 7.
Model depicting the tandem action of PINK1-Parkin as a damaged-gated molecular switch for cell fate specification.
Stimuli specific degradation of substrates by PINK1-Parkin pathway
We found that valinomycin and CCCP treatment caused different cell fates via the same PINK1/Parkin activation. The levels and kinetics of Parkin mitochondrial recruitment are comparable (Supplementary Figure 1). This result suggests cell fate decisions are AND-gates since Parkin mitochondrial recruitment alone is insufficient. Indeed, we discovered that the endogenous Mcl-1 level set by Parkin recruitment followed by ubiquitination specifies mitophagy vs. apoptosis decision in response to CCCP or valinomycin. Therefore targeting Parkin to mitochondria does not lead indiscriminate substrate ubiquitination. There are additional stimuli specific changes that are in play. Our study identified a key determinant in Mcl-1 level and an AND-gate in cell fate decisions. Future studies are needed to uncover stimuli specific determinant that makes Mcl-1 susceptible to Parkin ubiquitination. Regardless the specificity factor of Mcl-1 degradation, our results suggest that PINK1-Parkin pathway serves as damage-gated molecular switch to specify distinct cell fate decisions (Figure 7).
An autocatalytic mechanism for the PINK1-Parkin cascade
Mechanistically we linked valinomycin induced apoptotic response to ubiquitination and degradation of Mcl-1 by Parkin. Mcl-1 is an unstable protein and its steady state levels are regulated by several ubiquitin E3 ligases including Mule [35], SCFFbw7[36] and SCFTrCP[37]. Here we provide an example of signal-regulated Mcl-1 E3 ligase in Parkin. Using a reconstituted in vitro system, we demonstrated that two PINK1-dependent phosphorylation events drive amplification of its auto-ubiquitination and transubiquitination activity. PINK1 directly phosphorylates Ser65 of Parkin in the Ubl domain and ubiquitin at Ser65. This sets off an autocatalytic cycle for activating Parkin auto-ubiquitination and trans-ubiquitination of Mcl-1. While we still do not know precisely how Ser65 phosphorylation to enable Parkin to adopt an active conformation, we can speculate that phosphorylation in Ubl domain of Parkin may unmask the E2 docking site [23]. It is a mystery why phosphorylated ubiquitin can promote its activity. In essence, Ser65 phosphorylated ubiquitin could be derived from phosphorylation of auto-ubiquitinated Parkin by PINK1 or conjugation of free ubiquitin phosphorylated by PINK1 or both. Future experiments are needed to determine to what degree ubiquitin is phosphorylated in cells upon PINK1 activation. We show that a Parkin mutant defective in PINK1 phosphorylation can stimulate wild type Parkin E3 activity in trans. This result suggests that Parkin is a preferred substrate for itself and a small fraction of Parkin phosphorylation by PINK1 can trigger nonlinear amplification of E3 ligase activity. The positive feedback generated by Parkin auto-ubiquitination and autocatalysis is likely an underlying mechanism for irreversible cell fate determination. This model provides one explanation why wild type Parkin can rescue the defects of Parkin Ser65 mutant and robust Parkin accumulation on mitochondria. It will be interesting to determine whether this mechanism can operate locally in a spatial constrained manner.
Experimental Procedures
Cell culture, constructs and antibodies
The source of cell lines, recombinant DNA constructs and antibodies are listed in supplemental experimental procedures.
In vitro kinase assay
Recombinant wild-type and D359A (kinase-dead) in the background of MBP-TcPINK1 and the substrate His6-Sumo-Parkin (1-108) were purified and the kinase reaction were performed as described [11].
In Vitro ubiquitination assay
Purification of ubiquitin E1, E2 (UbcH7) and reagents for in vitro ubiquitination assay was described previously [38]. Wild-type or mutant Parkin proteins were purified from E.coli as described previously [23]. Human Mcl-1 (1-327 aa) was subcloned in pET15b and purified by Ni-NTA affinity chromatography. The ubiquitination assays were performed by incubating 1 μM His6-Mcl-1 with 0.5 μM E1, 5 μM UbcH7 (E2), 0.7 μM rParkin E3 complex, 20 μM ubiquitin (Sigma), and 1 μl 20 × energy regeneration system (10 mM ATP, 20mM HEPES pH 7.4, 10 mM MgOAc, 300 mM creatine phosphate, 0.5 mg/ml creatine phosphokinase) in a final volume of 20 μl. The reactions were incubated at 37°C for indicated time, terminated by boiling in 2×SDS protein sample buffer and analyzed by SDS-PAGE and immunoblotting with indicated antibodies.
Cell death assays
Three independent cell death assays were used. Detailed methods of RFP-Smac release, NucView caspase-3 biosensor and Hoechst 33258 staining as well as live cell imaging are described in supplementary experimental procedures.
Supplementary Material
Highlights.
PINK1-Parkin operates as a damaged gated molecular switch for cell fate decisions.
Parkin catalyzes ubiquitination of selective substrates in response to specific stimuli.
Parkin is the E3 ligase for Mcl-1 and mediates valinomycin-induced apoptosis.
PINK1 triggers autocatalytic activation of Parkin by phosphorylating Ser65 of Parkin.
Acknowledgments
We thank Drs. Kevan Shokat, Stefan Constantinescu, Nick Hertz and Michael Lazarus for providing critical reagents. We thank Kevin Dean, Yan Qin, Junglim Lee and Genevieve Park for technical assistance. We also want to thank Dr. James Goodrich for critical reading of the manuscript; Drs. Natalie Ahn, James Ferrell, Tobias Meyer and Amy Palmer and members of Liu laboratory for discussion and Drs. Miratul Muqit, Helen Walden, Hiroyuki Miyoshi, Xinde Zheng, Tony Hunter and Sabrina Spencer for expression vectors. This work was supported by grants from a Butcher Award from the University of Colorado, Cancer League of Colorado, and National Institutes of Health CA107098 to Xuedong Liu. The ImageXpress MicroXL was supported by a NCRR grant S10 RR026680 from NIH. This work was also in part sponsored by the U.S. Army Research Office and the Defense Advanced Research Projects Agency and was accomplished under Cooperative Agreement Number W911NF-14-2-0019.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: C.Z. and X. L designed the study; C.Z., S.L., Y.P., S.M. and Z.Z. performed experiments; C.Z., S.L., E.B. and X.L. analysed data; E.G. and J.S. provided PINK1 and Parkin null cell lines; C.Z., S.L. and X.L. wrote the manuscript.
While this manuscript was under review, four publications Koyano et al. Nature 510:162-6, Kazlauskaite et al. Open Biol 4:130213 2014; Kazlauskaite et al. Biochem J 460(1):127-39; Kane et al. JCB 205:143-53 independently reported the finding of phosphorylation of ubiquitin and Parkin by PINK1.
References
- 1.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 2.Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson's disease. Mov Disord. 2010;25(Suppl 1):S32–39. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cookson MR. The biochemistry of Parkinson's disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 4.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winklhofer KF. Parkin and mitochondrial quality control: toward assembling the puzzle. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narendra DP, Jin SM, Tanaka A, Suen D-F, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2008;8 doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu W, Ko HS, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080–120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou YS, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 15.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 2013;23:886–897. doi: 10.1038/cr.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S, Youle RJ. PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olzmann JA, Li L, Chudaev MV, Chen J, Perez FA, Palmiter RD, Chin LS. Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol. 2007;178:1025–1038. doi: 10.1083/jcb.200611128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka A, Cleland MM, Xu S, Narendra DP, Suen D-F, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, et al. Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem. 2011;286:11649–11658. doi: 10.1074/jbc.M110.144238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler S, Holmstrom KM, Treis A, Skujat D, Weber SS, Fiesel FC, Kahle PJ, Springer W. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–878. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 22.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 24.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, et al. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- 26.Narendra D, Tanaka A, Suen D-F, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narendra DP, Jin SM, Tanaka A, Suen D-F, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci U S A. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodroof HI, Pogson JH, Begley M, Cantley LC, Deak M, Campbell DG, van Aalten DM, Whitworth AJ, Alessi DR, Muqit MM. Discovery of catalytically active orthologues of the Parkinson's disease kinase PINK1: analysis of substrate specificity and impact of mutations. Open Biol. 2011;1:110012. doi: 10.1098/rsob.110012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P, Cilia E, Jonas EA, Valente EM. PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ. 2013;20:920–930. doi: 10.1038/cdd.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hertz NT, Berthet A, Sos ML, Thorn KS, Burlingame AL, Nakamura K, Shokat KM. A Neo-Substrate that Amplifies Catalytic Activity of Parkinson's-Disease-Related Kinase PINK1. Cell. 2013;154:737–747. doi: 10.1016/j.cell.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klinkenberg M, Thurow N, Gispert S, Ricciardi F, Eich F, Prehn JHM, Auburger G, Kogel D. Enhanced vulnerability of PARK6 patient skin fibroblasts to apoptosis induced by proteasomal stress. Neuroscience. 2010;166:422–434. doi: 10.1016/j.neuroscience.2009.12.068. [DOI] [PubMed] [Google Scholar]
- 33.Haque ME, Thomas KJ, D'Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci U S A. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 35.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, Lee DF, Liu JC, Zhong Q, Wang X, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ungermannova D, Gao Y, Liu X. Ubiquitination of p27Kip1 requires physical interaction with cyclin E and probable phosphate recognition by SKP2. J Biol Chem. 2005;280:30301–30309. doi: 10.1074/jbc.M411103200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.